Abstract
A major base lesion resulting from oxidative stress is 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxoG) that has ambiguous coding potential. Error-free DNA synthesis involves 8-oxoG adopting an anti-conformation to base pair with cytosine whereas mutagenic bypass involves 8-oxoG adopting a syn-conformation to base pair with adenine. Left unrepaired the syn-8-oxoG/dAMP base pair results in a G–C to T–A transversion. During base excision repair of this mispair, DNA polymerase (pol) β is confronted with gap filling opposite 8-oxoG. To determine how pol β discriminates between anti- and syn-8-oxoG, we introduced a point mutation (R283K) to alter insertion specificity. Kinetic studies demonstrate that this substitution results in an increased fidelity opposite 8-oxoG. Structural studies with R283K pol β show that the binary DNA complex has 8-oxoG in equilibrium between anti- and syn-forms. Ternary complexes with incoming dCTP resemble the wild-type enzyme, with templating anti-8-oxoG base pairing with incoming cytosine. In contrast to wild-type pol β, the ternary complex of the R283K mutant with an incoming dATP-analogue and templating 8-oxoG resembles a G–A mismatched structure with 8-oxoG adopting an anti-conformation. These results demonstrate that the incoming nucleotide is unable to induce a syn-8-oxoG conformation without minor groove DNA polymerase interactions that influence templating (anti-/syn-equilibrium) of 8-oxoG while modulating fidelity.
INTRODUCTION
Reactive oxygen species (ROS) are generated during normal metabolic processes and their levels increase in response to environmental stress. Exposure of DNA to ROS results in oxidative DNA damage and the generation of multiple types of DNA lesions. Left unrepaired these lesions contribute to mutagenesis, cancer and human disease (1). One of the most abundant lesions caused by oxidative DNA damage in both the DNA and nucleotide pools is 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxoG). At physiological pH, 8-oxoG has a carbonyl at C8 and is protonated at N7 (2). These modifications to guanine lead to additional hydrogen bonding interactions at the Hoogsteen edge and promote rotation about its glycosidic bond thereby encouraging both anti- and syn-conformations. This rotation about the glycosidic bond to the syn-conformation is further facilitated by a backbone clash between the adducted oxygen (O8) and the sugar–phosphate backbone in the anti-8-oxoG conformation (3).
The coding potential of 8-oxoG is dictated by the anti- or syn-conformation of the modified base. Similar to unmodified deoxyguanine, anti-8-oxoG base pairs with cytosine through traditional Watson–Crick hydrogen bonding interactions forming a non-mutagenic DNA lesion. In contrast, the mutagenic syn-8-oxoG conformation is able to base pair with adenine through its Hoogsteen edge. Kinetic studies have shown that DNA polymerases insert adenine opposite 8-oxoG frequently and with enhanced catalytic efficiency when compared with guanine (4). When compared with guanine, the catalytic efficiency of adenine insertion opposite 8-oxoG increases almost three orders of magnitude. If left unrepaired, the pro-mutagenic syn-8oxoG-dA base pair results in a G–C to T–A transversion mutation following DNA replication (5). This highlights the importance of the dual-coding properties of 8-oxoG being either error free or mutagenic when it is in the anti- or syn-conformations, respectively.
The prevalence of oxidative stress and subsequent mutagenic properties of 8-oxoG has resulted in an elegant cellular defense mechanism against 8-oxoG (4). This lesion is removed from DNA by a glycosylase that generates a gap in the DNA following removal of the damaged base and subsequent initiation of base excision repair (BER). DNA polymerase (pol) β is required to fill the single-nucleotide-gapped DNA intermediate. In the repair of the 8oxoG-C base pair, OGG1 glycosylase removes 8-oxoG resulting in a gapped DNA substrate with a templating cytosine that will be filled with high fidelity by pol β. When 8-oxoG escapes repair, there is a high probability that replication will result in adenine insertion. Thus, the cell also codes for a glycosylase, MYH, which removes adenine paired with 8-oxoG and initiates BER. In this situation, pol β encounters 8-oxoG as the templating nucleotide and will insert dCMP or dAMP opposite 8-oxoG resulting in error-free or mutagenic gap filling, respectively. Kinetic studies with a templating 8-oxoG have shown that pol β only incorporates dCTP over dATP by ∼2-fold (6). This poor 8-oxoG fidelity results in futile mutagenic ‘repair’ following insertion of dATP opposite 8-oxoG. Accordingly, mutagenic repair would be a cellular burden during times of elevated metabolic or environmental stress. These examples emphasize the importance of BER and pol β in modulating the repair of oxidative DNA damage and lesion bypass.
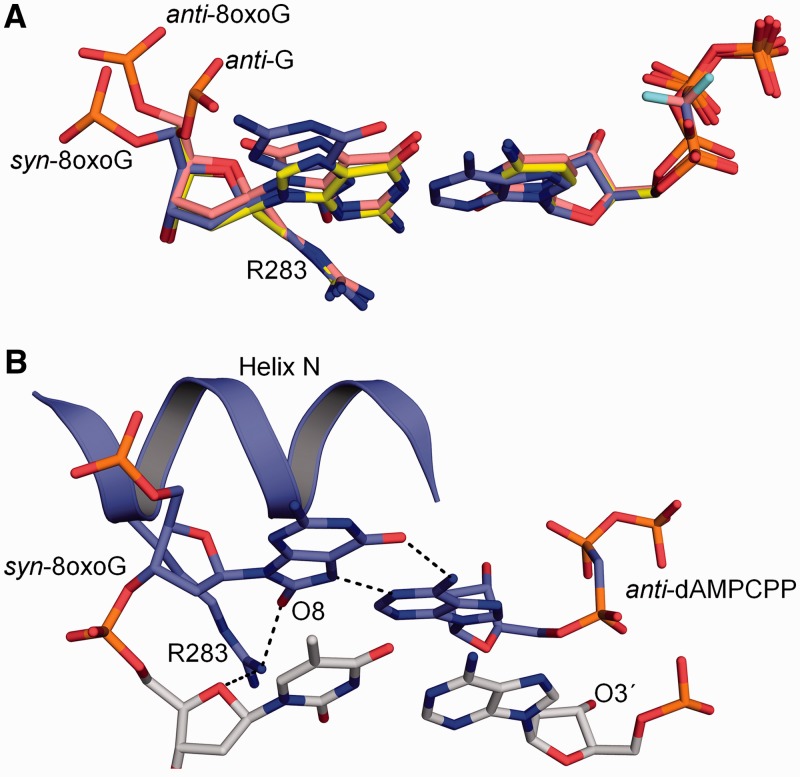
Currently, pol β is the only wild-type DNA polymerase that has been structurally characterized with a templating 8-oxoG base paired with either an incoming dCTP or dATP (7,8). Overall these structures indicate that the 8-oxoG lesion paired with dCTP or dATP is well tolerated in the polymerase active site. The structure with an incoming dCTP paired with anti-8-oxoG shows the greatest structural change at the phosphate backbone of the anti-8-oxoG, shifting 2.9 Å and rotating 200° to accommodate the carbonyl oxygen at C8 position (Figure 1A). This movement is a result of a steric clash between O8 and the phosphate backbone. Recently, the structure of pol β with a templating syn-8-oxoG Hoogsteen base pairing with dATP was determined and indicates that it is similar to that with an incoming dCTP (7). With the syn-8-oxoG structure, however, Arg283 is observed to stabilize the syn-conformation through a hydrogen bond to O8 in the DNA minor groove (Figure 1B). With guanine or anti-8-oxoG as the templating base, Arg283 interacts with the templating nucleotide upstream of the coding base. Importantly, Arg283 only interacts with the templating strand after nucleotide binding that induces subsequent polymerase subdomain repositioning.
Figure 1.
