Abstract
The transcription factors Signal Transducer and Activator of Transcription (STAT) 5A/B mediate prolactin-induced mammary development during pregnancy. However, it is not clear how the different processes, expansion and maturation of alveolar precursor cells and the differential induction of milk protein genes are regulated on a molecular level. We have used mouse genetics and genome-wide analyses to determine how altering concentrations of STAT5A and STAT5B impacts mammary epithelial development during pregnancy and the regulation of target genes. The presence of only a single Stat5a or Stat5b allele was sufficient for the establishment of histologically undifferentiated alveolar units and two alleles permitted the execution of a differentiation program similar to that found with all four alleles. While one copy of Stat5 induced limited expression of target genes, two copies activated a lactation-like gene signature. Using ChIP-seq analyses on intact tissue under physiological conditions, we found that highly expressed and regulated genes were bound by STAT5 in their promoter proximal regions, whereas upstream binding had minor biological consequences. Remarkably, 80% of the genes bound by STAT5 in vivo were not under STAT5 control. RNA polymerase II intensity was directly proportional to STAT5 concentration only on STAT5 regulated genes providing mechanistic insight by which STAT5 activates mammary specific genes.
INTRODUCTION
The lactating mammary gland consists of two histologically distinct structures, ducts and alveoli. While the establishment of ducts occurs mainly during puberty and is under the control of ovarian steroid hormones, the formation of alveoli during pregnancy is under the influence of progesterone and prolactin, the latter signaling mainly through the homologous transcription factors Signal Transducer and Activator of Transcription 5A (STAT5A) and STAT5B (referred to as STAT5) (1,2). Alveoli are specialized structures whose sole purpose is the production and secretion of milk during lactation. Alveolar progenitor cells respond to pregnancy signals with an initial burst of cell proliferation to establish immature alveolar units, which subsequently undergo differentiation culminating in the production of milk.
The absence of both Stat5 genes from mammary stem cells causes a failure in the formation of alveoli during pregnancy (3) demonstrating essential roles in the progression of the alveolar lineage (4). Ablation of Stat5 from mammary epithelium late in pregnancy causes these cells to die (3). This suggests that STAT5 controls distinct biological programs at different stages of pregnancy. Throughout pregnancy STAT5 concentrations rise in mammary epithelium, which is paralleled by an elevation of pSTAT5 levels, indicative of the active protein (5). One hallmark of mammary alveolar development is the sharp induction of some milk proteins around mid-pregnancy while others are induced just prior to parturition.
STAT5A and STAT5B are highly conserved and they can compensate, at least partially, for each other’s absence as shown by the deletion of the individual genes (6,7). In mammary tissue, STAT5A is more abundant than STAT5B and constitutes ∼70% of STAT5 levels (8). Mammary development in outbred Stat5a-null mice is overtly normal and dams can nurse their pups, suggesting that ∼30% STAT5 is sufficient for mammary tissue to function. STAT5 can be activated by a plethora of cytokines and both cell-specific and general target genes have been identified. While transcription of the majority of target genes, such as Igf1 and Socs2, is activated by STAT5 others, including Bcl6 (9), are suppressed. ChIP and ChIP-seq experiments have revealed that STAT5, in addition to recognizing promoter sequences, also binds to more distant and intronic sequences (9–14) suggesting a complex regulation of these genes.
Although it has been established that STAT5 is a driver of mammary development, key questions, some of which are equally relevant to other cell types under cytokine-STAT5 control, remain to be answered. First, to what extent is the sequential biphasic mammary epithelial program during pregnancy, epithelial cell proliferation followed by functional differentiation, triggered by distinct STAT5 concentrations? Second, are discrete gene expression programs in mammary epithelium activated by distinct STAT5 levels? Third, to what extent are genes bound by STAT5 in mammary tissue also expressed and regulated during pregnancy? Fourth, to what extent does cell-specificity modulate global STAT5 binding? To address these questions we have generated mice carrying different combinations of Stat5 alleles, thus harboring STAT5 concentrations between 0 and 100%. We analyzed development of mammary tissue from these mice during pregnancy and at lactation using histology and RNA-seq. Moreover, we performed genome-wide ChIP-seq studies to explore binding of STAT5A, STAT5B and RNA polymerase II as well as the distribution of histone H3K4 trimethylation (H3K4me3) in mammary tissue expressing different STAT5 levels. To our knowledge this is the first investigation in which STAT5 levels were manipulated by genetic methods to study the role of this transcription factor in a physiological setting during a developmental program.
MATERIALS AND METHODS
Generation of mice with different Stat5 alleles
Animals were handled and housed in accordance with the guidelines of NIH and all experiments were approved by the Animal Care and Use Committee of NIDDK. By mating Stat5ab+/− mice (3) with Stat5a−/− mice (6) and Stat5b−/− mice (7), we generated mice carrying a null allele of the Stat5ab locus and one functional allele of either Stat5a (Stat5ab−/Stat5b−) or Stat5b (Stat5ab−/Stat5a−). Stat5abf/f mice (3) were mated with the MMTV-Cre transgenic mouse line A (15) to generate Stat5abf/f;MMTV-Cre mice. We refer to the different mutations based on the allele that was retained in these mice. Wild-type mice and Stat5abfl/fl mice are referred to as AABB mice; Stat5abfl/fl;MMTV-Cre (with Stat5ab-deficient mammary epithelial cells) as Null mice; Stat5a−/− mice as BB mice; Stat5b−/− mice as AA mice; Stat5ab+/null mice as AB mice. Mice carrying only a single functional allele of either Stat5a (Stat5abnull/Stat5b−) or Stat5b (Stat5abnull/Stat5a−) are referred to as A mice and B mice, respectively. For some experiments, mice were mated and checked for vaginal plugs. The day when a vaginal plug was found is day 0.
Mammary tissue transplantation
Athymic nude mice (3 weeks old) were anesthetized with an intraperitoneal injection of avertin, the proximal part of the inguinal gland containing the mammary epithelium was excised and a small piece of mammary tissue from a virgin donor mouse was inserted into the remaining cleared fat pad. To assess the completeness of clearing, the excised tissues were processed for whole mount staining as described later. Eight weeks after transplantation, fat pads were harvested either from virgin hosts or the hosts were mated and their tissue was harvested on day 6 (p6) or day 13 of pregnancy (p13) or the day of parturition within <12 h after delivery (L1).
Histology
Harvested mammary tissues were fixed in 10% formalin, dehydrated through ethanol and xylene, embedded in paraffin and sectioned. Sections were stained with hematoxylin and eosin (H&E) by standard methods. For immunostaining, antigen unmasking was performed in a Decloaking chamber (Biocare Medical, Concord, CA, USA) using BORG Decloaker solution pH 9.5 (Biocare Medical) at 125°C, 18–24 PSI (pounds per inch) for 5 min. Sections were blocked for 30 min in TBS-T containing 3% goat serum. Primary antibodies were incubated overnight at 4°C (anti-phosphorylated STAT5, Cell Signaling, 1:200; anti-NKCC1, a gift from Dr. Jim Turner National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD, USA, 1:1000; anti-smooth muscle actin #A2547, Sigma, St. Louis, MO, USA, 1:1000). Alexafluor488 or 594 conjugated secondary antibodies (Invitrogen) were used at a dilution of 1:400 for 30 min at room temperature.
Western blotting
Anti STAT5A and anti STAT5B antibodies from several vendors were tested and we determined that Santa Cruz antibodies L-20 and C-17 were specific for STAT5A and STAT5B, respectively.
RNA-seq data processing
Poly(A) RNA was purified twice from 1 µg total RNA and cDNA was synthesized using SuperScript II (Invitrogen) and TruSeq RNA Sample Preparation Kit (Illumina Inc., San Diego, CA, USA), and sequenced using HiSeq 2000 (Illumina). The single-end reads of biological triplicates obtained from each sample were aligned to the mouse reference genome (mm9 assembly) using the TopHat program (16,17). The total number of the mapped reads on each gene was calculated with the HTSeq program (http://www-huber.embl.de/users/anders/HTSeq/). Transcript abundance was estimated by means of fragment per kilobase of exon per million fragments mapped (FPKM) according to the mapped reads on exons as described earlier (18). We observed that few extremely abundant transcripts, which absorbed more than 5% of the total mapped reads, led to the biased estimation of the FPKM values named ‘dilution effect’ (Supplementary Figure S1). To correct for this bias and identify less abundant in vivo STAT5 target genes, we applied multiple filter criteria. First, the genes containing more than 5% of the total mapped reads were excluded from initial FPKM calculation (Csn1s2a, Csn1s1, Csn2, Glycam1 and Wap at L1; Scd1 at p6), and their FPKMs were calculated along with the other genes as a separate procedure. Then, the calculated FPKMs of extremely abundant genes were merged with the initially calculated FPKMs of the other genes. The FPKM values of biological triplicates per sample were further normalized using the quantile normalization method (19). Second, genes showing more than 2-fold up-regulation between AABB and Null samples were selected. Third, genes showing more than five FPKM in the wild type (AABB) were finally used to eliminate potential false-positive detection. Totals of 370 (p6) and 750 (L1) genes were identified as STAT5 target genes. Clustering analyses was performed using Cluster 3.0 program (20). The RNA-seq data are deposited in GEO under accession number GSE37646.
