Abstract
AtCyp59 is a multidomain cyclophilin containing a peptidyl-prolyl cis/trans isomerase (PPIase) domain and an evolutionarily highly conserved RRM domain. Deregulation of this class of cyclophilins has been shown to affect transcription and to influence phosphorylation of the C-terminal repeat domain of the largest subunit of the RNA polymerase II. We used a genomic SELEX method for identifying RNA targets of AtCyp59. Analysis of the selected RNAs revealed an RNA-binding motif (G[U/C]N[G/A]CC[A/G]) and we show that it is evolutionarily conserved. Binding to this motif was verified by gel shift assays in vitro and by RNA immunopreciptation assays of AtCyp59 in vivo. Most importantly, we show that binding also occurs on unprocessed transcripts in vivo and that binding of specific RNAs inhibits the PPIase activity of AtCyp59 in vitro. Surprisingly, genome-wide analysis showed that the RNA motif is present in about 70% of the annotated transcripts preferentially in exons. Taken together, the available data suggest that these cyclophilins might have an important function in transcription regulation.
INTRODUCTION
Cyclophilins are ubiquitous proteins with a peptidyl-prolyl cis/trans isomerase (PPIase) activity and have important functions in protein folding (1). Typically they are small single-domain proteins but some of them have accessory domains. AtCyp59 is a member of the cyclophilin family which consists of 29 genes in Arabidopsis (2). AtCyp59 is unusually complex as it consists of a PPIase domain, an RRM motif, a Zn-knuckle and a charged C-terminal domain with RS/RD repeats (arginine/serine and arginine/aspartate) (3). It is an evolutionarily highly conserved protein present from Schizosaccharomyces pombe to humans, but the Zn-knuckle is a plant-specific addition. It was first described in Paramecium tetraurelia as a protein involved in cell morphogenesis (4).
The Arabidopsis protein Cyp59 was isolated in a yeast two-hybrid screen with plant SR (serine, arginine) proteins which are an important and conserved family of splicing factors (3). Deletion analysis showed that the C-terminal domain is indispensable for interacting with the SR proteins in vitro. AtCyp59 is a nuclear protein, but it does not significantly co-localize with SR proteins in nuclear speckles. Instead, its punctuate localization pattern resembles transcription initiation sites. In line with these observations, it was shown that AtCyp59 resides within a complex with the C-terminal repeat domain (CTD) of the largest subunit of the RNA polymerase II. These results suggested a possible function for AtCyp59 at the interface of transcription and splicing (3). It is now widely accepted that most splicing events occur co-transcriptionally whereby the CTD domain of RNA polymerase II plays very important roles in both transcription and RNA processing (5) and recently reviewed in (6). In general, the CTD acts as a binding platform for various protein factors during transcription, and at the same time recruits pre-mRNA processing proteins to the nascent transcripts (7,8). The CTD of most eukaryotes consists of a variable number of heptapeptide repeats (YSPTSPS) which undergo dynamic phosphorylation/dephosphorylation events on serine residues and thereby determine the course of transcription and binding of RNA processing factors. These include capping, splicing and polyadenylation factors whose dynamics are tightly coordinated with transcription (9,10). The severe growth effect upon overexpression of AtCyp59 and the fact that no mutants are available point to an essential function for this protein (3). The involvement of this class of cyclophilins in the transcription process was corroborated by experiments with the S. pombe orthologue Rct1, which showed that Rct1 is an essential gene, is recruited to actively transcribed genes and its deregulation affected CTD phosphorylation and RNA polymerase II transcription (11). However, the mechanism of how these cyclophilins function in the transcription cycle is unknown. Possible scenarios are that they might act directly on the CTD structure and therefore influence phosphorylation/dephosphorylation of the heptapeptide or they might act on kinases/phosphatases which are regulating the CTD. It is worth mentioning that other smaller PPIases have been shown to be important for correct CTD conformation and phosphorylation thereby influencing both transcription and RNA processing (12–16).
One of the most interesting features of AtCyp59 is its complex domain structure. PPIases are usually small proteins, but a few other complex cyclophilins have been described. For example, in the splicing complex several RS-domain-containing cyclophilins have been investigated in Arabidopsis and mammals suggesting a role in RNA processing (12,17,18). Other RRM-containing cyclophilins have been found but their functions are mostly not determined. Among them, the best described protein is hCyp33, a regulator of a histone acetyl transferase; however, the function of RNA binding is not well defined (19,20). Interestingly, the RRM domain of the multidomain AtCyp59 is evolutionarily highly conserved and is in fact the most conserved feature of this protein. This RRM has been shown to bind RNA with preferences to G and C bases (3). These data indicated an important contribution of the RRM to the activity of AtCyp59 raising the question of what are its RNA target(s) and what influence RNA binding has on cyclophilin activity. Approaches to these questions are not trivial as the tight regulation of AtCyp59 strongly hindered in vivo approaches for determining RNA targets.
In this article, we describe the identification of an RNA-binding motif for AtCyp59 by a genomic SELEX method using an Arabidopsis genomic RNA library. Binding to this motif was verified by electrophoretic mobility shift analyses (21) in vitro and by RNA immunoprecipitation experiments in vivo. Interestingly, the identified motif is present in about 70% of the annotated genes in Arabidopsis indicating that binding of AtCyp59 to mRNA might be a general feature of most RNA polymerase II transcripts. This is supported by the conservation of this motif in S. pombe. In addition, we have shown activity of the PPIase domain of AtCyp59 and importantly its regulation by binding to specific RNAs. Considering the evolutionary conservation of the RNA-binding motif and the known characteristics of this protein, these data indicate a function for AtCyp59 in the transcription cycle.
MATERIALS AND METHODS
Genomic SELEX
Using random priming, a representative library of the Arabidopsis thaliana genome was constructed containing overlapping sequences from 50 to 300 nt in length. Library fragments were generated using the method and adaptors described in (22). Each library fragment contained fixed primers suitable for PCR amplification and preceding T7 promoter sequence at 5′-end for in vitro RNA transcription.
For selection of binding RNAs, we incubated recombinant AtCyp59_RRM_Zn domain protein with the in vitro transcribed genomic RNA pool for 30 min at room temperature using neutral PBS buffer conditions (2 mM MgCl2, 0.5 mM DTT and 0.135 M NaCl, 27 mM KCl, 8 mM Na2HPO4, 2 mM NaH2PO4, pH = 7.5). Separation of bound and unbound fractions was performed via GST-tagged AtCyp59_RRM_Zn on 4B glutathione sepharose blocked with 100 µg tRNA (Sigma-Aldrich). Recovery of AtCyp59_RRM_Zn-binding RNAs was performed via urea-mediated denaturation followed by phenol/chloroform extraction. Selected sequences were amplified via reverse transcriptase-polymerase chain reaction (RT–PCR) as described (23). For the next SELEX cycle, obtained PCR products were again in vitro transcribed into RNA. In total, 10 cycles of in vitro selection were performed using molar ratio RNA:protein = 3:1 (or 10:1 on later cycles). On the ninth cycle the control (anti-GST selection) was performed using 10:1 molar ratio of recombinant GST over RNA library. Binding reaction was done as described above with the only change of keeping the RNA fraction unbound to the beads instead of beads fraction. Next, 1/10 of the selected library was cloned via T/A cloning to the pGEM-T easy vector and 386 clones were sequenced using Sanger sequencing method. The rest of the selected library was sequenced using 454 sequencing technology. Plant material preparation, plasmids and protein purification are described in Supplementary Methods.
Computational analysis of the SELEX library
The raw sequencing reads have been submitted to analysis with APART pipeline (24). For the adaptor filtering step, the following sequences have been used: AGGGGAATTCGGAGCGGGGCAGC (5′-adaptor), CATCCCAGCCCCGAGGAT (3′-adaptor). For downstream steps, only sequences with both adaptors were analyzed. All parameters of APART have been set to default, except the minimum number of reads per contig which has been changed to 2. For identification of the AtCyp59-binding motif consensus sequences, the seven contigs with the highest read number (accounting for 79.5% of all mapped reads) have been used. The motif identification has been performed using the MEME program (25) with the following non-default settings: motif width 7, minimum number of sites 4 and maximum number of sites 15.