Structural overview of wild-type pol β with templating 8-oxoG in the active site. (A) Overlay of the templating anti-G:dCMPCPP (PDB ID 2FMP), anti-8-oxoG:dCMP(CF2)PP (PDB ID 3RJI) and syn-8-oxoG:dAMPCPP (PDB ID 3RJF) in the pol β active site are shown in yellow, salmon and purple, respectively. The position of the templating phosphate backbone is indicated for each conformation. (B) Structure of the closed ternary pol β complex with a templating syn-8-oxoG Hoogsteen base pairing with dAMPCPP (PDB ID 3RJF) (7). The primer terminus (O3′) is indicated. Key hydrogen bonding interactions are shown as black dashes.
Combining rational site-directed mutagenesis with kinetic and structural approaches, we are able to identify DNA minor groove interactions that modulate the coding potential of 8-oxoG. These results provide mechanistic insights into substrate specificity, lesion bypass and the role that the polymerase and incoming nucleotide have on the anti-/syn-equilibrium of 8-oxoG. In addition to the mechanistic information obtained, we have also gained insight concerning the impact the incoming nucleotide has on the conformation of the templating 8-oxoG and the point at which the 8-oxoG switches between the syn- or anti-conformations.
MATERIALS AND METHODS
Human R283K pol β protein was overexpressed in Escherichia coli and purified as previously described (9). Binary complex crystals with a templating 8-oxoG in a 1-nt-gapped DNA were grown in a solution containing 50 mM imidazole, pH 7.5, 17% PEG3350 and 350 mM sodium acetate, as previously described (7,10). The binary complex crystals were transferred to a cryosolution containing 12% ethylene glycol, 50 mM imidazole, pH 7.5, 20% PEG3350, 90 mM sodium acetate, 5 mM non-hydrolyzable dNTP analogues (dCMP(CF2)PP or dAMP(CH2)PP) and 200 mM MnCl2. This resulted in formation of the ternary closed complex. Data for the binary complex crystal structure were determined using the same cryosolution described above, but lacking the non-hydrolyzable dNTP analogues and MnCl2. The dAMP(CH2)PP was purchased from Jena Biosciences.
Data were collected at 100 K on a SATURN92 CCD detector system mounted on a MiraMax-007HF rotating anode generator. Data were processed and scaled using the HKL2000 software package (11). Initial models were determined using molecular replacement with the previously determined open (3ISB), closed (2FMS), or mismatch (3C2M) structure of pol β as a reference (12). All Rfree flags were taken from the starting model. Refinement was performed using PHENIX and model building using Coot (13,14). The figures were prepared in PyMOL (Schrödinger LLC). Ramachandran analysis determined 100% of non-glycine residues lie in the allowed regions and at least 98% in favoured regions. The R283K pol β 8-oxoG binary structure was refined with the templating 8-oxoG base in alternate conformations. After refinement, the occupancies were determined to be 0.5 for each conformation.
A 34-mer oligonucleotide DNA substrate containing a single-nucleotide gap with a templating guanine was created as previously described with a templating guanine or 8-oxoG in the coding position (15). Steady-state kinetic parameters for single-nucleotide gap filling reactions were determined as previously described (16).
RESULTS
To probe the influence of pol β on the anti-/syn-conformation of a templating 8-oxoG, we have utilized a combination of site-directed mutagenesis, kinetic and structural studies to gain mechanistic insights for 8-oxoG lesion bypass during BER. Upon polymerase subdomain closure, Arg283 in α-helix N forms a hydrogen bond with the oxygen of the nucleotide sugar (O4′) upstream of the templating base, stabilizing the closed state of the polymerase. When 8-oxoG serves as the templating (coding) nucleotide, Arg283 is also within hydrogen bonding distance of the adducted oxygen (O8) in the closed conformation (Figure 1B). These observations indicate that Arg283 has two roles during pol β catalysis when an 8-oxoG is in the templating position, stabilizing both the closed polymerase state and the syn-conformation of 8-oxoG. These roles make Arg283 an ideal target for studying the mechanism of 8-oxoG lesion bypass.
R283K pol β incorporation opposite 8-oxoG
To analyse the impact that Arg283 has on the syn-conformation of 8-oxoG, we replaced arginine with lysine generating R283K pol β. We chose to utilize the conservative lysine substitution because it has only a moderate impact on polymerase insertion efficiency with non-damaged guanine in a 1-nt-gapped substrate (Figure 2A and Supplementary Figure S1). In comparison, previous studies have shown that alanine substitution results in a dramatic loss (>30 000-fold) in catalytic efficiency, likely due to the failure of the polymerase to undergo closure (17). Steady-state kinetic studies where MgCl2 was substituted with MnCl2 as a divalent cofactor were also undertaken (Figure 2). Manganese was necessary for crystallographic characterization of the closed substrate complex of the R283K mutant enzyme (see below). This ensured that the solution studies correlated with the structural analysis. Importantly, although the catalytic efficiencies with MnCl2 are greater than that observed with MgCl2, the specificities (i.e. discrimination) are similar with these alternate divalent metals (Figure 2A and Supplementary Figure S1).
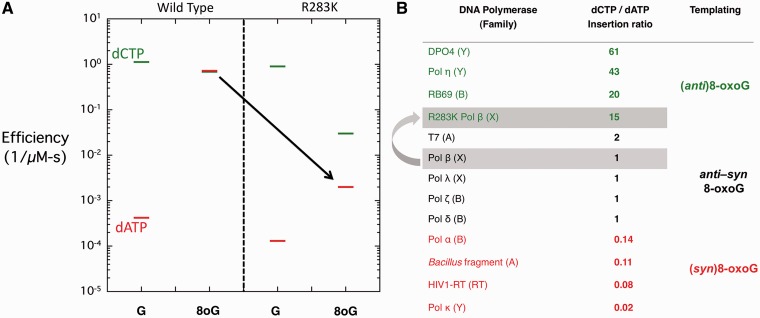
Figure 2.
Steady-state kinetic analysis of wild-type and R283K pol β. (A) A discrimination plot of 1-nt gap filling by wild-type and R283K pol β opposite non-damaged guanine (G) and 8-oxoG (8oG) with dCTP or dATP in the presence of MnCl2. The dCTP and dATP incorporation is shown with a green and red bar, respectively. The black arrow highlights the impact of the R283K mutation on dATP incorporation opposite 8-oxoG. (B) Table of select DNA polymerases (polymerase family) that have been characterized kinetically for incorporation opposite 8-oxoG (6,18,19,20–32). The dCTP/dATP ratio shows the preference for incorporation of dCTP relative to dATP. A value of 1 indicates no preference of incorporation, a value >1 indicates a preference for dCTP and a value <1 indicates a preference for dATP incorporation. The likely preferred glycosidic conformation of the templating 8-oxoG is indicated.
Figure 2A shows a discrimination plot with the catalytic efficiencies for dCTP and dATP incorporation opposite guanine (G) or 8-oxoG (8oG) by wild-type and R283K pol β with MnCl2. In this type of plot, the magnitude of discrimination or fidelity is represented by the distance between the log of the catalytic efficiencies of the alternate substrates. Opposite a non-damaged guanine, both the wild-type and R283K pol β have similar catalytic efficiencies, preferentially inserting dCTP by 3000- and 7000-fold, respectively. The similar efficiencies for dATP insertion opposite guanine suggest that wild-type and mutant enzymes are forming structurally similar complexes in the presence of an incorrect nucleotide. In contrast, dCTP and dATP incorporation opposite 8-oxoG by wild-type and R283K is very different (Figure 2A). Wild-type pol β incorporates both dCTP and dATP opposite 8-oxoG with nearly the same efficiency. This results from the enhanced incorporation efficiency of dATP opposite 8-oxoG because of the Hoogsteen hydrogen bonding interactions between the incoming dATP and syn-8-oxoG (Figure 1A). In contrast, the catalytic efficiency of R283K is reduced opposite 8-oxoG. With an incoming dATP, R283K fails to show the dramatically enhanced efficiency opposite 8-oxoG observed with wild-type enzyme (highlighted by a black arrow in Figure 2A). This suggests that it is unable to form a stable Hoogsteen base pair with 8-oxoG.