Chromatin immunoprecipitation coupled by parallel sequencing (ChIP-seq) data processing
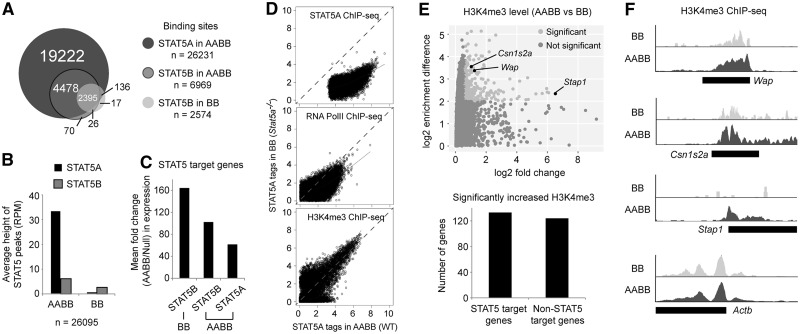
Frozen-stored mammary tissues were broken into powder with mortar and pestle and then cross-linked with 1% formaldehyde for 10 min. Nuclei were fractionated by sucrose density gradient centrifugation. Chromatin was fragmented to 200–300 bp by sonication using a MISONIX Sonicator 3000 (QSonica, Newtown, CT, USA). Antibodies against STAT5A (# sc-1081, Santa Cruz, CA, USA), STAT5B (# sc-835, Santa Cruz), RNA polymerase II (# ab5408, Abcam), and histone H3K4me3 (# 17-614, Millipore, Temecula, CA, USA) were used for ChIP. The ChIP DNA fragments were blunt-ended and ligated to the Illumina Indexed DNA adaptors using NEBNext ChIP-Seq Library Prep Master Mix Set for Illumina (# E6240, New England BioLabs, Ipswich, MA, USA), and sequenced using the Illumina HiSeq 2000. The single-end reads were aligned to the mouse reference genome (mm9 assembly) using the BWA program (21). The mapped reads of samples and respective input controls were analyzed using the HOMER peak calling program with default parameters (false discovery rate cutoff—0.001). Totals of 26 231 STAT5A and 6969 STAT5B peaks in wild type (AABB) and 2574 STAT5B peaks in Stat5a-null (BB) were identified as STAT5 binding sites (Figure 5). For visualization, total number of reads in each sample was normalized to 10 million. The ChIP-seq data are deposited in GEO under accession number GSE40930.
Figure 5.
Summary of genome-wide STAT5 binding sites at L1. (A) The Venn diagram shows the number of identified STAT5A and STAT5B sites (peaks) in AABB tissue and STAT5B sites in BB tissue. (B) Average peak heights of STAT5A and STAT5B in AABB and BB tissues were estimated after library size normalization (RPM, reads per 10 million, input subtracted). (C) Mean fold changes of STAT5A and STAT5B target genes in AABB tissue and STAT5B target genes in BB tissue were calculated. The genes containing STAT5 peaks within ±1 kb around TSSs were regarded as STAT5 target genes. (D) Normalized tag counts (RPM) of STAT5A, RNA polII and H3K4me3 from 200 bp around STAT5A peak centers at L1 were calculated and compared between AABB and BB. Log2-transformed values were used (x and y axes). (E) Normalized tags of H3K4me3 at positions 1 kb upstream and 2 kb downstream of TSS were summed up and divided by the size (3 kb) and then quantile normalized for comparison (top). The scatter plot shows the fold change (x-axis) and difference (y-axis) of H3K4me3 average enrichment between genes (spot) in AABB and BB. Cutoffs for significant changes were set as follows: 1.5-fold change (x-axis, AABB/BB) and four average tag difference (y-axis, AABB/BB). Among the genes showing significant changes of H3K4me3 enrichment, the number of STAT5 target and non-target genes was counted (bottom). (F) Genome browser views represent three gene loci (Wap, Csn1s2a and Stap1) showing changes of H3K4me3 level and one housekeeping gene locus (Actb).
Estimation of empirical P values
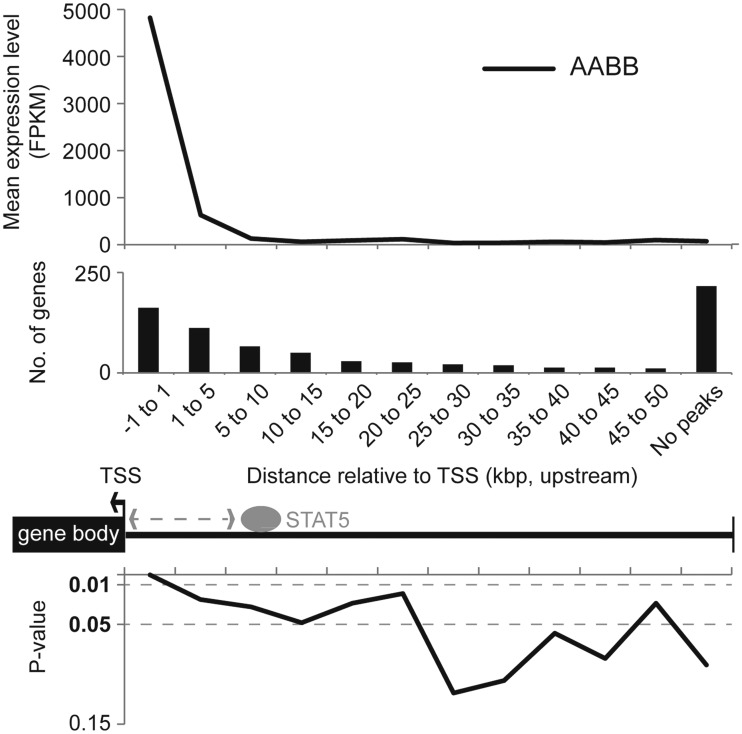
To estimate the significance of STAT5 binding distance to gene expression (Figure 6), we randomly resampled genes among all genes with replacement (the size of the resample was equal to the size of the given STAT5 target genes) and the mean expression values of the resampled set were calculated. This procedure was repeated 10 000 times for each category (defined by the distance of STAT5 binding site relative to TSS). Then, P values were empirically computed as the number of times the mean value of a randomly resampled set was greater than or equal to the observed mean expression value (22).
Figure 6.
Correlation between gene expression and STAT5 binding distance. Mean expression level of STAT5 target genes, which contained STAT5 peaks within the given position and expressed more than 2-fold (AABB/Null) is shown (top). The number of genes in each criterium is shown (middle). P values were calculated by comparing with random sets that have the same number of genes (10 000 iteration) (bottom). P values were empirically computed using the Monte Carlo algorithm (see ‘Materials and Methods’ section).
RESULTS
Generation of mammary epithelium with different Stat5a/b genotypes
This study investigated the extent to which STAT5 concentrations control mammary alveolar expansion and differentiation throughout pregnancy and the degree to which genomic STAT5A/B binding determines expression of nearby genes. Toward this end we generated mice carrying any combination of Stat5a and Stat5b alleles, thus expressing STAT5 at levels ranging from 0 to 100% (see ‘Materials and Methods’ for the nomenclature used to indicate the active Stat5 genes in these mice) and evaluated the mRNA and protein levels of STAT5 isoforms (Supplementary Figure S2). Since in mammary epithelium Stat5a mRNA levels are approximately twice as high as Stat5b levels (Supplementary Figure S2A), Stat5a-null (BB) tissue contains only ∼30% of total Stat5. Moreover, STAT5 concentrations in total mammary tissue of virgin mice in the presence of only one Stat5 allele were lower than expected (Supplementary Figure S2B), which is probably the result of blunted auto regulation (STAT5 binds to the Stat5a/b promoters as shown later). While mice with a germline deletion of the entire Stat5a/b locus displayed severe anemia and panleukopenia and died at birth (3,23), mice with only one functional allele of either Stat5a (A) or Stat5b (B) were viable and overtly normal. Mice lacking both Stat5b alleles (AA) are infertile (7), which prohibited studies on mammary tissue during pregnancy. To bypass this impediment, and to ensure that only epithelial-autonomous functions of STAT5 were investigated, mammary epithelium from mice carrying different combinations of Stat5 alleles was transplanted into wild-type hosts. Mammary tissue from these mice was analyzed at day 6 of pregnancy (p6) and at parturition (L1).
