Abstract
2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD)-inducible poly(ADP-ribose) polymerase (TiPARP/ARTD14) is a member of the PARP family and is regulated by the aryl hydrocarbon receptor (AHR); however, little is known about TiPARP function. In this study, we examined the catalytic function of TiPARP and determined its role in AHR transactivation. We observed that TiPARP exhibited auto-mono-ADP-ribosyltransferase activity and ribosylated core histones. RNAi-mediated knockdown of TiPARP in T-47D breast cancer and HuH-7 hepatoma cells increased TCDD-dependent cytochrome P450 1A1 (CYP1A1) and CYP1B1 messenger RNA (mRNA) expression levels and recruitment of AHR to both genes. Overexpression of TiPARP reduced AHR-dependent increases in CYP1A1-reporter gene activity, which was restored by overexpression of AHR, but not aryl hydrocarbon receptor nuclear translocator. Deletion and mutagenesis studies showed that TiPARP-mediated inhibition of AHR required the zinc-finger and catalytic domains. TiPARP and AHR co-localized in the nucleus, directly interacted and both were recruited to CYP1A1 in response to TCDD. Overexpression of Tiparp enhanced, whereas RNAi-mediated knockdown of TiPARP reduced TCDD-dependent AHR proteolytic degradation. TCDD-dependent induction of AHR target genes was enhanced in Tiparp−/− mouse embryonic fibroblasts compared with wildtype controls. Our findings show that TiPARP is a mono-ADP-ribosyltransferase and a transcriptional repressor of AHR, revealing a novel negative feedback loop in AHR signalling.
INTRODUCTION
ADP-ribosylation is a ubiquitous post-translational protein modification that is important for many cellular activities, including DNA repair, transcription, apoptosis, proliferation and cell death (1). The synthesis of poly-ADP-ribose polymers from nicotinamide adenine dinucleotide (NAD+) is catalysed by members of the poly(ADP-ribose) polymerase (PARP) family (1).
PARP-1 (also known as ADP-ribosyltransferase diphtheria-like toxin 1 [ARTD1]) is the founding and the most extensively studied member of the PARP family. PARP-1 is activated by DNA strand breaks to modify target proteins with poly-ADP-ribose (2). PARP-1 ribosylates itself (referred to as auto-ribosylation) as well as other nuclear proteins, such as histones (referred to as hetero-ribosylation) (3). Ribosylation of histones has been suggested to be an additional component of the histone code (4). To date, 17 different proteins have been identified that share the PARP catalytic domain and thus potentially exhibit poly(ADP-ribose) polymer synthase activity (5,6). PARP activity is dependent on the presence of a well-conserved glutamate residue within the histidine–tyrosine–glutamate (HYE) catalytic triad motif (5). However, the glutamate residue (E988 in PARP-1) and other important characteristics for poly(ADP-ribose) polymer synthase activity are not conserved or absent in 11 PARP family members (5,7). These findings led to the suggestion that the majority of the PARP family members may exhibit mono-ADP-ribosyltransferase (mART) activity and not poly(ADP-ribose) polymer synthase activity (5). These data prompted some investigators to suggest that the current nomenclature and classification of PARPs are inaccurate (6).
Mono-ADP-ribosylation is the pathogenic mechanism of several different bacterial toxins (8). Diphtheria, cholera and pertussis toxins are mARTs that transfer a single ADP-ribose moiety to their target proteins (9). Extracellular membrane-associated ecto-mARTs are a family of structurally related proteins expressed on the cell surfaces or secreted into the extracellular space, and thus do not contribute to intracellular ADP-ribosyltransferase activity (9). The sirtuin family of NAD+-dependent deacetylases (SIRTs) also exhibits mART activity (10); however, sirtuins are structurally distinct from PARPs and they function primarily as NAD+-dependent protein deacetylases. The identification of the key proteins regulating intracellular and nuclear mART activity has not been determined.
2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-inducible poly(ADP-ribose) polymerase (TiPARP, also known as ARTD14/PARP-7) is an uncharacterized member of the PARP family (1). TiPARP lacks the equivalent catalytic E988 residue of PARP-1 and is predicted to have mART rather than poly(ADP-ribose) polymer synthase activity (7). TiPARP also contains a C-terminal PARP catalytic domain, a centrally located WWE (tryptophan-tryptophan-glutamate) protein interaction domain and a single CCCH-type zinc-finger domain (1,11). The roles of the different domains, the cellular function and molecular targets of TiPARP are not known. Although, increased histone ADP-ribosylation was reported when core histones were incubated with in vitro translated TiPARP, its auto-ribosylation and mono- or poly-ADP-ribosylation activity and the specific histones modified by this enzyme have not been described (12).
TiPARP expression is regulated by TCDD via activation of the aryl hydrocarbon receptor (AHR) (12). The AHR is a ligand-activated transcription factor and member of the basic helix-loop-helix-Per-ARNT-Sim family of transcriptional regulators. The AHR mediates the toxic effects of environmental contaminants, such as TCDD (13). Once activated by ligand, AHR translocates from the cytoplasm into the nucleus where it associates with its obligatory heterodimerization partner the aryl hydrocarbon receptor nuclear translocator (ARNT). The AHR/ARNT heterodimer recognizes a DNA element designated the AHR response element (AHRE) located within the promoter and distal enhancer regions of AHR target genes, such as cytochrome P450 1A1 (CYP1A1), CYP1B1, aryl hydrocarbon receptor repressor (AHRR) and TiPARP (14,15). AHR molecules that fail to interact with ARNT are ubiquitinated and proteolytically degraded (16). TCDD-dependent increases in AHRR levels have been proposed to be part of an autoregulatory feedback loop involving AHR, ARNT and AHRR (17). In this model, AHRR binds and sequesters ARNT to reduce AHR transactivation. However, the validity of this model has been questioned after a recent study reported that AHRR directly interacted with AHR and that ARNT overexpression failed to rescue AHRR-dependent repression of AHR transactivation (18).
The consistent ligand-activated AHR-dependent increases in TiPARP mRNA expression levels across different cell and animal models, as well as the prominent role that PARP family members play in cell function prompted us to investigate the functional properties of TiPARP as well as its potential role in AHR transactivation. Our findings show that TiPARP is a nuclear mART, which exhibits auto- and hetero-ribosylation activities. We also show that TiPARP is a repressor of AHR transactivation, revealing a new mechanism of negative feedback control in AHR signalling.
MATERIALS AND METHODS
Chemicals
TCDD was purchased from Wellington Laboratories (Guelph, ON, Canada). Dimethyl sulfoxide (DMSO) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Cell culture media, foetal bovine serum (FBS) and trypsin were purchased from Wisent (Bruno, QC, USA). All other chemicals and biochemicals were of the highest quality available from commercial vendors.