The specificity for insertion of dCTP and dATP opposite 8-oxoG for select members from different DNA polymerase families is illustrated in Figure 2B. This insertion ratio provides insight into the templating base and preferential incorporation of either dATP or dCTP opposite 8-oxoG. Wild-type pol β has an insertion ratio of ∼1 suggesting that the templating 8-oxoG is stable in either the anti- or syn-conformation. In comparison, the Bacillus fragment (A-family) preferentially incorporates dATP opposite 8-oxoG (dCTP/dATP = 0.11), suggesting a preferential syn-conformation of 8-oxoG in the active site (18), whereas RB69 DNA polymerase (B-family) preferentially incorporates dCTP (dCTP/dATP = 20), suggesting an anti-conformation of 8-oxoG is preferred in the nascent base pair binding pocket (19). For pol β, the single lysine substitution at Arg283 results in a mutant enzyme that is less error prone (dCTP/dATP = 15), preferentially incorporating dCTP opposite 8-oxoG.
Structural characterization of the R283K pol β binary complex with templating 8-oxoG
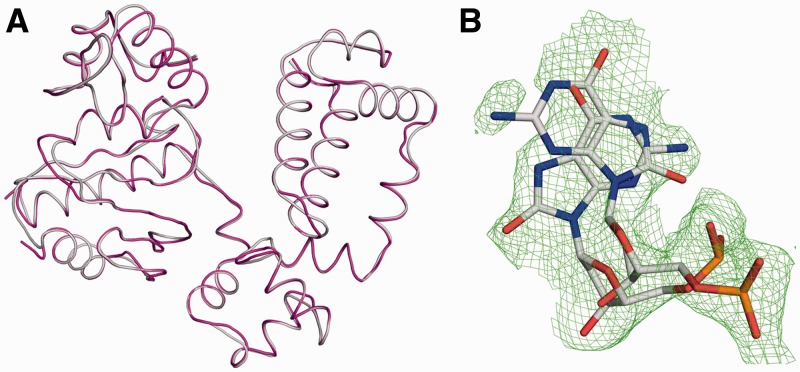
Structural studies with the open binary DNA complex of wild-type enzyme with a templating 8-oxoG indicate that 8-oxoG can adopt both the anti- and syn-conformations (7). To determine whether a templating 8-oxoG can adopt multiple conformations in the binary complex of the R283K mutant enzyme, we determined the crystallographic structure to 1.95 Å (Table 1). Figure 3A shows an overlay of the binary R283K pol β complex and the wild-type binary complex (PDB code 3ISB; root mean square deviation (RMSD) of 0.17 Å over 307 Cα atoms). The low RMSD indicates that the R283K mutation does not have an appreciable impact on the open conformation of the protein and inspection of the active site indicates no localized structural perturbations when compared with wild-type enzyme.
Table 1.
Data collection and refinement statistics
| R283K pol β 8-oxoG | R283K pol β 8-oxoG:dCMP(CF2)PP | R283K pol β 8-oxoG:dAMPCPP | |
|---|---|---|---|
| Data collection | |||
| Space group | P21 | P21 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 54.8,79.3,54.3 | 55.1,77.4,55.1 | 54.9,78.7,55.1 |
| α, β, γ (°) | 90,105.6,90 | 90,114.4,90 | 90,112.6,90 |
| Resolution (Å) | 50-1.95 | 50-2.20 | 50-2.0 |
| Rsym or Rmergea (%) | 11.4 (50.0) | 9.7 (57.9) | 6.7 (41.1) |
| I/σI | 11.2 (2.3) | 14.7 (2.9) | 22.0 (2.4) |
| Completeness (%) | 98.3 (86.3) | 99.9 (100) | 99.9 (100) |
| Redundancy | 4.7 (3.4) | 5.8 (5.7) | 4.8 (2.9) |
| Refinement | |||
| Resolution (Å) | 1.95 | 2.20 | 2.0 |
| No. reflections | 29 554 | 36 585 | 27 692 |
| Rwork/Rfree | 20.2/25.8 | 23.1/29.2 | 21.9/27.7 |
| No. atoms | |||
| Protein | 2631 | 2524 | 2522 |
| DNA | 658 | 634 | 634 |
| Water | 363 | 109 | 193 |
| B-factors (Å2) | |||
| Protein | 26.6 | 42.5 | 37.8 |
| DNA/dNTP/8OG | 28.3/–/49.3(47.1) | 51/39.8/64.8 | 42.3/35.8/69.3 |
| Water | 29.6 | 34.4 | 36.2 |
| RMS deviations | |||
| Bond lengths (Å) | 0.008 | 0.008 | 0.009 |
| Bond angles (°) | 1.123 | 1.153 | 1.192 |
| RCSB ID code | 4GXI | 4GXJ | 4GXK |
aHighest resolution shell is shown in parentheses.
Figure 3.
R238K pol β binary complex with a templating 8-oxoG in the active site. (A) Overlay of the wild-type (PDB ID 3ISB) and R283K (PDB ID 4GXI) pol β binary complex structures in grey and purple, respectively. (B) A Fo − Fc omit map (green) contoured at 2.5σ is shown for the templating 8-oxoG in the R283K pol β binary structure. The templating 8-oxoG can be modelled in both the syn- and anti-conformations with the phosphate backbone in two orientations.
Examination of the 8-oxoG in the templating position indicates that 8-oxoG has an elevated B-factor (48 Å2) relative to the rest of the DNA (28 Å2). The high B-factor correlates with the globular density for this templating base. As shown in Figure 3B, 8-oxoG can be modelled into an omit map in either the anti- or syn-conformations and likely adopts multiple orientations in the active site when pol β is in an open conformation. The templating 8-oxoG was initially modelled and refined in either the syn- or anti-conformation (Supplementary Figure S2). However, omit map analysis of either conformer indicated the existence of multiple conformations. In addition, the phosphodiester backbone of the templating nucleotide can be modelled into two stable conformations. These observations are consistent with the templating 8-oxoG in the R283K pol β open binary conformation being in equilibrium between the anti- and syn-conformations prior to binding the incoming nucleotide and undergoing polymerase subdomain closure.
Structure of R283K ternary 8-oxoG complexes
To determine the crystallographic structure of the ternary R283K pol β protein with a templating 8-oxoG, we utilized non-hydrolyzable dNTP analogues to prevent catalysis while maintaining the catalytically active primer terminus. Hydrolysis of the incoming nucleotide by an activated water molecule that infiltrates the active site has been observed in the absence of a primer O3′ (i.e. dideoxy-terminated primer sugar) (8). Importantly, these analogues are excellent mimics of dNTPs having similar metal affinities and coordination properties as native nucleotides (33). Since Arg283 is involved in stabilizing the closed conformation of pol β, it was not surprising that in the presence of MgCl2 the mutant enzyme remained in an open conformation (see below). This is consistent with the moderate loss of catalytic efficiency (Figure 2A and Supplementary Figure S1). As demonstrated previously (34), we utilized MnCl2 to promote polymerase closure to overcome the weak ability of R283K pol β to form a closed complex.