Stat5 dose-dependent mammary development
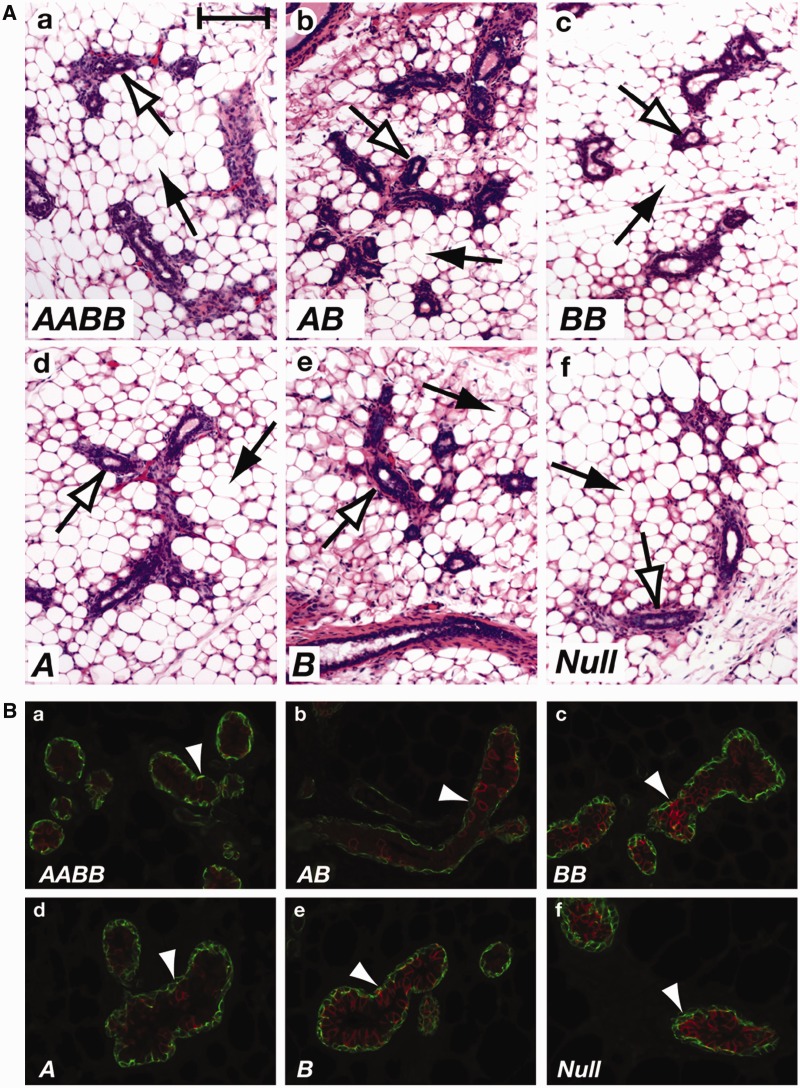
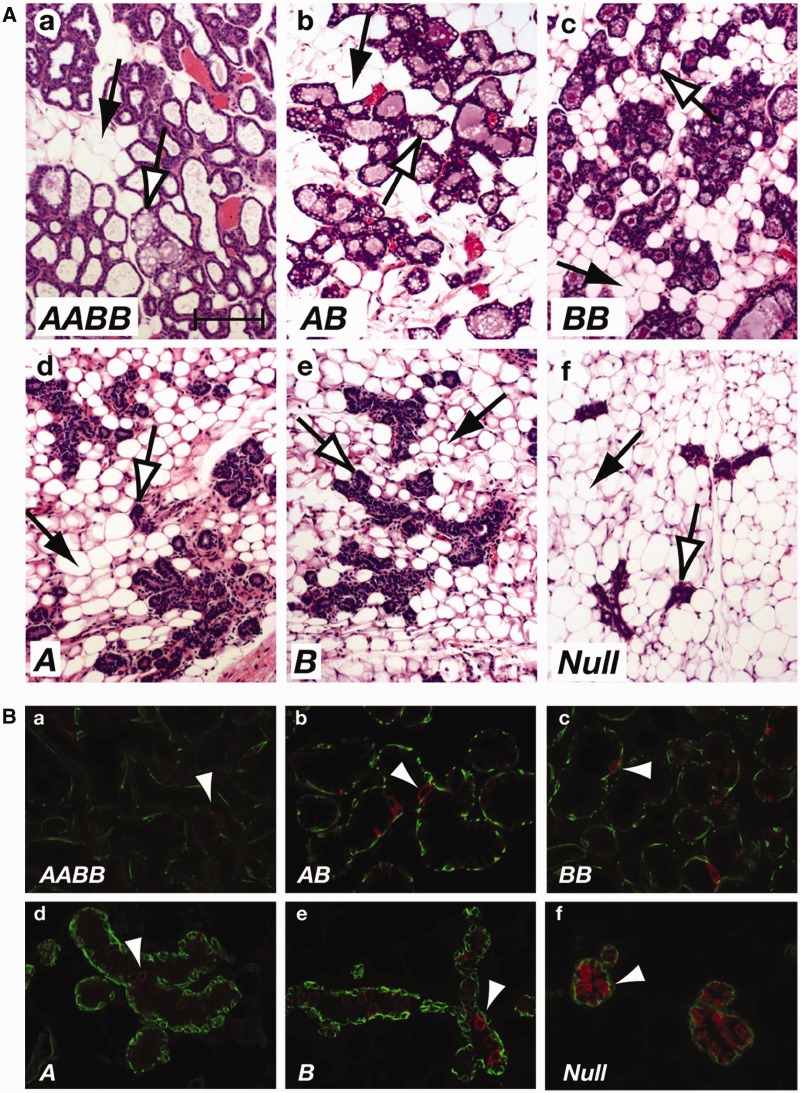
In addition to a surge in STAT5 activation, STAT5 levels increase in mammary epithelium during pregnancy (5) suggesting a specific dose requirement for progression of normal mammary development. To study dose dependency we determined the extent of epithelial development in the presence of different numbers of Stat5 alleles by assessing histological appearance and differentiation of mammary epithelium at parturition. While no bona fide alveoli developed in the complete absence of STAT5, one copy of either Stat5a or Stat5b was sufficient to promote development of small, unexpanded and histologically immature alveoli (Supplementary Figure S3; Figure 1A). The presence of two Stat5b alleles resulted in a further increase of alveolar units with small lumina. Overt differentiation, as judged by the presence of milk fat globules in secretory cells and milk secretion into the alveolar lumen, was observed in alveoli carrying two Stat5 alleles (AA, BB and AB) and complete differentiation was attained in the presence of all four Stat5 alleles. To determine to what extent STAT5 dose impacted mammary development at early pregnancy, a period of epithelial cell expansion, we analyzed tissue at day 6 of pregnancy (p6) (Figure 2A). At this stage small alveoli started to develop in all samples that expressed any level of STAT5 while Stat5-null tissue only contained undecorated ducts.
Figure 2.
Histology and IF staining for NKCC1 of mammary tissues of mice with various STAT5 dosages in early pregnancy. (A) Transplanted tissues from mice expressing Stat5a or Stat5b at various levels as indicated were harvested on day 6 of pregnancy and stained with H&E. At this stage alveolar development in all samples is sparse in all epithelial cells expressing Stat5 and is even more reduced in Null cells (f). Black arrows indicate stromal adipocytes and white arrows indicate alveolar epithelial cells. Scale bar = 80 µm. (B) Staining of the membrane transporter molecule NKCC1, which is downregulated as epithelial cells differentiate, indicates a more mature developmental stage in wild type (a) cells, intermediate maturity in cells with two or one Stat5 alleles (b and c) and strong staining in Null cells (f). Arrowheads indicate NKCC1-positive cells stained in red. Myoepithelial cells are visualized with antibodies against smooth muscle actin (green).
Next, we analyzed the presence of the sodium-potassium transporter NKCC1, a membrane protein found in ductal luminal cells in virgin mice that is down regulated during pregnancy concomitant with alveolar differentiation to gauge the differentiation status of mammary epithelia (24). At parturition NKCC1 was observed sporadically in the presence of all four Stat5 alleles or two alleles (Figure 1B), which is typical for cells that have attained a differentiation status equivalent to late pregnancy. In contrast, epithelial cells expressing only one Stat5 allele retained expression of NKCC1 in clusters of cells, indicative of an immature developmental state. Strong NKCC1 staining in the absence of all Stat5 alleles is indicative of a complete lack of differentiation. In contrast, at p6 the majority of alveolar cells in the samples with less than four Stat5 alleles contained higher levels of NKCC1 than wild-type AABB tissue (Figure 2B).
Figure 1.
Histology and IF staining for NKCC1 of mammary tissues of mice with various STAT5 dosages at parturition. The nomenclature of mice with the different genotypes is based on the alleles they have retained. We refer to wild-type mice and Stat5abfl/fl mice as AABB mice; Stat5abfl/fl;MMTV-Cre (with Stat5ab-deficient mammary epithelial cells) as Null mice; Stat5a−/− mice as BB mice; Stat5b−/− mice as AA mice; Stat5ab+/null mice as AB mice. Mice carrying only a single functional allele of either Stat5a (Stat5abnull/Stat5b−) or Stat5b (Stat5abnull/Stat5a−) are referred to as A mice and B mice, respectively. (A) Transplanted mammary tissues obtained from mice of different genotypes were collected on the day of parturition and analyzed by histology. Alveoli are expanded and filled with milk in the presence of four (a) and two (b and c) Stat5 alleles. Epithelial cells with only one active Stat5 allele (d and e) form dense alveoli lacking signs of secretory activity. Black arrows indicate stromal adipocytes and white arrows indicate alveolar epithelial cells. Scale bar = 80 µm. (B) Mammary tissues of transplanted epithelia obtained from mice of different genotypes were collected at parturition and sections were stained with anti-NKCC1 antibody (red) and α-smooth muscle actin (green). Arrowheads indicate NKCC1-positive cells stained in red. Myoepithelial cells are visualized with antibodies against smooth muscle actin (green).