Genome-wide distribution of the AtCyp59-binding motif
For the analysis of the AtCyp59-binding motif at a genome-wide scale, a motif descriptor based on experimentally verified motif variants has been constructed. The search has been performed using the glam2scan (26) software with the default parameter set and score cut-off 7. The sequences mapped to A. thaliana correspond to TAIR10 genome assembly (27). The occurrence of the 17 motifs experimentally validated in Arabidopsis was analysed in the transcription units (TUs). If more than one TU was annotated for a gene, the longest one was chosen for the motif search. Only the TUs of protein-coding genes were used in the analysis (TAIR10). As a control, the distribution of two scramble motifs (‘TAGCGTC’ and ‘CATGTGC’) was analyzed in the protein-coding genes of Arabidopsis. The same analysis was done for the TUs in S. pombe.
Additionally, a motif search was performed in TUs of different sizes in Arabidopsis. Small TUs were defined as TUs with a size between 500 and1000 nt. Medium TUs were defined with a size range between 1001 and 2000 nt and big TUs with a size range between 2001 and 4000 nt. Binomial tests were used for the P-value calculations.
Electrophoretic mobility shift assays
Synthetic 7 nt RNA oligonucleotide (Sigma-Aldrich) (see Table 1) (100 nM) or longer RNAs (see Supplementary Methods and Supplementary Table S3) was incubated on ice for 20 min in 10 mM HEPES-KOH, pH 7.9, 10 mM MgCl2, 50 mM KCl, 1 mM DTT, 0.025% Nonindet P-40, supplemented with protease inhibitor cocktail (Roche) and RNAse inhibitor (Promega) with various amounts of recombinant AtCyp59, AtCyp59_RMM_Zn, AtCyp59_3M_RRM_Zn, Rct1 protein as indicated. RNA–protein complexes were separated on 6–10% native polyacrylamide gels at 3 V/cm. Gels were stained with SYBR GREEN II dye and visualized on a Phosphoimager (Thyphoon 900) and quantified using ImageQuant 1.4 software. Assay was performed three times with independent protein purifications.
Table 1.
Range of dissociation constants of 17 different 7-nt RNA variants of the binding motif found in genomic SELEX and AtCyp59_RRM_Zn domain based on gel shift assay
| Sequence | Kd AtCyp59_RRM_Zn (nM) | Kd AtCyp59 (nM) |
|---|---|---|
| GGUGCCG | 40 ± 10 | 120 ± 25 |
| GUGGCCG | 40 ± 10 | 120 ± 25 |
| GUCGCCG | 40 ± 10 | 120 ± 25 |
| GUAGCCA | 105 ± 25 | 120 ± 25 |
| GCUGCCG | 105 ± 25 | 120 ± 25 |
| GAUGCCA | 105 ± 25 | 120 ± 25 |
| GACGCCA | 105 ± 25 | 120 ± 25 |
| GCCGCCG | 200 ± 35 | 120 ± 25 |
| GUUGCCG | 200 ± 35 | 120 ± 25 |
| GUAGCCG | 200 ± 35 | 120 ± 25 |
| GCGGCCG | 200 ± 35 | 120 ± 25 |
| GUCGCCA | 300 ± 40 | 120 ± 25 |
| GCCGCCA | 300 ± 40 | 120 ± 25 |
| GAUGCCG | 300 ± 40 | 120 ± 25 |
| GUGGCCA | 300 ± 40 | 120 ± 25 |
| GGAGCCA | 300 ± 40 | 120 ± 25 |
| GCGGCCA | 300 ± 40 | 120 ± 25 |
Preparation of whole-cell extracts from protoplasts and immunoprecipitation
Arabidopsis cell suspension protoplasts were isolated and transformed with pDEDH-AtCyp59-HA, pDEDH-Cyp59_3M-HA, pDEDH-GFP or pGREEN-MAPK6-HA as described (17). For double transformation experiments, Arabidopsis protoplasts were transformed with equal concentration of the following plasmid pairs: pDEDH-AtCyp59-HA/pDEDH-atSR34a; pDEDH-AtCyp59-HA/pDEDH-Mut_atSR34a; pDEDH-AtCyp59-HA/pDEDH-atRS2Z32 and pDEDHAtCyp59-HA/pDEDH-Mut_atRS2Z32. Transformed protoplasts were collected 24 h after transformation (15 min, 700g), frozen in liquid nitrogen, and resuspended in 300 μl (per 10 million protoplasts) protoplasts extraction buffer [50 mM HEPES-KOH pH 7.9, 2.5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 1% sodium dodecyl sulphate (SDS)], supplemented with EDTA-free protease inhibitor cocktail (Roche Diagnostics) and RNase inhibitor (Roche Diagnostics). Suspension was sonicated three times for 6 s and incubated on ice for 20 min with occasional mixing. After centrifugation (15 min, 14 000 rpm, 4°C), concentration of SDS in extracts was adjusted to 0.1% with SDS-free lysis buffer. Extracts were incubated for 1 h with magnetic Dynal beads (m-270 epoxy; Invitrogen), coupled with anti-HA antibodies produced in mouse (HA-7, monoclonal; Sigma-Aldrich) on rotary shaker at 4°C and then were washed three times with protoplast extraction buffer without SDS and three times with washing buffer (10 mM HEPES-KOH, pH 7.9). RNA was extracted via protein digest with 100 μg proteinase K (Sigma-Aldrich) and 5 μl 10% SDS in 400 μl washing buffer for 30 min at 55°C followed by phenol/chloroform extraction and ethanol precipitation. RNA was purified by DNAse I (Qiagen) treatment followed by Qiagen RNeasy Plant Mini kit. Purified RNA was used directly in RT–PCR (33 PCR cycles) or stored at −80°C. RT reaction was performed with equal volume of RNA sample, 15-mer oligo-dT or pre-mRNA primers targeting a nascent pre-mRNA after the polyA signal and M-MLV reverse transcriptase (Promega) according to the manufacturing instruction and then was used in standard PCR with Phusion polymerase (Finnzyme). Primers for target genes used in PCR are listed in the Supplementary Table S3.
PPIase activity assay
The PPIase activity of recombinant GST-AtCyp59 or GST-AtCyp59_3M was performed as described (28) by using the tetrapeptide substrate Suc-AAPF-pNA (N-succinyl-Ala-Leu-Pro-Phe p-nitroanilide; Sigma-Aldrich). All reagents were pre-equilibrated until the temperature reached 4°C. In a 1-ml glass cuvette, 800 nM GST-AtCyp59 or GST-AtCyp59_3M was mixed with 100 μl of α-chymotrypsin (Sigma-Aldrich; 60 mg/ml in 1 mM HCl), and the volume was adjusted to 975 μl with assay buffer (50 mM Hepes-KOH, pH 8.0 at 0°C, 100 mM NaCl, 2 mM MgCl2, 1 mM EDTA). The reaction was initiated by the addition of 25 μl of substrate (4 mM tetrapeptide in 470 mM anhydrous LiCl dissolved in trifluoroethanol). Changes in absorbance due to released p-nitroaniline were monitored at 390 nm at 0°C over a 3-min period in a PerkinElmer Lambda 35 UV/VIS spectrophotometer with a thermostatically controlled cuvette holder. To check PPIase activity of proteins in presence of RNA, 800 nM GST-AtCyp59 or GST-AtCyp59_3M was pre-incubated in reaction buffer with equal concentration of 7 nt RNA (GUGGCCG), polyA+ fraction of total RNA (800 nM) isolated from 21-day-old wt Col-0 plants using Micropoly(A) purist kit (Ambion). Pre-incubated protein was similarly used in the assay above. The experiments were performed five times with different preparations of proteins.
RESULTS
RNA targets of AtCyp59 identified by Genomic SELEX
The systematic evolution of ligands by exponential enrichment (SELEX) is an approach to isolate high-affinity binding partners for a given molecule and usually uses a random nucleic acid library (29,30). In contrast, Genomic SELEX has the advantage of selecting only from the sequences available in a given genome which enhances the possibility of isolating natural RNA targets (31,22). In vivo studies on AtCyp59 are hindered by its tight gene regulation with the consequence that Arabidopsis overexpression and mutant lines are currently not available. We therefore used Genomic SELEX as an in vitro method to identify RNAs bound by AtCyp59 and to discover RNA motif(s) for this protein.