Plasmids
Human and mouse TiPARP cDNAs were amplified by polymerase chain reaction (PCR) using pCMV6-XL4-hTiPARP or pCMV6-kan/neo-mTiparp as templates, respectively (Origene, Rockville, MD, USA). Chicken TiPARP was PCR amplified with primers previously described (19) using total Gallus gallus liver RNA as template (Zyagen, San Diego, CA, USA). Following amplification, the sequences were cloned into EcoRI and XhoI sites of pcDNA3.1 (Invitrogen, Carlsbad, CA, USA) or EcoRI and SalI sites of pEGFP-C2 (Clontech, Mountain View, CA, USA).
Cell culture
T-47D human breast carcinoma and HuH-7 human hepatoma were purchased from ATCC (Manassas, VA, USA). TREx-FLP-IN 293 human embryonic kidney cells were purchased from Invitrogen. T-47D cells were cultured in 1:1 F12 Ham’s nutrient mixture medium and Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS (v/v) and 1% penicillin-streptomycin. HuH-7 and TREx-FLP-IN 293 cells were cultured in high glucose (4500 mg/l) DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. All cell lines were maintained at 37°C and 5% CO2 and were subcultured every 2–3 days or when they reached 80% confluency.
Derivation of mouse embryonic fibroblasts
Wildtype, Tiparp-deficient (Tiparp−/−) and Tiparp+/− fibroblasts were prepared from E14.5 embryos derived from matings of mice heterozygous for disruption of the Tiparp allele (20) as previously described (21). Mice heterozygous for disruption of the Tiparp allele were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). Primary fibroblasts from wildtype, Tiparp−/− and Tiparp+/− siblings were immortalized at passage 2 after transfection with Simian virus large T antigen (SV40gp6) in pSG5 (Stratagene) and a puromycin resistance plasmid, and selected in puromycin-containing medium. The genotypes of the mouse embryonic fibroblasts (MEFs) were verified by PCR as previously described (20).
Reporter gene assays
HuH-7 cells were transfected with 50–1500 ng pcDNA-TiPARP and/or 250–750 ng pRc-CMV-hAHR (a generous gift from Dr. Patricia Harper) and/or 250–750 ng pcDNA4-hARNT (a generous gift from Dr. Kevin Gardner) or 200 ng pcDNA-mTiparp, pcDNA-chTiPARP, 200–1500 ng pCMV-XL-PARP-1 or pCMV-XL-PARP-12 (Origene) or 200 ng pcDNA-TiPARP truncation or point mutant plasmids and 200 ng of pCYP1A1-luc plasmid using Lipofectamine LTX. All reactions included 50 ng of pCH110-β-Gal (Pharmacia) to normalize for transfection efficiency. After 24 h, cells were treated with TCDD or DMSO for 24 h before luciferase and β-galactosidase assays were performed.
Transient transfection and RNAi
TiPARP (siTiPARP-12, J-013948-12-0005; siTiPARP-13, J-013948-13-0005) ON-TARGETplus siRNAs and DharmaFECT1 transfection reagent were purchased from Dharmacon (Lafayette, CO, USA). Briefly, T-47D cells were transfected with 100 nM of each siRNA against TiPARP, or non-targeting (NT) siRNA #2 (D-001810-02-20) (Dharmacon) were transfected with 4 µl DharmaFECT1. After 48 h, cells were treated with 10 nM TCDD or DMSO (0.1%) for 1 h for chromatin immunoprecipitation (ChIP) analysis (described below) or 1.5–24 h for mRNA analysis.
Transient transfection for RNA expression
HuH-7 cells were transiently transfected with 3 µg pcDNA or pcDNA-TiPARP using Lipofectamine LTX as stated above. After 24-h overexpression, cells were treated with 10 nM TCDD or DMSO (0.1%) for 6 h, and RNA isolation was performed.
RNA isolation and qPCR
T-47D cells were seeded in six-well plates at a density of 300 000 cells/well. After 24 h, cells were treated with 1 µg/ml actinomycin D (Sigma) for 1–24 h. MEFs were seeded in six-well plates at a density of 250 000 cells/well. The following day, cells were treated with 10 nM TCDD for 24 h or pre-treated with 10 µg/ml cycloheximide (CHX, Sigma) for 1 h and then treated with 10 nM TCDD for 6 h. RNA was isolated with RNeasy spin columns (Qiagen). For cDNA synthesis, 500 ng of extracted RNA was reversed transcribed using SuperScriptII (Invitrogen) and random hexamer primers. qPCR was performed with 1 µl of the cDNA synthesis reaction using Kapa SYBR FAST qPCR Master Mix (KapaBiosystems). Primers used to amplify CYP1A1, CYP1B1 and TiPARP mRNAs have been previously described (22,23). All target transcripts were normalized to β-actin and analysed using the comparative CT (ΔΔ CT) method. RNA data were presented as mean and standard error of the mean of three independent experiments.
32P-NAD+ Auto-ADP-ribosylation and Hetero-ribosylation assays
Glutathione S-transferase (GST)-tagged fusion proteins were purified according to standard procedures. Purified PARP-1 was purchased from Trevigen (Gaithersburg, MD, USA). Auto-ribosylation assays were carried out in a 30 µl reaction volume at 30°C in 1× PARP buffer, 2 µCi 32P-NAD+ (Perkin-Elmer, Woodbridge, ON, Canada) and/or 500 µM β-NAD+ (Sigma), 1 µg activated DNA (Sigma) or RNA. Approximately 5 µg of GST fusion partially purified protein, or 1 µg of PARP-1 was used. For hetero-ribosylation experiments, 8 µg of core histones (H2A, H2B, H3, H4) (New England Biolabs, Pickering, ON, Canada) were added to the reaction. Reactions were stopped by addition of 1× sample buffer, separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by autoradiography.
Western Blot
T-47D cells were transfected with siTiPARP, NT, pcDNA or pcDNA-TiPARP as described above. TREx-FLP-IN 293 cells were co-transfected with 1 µg pEGFP-TiPARP and siRNA for 48 h. HuH-7 cells were transfected with 2 µg pEGFP-TiPARP for 24 h, then treated with 25 µM MG-132 (Sigma) for 6 h. Whole cell extracts were prepared from transfected cells treated with 10 nM TCDD or DMSO, and proteins were resolved by SDS-PAGE and transferred to nitrocellulose membrane. The membrane was blocked in 2% (w/v) ECL-Advanced blocking agent for 1 h at room temperature and then incubated with anti-AHR (H-211, Santa Cruz) or anti-β-actin antibody (Sigma) overnight at 4°C.
For GFP blots, membrane was blocked with 10% fat-free milk, phosphate buffered saline (PBS) + 0.1% Tween 20 for 1 h and then incubated with anti-GFP antibody (JL-8, Clontech) overnight. All membranes were then washed and incubated with appropriate secondary antibody for 1 h at room temperature. After washing, the bands were visualized using ECL-Advance chemiluminescent substrate (GE Healthcare, Baie d’Urfe, QC, Canada) or SuperSignal West Dura substrate (Pierce, Rockford, IL, USA) according to the manufacturer's instructions.