To assess the correlation between STAT5 activity at the cellular level and the number of Stat5 alleles, we performed immunofluorescence (IF) for pSTAT5 at different stages of mammary gland development. As we monitored the development of mutant mammary epithelium embedded in the stroma of wild-type mice, an analysis of total mammary tissue by western blotting would not have been appropriate as the stroma derived from the wild-type host also contributes to pSTAT5 and STAT5 signals. First, we tested the acute cytokine response of STAT5 in mammary tissue carrying different genotypes. Virgin mice were injected with prolactin and tissue was stained for pSTAT5 (Supplementary Figure S4A). As expected, the amount of nuclear pSTAT5 was reduced according to the number of Stat5 alleles. Compared with control (AABB) tissue, there was weaker staining in tissues from BB and AB mice, which was further reduced in A and B tissues. No pSTAT5 positive cells were present in Stat5-null tissue. At parturition strong nuclear pSTAT5 was observed in the vast majority of alveolar epithelial cells from mice carrying all four Stat5 alleles (Supplementary Figure S4B). Nuclear pSTAT5 was also observed in AB epithelium and to a lesser extent in BB epithelium. Notably, as shown in Figure 1A, alveolar development also occurred in BB tissue and RNA-seq data (shown later) further demonstrated that this level of active STAT5 was sufficient to induce differentiation. The relationship between Stat5 genotypes and pSTAT5 IF at p6 was similar to that observed in virgin mice injected with prolactin (Supplementary Figure S4C).
STAT5 dependent gene expression at lactation
Having observed that histologically immature mammary alveoli can form in the presence of only one Stat5 allele and that morphologically discernible differentiation was attained in the presence of two alleles, we next asked to what extent gene expression programs in mammary tissue are dependent on the concentration of STAT5, both at parturition (L1) and early pregnancy (p6). Toward this end we performed a global assessment of gene expression by Illumina high-throughput RNA-seq analyses of wild type and Stat5-mutant mammary tissues at L1 (GSE37646). The analysis of total mammary tissue from germline Stat5a-null mice displays the specificity and validity of this experimental approach (Supplementary Figure S5). While in wild type tissue sequence tags were obtained for all exons, no tags were found over deleted exons in Stat5a-null tissue (BB). The sequence tags in Stat5-null mammary epithelium embedded in control mice stem from the wild-type stroma. To avoid any confounding systemic influences induced by the absence of STAT5, such as ovarian insufficiency, we again used mutant tissue that had been transplanted into cleared fat pads of wild-type hosts. As this study examined mutant mammary epithelium embedded in control stroma, differences in gene expression can be attributed to changes in the epithelial component. We analyzed total mammary tissue and decided not to enrich mammary epithelium as this requires extended enzymatic treatments that would alter the mRNA composition and thus not reflect the in situ situation.
Milk and its components are the defining characteristic of lactation. We found that at least 70% of mRNAs in differentiated mammary alveoli encode <10 milk protein species. Therefore, the presence of a few abundant mRNAs dilutes mRNA levels of other genes in fully differentiated control tissue compared with undifferentiated tissues expressing only one or no Stat5 allele (Supplementary Figure S1). To correct for the dilution effect, we recalculated the FPKM values after filtering out genes contributing more than 5% of total reads in each sample (see ‘Materials and Methods’ for a detailed description). Upon applying additional filters we identified a set of 750 genes under STAT5 control, i.e. genes, whose expression at parturition (L1) in the presence of all four Stat5 alleles was at least 2-fold higher than in the absence of STAT5 (Supplementary Table S2). This set includes genes encoding milk proteins and proteins controlling cell metabolism and secretion. Their expression range extended over more than four orders of magnitude and STAT5-dependent induction was between 2- and 700-fold. However, this number of STAT5-dependent genes is probably somewhat overestimated as the ratio of aveolar and ductal epithelium differs between mammary tissues with those two genotypes contributing to differences in gene expression. On the other hand, expansion of immature alveoli occurs in mammary tissue carrying only one Stat5b allele (B), which translates to ∼5–10% STAT5 levels, making it a better comparison to wild-type tissue. Expression of ∼400 genes was elevated at least 3-fold in AABB tissue compared with B tissue.
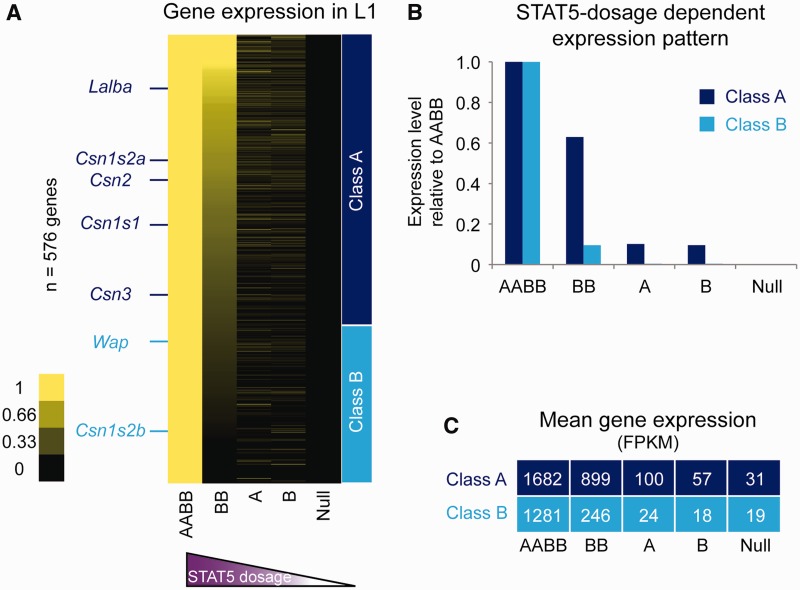
We next asked to what extent expression of these genes was dependent on the Stat5 genotype and thus on the concentration of STAT5. In addition to mammary tissue with all four Stat5 alleles (AABB) and Stat5-null tissue (null) we analyzed tissue with two STAT5 alleles (AB, BB) and with one allele (A, B) at L1 using RNA-seq. Of particular importance was tissue from Stat5a-null (BB) mice as they exhibit overtly normal mammary function. Clustering analysis revealed distinct gene classes that differentially responded to various STAT5 dosages (Supplementary Table S2; Figure 3). Class A includes the majority of milk protein genes, such as Csn1s1, Csn1s2a, Csn2, Csn3 and Lalba. Their expression was detectable in the absence of STAT5 and surged with increasing STAT5 levels. While absolute expression of these genes ranged between 1000 and 100 000 FPKM, induction by a full complement of STAT5 was between 10- and 700-fold. Notably, a cutback of STAT5 by ∼70% (BB tissue) resulted in a ∼50% reduced expression of these genes, and a drop by ∼90% (B tissue) yielded reduced expression by ∼98%. This demonstrates the presence of a threshold level of STAT5 permitting the highly induced expression during pregnancy of these genes. The much smaller class B that includes Wap and Csn1s2b was more strongly dependent on the levels of STAT5 for their expression. A decrease of STAT5 by ∼70% (BB tissue) resulted in an ∼85 and 99.5% reduced expression of the Wap and Csn1s2b genes, respectively (Figure 3B). In general, expression of class B genes was lower than that of class A genes (Figure 3C).
Figure 3.
STAT5 dose-dependent gene expression. (A) A total of 750 significantly induced genes by STAT5 were initially clustered according to gene expression patterns. Genes (174) that showed irrelevant expression patterns were not shown and 576 genes were categorized into two classes. (B) Mean gene expression of the classified genes was calculated at different STAT5 dosages and normalized relative to that of AABB tissue (wild type, set to 1). (C) Mean gene expressions of class A and class B at different STAT5 dosages were shown.
Detectable expression of most, if not all, mammary specific genes in the absence of STAT5 demonstrated that STAT5 does not convey their cell specificity but rather augments their expression during pregnancy. Although histologically mammary alveoli in the presence of only one copy of Stat5a or Stat5b appeared to be undifferentiated, gene expression patterns obtained from these tissues indicated the emergence of a partial differentiation signature (Supplementary Table S2). Genes in this group encode several transcription factors, including ELF5 and ID2, membrane transporters, gap junction and milk proteins. Several mammary-specific genes, such as Csn2, were induced up to 5-fold in the presence of one Stat5a allele, which amounts to ∼10 –15% of total STAT5. In contrast, other target genes, such as Csn1s2b and Wap, did not respond to this STAT5 concentration.