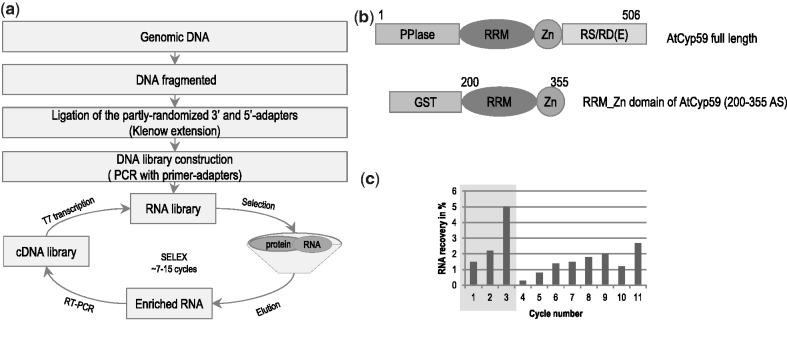
To create a genomic library 2 g of 3-week-old wild-type A. thaliana (Col-0) leaves were used to isolate genomic DNA. Thirty micrograms of this DNA was fragmented and used as template to create a DNA library by Klenow fragment reaction. The addition of primers with constant regions and T7 promoter sequences allowed the synthesis of an RNA library (Figure 1a). The sequences of primer adaptors were not present in the Arabidopsis genome. Library construction was performed in such a way as to create an RNA library with sizes of 30–300 nucleotides. Library construction and the selection of RNA targets were essentially done as described (22) and a more specific description for Arabidopsis is published elsewhere (32). For the selection procedure, the region encoding the RRM and Zn-knuckle domain of AtCyp59 was cloned and expressed as a recombinant GST-fusion protein. Binding to the RNA library was performed in a 3:1 molar excess of RNA over protein for the initial first three cycles of selection and then the ratio was changed to 10:1 to increase the stringency. Bound RNAs were separated on protein-GST beads and eluted with glutathione. To control for RNAs binding to the GST tag, a counter selection with GST protein was performed after the ninth cycle (Figure 1b). The selection was stopped after 11 rounds of SELEX when the recovery rate of RNA was about 3% of the input RNA. As a 10-fold molar excess of RNA over protein was used for competition, this value suggests that about 30% of the selected RNA pool is able to bind to AtCyp59.
Figure 1.
Genomic SELEX experiment with the multidomain cyclophilin AtCyp59_RRM_Zn domain. (a) Scheme of Arabidopsis RNA library preparation and selection process. (b) Schematic representation of the domain organization of the full-length AtCyp59 and GST-tagged construct of the RRM+Zn motif of AtCyp59. PPIase, peptidyl/prolyl cis/trans isomerase domain; RRM, RNA recognition motif; Zn, zinc finger motif class CCHC; RS/RD(E), domain enriched in arginine, serine, aspartate, glutamate; GST, glutathione S-transferase. (с) Enrichment of the RNA sequences from the Arabidopsis thaliana genome which bind AtCyp59. The RNA recovery was calculated as a percentage of the RNA material that was bound to the protein relatively to the amount of the initial RNA material in each cycle. During Cycles 1–3, the molecular ratio of the RNA to protein was 3:1, Cycles 4–9, 11 the ratio was 10:1, and Cycle 10 anti-GST selection.
To control the library construction and SELEX procedure, selected RNAs were reverse transcribed, cloned and sequenced by conventional sequencing. Most of the reads contained Arabidopsis sequences of about 40–50 nucleotides (Supplementary Figure S1b). Therefore, the selected library was sequenced by the 454 deep sequencing methods. We obtained a total of about 13 375 trimmed reads and about 70% mapped to the Arabidopsis genome (Supplementary Table S1). From these sequences almost 60% mapped uniquely. The sequences that are more than once mapped are probably affiliated to duplicated genomic regions. We also sequenced the genomic library with 454 technology and compared the abundance of genomic elements to the Arabidopsis genome and the SELEX library (Supplementary Figure S1c). All genomic elements were present in the genomic library albeit with a slightly different abundance, whereas the SELEX library showed a clear increase in exonic sequences and a decrease in intronic sequences.
Statistical examination of the selected sequences reads revealed that most of the targets of AtCyp59 reside in protein-coding genes (Figure 2a). Interestingly, the majority of reads were observed to map in antisense orientation to annotated genes, mostly due to the genomic location of two contigs with the highest read number (accounting for 50% of all reads). When reads were assembled into contigs, the ratio between contigs in sense and antisense annotations was 3:4 (Figure 2b). By investigating the gene structure of genes which contain selected sequences in the sense orientation, we discovered that binding occurs preferentially to exons of the annotated transcripts and only few hits corresponded to introns (Figure 2c). As the Genomic SELEX method samples the entire sequence of a given genome in an unbiased manner, the obtained data clearly show that the potential targets of AtCyp59 are mainly located in protein-coding genes with a high preference for exonic regions.
Figure 2.

Bioinformatics analysis of the selected library. (a) Genomic distribution of the mapped reads generated after selection with AtCyp59_RRM_Zn domain. (b) Strand distribution of the aligned contigs according to the annotated gene. (c) Localization of the reads within the annotated gene in sense orientation. (d) Predicted binding motif based on sequences alignment of the seven most abundant clusters in the SELEX library using the MEME program tool.
Bioinformatics analysis reveals an RNA-binding motif for AtCyp59
One of the purposes of any SELEX experiment is to find a common binding motif for a protein of interest within the pool of selected sequences. The seven most abundant clusters (accounting for 79.5% of all aligned reads) in our selection were the basis for the alignment with the MEME program, which searches for consensus binding motifs (33). The obtained consensus sequence G[U/C]N[G/A]CC[A/G] (Figure 2d) is clearly GC-rich which is in line with previously published data indicating that AtCyp59 prefers either G- or C-rich sequences (3). This G[U/C]N[G/A]CC[A/G] motif was highly enriched in the selected sequences (4.5 times/per 1000 nt) and about 50% of the sequenced reads contained the motif (Supplementary Table S2). As the motif discovery was based on 79.5% of the reads, there might be a possibility of another potential binding motif. It is well established that the composition of exons in general is more GC-rich and in particular the Arabidopsis exons of pre-mRNAs are biased towards a higher GC content (34). Therefore, our findings fitted well to the observation that sequencing reads were preferentially aligned to exons of protein-coding genes. Taken together, these results support the notion that the RNA targets of AtCyp59 are located within the exons of protein-coding genes.
In vitro verification of AtCyp59 binding to the selected motif
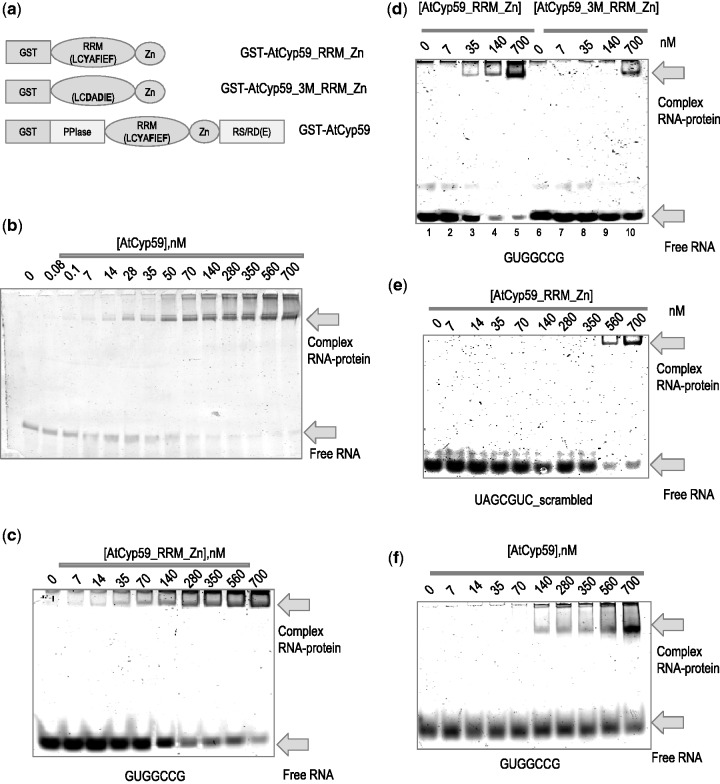
To verify the binding of AtCyp59 to its RNA targets, electrophoretic mobility shift assay with the GST-tagged version of the RRM and Zn-knuckle domain used in the selection process were performed. To control for unspecific binding, the RNP1 motif of the RRM was mutated in three conserved aromatic amino acids which are indispensable for RNA recognition (Y286D, F289D, F291D) (35). In addition, the full-length protein was expressed, purified and used along with the truncated version (Figure 3a).