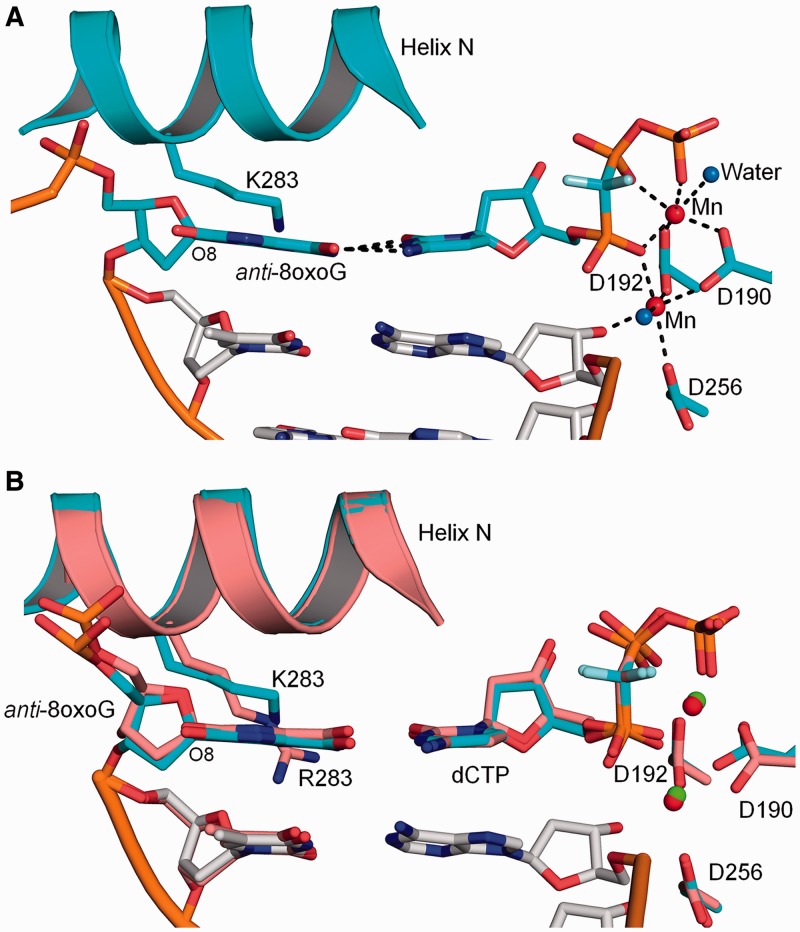
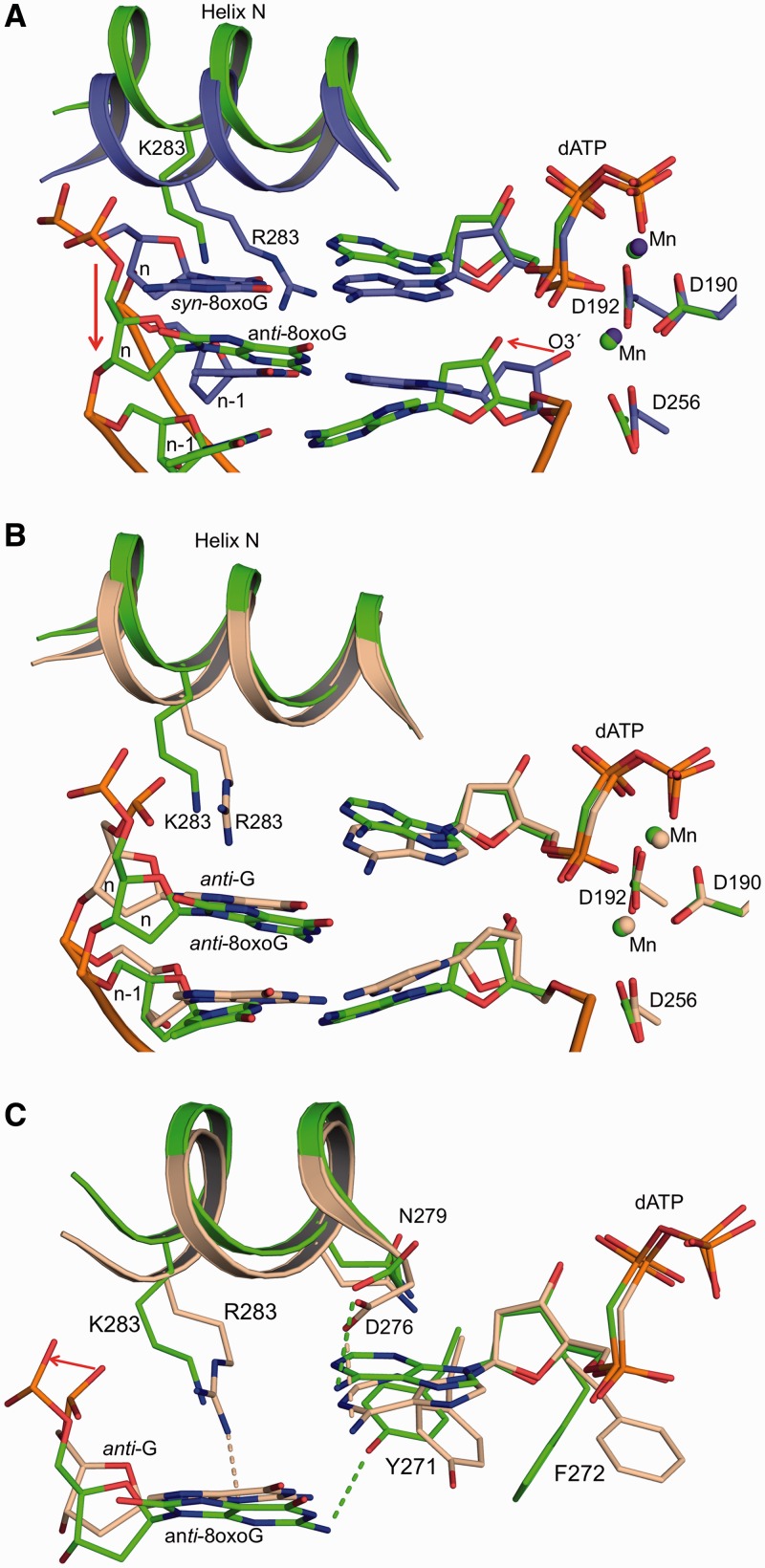
Crystals of the R283K ternary complex with a templating 8-oxoG (anti-conformation) base pairing with an incoming dCTP diffracted to 2.2 Å. This ternary complex is closed with α-helix N forming one face of the nascent base pair binding pocket (Figure 4). A superposition with the analogous wild-type complex indicates that the mutant structure is similar to that of the wild-type complex with an RMSD of 0.40 Å (305 Cα) (Figure 4B). In contrast to arginine at residue 283, Lys283 fails to form a hydrogen bond with the oxygen (O4′) of the sugar ring upstream of 8-oxoG. The incoming dCTP forms Watson–Crick hydrogen bonds with 8-oxoG, while the two Mn2+ ions are coordinated with octahedral geometry that include oxygens from the primer terminus (O3′), nucleoside triphosphate and active site aspartates (Asp190, 192 and 256). As observed in the wild-type ternary complex (7,8), 8-oxoG in the R283K complex adopts the anti-conformation and the phosphodiester backbone is displaced by the adducted oxygen (O8). The position of the 8-oxoG nucleotide (base and phosphodiester backbone) is nearly identical in both the wild-type and mutant ternary complexes (Figure 4B).
Figure 4.
R283K pol β ternary complex with a templating anti-8-oxoG:dCMP(CF2)PP base pair. (A) R283K pol β ternary complex active site with 8-oxoG in the anti-conformation base pairing with dCMP(CF2)PP. The key hydrogen bonding interactions are shown as black dashes. Mn2+ and water molecules are shown as red and blue spheres, respectively. The key active site residues and Lys283 (K283) are shown in stick format. (B) Structural overlay of the wild-type and R283K pol β ternary complexes are shown in salmon and cyan, respectively. Mn2+ from the R283K pol β structure is shown as red spheres and the Mg2+ from the wild-type pol β structure is shown in green.