For the immortalized MEFs, each cell line was plated at a density of 2.5 × 106 cells in 10-cm dishes. The following day, cells were treated with 10 nM TCDD or DMSO for 24 h, whole cell extracts were prepared and proteins resolved as described above. Membranes were incubated with anti-AHR (Enzo Life Sciences Inc., SA-210) overnight and then incubated for 1 h with anti-rabbit horseradish peroxidase–conjugated secondary antibody.
Chromatin immunoprecipitation assays
T-47D cells were plated in six-well plates in culture media and transfected for RNAi as described above. After 48-h TiPARP knockdown, cells were treated with either 10 nM TCDD or DMSO (0.1%) for 1 h, and ChIP assays were preformed as previously described (24), using 1 µg of anti-AHR, H-211, anti-ARNT or H-172 from Santa Cruz (Santa Cruz, CA, USA). Isolated DNA was quantified by qPCR using Kapa SYBR FAST qPCR Master Mix (KapaBiosystems, Woburn, MA, USA).
For transient overexpression experiments, 3 µg total DNA of pEGFP-TiPARP or pEGFP vector were transfected into HuH-7 cells using Lipofectamine LTX according to manufacturer’s recommendations (Invitrogen, Carlsbad, CA). After 24 h, cells were treated with 10 nM TCDD for 45 min and ChIP assays were performed using anti-GFP (632460, Clontech) at a dilution of 1:100. Primers used to amplify CYP1A1 and CYP1B1 enhancer regions have been previously described (22). Data were reported as percentages relative to 100% total input chromatin.
For immortalized MEFs, each of the cell lines were plated at a density of 2.5 × 106 cells in 10-cm dishes. The following day, cells were treated with 10 nM TCDD or DMSO for 45 min, and ChIP assays were performed using 1 µg of anti-Ahr (SA-210) antibody.
Indirect immunofluorescence
HuH-7 cells were seeded onto coverslips (250 000 cells/coverslip) and transfected with pEGFP-TiPARP, pRc-CMV-AHR and pcDNA-ARNT 24 h after plating. Twenty-four hours after transfection, cells were treated with 10 nM TCDD or DMSO for 1.5 h by addition of ligand directly to culture media, and cells were fixed in 4% paraformaldehyde and permeabilized with 0.4% Triton-X in PBS for 20 min. Cells were blocked in 1% goat serum/0.2% Triton-X for 1 h and incubated with anti-AHR overnight at 4°C. Cells were then incubated with Alexa 568-conjugated secondary antibody (Invitrogen) for 1 h, counterstained with 4,6-diamidino-2-phenylindole (DAPI) and mounted with Vectashield (Vector Laboratories). For detection of endogenous TiPARP, HuH-7 cells were fixated with methanol/acetone and permeabilized with 0.4% Triton-X in PBS for 20 min. The cells were blocked and incubated overnight with anti-TiPARP (84664; Abcam). Cells were then incubated with Alexa 568-conjugated secondary antibody for 1 h, counterstained with DAPI and mounted with Vectashield. Images were acquired using an Imager.Z1 epifluorescence microscope and Axiovision software (Zeiss) following deconvolution or an Olympus Fluoview 1000 confocal microscope.
Co-immunoprecipitation
TREx-FLP-IN 293 cells seeded in six-well plates at a density of 400 000 cells/well were transfected with 650 ng pRc-CMV-AHR, pRc-CMV-AHR-1-425, pCMV-AHR-TAD (464–848) (all generous gifts from Dr. Patricia Harper), 200 ng pcDNA4-hARNT and 650 ng pEGFP-TiPARP-FL or 650 ng pEGFP-TiPARP truncation with Fugene HD (Roche, Laval, QC, Canada) according to the manufacturer’s instructions. Twenty-four hours following transfection, cells were treated with 10 nM TCDD or DMSO for 1.5 h and whole cell extracts were prepared. Two micrograms of anti-AHR antibody (H-211) or anti-GFP (632460, Clontech) 1:200 dilution were incubated with whole cell extracts for 1 h in 4°C with constant rotation followed by incubation with protein-A beads for 1 h. Beads were washed five times with NP-40 buffer (20 mM Tris Base pH 8.0, 150 mM NaCl, 10% glycerol, 1% nonidet P-40, 2 mM ethylenediaminetetraacetic acid) and eluted in 1X sample buffer, separated by SDS-PAGE and transferred to nitrocellulose membrane. Membranes were exposed to anti-GFP (JL-8, Clontech), anti-AHR (H-211) or anti-AHR (N-19, Santa Cruz). All membranes were then washed and incubated with the appropriate secondary antibodies for 1 h at room temperature. After washing, bands were visualized using SuperSignal West Dura substrate or ECL-Advance.
Statistical analysis
All data were presented as means and standard error of the mean. One-way Analysis of Variance (ANOVA) followed by Tukey’s multiple comparison tests or two-tailed Student’s t-tests were used to assess statistical significance (P < 0.05).