Figure 3.
Binding of the AtCyp59 to its RNA targets in vitro. (a) Schematic representation of AtCyp59 protein variants used in the in vitro studies. The amino acids involved in RNA binding to the RRM (RNP1 motif) domain are marked in bold. These amino acids have been mutated in AtCyp59_3M_RRM_Zn. (b) 6% native polyacrylamide gel electrophoresis of a 180-nt RNA (gene At3G53500) containing one of the binding motifs (GCUGCCG). (c) 10% PAA gel electrophoresis of a 7-nt RNA sequence (GUGGCCG) incubated with an increasing concentration of the AtCyp59_RRM_Zn. (d) Comparison of binding to the 7-nt RNA (GUGGCCG) between wild-type AtCyp59_RRM_Zn protein (lanes 1–5) and the mutated RRM domain (lanes 6–10). (e) 10% native polyacrylamide gel electrophoresis of a 7-nt scrambled RNA (based on consensus sequence) incubated with an increasing concentration of the of AtCyp59_ RRM_Zn. (f) RNA–protein complex formation upon full-length AtCyp59 binding to the 7-nt RNA (GUGGCCG) on 6% PAA native gel. (b–f) The free RNA and RNA–protein complexes are indicated by arrows. Gels were stained with SYBR GREEN II dye. The concentration of the protein is shown in the nM range.
An initial test used one of the sequences obtained in the genomic SELEX screen, a 180-nt RNA from the AT3G53500 gene (coding for the Arabidopsis SR protein At-RS2Z32) containing one of the binding motifs (GUCGCCG). Full-length AtCyp59 protein was added in increasing amounts and the binding reaction was separated on a native PAA gel. As shown in Figure 3b, prominent complex formation between AtCyp59 and the RNA with a KD of 50 nM was observed suggesting that the sequence contained a functional binding motif.
The binding motif for AtCyp59 G[U/C]N[G/A]CC[A/G] was used to search the sequencing reads and 17 sequence variations of the seven nucleotide motif were generated (Table 1, Supplementary Figure S1). All these 17 short RNA oligonucleotides were used for binding tests in an electrophoretic mobility shift assay on 10% native gels. Table 1 shows all tested RNA oligonucleotides listed according to their binding affinities with a KD in the range of 40–300 nM. No significant preference for a particular nucleotide at the three variable positions could be observed, except that a G at the last position is preferable. By using one of these motifs, GUGGCCG, and the RRM_Zn-knuckle domain of AtCyp59, we observed strong binding with a KD of 40 nM (Figure 3c). In contrast, when the mutated protein was used, binding dropped significantly to >700 nM (Figure 3d, lanes 6–10). This value is similar in range to that obtained with a scrambled RNA oligonucleotide (UAGCGUC) bound to the non-mutated AtCyp59_RRM_Zn protein (Figure 3e). Thus, the RNP1 motif of the RRM domain is at least partially responsible for binding RNA. As the RRM is the most conserved domain in this cyclophilin family, we tested the S. pombe orthologue Rct1 in this binding assay using the Arabidopsis motif variant GUGGCCG. Interestingly, we observe similar binding of this oligo RNA to the Rct1 protein as to the Arabidopsis protein (Supplementary Figure S2c and d). These experiments strongly support an evolutionarily conserved function for the RRM of this family of cyclophilins.
To investigate the possible influence of the other domains of AtCyp59 on binding, the same experiments were performed using the full-length AtCyp59 protein. Interestingly, all the tested motif variants now showed a very similar dissociation constant of about 120 nM (Table 1 and Figure 3f). This levelling of the binding affinities to the 7-nt oligos by the other domains of AtCyp59 suggests that the motif variants might bind equally well to the full-length protein. However, as can be observed in the case of the GUCGCCG motif in the context of the AT3G53500 (At-RS2Z32) gene (Figure 3b and Supplementary Figure S2a and b) yielding a KD of 50 nM, the RNA sequence context within the transcript might have an additional influence on binding to a particular motif. In summary, the in vitro experiments have verified the binding motif for AtCyp59 from sequences selected by the Genomic SELEX method and have shown that this motif is evolutionarily highly conserved.
RNA transcripts containing the selected binding motif are bound by AtCyp59 in vivo
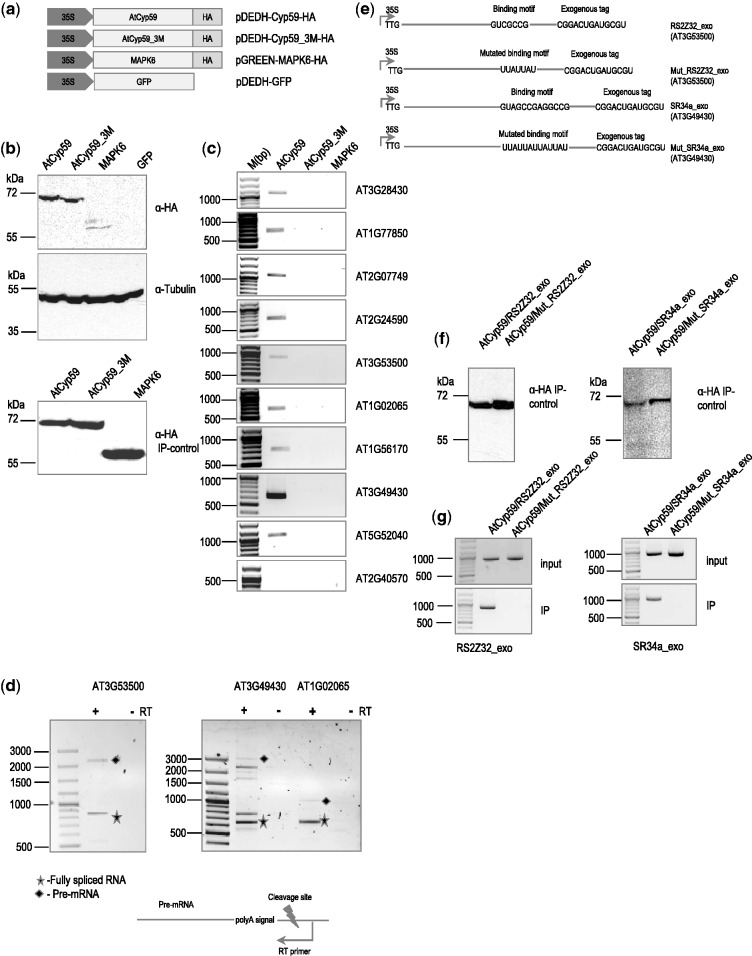
The in vivo testing of RNAs with a binding motif for AtCyp59 proved difficult. All our efforts to establish stably transformed plants or tissue culture cell lines failed as even small changes in the level of AtCyp59 are detrimental to cell growth (3, and our unpublished data). We therefore used a transient expression system in plant protoplasts where we used HA-tagged WT and mutated Cyp59_3M (3 M in RNP1) proteins (Figure 4a) for RNA immunoprecipitation to show specific binding of AtCyp59 to endogenous mRNAs containing the selected motif. A plasmid expressing HA-tagged MAPK6 kinase was used as control and transformation efficiency was monitored by a 35S-GFP construct. Protoplasts isolated from an Arabidopsis cell suspension culture were transformed with DNA constructs and after 24-h extracts were prepared and used for immunoprecipitation with anti-HA antibody. Figure 4b shows a western blot of the total protein isolated from transformed plant protoplasts with anti-HA antibodies demonstrating that both the AtCyp59 and the mutated AtCyp59_3M were expressed at similar levels (top panel) and were efficiently immunoprecipitated with anti-HA antibody (bottom panel). To test if the transformation of the HA-tagged protein itself affects the levels of the tested endogenous RNAs, control RT–PCR experiments were performed. None of the transfected proteins influenced the mRNA expression level of any of the tested endogenous RNAs (Supplementary Figure S3a). RNAs co-precipitated with AtCyp59, Cyp59_3M and MAPK6 were isolated and analysed by RT–PCR using oligonucleotides corresponding to endogenous mRNAs containing the selected motif. The majority of the motif-containing RNAs (10 of 13) could be recovered by IP of the HA-tagged AtCyp59 but not by the mutated AtCyp59_3M or the MAPK6 kinase control (Figure 4c and Supplementary Figure S3b).
Figure 4.