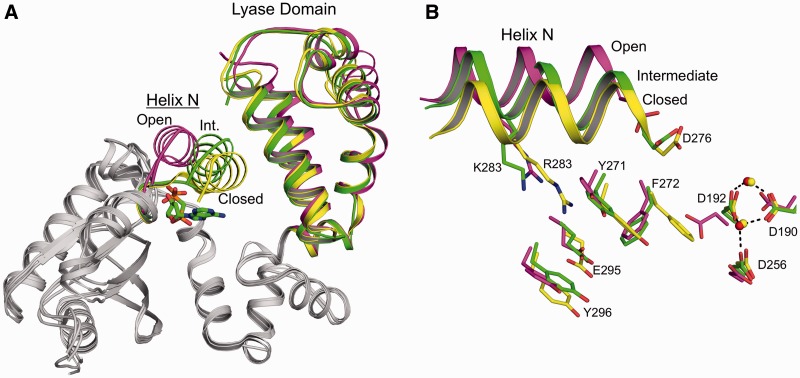
To provide molecular insight into the kinetic observation of the reduced dATP incorporation opposite 8-oxoG by the R283K mutant (Figure 2A), we solved the crystallographic structure of the R283K pol β ternary complex with an incoming dATP analogue and templating 8-oxoG to 2.0 Å (Table 1). Surprisingly, the structure shows that the R283K mutant failed to undergo complete closure and is trapped in an intermediate conformation (Figure 5). Structural studies with wild-type pol β indicate that during polymerase closure, a moderate structural repositioning occurs at the lyase domain whereas the largest structural change occurs with the pol β N-subdomain (i.e. fingers of right-handed DNA polymerases). This repositioning results in the nascent base pair sandwiched between the primer terminal base pair and α-helix N. As shown in Figure 5A, α-helix N of R283K is in an intermediate position between the open and closed states. The lyase domain has undergone complete closure, indicating that this partial closure event is likely due to the R283K mutation in α-helix N. A signalling cascade between the active site and α-helix N during polymerase closure involves Asp192, Arg258, Tyr271, Phe272, Arg283, Glu295 and Tyr296 (35–37). Figure 5B illustrates the relative position of these key residues for the open (PDB code 3ISB), intermediate (R283K) and closed (PDB code 2FMS) states of pol β. The aspartate residues involved in binding the active site metal ions are shown coordinating the MnCl2 ions in the intermediate and closed conformations. In comparison, other key residues for the intermediate R283K ternary complex are in alternative conformations between the open and closed states. Lys283 is in an intermediate location due to the loss of the hydrogen bond in the DNA minor groove. Tyr271 is positioned in an intermediate conformation and has not fully moved to its closed position, thus failing to promote the movement of Phe272 to its ‘closed’ conformation. The intermediate conformation of Tyr271 is likely stabilized by a hydrogen bond between its side chain hydroxyl and N2 of 8-oxoG (Figure 6B, discussed below).
Figure 5.
Intermediate conformation of the R283K pol β ternary complex with templating anti-8-oxoG and incoming dAMPCPP. (A) Structural overlay of the R283K anti-8-oxoG:dAMPCPP ternary complex with the wild-type open binary (PDB ID 3ISB) and closed ternary (PDB ID 2FMS) pol β complexes shown in green, purple and yellow, respectively. The lyase domain and α-helix N (Helix N) are indicated. The position of α-helix N is designated as open, intermediate (Int.) or closed for each structure. The incoming nucleotide for the R283K ternary complex is coloured green in the active site. (B) Key amino acids that are repositioned during subdomain closure are shown for the open (PDB ID 3ISB), intermediate (R283K 8-oxoG:dATP) and closed (PDB ID 2FMS) pol β complexes. Helix N is shown in a ribbon representation and key amino acids are in stick format. Mn2+ and Mg2+ are shown as red and yellow spheres, respectively.
Figure 6.
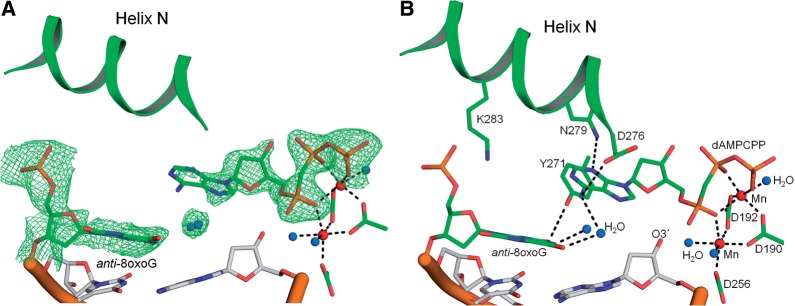
R283K pol β ternary complex with a templating anti-8-oxoG and incoming dAMPCPP. The templating anti-8-oxoG, incoming dAMPCPP and α-helix (Helix) N are shown in green. Mn2+ and water molecules are shown as red and blue spheres, respectively. (A) Active site of the R283K pol β anti-8-oxoG:dAMPCPP with a Fo − Fc map (green) contoured at 3σ. (B) Key active site hydrogen bonding interactions between the templating anti-8-oxoG and incoming dATP are shown with black dashes. The key amino acids are shown in stick format and the octahedral geometry for each metal ion is illustrated.
In the wild-type pol β syn-8-oxoG:dATP ternary complex, Arg283 stabilizes both the syn-conformation of 8-oxoG and the closed polymerase complex (7). By putting a lysine residue at this position, the kinetic studies suggest that the syn-conformation of 8-oxoG in the active site was not as common as with wild-type enzyme, indicated by the decreased dATP insertion efficiency (Figure 2A). Consistent with the kinetic studies, the structure of the corresponding ternary complex of the mutant indicates that the dATP analogue fails to base pair with the templating 8-oxoG. Figure 6A shows an omit map for the templating anti-8-oxoG and incoming dATP in the R283K active site. The incoming dATP analogue shows clear density for the sugar and phosphate moieties, while the base is more diffuse indicative of multiple positions and is consistent with previous structures of mismatches in the confines of the pol β active site (38). Importantly, the templating 8-oxoG adopts an anti-conformation with the phosphate backbone shifted away from the adducted oxygen. In addition, the inability of the incoming nucleotide to be stabilized with Hoogsteen hydrogen bonds suggests that the syn-conformation of 8-oxoG is not significantly populated. Figure 6B shows key hydrogen bonding interactions in the R283K active site. Lys283 fails to form hydrogen bonding interactions with 8-oxoG or the sugar of the upstream template nucleotide. The templating 8-oxoG forms hydrogen bonding interactions with two water molecules that have infiltrated the active site, likely due to the partially open intermediate state of α-helix N. These water molecules are also within hydrogen bonding distance of N6 of the incoming dATP. The N2 of anti-8-oxoG is hydrogen bonding to the hydroxyl of Tyr271. The incoming dATP analogue is within hydrogen bonding distance of Asp276(Oγ) and Asn279(Nδ2) with N6 and N3 of dATP, respectively. The triphosphate of the incoming nucleotide participates in the octahedral coordination of the two active site MnCl2 ions. Interestingly, the primer terminus is not coordinating the catalytic metal ion and has shifted away from the triphosphate permitting a water molecule to occupy this coordination position.
Overlaying the ternary substrate complexes of wild-type and mutant enzymes with 8-oxoG/dATP shows significant differences in the active site with an RMSD of 0.8 Å over 291 Cα atoms. Localized analysis of the active site of the two proteins (Figure 7A) shows that the R283K ternary complex is not fully closed in contrast to wild-type enzyme. The templating 8-oxoG in the R283K pol β active site has undergone major adjustments shifting upstream by 3.7 Å at the sugar moiety. This upstream template shift results in movement of the primer terminus away from the catalytically competent orientation. Overlaying the mismatch structure of wild-type pol β containing a templating guanine and incoming dATP analogue with the mutant anti-8oxoG:dATP structure has an RMSD of 0.34 Å over 277 Cα atoms, highlighting the similarities between the two structures. Figure 7B shows that while α-helix N of the R283K structure has not undergone complete closure, the templating strand, incoming nucleotide and primer terminus are in a similar orientation as the wild-type mismatch structure. Comparing key amino acid interactions indicates that Asp276 and Asn279 form similar contacts in the mismatch and mutant structures (Figure 7C); the major differences are at residues Lys283, Tyr271 and Phe272. The lysine at 283 in the mutant fails to interact with N3 of the templating guanine as observed in the wild-type mismatch structure (38); the loss of this interaction probably prevents complete closure of the N-subdomain. As a result of the partial closing of the mutant, Tyr271 and Phe272 are in intermediate positions. The similarities between the wild-type mismatch and R283K/8oxoG:dATP structures are consistent with the kinetic characterization (Figure 2A). The incorporation efficiency of dATP insertion opposite 8-oxoG with the R283K mutant is similar to that observed for wild-type enzyme opposite a non-damaged guanine. Thus, the mutant discriminates against dATP insertion opposite 8-oxoG through a similar conformational strategy as wild-type enzyme to deter insertion of dATP opposite guanine.