STAT5 controlled gene expression in early pregnancy
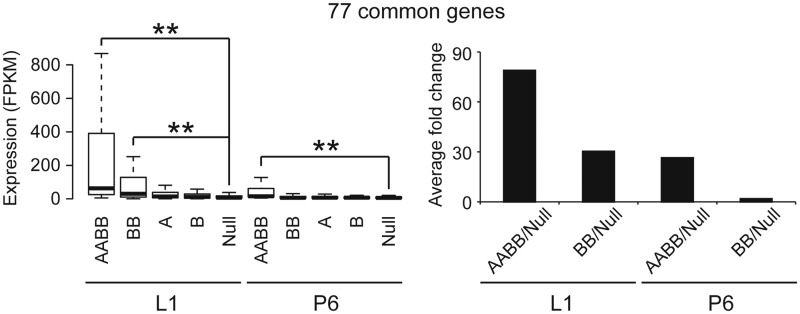
While at the end of pregnancy the majority of STAT5 target genes is already highly expressed in mammary tissue with two Stat5b alleles (30% STAT5) the STAT5 dose requirements at early pregnancy, when alveolar epithelium commences to differentiate, is not known. In addition, while most, if not all, STAT5-induced genes at parturition could be linked to milk production, lipid metabolism and secretion, the identification of STAT5 target genes at early pregnancy might unveil candidates for the regulation of cell proliferation and initial stages of differentiation. Therefore, we established the transcriptome of mammary tissues obtained from mice at day 6 of pregnancy (p6). Upon applying the same filtering criteria a set of ∼370 genes was induced more than 2-fold in control (AABB) compared with Stat5-null tissue (Supplementary Table S3). 77 of these genes were shared between the p6 and L1 samples and encoded mainly milk proteins and other mammary-specific or mammary-enriched differentiation markers. While at L1 many of these 77 common genes were induced more than 30-fold in the presence of only two Stat5b alleles, a full set of Stat5 was required at p6 (Supplementary Table S3; Figure 4). Moreover, expression of these genes at p6 reached only ∼2–3% of the levels seen at the onset of lactation (Supplementary Table S1). This demonstrates that while STAT5 levels during early pregnancy are a limiting factor in the activation of differentiation-specific genetic programs, high STAT5 concentrations at the onset of lactation result in a more widespread activation of target genes. Of note, the degree of pSTAT5 parallels the number of Stat5 alleles in mammary epithelium (Supplementary Figure S4).
Figure 4.
Expression pattern of STAT5-induced genes common to day 6 of pregnancy and day 1 of lactation. Expression levels (measured by FPKM) of 77 common genes at each STAT5 dosage in P6 and L1 are shown in box plots. The minimum, first quartile, median, third quartile and mean values were used (left panel). Average fold change of gene expression was calculated as the expression level at full dosage of STAT5 (AABB) over the expression level in the absence of STAT5 (Null). P values were calculated using the Wilcoxon–Mann–Whitney test. **P-value < 0.001.
A set of ∼250 genes under apparent STAT5 control was unique to mammary tissue at p6 (Supplementary Table S3). This group included several transcription factors, such as GATA3, MYC, RELB and CITED1, some of which have been linked to mammary development. Thus, analysis of differentially activated genes at early pregnancy probably enriches for proteins controlling the expansion and early differentiation steps of mammary epithelium.
Linking genome-wide STAT5 binding and RNA polymerase II to gene expression
To identify STAT5-regulated genes induced during pregnancy that were also bound by STAT5 we performed ChIP-seq analyses for STAT5A and STAT5B at parturition in wild-type (AABB) mammary tissue. We also included Stat5a-null (BB) tissue in this analysis as the presence of only STAT5B permits overtly normal mammary development, lactation appears to be unaffected in outbred mouse strains and gene expression was similar to AABB tissue. Thus, genes bound by STAT5B in BB mammary tissue should reflect genuine STAT5 targets required for normal development and differentiation. At parturition mammary epithelium has been subjected to a full complement of pregnancy signals for 19 days, including prolactin as a key activator of STAT5, and saturated STAT5 binding can be inferred. In total, more than 26 000 STAT5A peaks were identified in control but not in Stat5a-null mammary tissue (Figure 5). The ∼6873 STAT5B peaks almost completely overlapped with the STAT5A peaks in wild-type tissue suggesting that there are no high affinity binding sites restricted to STAT5B. In general, the average STAT5A peak height was 5-fold higher than that of STAT5B (Figure 5B), which is probably the result of a greater abundance of STAT5A and possibly higher antibody affinity. This difference would explain the excess of STAT5A over STAT5B peaks. Up to 15% of the genes bound by STAT5A and 36% of the genes bound by STAT5B were under STAT5 control as measured by RNA-seq of wild type and Stat5-null mammary tissue (Supplementary Figure S6). This demonstrates that STAT5 binding is no definitive predictor of the associated gene being under STAT5 control. The 2395 STAT5B binding sites in Stat5a-null and 6873 STAT5B binding sites in wild-type mammary tissues, which probably reflect high affinity STAT5 binding sites, were a better predictor for gene activation. Indeed, mean expression fold changes of the genes targeted by STAT5B in Stat5a-null mammary tissue were higher than those of the others (Figure 5C).
We also analyzed genome-wide RNA polymerase II (RNA polII) and H3K4me3 marks. In general, genes bound by STAT5 were also marked by both RNA polII and H3K4me3 indicating that they were poised for expression. The level of STAT5 binding was positively correlated with the enrichment level of RNA PolII and H3K4me3 near promoter regions (Supplementary Figure S7). Looking in more detail, a cutback of STAT5 by ∼70% (BB) resulted in a decreased level of RNA polII but not H3K4me3 near the STAT5 binding sites as overall spots shifted toward AABB (Figure 5D). However, comparison of H3K4me3 enrichment on promoter regions between AABB and BB tissues revealed that some STAT5 target and non-target genes showed a significant decrease of H3K4me3 in BB tissue (Figure 5E; Supplementary Table S4). Manual evaluation of several loci including STAT5 target genes (Wap and Csn1s2a), and a non-STAT5 target gene (Stap1) confirmed the decrease of H3K4me3 marks, which did not occur over the housekeeping gene, Actb (Figure 5F). The relationship between STAT5 and histone modifications remains to be investigated.
Out of the 750 genes identified as being regulated during pregnancy and at least partially dependent on the presence of STAT5, 532 genes were bound by STAT5A at 1218 positions in their flanking regions (+1 kb ∼ TSS ∼ −50 kb) of transcription start sites (TSSs) (Supplementary Table S1). In total, 50% of these STAT5A peaks coincided with gamma interferon activated sequence (GAS) motifs (TTCnnnGAA). In 31, 21 and 48% of these genes STAT5A binding was obtained within 1, 5 and 50 kb upstream regions, respectively. In general, STAT5A peaks in these genes coincided with weaker STAT5B peaks. Next, we determined whether the position of STAT5 binding within these gene loci was correlated with their absolute expression and/or the degree of STAT5 dependence. The vast majority of genes with the highest expression were bound by STAT5 within promoter proximal sequences (Figure 6). Genes expressed at low levels were bound by STAT5 at sequences beyond −10 kb and these levels were similar to genes that were not recognized by STAT5. Since promoter proximal, but not distal, STAT5 binding induced strong gene induction we propose that STAT5 is a promoter-centric transcription factor and probably less of an enhancer component. Moreover, genes with the highest induction levels bound STAT5 preferentially within the promoter proximal region. Out of the 42 genes, whose expression was induced more than 100-fold in the presence of STAT5, 21 were recognized by STAT5 within 1 kb of promoter sequence. In contrast, out of the 506 genes induced between 2 - and 5-fold <14% fulfilled these criteria (Supplementary Table S5).
Stat5-null mammary epithelium fails to form differentiated alveoli during pregnancy and it could thus be argued that a comparison with AABB tissue is not completely representative. Therefore, we identified genes differentially expressed between AABB (100% STAT5) and B (5–10% STAT5) mammary tissue at L1 with the same approach as described and analyzed their ability to be recognized by STAT5. Approximately 632 genes were differentially expressed between AABB and B tissue at L1 (Supplementary Table S6). Again, the ones with the highest expression and greatest induction were bound by STAT5 within promoter proximal sequences.