Transiently expressed AtCyp59 HA-tagged proteins in the A. thaliana protoplasts bind to endogenous mRNAs in vivo. (a) Schematic representation of the AtCyp59 constructs used in the transient transformation experiments. 35S, CaMV 35S RNA promoter sequence; HA, hemagglutinine antigen; 3M; mutations in the RRM domain (the same as in Figure 3a); GFP, green fluorescence protein; MAPK6, mitogen-activated protein kinase 6. (b) Western blot analyses of cells overexpressing the depicted constructs were performed using anti-HA antibody (upper panel) or anti-tubulin antibody (middle panel). Bottom panel is a western blot analysis of an HA immunoprecipitation used for RNA analysis shown in (c). Molecular weight markers are indicated on the left side. (c) Analysis of RNAs immunoprecipitated with the indicated proteins. RT–PCR with primers to the genes indicated on the right side (33 PCR cycles; primers used are listed in the Supplementary Table S3). Molecular weight marker is shown on the left side in bp range. (d) Analysis of selected pre-mRNAs immunoprecipitated with AtCyp59. Primers for RT reaction are designed to tag a pre-mRNA after the polyA signal; primers in the PCR are the same as in (c). +RT, PCR with reverse transcriptase added; -RT, PCR without reverse transcriptase; filled diamond, pre-mRNA product; star, fully spliced mRNA product. Other bands in the AT3G4930 lane are partially spliced products. (e) Schematic representations of the RNA constructs used for double transformation experiment with AtCyp59. TTG, mutated translational start codon; line, coding sequence of the gene; exogenous tag, partial sequence of the hemagglutinine antigen. (f) Western blot analyses of immunoprecipitation experiments with cells overexpressing the depicted combinations of constructs. The molecular weight marker is displayed on the left side. (g) RT–PCR with primers to the genes indicated at the bottom (primers used are listed in the Supplementary Table S3). Molecular weight marker is shown on the left side in bp range. Upper panel is a RT–PCR from total RNA isolated from protoplasts double transformed with the depicted constructs. Lower panel is an RT–PCR of RNA isolated after IP. Bands shown are amplified using the primers to the synthetic RNA construct which were designed to differentiate from endogenously expressed RNA.
As AtCyp59 is involved in transcription regulation, we argued that it should bind to the nascent transcript. Therefore, we repeated the RT–PCR experiment for three target transcripts using an RT primer downstream of the polyA addition site. This should only capture RNAs before 3′-end cleavage and polyA addition. In the PCR analysis in Figure 4d, we observe recovery of pre-mRNAs, partially spliced RNAs as well as of spliced RNAs showing that binding of AtCyp59 must have occurred on an unprocessed transcript. To control for DNA contamination, PCRs were carried out without prior reverse transcription. Additionally, these experiments indicate that splicing can occur before 3′-end processing as suggested previously.
To investigate if binding indeed occurred to the established RNA-binding motif, we designed an experiment where we used wild-type AtCyp59 and transformed it together with a construct containing one of the IP mRNAs which was mutated at the ATG (TTG) to avoid protein overexpression and with a tag at the 3′-end to distinguish it from the endogenously expressed mRNA. In addition, we mutated the AtCyp59-binding motif to a U/A-stretch in this mRNA and used this construct in co-transformation experiments with AtCyp59. This was done for the mRNAs of AT3G49430 (At-SR34a) and AT3G53500 (At-RS2Z32) (Figure 4e). Figure 4f (upper panels) shows a western blot with anti-HA antibodies from the immunoprecipitation of the co-transformation experiments to control for AtCyp59 expression. Figure 4g (upper panels) shows RT–PCR of total RNA of the co-transformation extract indicating that all transfected target RNAs were expressed in equivalent amounts. RT–PCR analyses of the IP (Figure 4g, lower panels) show that the RNAs containing the selected motif are present in the IP in contrast to the mRNAs with the mutated binding motif.
Taken together, these experiments show that AtCyp59 binds to the selected binding motif in vivo and most importantly that binding likely occurs co-transcriptionally.
AtCyp59 is an active PPIase and its activity is inhibited by RNA binding
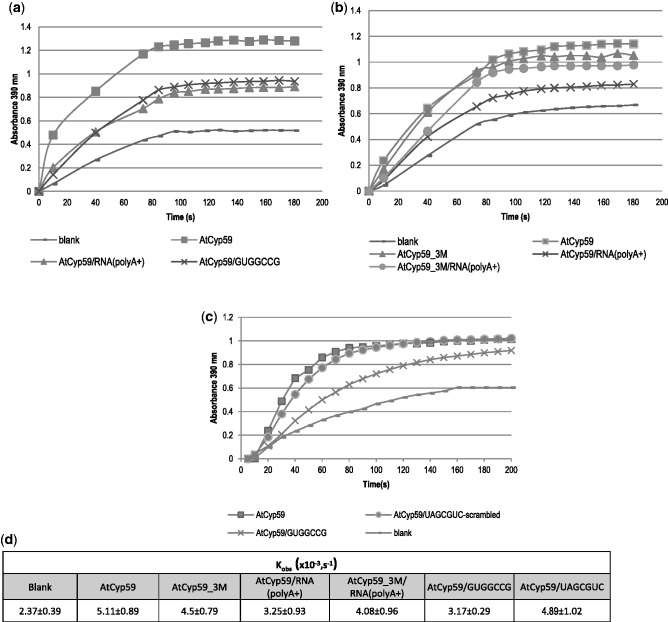
Having demonstrated that AtCyp59 indeed binds specific RNA sequences in vivo, it was of great interest to investigate if RNA binding could have an influence on the enzymatic activity (PPIase) of AtCyp59. However, it has never been shown that either the A. thaliana or the S. pombe protein possesses PPIase activity. We therefore used an in vitro PPIase assay and tested recombinant AtCyp59. This assay uses a substrate that has the peptide bond in cis conformation adjacent to a proline (Suc-Ala-Ala-Pro-Phe-p-nitroanilide) (36,28). This proline is followed by a p-nitro-Phe group which can be cleaved off by chymotrypsin if the peptide bond adjacent to the proline is in trans conformation. Conversion of substrate from cis to trans was measured by reading the absorbance of the released p-nitroaniline at 390 nm. Observed reaction rates were calculated as an average from four independent experiments. Figure 5a shows that addition of recombinant AtCyp59 to the substrate accelerated the spontaneous cis/trans isomerization considerably (blank: Kobs = 2.37 ± 0.39;+Cyp59: Kobs = 5.11 ± 0.89; Figure 5d). Addition of AtCyp59_3M in this assay resulted in only a small reduction in activity (Kobs = 4.50 ± 0.79), indicating that the mutations in the RRM do not affect the PPIase activity of AtCyp59 significantly (Figure 5b and d). To test if binding of RNA influences PPIase activity, we first pre-incubated a polyA+ mRNA from total Arabidopsis RNA with wild-type AtCyp59 before adding it to the PPIase reaction. As shown in Figures 5a and d, we see a significant effect of reduced PPIase activity (Kobs = 3.25 ± 0.93) on the polyA+ fraction. The effect on polyA+ RNA is in line with our observation that the binding motif of AtCyp59 mainly occurs in protein-coding genes. Furthermore, incubation with a 7-nt motif sequence (GUGGCCG) similarly decreased the activity of AtCyp59 PPIase (Kobs = 3.17 ± 0.29) (Figure 5a, c and d), whereas incubation with a scrambled oligo (UAGCGUC) did not affect the activity (Kobs = 4.89 ± 1.02). We also used the mutated AtCyp59_3M protein pre-incubated with polyA+ RNA. We observed a slight reduction of the PPIase activity compared with the AtCyp59_3M without RNA (Kobs = 4.50 ± 0.79 versus Kobs = 4.08 ± 0.96; Figure 5b and d). This observation corresponds to results from in vitro RNA-binding assays showing that the mutated protein still has a residual binding capacity for the RNA (Figure 3d and e). Taken together, these data show that AtCyp59 possesses an active PPIase domain and that specific binding of RNA to the RRM of AtCyp59 reduces the PPIase activity in vitro.
Figure 5.