Figure 7.
Structural comparison of R283K anti-8-oxoG:dAMPCPP and wild-type pol β ternary complexes. Active site aspartate residues (D190, D192 and D256), residue 283 (K283 or R283), templating 8-oxoG, incoming nucleotide and the primer terminus (O3′) are indicated. (A) Structural overlay of the R283K anti-8-oxoG:dAMPCPP and wild-type syn-8-oxoG:dAMPCPP pol β ternary complexes are shown in green and purple, respectively. The key movements of the primer terminus and templating strand are highlighted with red arrows. The templating nucleotide is labelled n and its upstream neighbour is n − 1. (B) Structural overlay of the R283K anti-8-oxoG:dATP and mismatch wild-type G:dATP pol β (PDB ID 3C2M) ternary complexes are shown in green and brown, respectively. (C) Key amino acid contacts between the active site residues and the templating base or incoming nucleotide of the R283K anti-8-oxoG:dATP and mismatch wild-type G:dATP pol β ternary complexes are shown with green and brown dashes, respectively. The movement of the phosphate backbone as a result of the adducted oxygen on 8-oxoG templating base is highlighted with a red arrow.
DISCUSSION
The oxidized purine 8-oxoG is able to adopt either an anti- or syn-conformation that will promote error-free or mutagenic replication, respectively. The alternate coding potential of 8-oxoG is modulated by the glycosidic torsion angle that controls whether its Watson–Crick or Hoogsteen edges are used for base pairing with cytosine or adenine, respectively. Structural characterization of duplex DNA containing 8-oxoG indicates that the glycosidic preference of 8-oxoG is determined by its base-pairing partner, inducing the syn-conformation when paired with adenine (39,40) and the anti-conformation when paired with cytosine (41,42). The 8-oxoG nucleoside favours the syn-conformation due to steric repulsion with the sugar. In this context, 8-oxodGTP is preferentially inserted opposite adenine since the anti-conformation is excluded from the dNTP-binding pocket due to steric and electrostatic barriers between O8 and Pα (43).
While the structural studies provide insight into the inherent conformations of 8-oxoG when in solution or duplex DNA, kinetic analysis of polymerase specificity tabulated in Figure 2B indicates that the polymerase active site influences the conformation of 8-oxoG during replication. To date only a select number of polymerases have been structurally characterized with a templating 8-oxoG in the active site, these include RB69 (B-family) (19,44,45), Dpo4 (Y-family) (20), pol κ (Y-family) (21,22), pol ι (Y-family) (46), pol η (Y-family) (47), T7 (A-family) (48) and pol β (X-family) (7,8). Structures of ternary substrate complexes indicate that with an incoming dCTP-positioned opposite anti-8-oxoG, very little structural changes are required to accommodate the lesion in the active site. In the case of pol β, the only appreciable change is the repositioning of the phosphate backbone of 8-oxoG to accommodate O8 of the modified guanine residue (7,8) (Figure 1). In other cases (e.g. RB69 and T7), the phosphate backbone does not sterically interfere with O8 due to a 90° bend in the template strand at the active site permitting the polymerase to modify the conformation of the sugar–phosphate backbone (19,48). There are however specific active site contacts with the templating 8-oxoG base. Both T7 (48,49) and Dpo4 DNA polymerases (20) form stabilizing hydrogen bonds with the anti-8-oxoG at the adducted oxygen (O8). RB69 has a strong preference for inserting dCTP opposite anti-8-oxoG due to interactions with the adducted oxygen (O8), discouraging the syn-8-oxoG conformation (44). Altering these residues enhances 8-oxoG mutagenesis (20,44,49). Together these studies highlight the impact that the active site has on conformational selection of the anti-/syn-conformation of 8-oxoG in both the templating and incoming nucleotide positions.
Active site contacts regulate mutagenic or error-free bypass of 8-oxoG
The kinetics of nucleotide incorporation opposite 8-oxoG by wild-type pol β indicates a lack of discrimination for dCTP and dATP incorporation (Figure 2). This implies that in solution the active site easily accommodates both the anti- and syn-conformations of 8-oxoG and that the incoming nucleotide may dictate the orientation of the templating base in the pol β active site. This is also consistent with the open binary pol β structure displaying 8-oxoG in multiple conformations based on diffuse electron density and high B-factors (Figure 3 and Supplementary Figure S2). However, structural studies of pol β with a templating syn-8-oxoG Hoogsteen base pairing to dATP indicate that Arg283 forms a stabilizing hydrogen bond with O8 in the closed ternary complex (7). This contact is unique to syn-8-oxoG and not observed with non-damaged guanine and anti-8oxoG; in these latter cases, Arg283 interacts with the nucleotide upstream of the templating nucleotide. Thus, the pol β active site appears to influence the anti-/syn-conformation of 8-oxoG. To test whether this contact influences 8-oxoG specificity, we generated a R283K mutant that resulted in pol β favouring error-free dCTP incorporation opposite anti-8oxoG (Figure 2). This indicates that the nucleotide alone is not able to dictate the orientation of the 8-oxoG in the active site of pol β and that active site contacts play a key role in regulating the error-free and mutagenic bypass of 8-oxoG. Surprisingly, the loss of Arg283 interactions resulted in pol β utilizing a mismatch strategy to avert misinsertion of dATP opposite 8-oxoG. This strategy utilizes the formation of a mismatch ternary complex in which the templating strand is shifted upstream, repositioning of the primer terminus away from the catalytic metal, and the subsequent generation of an apparent abasic site (i.e. the templating pocket lacks a base) (38). Together these studies infer that the base pairing of the incoming dATP to the templating 8-oxoG is not strong enough to maintain or induce the syn-conformation, and that the closed polymerase active site has an inherent preference for the anti-conformation that can only be overcome by Arg283 stabilizing the syn-conformation of 8-oxoG.
Previous computational studies with wild-type pol β containing a templating syn-8-oxoG Hoogsteen base pairing with dATP resulted in the polymerase having a partially open active conformation (50). This was a consequence of α-helix N failing to undergo complete closure. In failing to undergo closure, key amino acids that are involved in facilitating polymerase closure and enzyme activation fail to reach the fully closed conformation. These studies also observed that an anti-8oxoG failed to hydrogen bond with an incoming dATP and resulted in a mismatched ternary complex in the active site. The intermediate state of the N-subdomain results in additional water molecules penetrating the active site (Figure 6). This is consistent with other structural studies using Bacillus fragment Pol I where incomplete subdomain (i.e. fingers) closure permitted additional water molecules to penetrate the active site (51).
Stabilization of 8-oxoG during polymerase closure
The diffuse electron density for the templating 8-oxoG in the binary pol β active site indicates that 8-oxoG is in equilibrium between anti- and syn-conformations prior to polymerase closure (Figure 3). In comparison, 8-oxoG in the closed ternary complex of wild-type pol β exhibits a single glycosidic conformation indicating that the polymerase has captured 8-oxoG in a stable anti- or syn-orientation depending on the incoming nucleotide (7,8). This raises the question of when does 8-oxoG form a stable configuration in the polymerase active site, during or after polymerase closure. Structural analysis of the wild-type and R283K pol β active site in the closed ternary complex indicates the templating 8-oxoG is unable to accommodate glycosidic rotation between the anti- and syn-configurations after closure. Supplementary Figure S3 indicates that the nascent base pair binding pocket is spatially limited in the closed conformation and would not permit free rotation around the glycosidic bond of the templating nucleotide. The inability to stabilize the syn-8oxoG conformation in the R283K active site results in a mismatch ternary complex inferring that the polymerase requires Arg283 to stabilize the error-prone syn-conformation. For Arg283 to contact the templating 8-oxoG, it must move ∼6 Å towards the active site during polymerase closure. This large translation necessary for Arg283 to stabilize the syn-8-oxoG indicates the polymerase captures the 8-oxoG conformation during polymerase closure and the lack of this contact during closure results in a mismatch ternary complex. This type of mechanism allows the polymerase to scan the templating base during subdomain closure and capture either the anti- or syn-conformation based on the incoming nucleotide together with the energetically stable conformation of the templating base.