RESULTS
TiPARP is a mono-ADP-ribosyltransferase
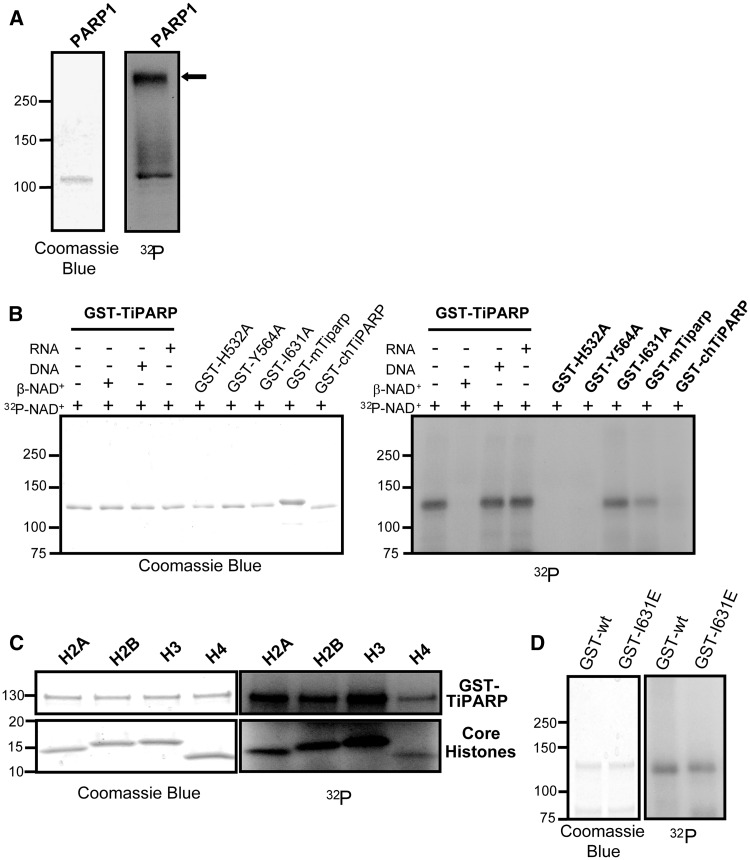
The catalytic activities of PARPs are dependent on a well-conserved glutamate residue within the active site that is a critical residue within the HYE triad sequence (25,26). Sequence alignment of the PARP catalytic core sequences of the 17 PARP family members showed that H532, Y564 and I631 of TiPARP are equivalent to the PARP catalytic HYE triad. Because TiPARP has an isoleucine in place of the catalytic glutamate and a shortened connecting loop, it is predicted to possess mART rather than PARP activity (7); however, this has not been experimentally determined. To examine the catalytic activity of TiPARP, we compared the enzymatic properties of TiPARP with that of PARP-1 using an auto-ADP-ribosylation assay. PARP-1 demonstrated auto-modification with the incorporation of 32P-NAD+ and a marked shift in mobility and smearing pattern, which was indicative of poly(ADP-ribose) polymer formation (Figure 1A). Partially purified GST-TiPARP also showed evidence of auto-modification but no apparent change in mobility, suggesting that TiPARP exhibited mART rather than poly(ADP-ribose) polymer synthase activity (Figure 1B). This activity was competed away with unlabelled β-NAD+. We cannot exclude the possibility that TiPARP generates short oligo-ADP-ribose units because they would not alter the migration of the protein. Because PARP-1 activity was known to be stimulated by its interaction with DNA (27), and TiPARP contained a zinc-finger domain with putative RNA binding capacity (28), we tested whether the presence of DNA or RNA modified TiPARP catalytic activity. Addition of activated DNA or DNAse-treated total RNA did not alter the migration pattern of GST-TiPARP (Figure 1B). The catalytically inactive mutants GST-TiPARP-H532A and GST-TiPARP-Y564A did not show any evidence of auto-modification or shift in mobility (Figure 1B). Surprisingly, GST-TiPARP-I631A demonstrated similar auto-ribosylation activity to that of wildtype, suggesting that this residue was not required for auto-ribosylation activity (Figure 1B).
Figure 1.
TiPARP is a mART with auto- and hetero-ribosylation activities. (A) Auto-ADP-ribosylation of PARP1, purified PARP1 was incubated with 32P-NAD+ and 1 µg activated DNA then analysed the SDS-PAGE and autoradiography (B) Auto-ADP-ribosylation of GST-TiPARP, GST-TiPARP catalytic mutants (GST-TiPARP-H532A, –Y564A and –I631A), GST- and mouse Tiparp (GST-mTiparp) and GST-chicken TiPARP (GST-chTiPARP). Auto-ADP-ribosylation reactions were carried out with semi-purified GST-TiPARP fusion proteins and 32P-NAD+ or co-incubated 500 µM β-NAD+ or 1 µg activated DNA or RNA. Automodified GST-TiPARP fusion proteins were analysed by SDS-PAGE and autoradiography. (C) Hetero-ADP-ribosylation of core histones by GST-TiPARP. GST-TiPARP was incubated with 32P-NAD+ and histones H2A, H2B, H3 or H4 and analysed by SDS-PAGE and autoradiography. (D) Comparison of auto-ADP-ribosylation of GST-TiPARP-I631E (GST-I631E) to wildtype GST-TiPARP (GST-wt). All figures represent data from three independent experiments. The closed arrow denotes mobility shifted PARP1.
Next we evaluated the catalytic activity of mouse and chicken (G. gallus) TiPARP as GST fusion proteins. GST-mouse Tiparp (GST-mTiparp) demonstrated a similar pattern of auto-ribosylation to that of GST-TiPARP (Figure 1B). GST-chicken TiPARP (GST-chTiPARP) demonstrated weak auto-ribosylation activity compared with human, despite the loading of similar amounts of protein (Figure 1B). Addition of activated DNA or DNAse-treated RNA did not alter the migration patterns or auto-ribosylation abilities of GST-mTiparp or GST-chTiPARP (Supplementary Figure S1). We then compared the ability of GST-TiPARP and PARP-1 to ribosylate core histones. In agreement with a previous report (29), PARP-1 ADP-ribosylated each of the core histones (data not shown). GST-TiPARP also mono-ADP-ribosylated each of the core histones as evidenced by the lack of a marked mobility shift in molecular weight of the histones (Figure 1C). No ADP-ribosylation of core histones was observed in the presence of GST-TiPARP-H532A and GST-TiPARP-Y564A (data not shown). Similar to wildtype, GST-TiPARP-I631A also ribosylated core histones in the presence of 32P-NAD+ (Supplementary Figure S2).
To determine if the mutation of isoleucine 631 to a glutamate would convert TiPARP mART activity to polymerase activity, we analysed the auto-ribosylation activity of GST-TiPARP-I631E mutant. The auto-ribosylation activity of GST-TiPARP-I631E was similar to that of wildtype (Figure 1D), exhibiting mART but not poly(ADP-ribose) polymer synthase activity.