PPIase activity of AtCyp59 decreases upon binding to the mRNA (polyA+) or to a 7-nt RNA binding motif. (a) PPIase activity in the absence (−) or in the presence (filled square) of AtCyp59. PPIase activity is inhibited upon binding either to the polyA+ fraction of RNA (filled triangle) or to an RNA binding motif (×). (b) PPIase activity in the absence (−) or in the presence of AtCyp59 (filled square), or in the presence of AtCyp59_3M (filled triangle). PPIase activity of AtCyp59 upon binding to the polyA+ fraction of RNA (×). PPIase activity of AtCyp59_3M upon binding to the polyA+ fraction of RNA (filled circle). (c) PPIase activity in the absence (−) or in the presence (filled square) of AtCyp59. PPIase activity is inhibited upon binding to the specific RNA binding motif (×) but is not inhibited upon binding to the scramble RNA oligonucleotide (filled circle). (d) Kobs of four independent measurements of the PPIase activity. Kinetics is calculated as first-order Michaelis–Menten reaction.
The AtCyp59-binding motif is present in most RNA polymerase II transcripts
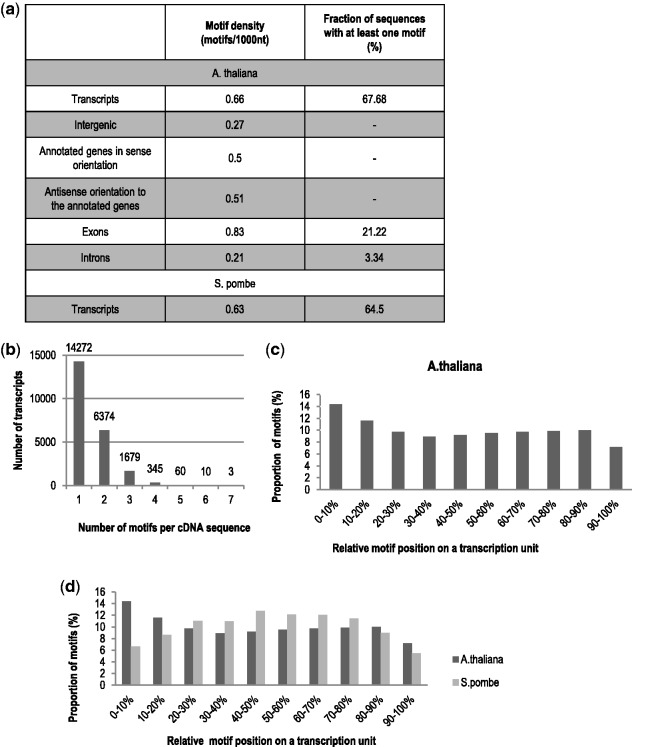
Having verified the binding motif by in vitro and in vivo methods, it was interesting to get a more genomic view of the distribution of the G[U/C]NGCC[A/G] motif in the Arabidopsis genome. The 17 motif variants which were tested in vitro (Table 1) were used to construct an experimentally validated version of the binding motif descriptor. The analysis of the Arabidopsis genome (TAIR10) reveals that about 70% of the Arabidopsis protein-coding genes contained one of the AtCyp59-binding motifs (Figure 6a). Considering that we did not test all possible variants for this motif, the number of RNA polymerase II transcripts possessing this motif might be much higher. It is very interesting that the motif density (number of the motifs per 1000 nt) is equally distributed to sense and antisense transcripts of the annotated genes (Figure 6a, 0.5 in sense and 0.51 in antisense orientation), while for intergenic regions the number is much lower (0.27). Even more drastic changes have been observed comparing exons and introns of protein-coding genes. The motif densities varied significantly, from 0.83 in exons to 0.21 in introns. These data show that the predominant location of the binding motif is within exons of protein-coding genes. Furthermore, we aligned the motif to the cDNA sequences (corresponding to mature mRNAs) and found that the majority of the transcripts contain the binding motif only once or twice in a given transcript body (Figure 6b). The analysis of the 17 motifs in the S. pombe genome revealed a similar distribution as in Arabidopsis (Figure 6a). About 64% of protein coding transcripts possess a motif mostly in the coding region.
Figure 6.
Computational analysis of the binding motif distribution in the A. thaliana genome. (a) Distribution of the AtCyp59 binding motif variants in the A. thaliana genome according to TAIR 10 annotation. (b) Frequency of occurrence of the binding motif variants per cDNA sequence in the A. thaliana genome. Each bar on the diagram represents the number of genes with the indicated number of the binding motifs per transcript sequence. (c) Arabidopsis AtCyp59 motif distribution in the TUs. Distribution of 17 motifs experimentally validated in A. thaliana is shown in 10 windows of the transcription units. (d) A. thaliana and S. pombe motif distribution. Distribution of the 17 motifs experimentally validated in A. thaliana is shown for Arabidopsis (dark grey) and S. pombe transcription units (light grey bars).
To examine if the motif had any general tendency for a location within the TU, we normalized motif occurrence to transcript length. Taking all 17 verified motif variants into account, we found a slightly higher proportion of motifs in the beginning and a decrease towards the end of the Arabidopsis transcript (Figure 6c). A similar analysis of the 17 motifs on the S. pombe genome showed a more prominent accumulation of the motifs towards the gene body (Figure 6d, grey bars). To determine a possible influence of gene length on the distribution of the motif, we performed the same analysis on small (750 ± 50 bp), medium (1500 ± 100 bp) and long (3000 ± 200 bp) genes (Supplementary Figure S4). Interestingly, the small genes profiles are similar to the S. pombe genomic distribution which is coherent with the smaller gene sizes in yeast. The bell–shaped curve is probably due to the 5′- and 3′-UTR having less motif variants. The medium- and long-size genes behave similar as in the genomic analysis (Figure 6c and d and Supplementary Figure S4) indicating that the observed motif peak at the beginning of genes comes from the lager genes as they contain more coding sequences in the first 10% of the nucleotide sequence.
Taken together, the fact that the AtCyp59-binding motif occurs in almost 70% of the protein-coding genes implicates that it might be important for the majority of RNA polymerase II TUs. This is supported by the bioinformatics analysis showing that it is equally abundant in antisense transcripts. In addition, the evolutionarily conserved abundance and location of this motif points to an important function of this protein in the transcription process.
DISCUSSION
In this article, we determined the RNA targets of an RRM-containing cyclophilin AtCyp59 by using a Genomic SELEX method. The selected sequences enabled us to identify the RNA-binding consensus motif for the evolutionarily highly conserved RRM domain of this multidomain protein. The binding of AtCyp59 to its RNA targets was confirmed in vitro by mobility shift assays and in vivo by RNA immunoprecipitation from protoplasts transiently expressing the HA-tagged protein. In addition, we showed that mutations in either the RRM domain of AtCyp59 or in the RNA motif sequence decrease the binding specificity of the AtCyp59 to its RNA targets. Furthermore, we show that recombinant AtCyp59 exhibits PPIase activity and that this activity is decreased upon binding to the specific RNA-binding motif. Genome-wide analysis of the RNA binding motif showed its presence in >70% of the A. thaliana mRNAs and its prevalent localization in the coding region. We also demonstrated that the RNA-binding motif and its genome-wide distribution are evolutionarily conserved.
Genomic SELEX is a method for the identification of an RNA-binding motif for a given protein. In our selection, the identified motif which consists of a 7-nt-long RNA consensus sequence G[U/C]N[G/A]CC[A/G], was consistent with previous data, which showed that AtCyp59 interacts with C- and G-rich RNA oligonucleotides in vitro (3). Sequence variations of the motif which were present in the SELEX sequences were tested in vitro. Interestingly, the binding motif variants showed variable binding affinities to the RRM-Zn domain of AtCyp59 which was used for selection. Furthermore, binding of the 7-nt binding motif variations to the full-length AtCyp59 showed an equal overall binding affinity to the motif variants, suggesting an influence of the other AtCyp59 domains in this interaction. AtCyp59 interacts with its RNA target sequence specifically, since mutations in the RRM domain of AtCyp59, which are known to be generally involved in RNA recognition (37), decreased binding activity. Similarly, mutations in the sequence of the 7-nt RNA consensus motif also decreased its affinity to the RRM_Zn domain of AtCyp59. In addition, when we used longer RNA transcripts containing the binding motif, we observed an improved binding to the full-length AtCyp59. As we have indications that AtCyp59 possesses an RNA chaperone activity, this effect might be due to an impact on local RNA structure. Thus, full-length AtCyp59 regulates binding to RNA targets and might also change RNA structure.