Comparative analysis of STAT5-bound genes
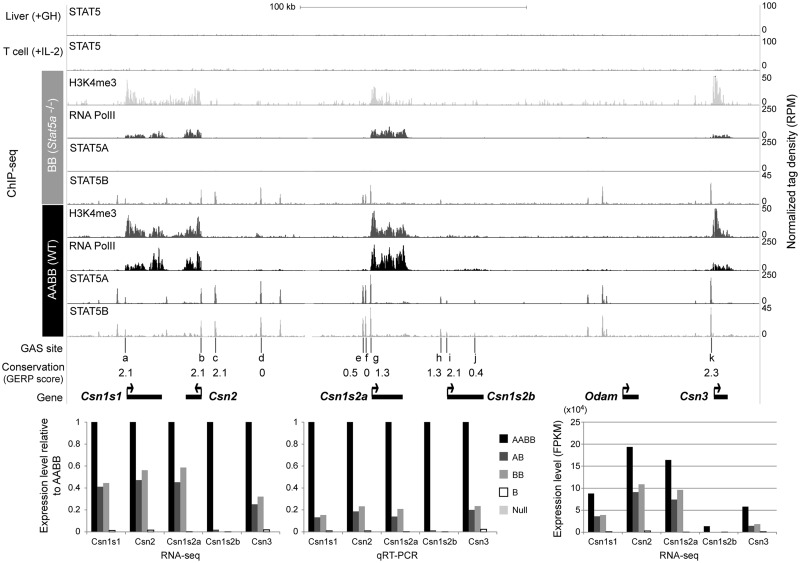
It is not clear to what extent STAT5 binding to specific genes controls their overall expression and the degree of activation in different cell types. To address this question we analyzed STAT5 bound genes expressed either specifically in mammary tissue or in several cell types (Table 1). We first focused on the casein locus, which encodes five major milk proteins, whose expression is largely restricted to differentiating mammary epithelium (Figure 7). STAT5 recognized 11 sequences within this 250 kb locus, all of which coincided with GAS motifs. Based on public databases, no STAT5 binding was observed in liver tissue (14) or T cells (12) where these genes are not expressed. Each casein promoter was marked by H3K4me3 marks and RNA polII binding. The Csn1s2b gene, which is expressed at ∼10% of other caseins, is characterized by smaller STAT5 and H3K4me3 peaks and less RNA polII binding. The Csn2 and Csn1s2a genes, which encode the two most abundant mRNAs in this locus, are separated by ∼75 kb and their transcription faces into opposite direction. STAT5 bound to seven sites in this region, two close to the Csn2 gene and three near the Csn1s2a gene. One or more STAT5 peaks that coincided with H3K4me3 marks and RNA polII loading were also detected over the other Casein gene promoters. In general, only promoter proximal GAS motifs bound by STAT5 were conserved between species, suggesting that other sites, although bound by STAT5 might have less physiological relevance (see ‘Discussion’). No STAT5A binding was detected in BB tissue and STAT5B peaks appeared to be unaltered. However, reduced intensity of H3K4me3 marks and RNA polII binding on the Csn1s1, Csn2, Csn1s2a and Csn3 genes in BB tissue reflected their slightly reduced expression. The expression levels determined by RNA-seq were confirmed by qRT-PCR (Figure 7). Although STAT5B binding was detected at normal levels, the Csn1s2b gene was silent in the absence of STAT5A, suggesting that STAT5B binding is below the threshold required to activate gene expression. These experiments demonstrate that within a given locus containing several STAT5 regulated genes, binding intensity directly reflects gene expression levels. In addition, the degree of H3K4me3 is dependent on STAT5 levels at least in part, suggesting that STAT5 may contribute to the establishment of histone modifications.
Table 1.
STAT5 regulation of and binding to known target genes
| Gene | Expression (FPKM)a |
Fold change | Sum of STAT5A peak heights (relative to TSS)b |
||||
|---|---|---|---|---|---|---|---|
| AABB | Null | AABB/Null | Mammary tissue |
T cells | Liver | ||
| +1 to −1 | −1 to −10 | +1 to −10 | +1 to −10 kb | ||||
| Csn2 | 193 548 | 558 | 347 | 154 | 175 | No | No |
| Csn1s2b | 136 10 | 12 | 1133 | 35 | 72 | No | No |
| Wap | 109 772 | 327 | 335 | 115 | 283 | No | No |
| Cish | 34 | 7 | 5 | 357 | No | Yes | Yes |
| Socs3 | 1 | 6 | 0 | 99 | 109 | Yes | Yes |
| Bcl6 | 3 | 33 | 0 | 81 | 37 | Yes | Yes |
| Stat5a | 56 | 8c | 7 | No | 97 | No | No |
aExpression level was measured by RNA-seq at day 1 of lactation.
bTotal number of mapped reads in each sample was normalized to 10 million (input subtracted).
cStat5a transcripts from wild-type stroma.
Figure 7.
STAT5 binding and chromatin features of the casein gene cluster. Genome browser tracks represent enrichment of STAT5A, STAT5B, H3K4me3 and RNA PolII in wild type (AABB) and Stat5a-null mammary tissues as well as STAT5 in liver and T cells. The liver and T-cell STAT5 ChIP-seq data sets were obtained from previous studies (GSE31578 and GSE36890). Conservation of GAS motifs (TTCnnnGAA) was calculated using the GERP score (the higher score means higher conservation) (47). Expression level of five milk protein genes was measured by both RNA-seq and qRT-PCR (bottom left). Absolute expression level of the milk protein genes is shown (bottom right).
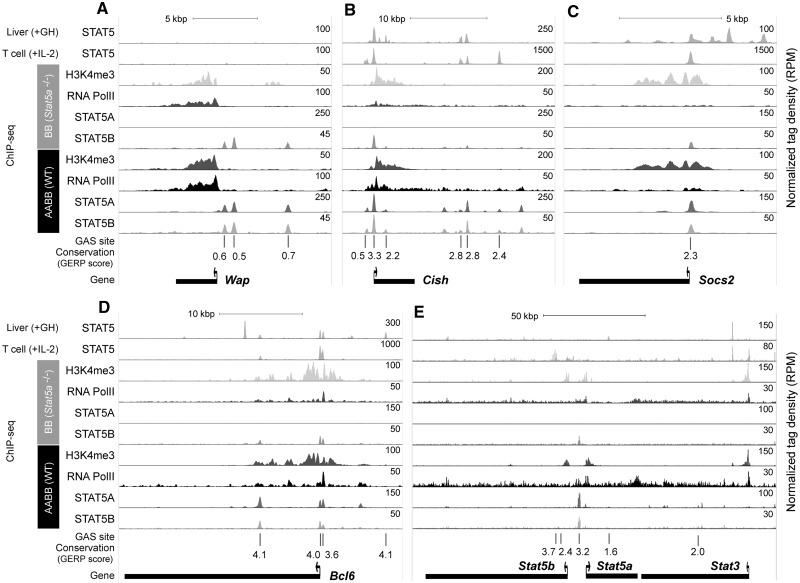
The Wap gene represents a class of STAT5 target genes that requires a STAT5 threshold of ∼30% to attain high expression during lactation. Three strong STAT5A binding peaks were detected in the promoter region, which also was decorated by extensive H3K4me3 marks (Figure 8A). As this gene is not expressed in T cells and in liver it was not surprising to see an absence of STAT5 binding in these tissues. Cish, a well-known STAT5 target genes in a diverse set of cell types, is expressed at fairly low levels in mammary tissue but induced 5-fold by STAT5. The strong STAT5 peak over the promoter in mammary tissue coincided with peaks observed in T cells and liver (Figure 8B). Additional binding, some of it cell-specific, was detected 3′ of the gene. Socs2, another well-established STAT5 target gene in many cell types, is expressed at very low levels in mammary tissue and no activation by STAT5 was observed. However, strong STAT5 peaks over GAS motifs in the promoter were detected in mammary tissue and other cell types (Figure 8C). As a representative of the rare class of genes, whose expression is apparently repressed by STAT5 we analyzed the Bcl6 gene (Figure 8D). Again, strong binding over the promoter was observed in mammary tissue, liver and T cells. The strong intronic binding site was conserved between mammary tissue and liver. Finally, we analyzed the STAT5 locus and identified STAT5A and STAT5B binding in the promoter region shared by both isoforms (Figure 8E). We propose that this binding is the cause of a STAT5 auto regulatory loop. Indeed, STAT5 levels in the presence of only one copy of Stat5a or Stat5b were less than predicted based on the genotype.
Figure 8.
STAT5 binding and chromatin features of STAT5 target genes. Genome browser tracks represent enrichment of STAT5A, STAT5B, H3K4me3 and RNA PolII in wild type (AABB) and Stat5a-null mammary tissues as well as STAT5 in liver and T cells. The liver and T-cell STAT5 ChIP-seq data sets were obtained from previous studies (GSE31578 and GSE36890). Conservation of GAS motifs (TTCnnnGAA) was calculated using the GERP score (the higher score means more conserved). (A) Wap gene, (B) Cish gene, (C) Socs2 gene, (D) Bcl6 gene and (E) Stat5a/b genes.
These examples emphasize that STAT5 binding to promoter sequences is neither an indicator for the overall expression of the respective genes nor does it reflect STAT5-inducibility (Table 1). However, it is clear from comparative studies that pregnancy-induced and STAT5-regulated genes expressed mainly in mammary tissue are not recognized by STAT5 in non-expressing cell types, suggesting a chromatin structure that prevents STAT5 access.
DISCUSSION
The question whether the concentration of a specific transcription factor and its binding location determine its ability to activate specific genetic and biological programs remains a central issue in biology. Using mouse genetics and genome-wide analyses we now demonstrate that a distinct number of Stat5 alleles, and therefore specific STAT5 concentrations, which are reflective of the corresponding nuclear pSTAT5 levels, elicit specific biological and genetic responses in mammary tissue during pregnancy. Thus, mammary development and function depend on prolactin-induced activation of STAT5 and on an increased concentration of STAT5 during pregnancy. Moreover, there is a close relationship between the number of Stat5 alleles and the levels of pSTAT5, and its ability to activate gene classes during pregnancy. We propose a two-stage model of STAT5 dose-dependent mammary gland development. While low concentrations of STAT5 ensure the expansion of alveolar epithelium, high levels are required for cells to proceed through a differentiation program that culminates in milk production. Moreover, our findings suggest that the concentration of STAT5 is a rate-limiting factor in the control of mammary development during pregnancy. If STAT5 levels were not a rate-limiting factor, reducing the number of Stat5 alleles should have had less of an impact on the development and differentiation of alveoli.