Identification of TiPARP as a novel negative regulator of AHR
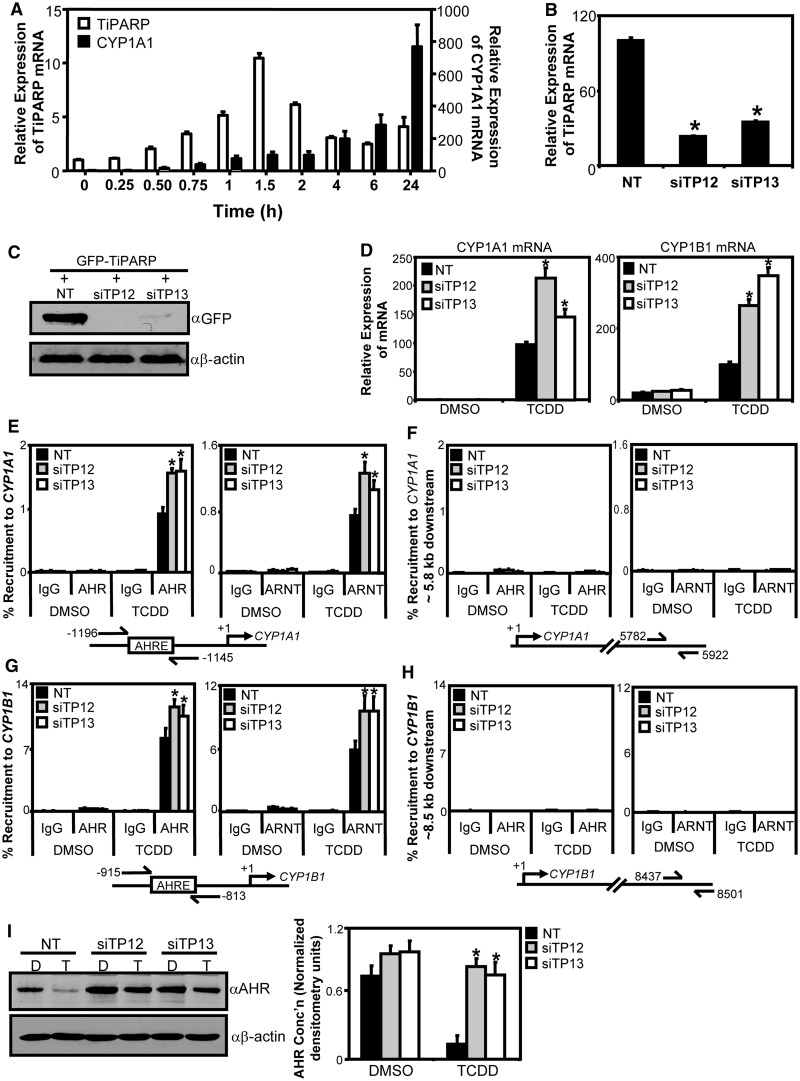
TiPARP was first reported to be a TCDD-responsive gene that was regulated by AHR in mouse hepatoma cells (12); however, little is known about the regulation of TiPARP in human cell lines. With this in mind, we determined the temporal changes in TiPARP expression after treatment of T-47D human breast carcinoma cells with TCDD. The results were compared with those obtained for CYP1A1. TCDD-dependent increases in TiPARP mRNA levels were rapid and peaked at 1.5 h, whereas CYP1A1 mRNA levels increased over the 24-h time course (Figure 2A). Similarly, Tiparp mRNA induction peaked after 1.5 h TCDD treatment in mouse Hepa1c1c7 cells (Supplementary Figure S3) (30). However, the TiPARP mRNA declined less rapidly compared with T-47D cells, suggesting possible cell type differences in TiPARP regulation or in mRNA stability. Similar kinetic differences between TCDD-induced TiPARP and CYP1A expression levels were observed in chick embryo hepatocytes (19), demonstrating that these regulatory differences between TiPARP and CYP1A genes are conserved across species. To test whether TiPARP modulates AHR activity, we then used RNAi-mediated knockdown of TiPARP in T-47D cells and examined TCDD-dependent changes in CYP1A1 and CYP1B1 mRNA levels. TiPARP mRNA levels were reduced to approximately 23–34% compared with NT control cells (Figure 2B). Endogenous TiPARP protein levels could not be assessed by immunoblot because of the lack of suitable antibody; however, co-transfection of GFP-tagged TiPARP with siRNA sequences demonstrated knockdown of overexpressed GFP-TiPARP protein (Figure 2C). RNAi-mediated knockdown of TiPARP resulted in significantly greater TCDD-dependent induction of CYP1A1 and CYP1B1 mRNA expression levels compared with NT control cells after 24-h exposure (Figure 2D). Similar results were obtained using HuH-7 human hepatoma and NCI-N87 human gastric carcinoma cell lines (Supplementary Figure S4).
Figure 2.
TiPARP knockdown increased TCDD-induced AHR transactivation. (A) Temporal analysis of TCDD-induced TiPARP and CYP1A1 mRNA expression levels. T-47D cells were treated with 10 nM TCDD at the times indicated. (B) TiPARP mRNA expression levels in T-47D cells following 48 h knockdown with single siRNAs against TiPARP (siTP12 and siTP13) and NT control. Expression levels significantly (P < 0.05) lower than NT were denoted with an asterisk. (C) Western blot analysis of GFP-TiPARP protein overexpression following RNAi-mediated TiPARP knockdown in TREx-FLP-IN 293 cells. (D) TiPARP knockdown increased TCDD-induced CYP1A1 and CYP1B1 mRNA expression levels in T-47D cells following 24-h treatment. Gene expression levels significantly (P < 0.05) greater than NT were denoted with an asterisk. TiPARP knockdown increased TCDD-induced AHR and ARNT recruitment to CYP1A1 (E and F) and CYP1B1 (G and H). AHR and ARNT to CYP1A1 regulatory region (E), CYP1A1 distal downstream region (F), CYP1B1 regulatory region (G) and CYP1B1 distal downstream region (H). T-47D cells with TiPARP knocked down were treated with 10 nM TCDD for 1 h and harvested for ChIP assays as described in ‘Materials and Methods’ section. Percent recruitment significantly greater (P < 0.05) than NT is denoted with an asterisk. (I) TiPARP knockdown reduced TCDD-induced AHR degradation in T-47D cells. T-47D cells following TiPARP knockdown were treated with TCDD (T) or DMSO (D) for 24 h and AHR protein expression was determined by Western blot analysis. Left panel: representative western blot from three independent experiments; right panel: Quantification of AHR protein levels, densitometry was performed using ImageJ analysis software (NIH). Asterisks denoted AHR protein levels significantly (P < 0.05, two-tailed Student’s t-test) greater than treatment-matched NT-transfected cells. All data were presented as means ± S.E.M for three independent experiments. Statistical analysis was determined by two-tailed Student’s t-test.
To examine the TiPARP-dependent mechanism of AHR repression, we performed ChIP assays in T-47D cells after RNAi-mediated knockdown of TiPARP and treated with TCDD for 1 h. In agreement with increased CYP1A1 and CYP1B1 mRNA levels, knockdown of TiPARP resulted in significantly greater recruitment of AHR and ARNT to the CYP1A1 and CYP1B1 enhancer regions compared with the NT control (Figure 2E and G). No significant differences in AHR or ARNT recruitment to downstream genomic control regions for both genes were observed (Figure 2F and H).
We next determined the effect of TiPARP knockdown on ligand-induced AHR protein degradation. Consistent with the gene expression and ChIP findings, knockdown of TiPARP also reduced AHR degradation following 24-h TCDD treatment (Figure 2I), suggesting that TiPARP may play a role in the proteolytic degradation of AHR.