The interaction between AtCyp59 and RNA targets was confirmed in vivo using transient expression of the AtCyp59 in protoplasts. We were restricted to transient expression for the experiments in vivo due to our inability to produce stable overexpression tagged lines of AtCyp59. This is most probably caused by the tight regulation of AtCyp59 levels in vivo (3,11). RNA immunoprecipitation analysis showed that wild-type AtCyp59 could immunoprecipitate endogenous RNA targets. Furthermore, mutations in the RRM domain of AtCyp59 abolished recovery of specific RNAs after immunoprecipitation. This suggests a direct recognition of RNA targets through the RRM domain of AtCyp59. When we performed a double transformation of the protoplasts with AtCyp59 and exogenous RNA targets, these RNAs could be also recovered by immunoprecipitation of AtCyp59. If the RNA-binding motif was mutated, AtCyp59 could no longer bind to the exogenous RNAs. These data show that AtCyp59 binds to motif containing mRNAs in vivo in a sequence-specific manner. Most importantly, these experiments also demonstrated that AtCyp59 binds to the transcript in the course of transcription as also unspliced or partially spliced RNAs were immunoprecipitated.
In addition, we could demonstrate PPIase activity of the recombinant AtCyp59 in vitro. The observed Kobs rates are in line with previously described PPIase activities of other isomerases (38). This also suggests that AtCyp59 could act as a PPIase enzyme in vivo. Most interestingly, the PPIase activity was reduced when the RRM domain of AtCyp59 was bound specifically to its RNA-binding motif or to polyA+ RNA but not to a scrambled RNA oligo. The mutations in the RRM domain of AtCyp59 slightly reduced its enzymatic activity similar to control mutations in the region between the RRM and the PPIase domain (data not shown). This minor decrease in activity might therefore come from changes in protein structure upon mutation in regions unrelated to the PPIase domain. However, if we added specific RNA to Cyp59_3M (mutated in the RRM), the PPIase activity did not change indicating that the decrease in activity upon RNA binding is exerted through the RRM domain. This suggests that binding of specific RNA to AtCyp59 causes structural changes that feed back to the PPIase domain resulting in a decrease in enzymatic activity. From the available data from AtCyp59 and spRct1 (3,11), we know that this protein is located in transcription/splicing complexes, but the mechanism of its action is unknown. There is one other RRM-containing cyclophilin, hCyp33, described in the literature which regulates a histone acetyl transferase. Contrary to AtCyp59, PPIase activity of hCyp33 is stimulated by binding AU-rich RNA (20) but neither a motif nor a natural RNA target has been identified (19). However, our results with AtCyp59 predict a possible negative feedback loop regulating PPIase activity upon RNA binding.
AtCyp59 is an interesting protein, because of its multidomain organization that includes a PPIase domain followed by an RRM domain, Zn knuckle and the C-terminal-charged domain. AtCyp59 is a nuclear protein and further analyses showed that AtCyp59 interacts in vitro and in vivo with the RNA polymerase II (3). Overexpression of the protein in cell culture was detrimental to cell growth and had an effect on the level of RNA polymerase II. Additional information about this highly conserved protein came from analysis of its S. pombe orthologue, Rct1, which was identified as an essential gene where overexpression of Rct1 led to a decrease in CTD phosphorylation. In contrast, lower levels of Rct1 upregulated CTD phosphorylation (11). This upregulation resulted in a decrease in RNA polymerase II activity as shown in run-on transcription assays. Furthermore, it was shown that Rct1 is associated with actively transcribed genes following the RNA polymerase II profile along the gene. These data suggested an important activity provided by this multidomain cyclophilin for transcription elongation. The RRM domain is evolutionarily the most conserved domain of this protein suggesting its important contribution to the overall activity of the protein. The experiments presented in this articles now show binding of AtCyp59 to the nascent transcript. In addition, they demonstrate that the selected binding motif, its abundance and genomic localization are also evolutionarily conserved.
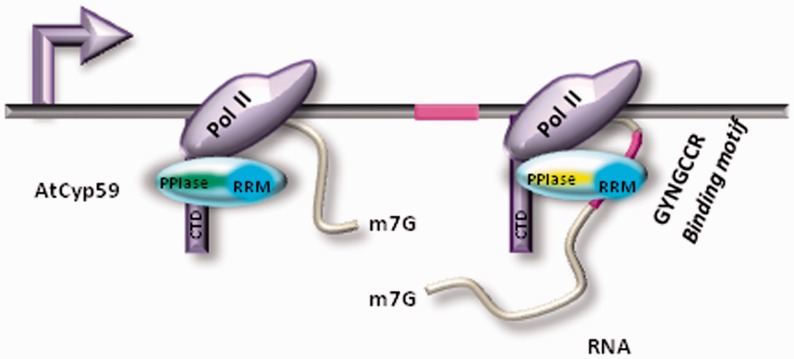
We predicted that AtCyp59 might bind a small regulatory RNA (similarly to the P-TEFb transcription factor regulated by 7SK RNA (39)) or alternatively might bind a subset of RNA polymerase II transcripts. Surprisingly, our genome-wide analysis of the AtCyp59 RNA-binding motif showed that this motif is present in the majority of RNA polymerase II transcripts in sense and antisense orientation (Figure 6a). If one assumes a universal function for this cyclophilin in transcription, the binding motif should be also present in antisense transcripts as antisense transcription of protein coding genes is pervasive in many genomes (40,41). Furthermore, the binding affinities to the motif variants should be similar which is what we observe when using full-length AtCyp59 in binding assays. These data suggest that AtCyp59 might bind to many endogenous transcripts (sense and antisense) with similar strength. Most importantly, we have shown that binding of specific RNAs or polyA+ RNAs reduces the PPIase activity of AtCyp59 in vitro. Therefore, binding of AtCyp59 to its natural RNA targets might also change PPIase activity in vivo and thus send a signal to the transcription machinery. As AtCyp59 interacts with the RNA polymerase II complex, a possible scenario could be that it re-structures the heptapeptide repeats on the CTD thus influencing phosphorylation similarly to the activity of the PPIase Pin1 in humans and Ess1 in Saccharomyces cerevisiae (12,16,42,43). Alternatively, it might influence kinase or phosphatase activities which act on the CTD repeats. As the position of the binding motif is distributed along the coding region of the gene, we suggest that AtCyp59 might bind to the nascent transcript in the elongation phase of transcription. The consequences of RNA binding on the function of this protein in vivo as well as on its temporal and spatial interaction with its partners are currently unknown. However, summarizing the available data for this cyclophilin, we propose a model (Figure 7) where in the course of transcription RNA-dependent inhibition of the PPIase activity of AtCyp59 influences RNA polymerase II activity. Our data indicate that this multidomain cyclophilin might have a role in transcription regulation, which is in line with the observation that it is an essential gene and its deregulation is detrimental to cell growth (3,11).
Figure 7.
Model of how binding of AtCyp59 to the transcript might influence transcription. When the RRM domain (dark blue) of AtCyp59 (light blue) is not engaged in RNA–protein interaction, activity of its PPIase domain is unaffected (green). However, upon binding of AtCyp59 to the binding motif (purple) on the RNA transcript, the PPIase activity of AtCyp59 decreases (yellow). These changes might serve as a signal to modulate RNA polymerase II activity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–4 and Supplementary Methods.
FUNDING
The Austrian Science Fund (FWF) [SFB 1710, 1711; DK W1207] and the Austria Genomic Program (GENAU III) [ncRNAs]; the EU FP6 Programme Network of Excellence on Alternative Splicing (EURASNET) [LSHG-CT-2005-518238]. Funding for open access charge: Austrian Science Fund, FWF.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Doris Chen for initial bioinformatics support and Zdravko Lorkovic, John WS Brown and Alwin Köhler for invaluable discussions. O.B. performed all experiments and analysed data; M.Z. and Y.M. did the computer analysis, motif finding and genome analysis; T.S. did the initial cycolphilin assays; M.K supervised experiments; A.B. designed and supervised the experiments; A.B, O.B. and M.K. wrote the article.