Based on the work presented here, we propose mechanistic steps of 8-oxoG lesion bypass by pol β. First, pol β binds to single-nucleotide-gapped DNA with a templating 8-oxoG forming an open binary state with the phosphate backbone and templating 8-oxoG in equilibrium between both the anti- and syn-conformations. Then, an incoming nucleotide binds to the open binary complex. In the case of an incoming dCTP, the equilibrium is shifted to the anti-8oxoG conformation and stabilized by both Watson–Crick hydrogen bonding interactions and the movement of the phosphodiester backbone. In the case of an incoming dATP, the nucleotide likely forms weaker Hoogsteen hydrogen bonds with the templating syn-8-oxoG in the open conformation, but does not appreciably shift the 8-oxoG equilibrium towards the syn-conformation. This is based on the inability of the incoming dATP to stably form a Hoogsteen base-pairing interaction in the absence of the Arg283 residue. In addition, recent studies by our group to structurally characterize nucleotide-binding states to the open conformation of pol β with an 8-oxoG templating base consistently captured the incoming dCTP base pairing with the anti-8oxoG in the open conformation, but never observed Hoogsteen base pairing with the incoming dATP (34). Following nucleotide binding, pol β undergoes closure with an incoming dCTP base pairing to anti-8oxoG, resulting in a stable ternary complex poised for insertion. In the case of an incoming dATP, wild-type pol β captures the syn-conformation through hydrogen bonding interactions between Arg283 and O8 of 8-oxoG during subdomain closure. This syn-conformation is further stabilized by the incoming dATP Hoogsteen base pairing with the templating base to form the final ternary complex poised for insertion. The lack of the Arg283 contact during subdomain closure results in the templating 8-oxoG having a preferential orientation for the anti-conformation resulting in a mismatch ternary complex, leading to reduced incorporation of dATP.
The ability of a polymerase to undergo mutagenic or error-free lesion bypass has important ramifications for the replication of the genome. In the case of pol β, the balance between the anti- and syn-conformations is not dictated solely by the incoming nucleotide and is partially controlled by active site residues that contact the templating base. In this sense, pol β actively promotes mutagenesis through stabilizing the syn-8oxoG conformations by Arg283 thereby promoting insertion of adenine. This is similar to the mutagenic interaction that Asn279 of pol β provides to stabilize the syn-conformation of an incoming nucleotide 8-oxodGTP while it Hoogsteen base pairs with adenine (6,43). These residues make wild-type pol β mutagenic during interaction with these oxidized substrates since they promote the mutagenic syn-8-oxoG conformation. Accordingly, mutating either the Arg283 or Asn279 residue in pol β results in increased specificity for dCTP incorporation opposite anti-8-oxoG. These results suggest that it might be possible to reduce the mutagenic response of cells to oxidative stress by expressing rationally engineered pol β modified at Asn279 and/or Arg283.
ACCESSION NUMBERS
4GXI, 4GXJ, 4GXK.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
Research Project Numbers [Z01-ES050158 and Z01-ES050159]; Division of Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences and was in association with the National Institutes of Health [1U19CA105010]. Funding for open access charge: Research Project [Z01-ES050158].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr C.E. McKenna for the α,β-difluoromethylene dCTP analogue. We acknowledge support from Lars Pedersen and the Collaborative Crystallography Group at NIEHS.
REFERENCES
- 1.Ames BN, Gold LS. Endogenous mutagens and the causes of aging and cancer. Mutat. Res. 1991;250:3–16. doi: 10.1016/0027-5107(91)90157-j. [DOI] [PubMed] [Google Scholar]
- 2.Culp SJ, Cho BP, Kadlubar FF, Evans FE. Structural and conformational analyses of 8-hydroxy-2′-deoxyguanosine. Chem. Res. Tox. 1989;2:416–422. doi: 10.1021/tx00012a010. [DOI] [PubMed] [Google Scholar]
- 3.Uesugi S, Ikehara M. Carbon-13 magnetic resonance spectra of 8-substituted purine nucleosides: characteristic shifts for the syn conformation. J. Am. Chem. Soc. 1977;99:3250–3253. doi: 10.1021/ja00452a008. [DOI] [PubMed] [Google Scholar]
- 4.Beard WA, Batra VK, Wilson SH. DNA polymerase structure-based insight on the mutagenic properties of 8-oxoguanine. Mutat. Res. 2010;703:18–23. doi: 10.1016/j.mrgentox.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 6.Miller H, Prasad R, Wilson SH, Johnson F, Grollman AP. 8-OxodGTP incorporation by DNA polymerase β is modified by active-site residue Asn279. Biochemistry. 2000;39:1029–1033. doi: 10.1021/bi991789x. [DOI] [PubMed] [Google Scholar]
- 7.Batra VK, Shock DD, Beard WA, McKenna CE, Wilson SH. Binary complex crystal structure of DNA polymerase β reveals multiple conformations of the templating 8-oxoguanine lesion. Proc. Natl Acad. Sci. USA. 2012;109:113–118. doi: 10.1073/pnas.1112235108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krahn JM, Beard WA, Miller H, Grollman AP, Wilson SH. Structure of DNA polymerase β with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure. 2003;11:121–127. doi: 10.1016/s0969-2126(02)00930-9. [DOI] [PubMed] [Google Scholar]
- 9.Beard WA, Wilson SH. Purification and domain-mapping of mammalian DNA polymerase®. Methods Enzymol. 1995;262:98–107. doi: 10.1016/0076-6879(95)62013-3. [DOI] [PubMed] [Google Scholar]
- 10.Batra VK, Beard WA, Shock DD, Krahn JM, Pedersen LC, Wilson SH. Magnesium induced assembly of a complete DNA polymerase catalytic complex. Structure. 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otwinowski Z, Minor W. Processsing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 12.Beard WA, Shock DD, Batra VK, Pedersen LC, Wilson SH. DNA polymerase β substrate specificity: side chain modulation of the ‘A-rule’. J. Biol. Chem. 2009;284:31680–31689. doi: 10.1074/jbc.M109.029843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 15.Cavanaugh NA, Beard WA, Wilson SH. DNA polymerase β ribonucleotide discrimination: insertion, misinsertion, extension, and coding. J. Biol. Chem. 2010;285:24457–24465. doi: 10.1074/jbc.M110.132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beard WA, Shock DD, Wilson SH. Influence of DNA structure on DNA polymerase β active site function: extension of mutagenic DNA intermediates. J. Biol. Chem. 2004;279:31921–31929. doi: 10.1074/jbc.M404016200. [DOI] [PubMed] [Google Scholar]
- 17.Beard WA, Shock DD, Vande Berg BJ, Wilson SH. Efficiency of correct nucleotide insertion governs DNA polymerase fidelity. J. Biol. Chem. 2002;277:47393–47398. doi: 10.1074/jbc.M210036200. [DOI] [PubMed] [Google Scholar]
- 18.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 19.Freisinger E, Grollman AP, Miller H, Kisker C. Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J. 2004;23:1494–1505. doi: 10.1038/sj.emboj.7600158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eoff RL, Irimia A, Angel KC, Egli M, Guengerich FP. Hydrogen bonding of 7,8-dihydro-8-oxodeoxyguanosine with a charged residue in the little finger domain determines miscoding events in Sulfolobus solfataricus DNA polymerase Dpo4. J. Biol. Chem. 2007;282:19831–19843. doi: 10.1074/jbc.M702290200. [DOI] [PubMed] [Google Scholar]