Correlating number of Stat5 alleles with STAT5 and pSTAT5 levels
In mammary tissue, ∼70% of total Stat5 mRNA consists of Stat5a and 30% of Stat5b. Although it can be anticipated that a similar ratio will exist for the respective protein levels, this cannot be fully verified as anti STAT5A and anti STAT5B antibodies have different affinities. Reducing the number of Stat5 alleles from four to two results in a slight reduction of the corresponding protein levels. However, STAT5 protein levels in the presence of only one Stat5 allele are lower than expected, which most likely is due to a positive feedback loop. In fact, ChIP seq experiments demonstrated that STAT5A binds GAS motifs in sequences in the shared Stat5a and Stat5b gene promoters.
Furthermore, pSTAT5 levels in mammary epithelial cells, as measured by IF on histological sections, correlated with the number of Stat5 alleles. The cellular heterogeneity of pSTAT5 staining during pregnancy likely reflects the unique differentiation status of individual cells. It is well established that differentiation of mammary epithelial cells during pregnancy is asynchronous (25).
STAT5 in pregnancy-dependent transcription programs
Transcription factor haploinsufficiencies have been reported as the underlying cause of several disorders, emphasizing that the concentration of transcription factors is critical for the establishment of biological programs. In mammary tissue, haploinsufficiency has been observed for the PRLR and the transcription factor ELF5, demonstrating critical threshold levels needed for normal development during pregnancy (26,27). In the presence of only one allele of either the Prlr or Elf5, mammary tissue fails to fully differentiate, which reflects the reduced expression of JAK2/STAT5 target genes (28–30). While insufficient PRLR levels could directly impact the extent of STAT5 activation, reduced ELF5 concentration might result in impaired alveolar epithelial development downstream from or in parallel with STAT5. Elf5 is bound by STAT5 and depends on it for expression suggesting that ELF5 is a downstream executor of STAT5.
Genomic responses to STAT5 largely depend on its concentration and the developmental stage of the tissue. While at lactation the overwhelming majority of STAT5 target genes encoded proteins related to the production of milk and secretion, genes activated during pregnancy were enriched for transcription factors and other regulatory proteins potentially regulating mammary epithelial cell expansion and differentiation. Our in vivo studies at normal and reduced STAT5 levels correlate well with in vitro overexpression studies using a constitutively active STAT5 that is able to force differentiation of several cell types in the absence of cytokine signaling. Specifically, ectopic expression of constitutively active STAT5 in mammary epithelial cells induced alveolar development and differentiation (31–36). In the hematopoietic system intermediate levels of a constitutively active STAT5 were sufficient to turn on genes driving long-term expansion, but maximal STAT5 levels were needed to ensure a shift toward erythroid differentiation (36). This demonstrates the concentration dependent ability of STAT5 to activate specific gene classes.
The expression levels of genes under STAT5 control span four orders of magnitude, with milk protein genes expressed at 5000-fold higher levels than non-mammary specific target genes, such as Cish. This difference might not be exclusively the result of transcriptional changes but also include post-transcriptional events, such as mRNA stabilization (37). Currently, the contribution of post-transcriptional events for different classes of STAT5 target genes is not understood. However, transgenes composed of milk protein gene promoters linked to heterologous sequences can be expressed at similar levels to endogenous milk protein genes (38), suggesting the fundamental contribution of promoter elements in determining expression levels.
Several direct STAT5 target genes encode transcription factors and other regulatory proteins. Gene knockout mice for some of them exist and their phenotypes are complete or partial phenocopies of STAT5 loss, suggesting that these genes execute STAT5 function. Among them are the transcription factors ELF5 (27,29,30), ID2 (39), GATA3 (40) and IRX2. CIDEA, an integral component of milk fat globules, also appears to be a transcription co-factor activating the Xdh gene that encodes xanthine dehydrogenase, an enzyme required for milk lipid secretion (41,42). Our study now reveals that both genes are genuine STAT5 targets.
Productive and opportunistic STAT5 binding
The strong correlation between the location of STAT5 binding with respect to the TSS and overall gene expression and induction levels suggests that STAT5 functions as transcription factor only when bound to promoter proximal sequences. Even strong STAT5 binding to sequences beyond −5 kb had only a small impact on the overall expression and regulation of the respective genes during pregnancy. Classically, the functional relevance of putative transcription factor binding sites, identified through sequence motifs, has been studied using in vitro systems in which TF binding sites were analyzed out of context. Since in vivo only those binding events close to the TSS have a disproportionally high impact, it is ambiguous to define the physiological significance of distal GAS motifs or even distal STAT5 binding site based on short sequence motifs. Our findings linking TF binding location with gene expression levels agree with the model developed by Fraenkel and colleagues that predicts and confirms that proximity of TF binding to the TSS greatly affects overall gene expression (43). We estimate that ∼80% of genes bound by STAT5 are not under its regulation and the biological significance of these in vivo interactions is not known. The lack of correlation between STAT5 binding and transcriptional activation of nearby genes is not unique to mammary epithelium as we have observed a similar phenomenon in an experimental system based on mouse embryonic fibroblasts overexpressing STAT5 (44).
Comparative analyses of STAT5 ChIP assays and ChIP-seq data sets from different cell types highlight both common and cell-specific STAT5 binding patterns. STAT5-regulated genes expressed exclusively in mammary epithelium were not recognized by STAT5 in non-mammary cell types (12,14), thus demonstrating cell-specific accessibility of these loci. Selective DNase I hypersensitivity patterns further emphasize cell-specific conformation of mammary loci (45). A common denominator of genes that are under STAT5 control in diverse cell types appears to be STAT5 binding to conserved promoter sequences. Additional cell-specific STAT5 islands were detected on several genes, including Cish, Bcl6 (14) and Igf1 (10,11,13,46) suggesting the possibility of modular regulation. Specifically, STAT5 bound to the promoter shared by Stat5a and Stat5b only in mammary cells demonstrating an auto regulatory loop as evidenced by the reduced STAT5 levels in cells expressing only one Stat5 allele.
This study has provided not only mechanistic insight into the fundamental role of STAT5 in mammary epithelial development during pregnancy and the activation of gene expression programs but has also opened new perspectives and questions. Most notably, the biological relevance, if any, of strong STAT5 binding to genes that are not under STAT5 control remains to be investigated. Moreover, some gene sets are bound by STAT5 across various cell types but are subject to cell-specific STAT5 control. Finally, our findings that the degree of RNA polymerase binding and H3K4me3 marks parallel STAT5 binding levels at genes under STAT5 control suggests that transcription factors have the capacity to influence promoters and not just access open chromatin.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–6 and Supplementary Figures 1–7.
FUNDING
The Intramural Research Programs (IRP) of NIDDK at the National Institutes of Health (NIH), USA. Funding for the open access charge: the World Class University Program, Ministry of Education, Science and Technology, through the National Research Foundation of Korea, South Korea [R31-10069]; WCU Research Center, Dankook University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the Intramural Research Program (IRP) of NIDDK, NIH. D.Y.: Experimental design, mouse experiments, RNA-seq and ChIP-seq experiments, data analysis, writing manuscript; K.K.: bioinformatics data analysis, writing paper; G.W.R.: experimental design, mouse experiments, data analysis, writing manuscript; L.H.: experimental design, data analysis, writing manuscript. We thank Dr. Harold Smith (NIDDK Genomics core) for sequencing RNA-seq and ChIP-seq libraries.