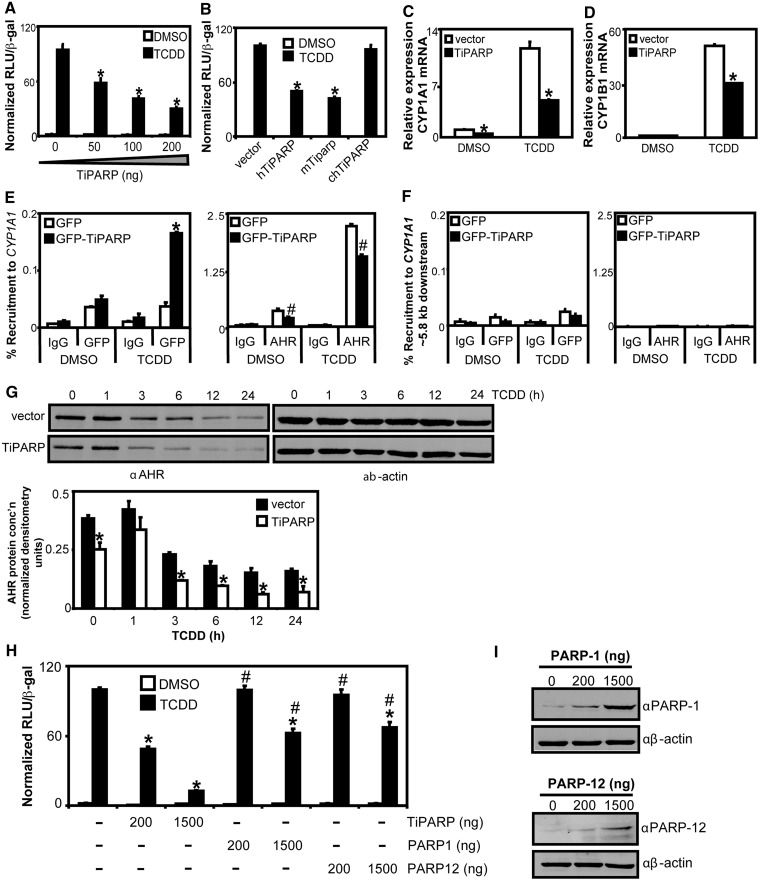
Because the RNAi experiments suggested that TiPARP may act as a negative regulator of AHR transactivation, we investigated the ability of TiPARP overexpression to repress AHR-dependent gene expression in HuH-7 cells transfected with a CYP1A1-regulated luciferase reporter plasmid (CYP1A1-luc). Transient transfection of increasing amounts of TiPARP resulted in a dose-dependent repression of TCDD-induced reporter gene activity (Figure 3A). Similar results were observed using a CYP1B1-regulated luciferase reporter plasmid (data not shown). Transfection of up to 1 µg DNA of the catalytic inactive mutant TiPARP-H532A did not repress reporter gene activity (Supplementary Figure S5). Mouse Tiparp, which shares 92.8% sequence homology with human TiPARP, also repressed AHR transactivation (Figure 3B). However, chicken TiPARP, which only exhibits 68.5% sequence homology with human TiPARP, did not. Similar findings were observed after transfection with GFP-tagged TiPARP fusion proteins. Transfection of equal amounts of GFP-TiPARP plasmid DNA resulted in similar protein expression levels for human and mouse GFP-tagged TiPARP, whereas higher protein levels of chicken TiPARP were observed (Supplementary Figure S6). Collectively, these data showed that TiPARP was a negative regulator of AHR transactivation, but this effect may exhibit species-specificity.
Figure 3.
TiPARP overexpression reduced TCDD-induced AHR signalling. (A) Concentration-dependent reduction of TCDD-induced CYP1A1-luc reporter gene activity with TiPARP overexpression. (B) Mammalian TiPARP inhibited TCDD-induced CYP1A1-luc reporter gene activity. HuH-7 cells were co-overexpressed with pCYP1A1-luc and 200 ng pcDNA-TiPARP, pcDNA-mTiparp or pcDNA-chTiPARP, and luciferase activity was determined. Values were normalized to 100% TCDD treatment. Reporter gene activity significantly lower than (P < 0.05) TCDD-treated empty vector is denoted with an asterisk. (C and D) TiPARP overexpression repressed TCDD induction of CYP1A1 (C) and CYP1B1 (D) mRNA in HuH-7 cells. Asterisks denote mRNA expression significantly lower (P < 0.05, two-tailed Student’s t-test) than treatment-matched empty vector transfected cells. (E) Overexpressed GFP-TiPARP recruitment to CYP1A1 regulatory region was induced by TCDD and repressed AHR recruitment. (F) Recruitment of GFP-TiPARP and AHR to CYP1A1 distal downstream region. HuH-7 cells were transfected with pEGFP empty vector or pEGFP-TiPARP and dosed with TCDD for 45 min and cells were harvested for ChIP assays. GFP-TiPARP recruitment, percent recruitment of transfection-matched cells significantly greater (P < 0.05, two-tailed Student’s t-test) than DMSO was denoted with an asterisk. Percent recruitment of AHR significantly lower (P < 0.05, two-tailed Student’s t-test) than treatment-matched GFP-transfected cells was denoted with a pound sign. Data shown were representative of three independent experiments. (G) TiPARP overexpression increased TCDD-induced AHR proteasomal degradation. T-47D cells were transfected with vector or pcDNA-TiPARP and dosed with TCDD for the indicated times and AHR protein levels were determined by western blot. Upper panel: representative western blots from three independent experiments; lower panel: Quantification of AHR protein levels, densitometry was performed using ImageJ analysis software (NIH). Asterisks denoted AHR protein levels significantly (P < 0.05, two-tailed Student’s t-test) lower than treatment-matched vector-transfected cells. (H) CYP1A1-luc reporter gene activity was preferentially inhibited by TiPARP. HuH-7 cells were co-overexpressed with 200 ng or 1500 ng of pcDNA-TiPARP, pCMV-PARP-1 or pCMV-PARP-12 and pCYP1A1-luc and treated with TCDD. (I) Western blot analysis of PARP-1 and PARP-12 protein overexpression in transfected HuH-7 cells. Reporter gene activity significantly lower (P < 0.05) than empty vector transfected cells was denoted with an asterisk. Reporter gene activity significantly (P < 0.05) greater than TiPARP-transfected cells (1500 ng) was denoted with a pound sign. All luciferase data were presented as means ± S.E.M from three independent experiments and statistical significance analysed by one-way ANOVA and Tukey’s multiple comparisons test.
To examine if TiPARP overexpression inhibited endogenous CYP1A1 and CYP1B1 mRNA expression, we transiently transfected HuH-7 cells with TiPARP, treated them with TCDD for 24 h before isolating RNA and performing qPCR. We observed a significant repression of TCDD-dependent increases in CYP1A1 and CYP1B1 mRNA levels in cells overexpressing TiPARP (Figure 3C and D).
We then determined if TiPARP was recruited to the enhancer regions of AHR target genes using ChIP assays. ChIP assays performed on extracts from HuH-7 cells transfected with GFP-TiPARP revealed a small but significant TCDD-dependent increase in GFP-TiPARP recruitment to CYP1A1 after 45 min of TCDD treatment (Figure 3E), but not to a downstream chromatin region that was not bound by AHR (Figure 3F). Interestingly, reduced AHR occupancy at CYP1A1 was also observed in cells overexpressing GFP-TiPARP (Figure 3E). Time course studies showed a reduction in AHR protein levels in cells overexpressing TiPARP both in the presence or absence of TCDD (Figure 3G), which was in support of the increased AHR protein levels observed after RNAi-mediated knockdown of TiPARP (Figure 2).