- 21.Irimia A, Eoff RL, Guengerich FP, Egli M. Structural and functional elucidation of the mechanism promoting error-prone synthesis by human DNA polymerase κ opposite the 7,8-dihydro-8-oxo-2′-deoxyguanosine adduct. J. Biol. Chem. 2009;284:22467–22480. doi: 10.1074/jbc.M109.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasquez-Del Carpio R, Silverstein TD, Lone S, Swan MK, Choudhury JR, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Structure of human DNA polymerase κ inseerting dATP Opposite an 8-oxoG DNA lesion. PLoS One. 2009;4:e5766. doi: 10.1371/journal.pone.0005766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown JA, Duym WW, Fowler JD, Suo Z. Single-turnover kinetic analysis of the mutagenic potential of 8-oxo-7,8-dihydro-2′-deoxyguanosine during gap-filling synthesis catalyzed by human DNA polymerases λ and β. J. Mol. Biol. 2007;367:1258–1269. doi: 10.1016/j.jmb.2007.01.069. [DOI] [PubMed] [Google Scholar]
- 24.Carlson KD, Washington MT. Mechanism of efficient and accurate nucleotide incorporation opposite 7,8-dihydro-8-oxoguanine by Saccharomyces cerevisiae DNA polymerase η. Mol. Cell. Biol. 2005;25:2169–2176. doi: 10.1128/MCB.25.6.2169-2176.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Einolf HJ, Guengerich FP. Fidelity of nucleotide insertion at 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase δ: steady-state and pre-steady-state kinetic analysis. J. Biol. Chem. 2001;276:3764–3771. doi: 10.1074/jbc.M006696200. [DOI] [PubMed] [Google Scholar]
- 26.Furge LL, Guengerich FP. Analysis of nucleotide insertion and extension at 8-oxo-7,8-dihydroguanine by replicative T7 polymerase exo(-) and human immunodeficiency virus-1 reverse transcriptase using steady-state and pre-steady-state kinetics. Biochemistry. 1997;36:6475–6487. doi: 10.1021/bi9627267. [DOI] [PubMed] [Google Scholar]
- 27.Haracska L, Prakash L, Prakash S. Role of human DNA polymerase κ as an extender in translesion synthesis. Proc. Natl Acad. Sci. USA. 2002;99:16000–16005. doi: 10.1073/pnas.252524999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haracska L, Prakash S, Prakash L. Yeast DNA polymerase ζ is an efficient extender of primer ends opposite from 7,8-dihydro-8-oxoguanine and O6-methylguanine. Mol. Cell. Biol. 2003;23:1453–1459. doi: 10.1128/MCB.23.4.1453-1459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8- oxoguanine by DNA polymerase η. Nat. Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 30.Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 2006;4:e11. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 32.Zang H, Irimia A, Choi J-Y, Angel KC, Loukachevitch LV, Egli M, Guengerich FP. Efficient and high fidelityincorporation of dCTP opposite 7,8-dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J. Biol. Chem. 2006;281:2358–2372. doi: 10.1074/jbc.M510889200. [DOI] [PubMed] [Google Scholar]
- 33.Blackburn GM, Kent DE, Kolkmann F. The synthesis and metal binding characteristics of novel, isopolar phosphonate analogues of nucleotides. J. Chem. Soc. Perkin Trans. 1984;1:1119–1125. [Google Scholar]
- 34.Freudenthal BD, Beard WA, Wilson SH. Structures of dNTP intermediate states during DNA polymerase active site assembly. Structure. 2012;20:1829–1837. doi: 10.1016/j.str.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawaya MR, Prasad P, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 36.Kirby TW, DeRose EF, Cavanaugh NA, Beard WA, Shock DD, Mueller GA, Wilson SH, London RE. Metal-induced DNA translocation leads to DNA polymerase conformational activation. Nucleic Acids Res. 2012;40:2974–2983. doi: 10.1093/nar/gkr1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase β. Chem. Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 38.Batra VK, Beard WA, Shock DD, Pedersen LC, Wilson SH. Structures of DNA polymerase β with active site mismatches suggest a transient abasic site intermediate during misincorporation. Mol. Cell. 2008;30:315–324. doi: 10.1016/j.molcel.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kouchakdjian M, Bodepudi V, Shibutani S, Eisenberg M, Johnson F, Grollman AP, Patel DJ. NMR structural studies of the ionizing radiation adduct 7-hydro-8- oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-Oxo-7H-dG(syn)•dA(anti) alignment at lesion site. Biochemistry. 1991;30:1403–1412. doi: 10.1021/bi00219a034. [DOI] [PubMed] [Google Scholar]
- 40.McAuley-Hecht KE, Leonard GA, Gibson NJ, Thomson JB, Watson WP, Hunter WN, Brown T. Crystal structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base pairs. Biochemistry. 1994;33:10266–10270. doi: 10.1021/bi00200a006. [DOI] [PubMed] [Google Scholar]
- 41.Oda Y, Uesugi S, Ikehara M, Nishimura S, Kawase Y, Ishikawa H, Inoue H, Ohtsuka E. NMR studies of a DNA containing 8-hydroxydeoxyguanosine. Nucleic Acids Res. 1991;19:1407–1412. doi: 10.1093/nar/19.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lipscomb LA, Peek ME, Morningstar ML, Verghis SM, Miller EM, Rich A, Essigmann JM, Williams LD. X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc. Natl Acad. Sci. USA. 1995;92:719–723. doi: 10.1073/pnas.92.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Batra VK, Beard WA, Hou EW, Pedersen LC, Prasad R, Wilson SH. Mutagenic conformation of 8-oxo-7,8-dihydro-2′-dGTP in the confines of a DNA polymerase active site. Nat. Struct. Mol. Biol. 2010;17:889–890. doi: 10.1038/nsmb.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beckman J, Wang M, Blaha G, Wang J, Konigsberg WH. Substitution of ala for Tyr567 in RB69 DNA polymerase allows dAMP to be inserted opposite 7,8-dihydro-8-oxoguanine. Biochemistry. 2010;49:4116–4125. doi: 10.1021/bi100102s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hogg M, Rudnicki J, Midkiff J, Reha-Krantz L, Doublié S, Wallace SS. Kinetics of mismatch formation opposite lesions by the replicative DNA polymerase from bacteriophage RB69. Biochemistry. 2010;49:2317–2325. doi: 10.1021/bi901488d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirouac KN, Ling H. Unique active site promotes error-free replication opposite an 8-oxo-guanine lesion by human DNA polymerase iota. Proc. Natl Acad. Sci. USA. 2011;108:3210–3215. doi: 10.1073/pnas.1013909108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silverstein TD, Jain R, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis for error-free replication of oxidatively damaged DNA by yeast DNA polymerase η. Structure. 2010;18:1463–1470. doi: 10.1016/j.str.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brieba LG, Eichman BF, Kokoska RJ, Doublie S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brieba LG, Kokoska RJ, Bebenek K, Kunkel TA, Ellenberger T. A lysine residue in the fingers subdomain of T7 DNA polymerase modulates the miscoding potential of 8-oxo-7,8-dihydroguanosine. Structure. 2005;13:1653–1659. doi: 10.1016/j.str.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Reddy S, Beard WA, Wilson SH, Schlick T. Differing conformational pathways before and after chemistry for insertion of dATP versus dCTP opposite 8-oxoG in DNA polymerase β. Biophys. J. 2007;92:3063–3070. doi: 10.1529/biophysj.106.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu EY, Beese LS. The structure of a high fidelity DNA polymerase bound to a mismatched nucleotide reveals an ‘ajar’ intermediate conformation in the nucleotide selection mechanism. J. Biol. Chem. 2011;286:19758–19767. doi: 10.1074/jbc.M110.191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.