REFERENCES
- 1.Wakao H, Gouilleux F, Groner B. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO J. 1995;14:854–855. doi: 10.1002/j.1460-2075.1995.tb07064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat. Rev. Mol. Cell. Biol. 2005;6:715–725. doi: 10.1038/nrm1714. [DOI] [PubMed] [Google Scholar]
- 3.Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, Robinson GW, Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell. Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamaji D, Na R, Feuermann Y, Pechhold S, Chen W, Robinson GW, Hennighausen L. Development of mammary luminal progenitor cells is controlled by the transcription factor STAT5A. Genes Dev. 2009;23:2382–2387. doi: 10.1101/gad.1840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X, Robinson GW, Hennighausen L. Activation of Stat5a and Stat5b by tyrosine phosphorylation is tightly linked to mammary gland differentiation. Mol. Endocrinol. 1996;10:1496–1506. doi: 10.1210/mend.10.12.8961260. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 7.Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, Waxman DJ, Davey HW. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc. Natl Acad. Sci. USA. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, Robinson GW, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc. Natl Acad. Sci. USA. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer RD, Laz EV, Su T, Waxman DJ. Male-specific hepatic Bcl6: growth hormone-induced block of transcription elongation in females and binding to target genes inversely coordinated with STAT5. Mol. Endocrinol. 2009;23:1914–1926. doi: 10.1210/me.2009-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chia DJ, Rotwein P. Defining the epigenetic actions of growth hormone: acute chromatin changes accompany GH-activated gene transcription. Mol. Endocrinol. 2010;24:2038–2049. doi: 10.1210/me.2010-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chia DJ, Varco-Merth B, Rotwein P. Dispersed Chromosomal Stat5b-binding elements mediate growth hormone-activated insulin-like growth factor-I gene transcription. J. Biol. Chem. 2010;285:17636–17647. doi: 10.1074/jbc.M110.117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin JX, Li P, Liu D, Jin HT, He J, Ata Ur Rasheed M, Rochman Y, Wang L, Cui K, Liu C, et al. Critical Role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function. Immunity. 2012;36:586–599. doi: 10.1016/j.immuni.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rotwein P. Mapping the growth hormone—Stat5b—IGF-I transcriptional circuit. Trends Endocrinol. Metab. 2012;23:186–193. doi: 10.1016/j.tem.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Laz EV, Waxman DJ. Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol. Cell. Biol. 2012;32:880–896. doi: 10.1128/MCB.06312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, Li M, Furth PA, Hennighausen L. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C, Salzberg SL. How to map billions of short reads onto genomes. Nat. Biotechnol. 2009;27:455–457. doi: 10.1038/nbt0509-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 19.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 20.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.North BV, Curtis D, Sham PC. A note on the calculation of empirical P values from Monte Carlo procedures. Am. J. Hum. Genet. 2002;71:439–441. doi: 10.1086/341527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, Li D, Durum SK, Jiang Q, Bhandoola A, et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc. Natl Acad. Sci. USA. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shillingford JM, Miyoshi K, Robinson GW, Bierie B, Cao Y, Karin M, Hennighausen L. Proteotyping of mammary tissue from transgenic and gene knockout mice with immunohistochemical markers: a tool to define developmental lesions. J. Histochem. Cytochem. 2003;51:555–565. doi: 10.1177/002215540305100501. [DOI] [PubMed] [Google Scholar]
- 25.Robinson GW, McKnight RA, Smith GH, Hennighausen L. Mammary epithelial cells undergo secretory differentiation in cycling virgins but require pregnancy for the establishment of terminal differentiation. Development. 1995;121:2079–2090. doi: 10.1242/dev.121.7.2079. [DOI] [PubMed] [Google Scholar]
- 26.Ormandy CJ, Camus A, Barra J, Damotte D, Lucas B, Buteau H, Edery M, Brousse N, Babinet C, Binart N, et al. Null mutation of the prolactin receptor gene produces multiple reproductive defects in the mouse. Genes Dev. 1997;11:167–178. doi: 10.1101/gad.11.2.167. [DOI] [PubMed] [Google Scholar]
- 27.Zhou J, Chehab R, Tkalcevic J, Naylor MJ, Harris J, Wilson TJ, Tsao S, Tellis I, Zavarsek S, Xu D, et al. Elf5 is essential for early embryogenesis and mammary gland development during pregnancy and lactation. EMBO J. 2005;24:635–644. doi: 10.1038/sj.emboj.7600538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris J, Stanford PM, Sutherland K, Oakes SR, Naylor MJ, Robertson FG, Blazek KD, Kazlauskas M, Hilton HN, Wittlin S, et al. Socs2 and elf5 mediate prolactin-induced mammary gland development. Mol. Endocrinol. 2006;20:1177–1187. doi: 10.1210/me.2005-0473. [DOI] [PubMed] [Google Scholar]
- 29.Naylor MJ, Oakes SR, Gardiner-Garden M, Harris J, Blazek K, Ho TW, Li FC, Wynick D, Walker AM, Ormandy CJ. Transcriptional changes underlying the secretory activation phase of mammary gland development. Mol. Endocrinol. 2005;19:1868–1883. doi: 10.1210/me.2004-0254. [DOI] [PubMed] [Google Scholar]
- 30.Oakes SR, Naylor MJ, Asselin-Labat ML, Blazek KD, Gardiner-Garden M, Hilton HN, Kazlauskas M, Pritchard MA, Chodosh LA, Pfeffer PL, et al. The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Genes Dev. 2008;22:581–586. doi: 10.1101/gad.1614608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong J, Tong T, Reynado AM, Rosen JM, Huang S, Li Y. Genetic manipulation of individual somatic mammary cells in vivo reveals a master role of STAT5a in inducing alveolar fate commitment and lactogenesis even in the absence of ovarian hormones. Dev. Biol. 2010;346:196–203. doi: 10.1016/j.ydbio.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iavnilovitch E, Cardiff RD, Groner B, Barash I. Deregulation of Stat5 expression and activation causes mammary tumors in transgenic mice. Int. J. Cancer. 2004;112:607–619. doi: 10.1002/ijc.20484. [DOI] [PubMed] [Google Scholar]
- 33.Iavnilovitch E, Groner B, Barash I. Overexpression and forced activation of stat5 in mammary gland of transgenic mice promotes cellular proliferation, enhances differentiation, and delays postlactational apoptosis. Mol. Cancer Res. 2002;1:32–47. [PubMed] [Google Scholar]
- 34.Schepers H, van Gosliga D, Wierenga AT, Eggen BJ, Schuringa JJ, Vellenga E. STAT5 is required for long-term maintenance of normal and leukemic human stem/progenitor cells. Blood. 2007;110:2880–2888. doi: 10.1182/blood-2006-08-039073. [DOI] [PubMed] [Google Scholar]
- 35.Vafaizadeh V, Klemmt P, Brendel C, Weber K, Doebele C, Britt K, Grez M, Fehse B, Desrivieres S, Groner B. Mammary epithelial reconstitution with gene-modified stem cells assigns roles to Stat5 in luminal alveolar cell fate decisions, differentiation, involution, and mammary tumor formation. Stem Cells. 2010;28:928–938. doi: 10.1002/stem.407. [DOI] [PubMed] [Google Scholar]
- 36.Wierenga AT, Vellenga E, Schuringa JJ. Maximal STAT5-induced proliferation and self-renewal at intermediate STAT5 activity levels. Mol. Cell. Biol. 2008;28:6668–6680. doi: 10.1128/MCB.01025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guyette WA, Matusik RJ, Rosen JM. Prolactin-mediated transcriptional and post-transcriptional control of casein gene expression. Cell. 1979;17:1013–1023. doi: 10.1016/0092-8674(79)90340-4. [DOI] [PubMed] [Google Scholar]
- 38.Devinoy E, Thepot D, Stinnakre MG, Fontaine ML, Grabowski H, Puissant C, Pavirani A, Houdebine LM. High level production of human growth hormone in the milk of transgenic mice: the upstream region of the rabbit whey acidic protein (WAP) gene targets transgene expression to the mammary gland. Transgenic Res. 1994;3:79–89. doi: 10.1007/BF01974085. [DOI] [PubMed] [Google Scholar]
- 39.Miyoshi K, Meyer B, Gruss P, Cui Y, Renou JP, Morgan FV, Smith GH, Reichenstein M, Shani M, Hennighausen L, et al. Mammary epithelial cells are not able to undergo pregnancy-dependent differentiation in the absence of the helix-loop-helix inhibitor Id2. Mol. Endocrinol. 2002;16:2892–2901. doi: 10.1210/me.2002-0128. [DOI] [PubMed] [Google Scholar]
- 40.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vorbach C, Scriven A, Capecchi MR. The housekeeping gene xanthine oxidoreductase is necessary for milk fat droplet enveloping and secretion: gene sharing in the lactating mammary gland. Genes Dev. 2002;16:3223–3235. doi: 10.1101/gad.1032702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W, Lv N, Zhang S, Shui G, Qian H, Zhang J, Chen Y, Ye J, Xie Y, Shen Y, et al. Cidea is an essential transcriptional coactivator regulating mammary gland secretion of milk lipids. Nat. Med. 2012;18:235–243. doi: 10.1038/nm.2614. [DOI] [PubMed] [Google Scholar]
- 43.MacIsaac KD, Lo KA, Gordon W, Motola S, Mazor T, Fraenkel E. A quantitative model of transcriptional regulation reveals the influence of binding location on expression. PLoS Comput. Biol. 2010;6:e1000773. doi: 10.1371/journal.pcbi.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu BM, Kang K, Yu JH, Chen W, Smith HE, Lee D, Sun HW, Wei L, Hennighausen L. Genome-wide analyses reveal the extent of opportunistic STAT5 binding that does not yield transcriptional activation of neighboring genes. Nucleic Acids Res. 2012;40:4461–4472. doi: 10.1093/nar/gks056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rijnkels M, Kabotyanski E, Montazer-Torbati MB, Hue Beauvais C, Vassetzky Y, Rosen JM, Devinoy E. The epigenetic landscape of mammary gland development and functional differentiation. J. Mammary Gland Biol. Neoplasia. 2010;15:85–100. doi: 10.1007/s10911-010-9170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chia DJ, Young JJ, Mertens AR, Rotwein P. Distinct alterations in chromatin organization of the two IGF-I promoters precede growth hormone-induced activation of IGF-I gene transcription. Mol. Endocrinol. 2010;24:779–789. doi: 10.1210/me.2009-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.