TiPARP selective repression of AHR activity
Because PARP-1 and other PARP family members can function as transcriptional repressors or activators (31), we examined the ability of PARP-1 and PARP-12 to modulate AHR transactivation. PARP-1 was selected because it is the most studied PARP, whereas PARP-12 was chosen because it shares the greatest sequence homology (46.8%) with TiPARP compared with other members of the PARP family (1). Transfection of HuH-7 cells with 200 ng of TiPARP caused ∼50% reduction in TCDD-dependent induction of CYP1A1-luc reporter gene activity, whereas transfection with the same amount of PARP-1 or PARP-12 had no effect (Figure 3H). Increasing the amount of transfected plasmid DNA to 1500 ng caused a >85% reduction in CYP1A1-luc reporter gene activity by TiPARP, but only ∼40% reduction by PARP-1 or PARP-12. Western blots confirmed the expression of PARP-1 and PARP-12 (Figure 3I). These results suggested that TiPARP exhibited selective repression of AHR transactivation compared with other PARP family members.
Repression of AHR by TiPARP requires the zinc-finger and catalytic domains
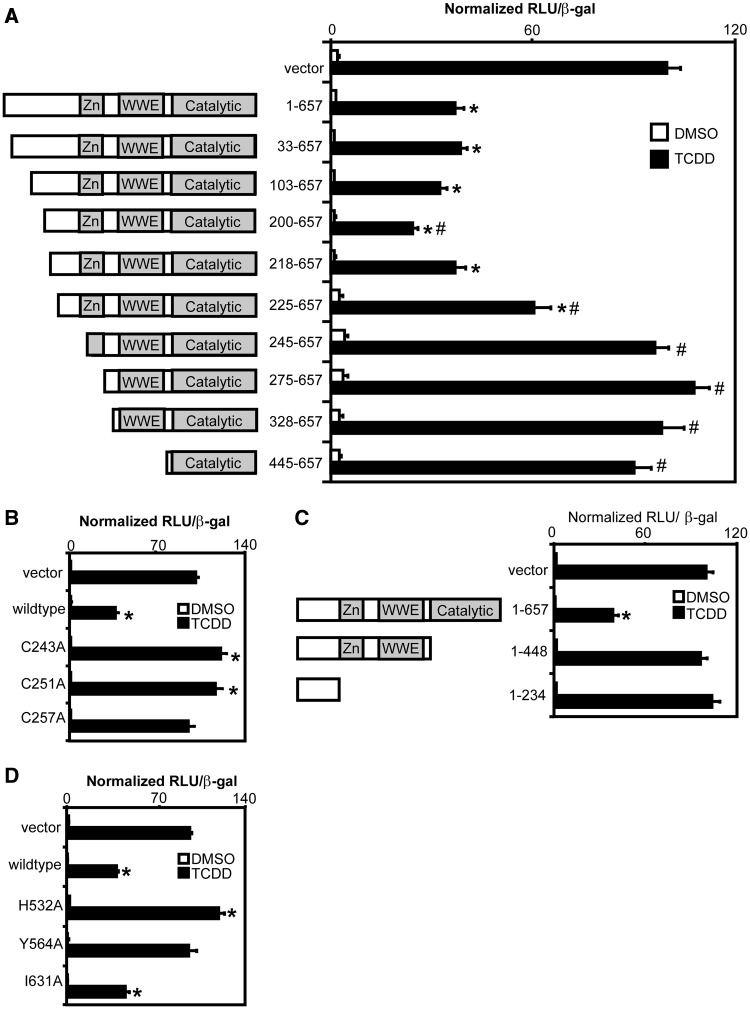
To determine the mechanism of TiPARP-dependent repression of AHR, we created a series of TiPARP deletions and site-directed point mutants, and determined their ability to repress AHR-dependent reporter gene activity. The expression of each of the TiPARP deletions and point mutants was confirmed by in vitro translation (data not shown). TiPARP N-terminal deletion constructs expressing amino acids 33–657 to 218–657 repressed CYP1A1-regulated reporter gene activity similar to that of wildtype, with the exception of 200–657, which exhibited greater repressor activity (Figure 4A). The 225–657 truncation construct exhibited a significantly reduced ability to repress reporter gene activity, whereas transfection of 245–657 to 445–657 truncations did not repress AHR-regulated reporter gene activity. Similar findings were observed using GFP-tagged TiPARP fusion proteins for all TiPARP deletion and mutant variants (Supplementary Figure S7). Western blot analysis of the GFP-TiPARP proteins showed that the lack of repression was not due to reduced protein expression of the different GFP-TiPARP variants (Supplementary Figure S8). Because the zinc-finger domain (residues 237–264) overlapped with the putative repressor domain (residues 218–245), we tested the importance of this domain in TiPARP-mediated repression by creating single point mutants of the cysteine residues of CCCH-type zinc-finger motif. All three TiPARP cysteine point mutants failed to inhibit TCDD-induced CYP1A1-luc reporter gene activity (Figure 4B). Similar findings were observed with GFP-tagged TiPARP zinc-finger variants (Supplementary Figures S7/S8).
Figure 4.
Inhibition of AHR signalling by TiPARP required zinc-finger and catalytic domains. Repression of TCDD induction of CYP1A1-luc reporter gene activity by (A) TiPARP N-terminal truncations, (B) Zinc-finger point mutants, (C) C-terminal truncations and (D) catalytic point mutants. Full-length TiPARP (1-657, wildtype), TiPARP truncation or point mutant constructs were co-overexpression with pCYP1A1-luc in HuH-7 cells and treated with TCDD. Data were presented as means ± S.E.M. from three independent experiments. Significance was analysed by one-way ANOVA and Tukey’s multiple comparisons test. Reporter gene activity significantly different than (P < 0.05) TCDD-treated empty vector or TiPARP (1-657) was denoted with an asterisk or pound sign, respectively.
To evaluate the importance of the TiPARP catalytic domain in mediating the repression of AHR transactivation, we created C-terminal deletion and catalytic point mutants, and evaluated them in our transient transfection reporter gene assay. Deletion of the TiPARP C-terminus containing the catalytic domain (residues 449–657) abolished the ability of TiPARP to repress AHR-dependent reporter gene activity (Figure 4C). The ability of TiPARP to repress AHR transactivation was prevented with the H532A and Y564A point mutants, but not I631A (Figure 4D), which retained catalytic activity (Figure 1B). Together, these data demonstrated that TiPARP-mediated repression of AHR transactivation required both the zinc-finger and catalytic domains. Similar results were observed with GFP-tagged TiPARP mutants, and immunoblotting confirmed that the lack of repression was not due to lack of protein expression (Supplementary Figures S7/S8).