REFERENCES
- 1.Nicholson LK, Lu KP. Prolyl cis-trans isomerization as a molecular timer in Crk signaling. Mol. Cell. 2007;25:483–485. doi: 10.1016/j.molcel.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Romano PG, Horton P, Gray JE. The Arabidopsis cyclophilin gene family. Plant Physiol. 2004;134:1268–1282. doi: 10.1104/pp.103.022160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gullerova M, Barta A, Lorkovic ZJ. AtCyp59 is a multidomain cyclophilin from Arabidopsis thaliana that interacts with SR proteins and the C-terminal domain of the RNA polymerase II. RNA. 2006;12:631–643. doi: 10.1261/rna.2226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krzywicka A, Keller AM, Cohen J, Jerka-Dziadosz M, Klotz C. KIN241: a gene involved in cell morphogenesis in Paramecium tetraurelia reveals a novel protein family of cyclophilin-RNA interacting proteins (CRIPs) conserved from fission yeast to man. Mol. Microbiol. 2001;42:257–267. doi: 10.1046/j.1365-2958.2001.02634.x. [DOI] [PubMed] [Google Scholar]
- 5.Bentley D. The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 2002;14:336–342. doi: 10.1016/s0955-0674(02)00333-2. [DOI] [PubMed] [Google Scholar]
- 6.Munoz MJ, de la Mata M, Kornblihtt AR. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem. Sci. 2010;35:497–504. doi: 10.1016/j.tibs.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Natalizio BJ, Robson-Dixon ND, Garcia-Blanco MA. The carboxyl-terminal domain of RNA polymerase II is not sufficient to enhance the efficiency of pre-mRNA capping or splicing in the context of a different polymerase. J. Biol. Chem. 2009;284:8692–8702. doi: 10.1074/jbc.M806919200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brody Y, Neufeld N, Bieberstein N, Causse SZ, Bohnlein EM, Neugebauer KM, Darzacq X, Shav-Tal Y. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 2011;9:e1000573. doi: 10.1371/journal.pbio.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 10.Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gullerova M, Barta A, Lorkovic ZJ. Rct1, a nuclear RNA recognition motif-containing cyclophilin, regulates phosphorylation of the RNA polymerase II C-terminal domain. Mol. Cell Biol. 2007;27:3601–3611. doi: 10.1128/MCB.02187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu YX, Hirose Y, Zhou XZ, Lu KP, Manley JL. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev. 2003;17:2765–2776. doi: 10.1101/gad.1135503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh N, Ma Z, Gemmill T, Wu X, Defiglio H, Rossettini A, Rabeler C, Beane O, Morse RH, Palumbo MJ, et al. The Ess1 prolyl isomerase is required for transcription termination of small noncoding RNAs via the Nrd1 pathway. Mol. Cell. 2009;36:255–266. doi: 10.1016/j.molcel.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu YX, Manley JL. Pin1 modulates RNA polymerase II activity during the transcription cycle. Genes Dev. 2007;21:2950–2962. doi: 10.1101/gad.1592807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poschmann J, Drouin S, Jacques PE, El Fadili K, Newmarch M, Robert F, Ramotar D. The peptidyl prolyl isomerase Rrd1 regulates the elongation of RNA polymerase II during transcriptional stresses. PLoS One. 2011;6:e23159. doi: 10.1371/journal.pone.0023159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma Z, Atencio D, Barnes C, Defiglio H, Hanes SD. Multiple roles for the Ess1 prolyl isomerase in the RNA polymerase ii transcription cycle. Mol. Cell Biol. 2012;32:3594–3607. doi: 10.1128/MCB.00672-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorkovic ZJ, Lopato S, Pexa M, Lehner R, Barta A. Interactions of Arabidopsis RS domain containing cyclophilins with SR proteins and U1 and U11 small nuclear ribonucleoprotein-specific proteins suggest their involvement in pre-mRNA Splicing. J. Biol. Chem. 2004;279:33890–33898. doi: 10.1074/jbc.M400270200. [DOI] [PubMed] [Google Scholar]
- 18.Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M, Will CL, Pena V, Luhrmann R, Stelzl U. Dynamic protein-protein interaction wiring of the human spliceosome. Mol, Cell. 2012;45:567–580. doi: 10.1016/j.molcel.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Song J, Milne TA, Wang GG, Li H, Allis CD, Patel DJ. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell. 2010;141:1183–1194. doi: 10.1016/j.cell.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Han R, Zhang W, Yuan Y, Zhang X, Long Y, Mi H. Human CyP33 binds specifically to mRNA and binding stimulates PPIase activity of hCyP33. FEBS Lett. 2008;582:835–839. doi: 10.1016/j.febslet.2008.01.055. [DOI] [PubMed] [Google Scholar]
- 21.Robertson JF, Semiglazov V, Nemsadze G, Dzagnidze G, Janjalia M, Nicholson RI, Gee JM, Armstrong J. Effects of fulvestrant 250mg in premenopausal women with oestrogen receptor-positive primary breast cancer. Eur. J. Cancer. 2007;43:64–70. doi: 10.1016/j.ejca.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 22.Lorenz C, von Pelchrzim F, Schroeder R. Genomic systematic evolution of ligands by exponential enrichment (Genomic SELEX) for the identification of protein-binding RNAs independent of their expression levels. Nat. Protoc. 2006;1:2204–2212. doi: 10.1038/nprot.2006.372. [DOI] [PubMed] [Google Scholar]
- 23.Zimmermann B, Gesell T, Chen D, Lorenz C, Schroeder R. Monitoring genomic sequences during SELEX using high-throughput sequencing: neutral SELEX. PLoS One. 2010;5:e9169. doi: 10.1371/journal.pone.0009169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zywicki M, Bakowska-Zywicka K, Polacek N. Revealing stable processing products from ribosome-associated small RNAs by deep-sequencing data analysis. Nucleic Acids Res. 2012;40:4013–4024. doi: 10.1093/nar/gks020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frith MC, Saunders NF, Kobe B, Bailey TL. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 2008;4:e1000071. doi: 10.1371/journal.pcbi.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40(Database issue):D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kofron JL, Kuzmic P, Kishore V, Colon-Bonilla E, Rich DH. Determination of kinetic constants for peptidyl prolyl cis-trans isomerases by an improved spectrophotometric assay. Biochemistry. 1991;30:6127–6134. doi: 10.1021/bi00239a007. [DOI] [PubMed] [Google Scholar]
- 29.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 30.James TD, Cashel M, Hinton DM. A mutation within the beta subunit of Escherichia coli RNA polymerase impairs transcription from bacteriophage T4 middle promoters. J. Bacteriol. 2010;192:5580–5587. doi: 10.1128/JB.00338-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S, Shi H, Lee DK, Lis JT. Specific SR protein-dependent splicing substrates identified through genomic SELEX. Nucleic Acids Res. 2003;31:1955–1961. doi: 10.1093/nar/gkg286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamm S, Smith C, Lührmann R. Alternative Pre-mRNA Splicing: Theory and Protocols. New York: Wiley; 2012. p. 660. [Google Scholar]
- 33.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 34.Carels N, Bernardi G. The compositional organization and the expression of the Arabidopsis genome. FEBS Lett. 2000;472:302–306. doi: 10.1016/s0014-5793(00)01476-9. [DOI] [PubMed] [Google Scholar]
- 35.Mayeda A, Munroe SH, Caceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer G, Bang H, Mech C. [Determination of enzymatic catalysis for the cis-trans-isomerization of peptide binding in proline-containing peptides]. (Translated from ger) Biomed. Biochim. Acta. 1984;43:1101–1111. (in Germany) [PubMed] [Google Scholar]
- 37.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 38.Wiborg J, O'Shea C, Skriver K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. 2008;413:447–457. doi: 10.1042/BJ20071568. [DOI] [PubMed] [Google Scholar]
- 39.Eilebrecht S, Benecke BJ, Benecke A. 7SK snRNA-mediated, gene-specific cooperativity of HMGA1 and P-TEFb. RNA Biol. 2011;8:1084–1093. doi: 10.4161/rna.8.6.17015. [DOI] [PubMed] [Google Scholar]
- 40.Chen HM, Neiman AM. A conserved regulatory role for antisense RNA in meiotic gene expression in yeast. Curr. Opin. Microbiol. 2011;14:655–659. doi: 10.1016/j.mib.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ni T, Tu K, Wang Z, Song S, Wu H, Xie B, Scott KC, Grewal SI, Gao Y, Zhu J. The prevalence and regulation of antisense transcripts in Schizosaccharomyces pombe. PLoS One. 2010;5:e15271. doi: 10.1371/journal.pone.0015271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang M, Wang XJ, Chen X, Bowman ME, Luo Y, Noel JP, Ellington FA, Zhang Y. Structural and Kinetic analysis of prolyl-isomerization/phosphorylation cross-talk in the CTD code. ACS Chem Biol. 2012;7:1462–1470. doi: 10.1021/cb3000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J. Biol. Chem. 2011;286:5717–5726. doi: 10.1074/jbc.M110.197129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.