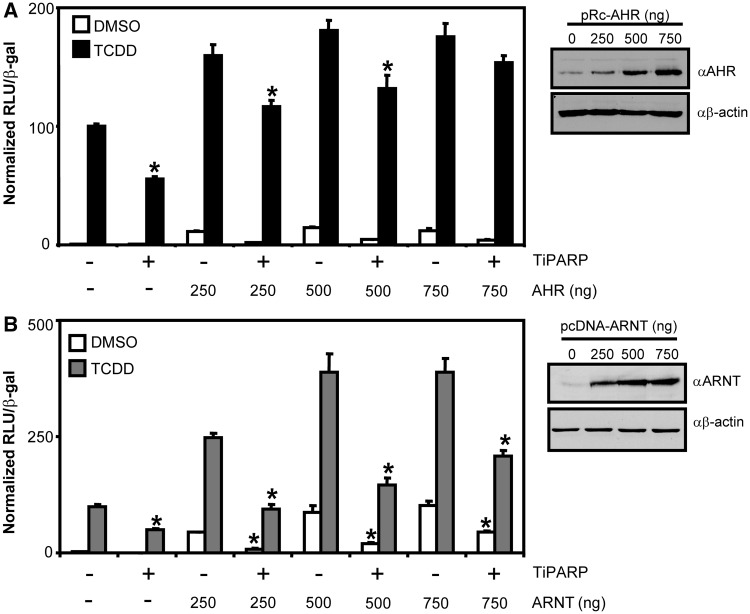
Ectopic AHR, but not ARNT, prevented TiPARP-mediated inhibition of AHR transactivation
The AHRR negatively regulates AHR transactivation by competing with AHR for ARNT dimerization and binding to the AHRE (17), but also through ARNT-independent mechanisms (18,32). To evaluate the role of ARNT in TiPARP-mediated repression of AHR transactivation, we assessed the ability of TiPARP to repress AHR activity in the presence of increasing amounts of AHR or ARNT. Transient co-transfection of AHR, but not ARNT, rescued the TiPARP-dependent repression of CYP1A1-regulated reporter gene activity (Figure 5). No repression or AHR or ARNT rescue was observed after transfection with the catalytically inactive TiPARP-H532A mutant (Supplementary Figure S9). These data suggested that the TiPARP-dependent repression of AHR transactivation was independent of ARNT.
Figure 5.
Overexpression of AHR but not ARNT prevented TiPARP-mediated inhibition of AHR transactivation. HuH-7 cells were co-transfected with pCYP1A1-luc, TiPARP (200 ng) and (A) AHR (250–750 ng) or (B) ARNT. Results represented the means ± S.E.M from three independent experiments and were analysed by one-way ANOVA and Tukey’s multiple comparisons test. Reporter gene activity of cells co-overexpressing TiPARP significantly lower than (P < 0.05) transfection- and treatment-matched control cells was denoted with an asterisk. Western blots confirming the presence of increased AHR and ARNT protein expression following transient transfection were presented to the right of the respective bar graphs.
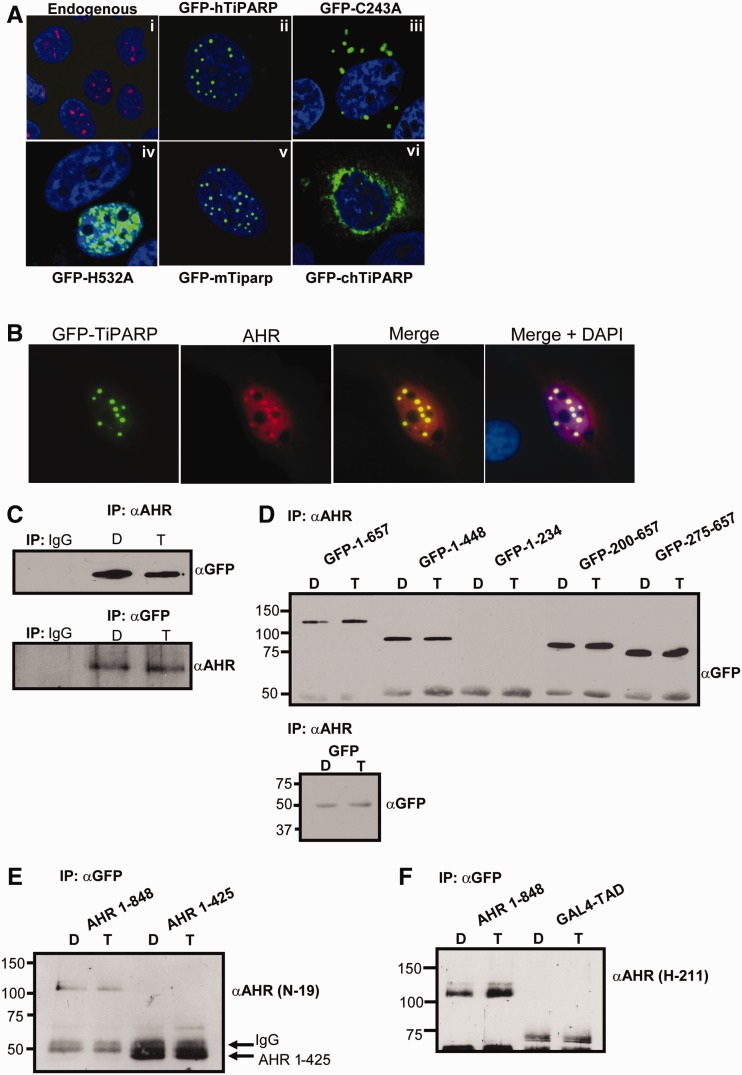
AHR and TiPARP interact and co-localize within the nucleus
To examine direct interactions between AHR and TiPARP, we performed co-localization and co-immunoprecipitation experiments. We first determined the subcellular localization of TiPARP. Endogenous TiPARP localized as small nuclear foci in HuH-7 cells (Figure 6A, panel i). Overexpressed GFP-TiPARP exhibited focal nuclear expression patterns similar to endogenous TiPARP (Figure 6A, panel ii). The GFP-tagged catalytic mutant H532A displayed a diffuse nuclear expression pattern, whereas GFP-tagged zinc-finger mutant C243A displayed cytosol foci, suggesting the zinc-finger domain was important for nuclear localization (Figure 6A, panel iii and iv). GFP-tagged mouse Tiparp expressed a similar nuclear focal pattern to human, while GFP-tagged chicken TiPARP was expressed primarily in the cytosol (Figure 6A, panel v and vi). These observations revealed that the nuclear and focal organization within the nucleus were important factors influencing the ability of TiPARP to repress AHR transactivation.
Figure 6.
TiPARP interacted with AHR in the nucleus. (A) Nuclear localization of endogenous TiPARP (40× magnification; panel i), overexpressed GFP-TiPARP (63×; panel ii), zinc-finger mutant GFP-TiPARP-C243A (63×; panel iii), catalytic mutant GFP-TiPARP-H532A (63×; panel iv), mouse GFP-Tiparp [GFP-mTiparp (63×; panel v)]. Images were acquired using an Olympus Fluoview 1000 confocal microscope. (B) Co-localization of overexpressed GFP-TiPARP and AHR in HuH-7 cells (63× magnification). Cells were treated with 10 nM TCDD for 1.5 h and fixed, immunostained for AHR, counterstained with DAPI and mounted using Vectashield. Images were acquired using an Imager.Z1 epifluorescence microscope and Axiovision software (Zeiss) following deconvolution. (C) Reciprocal co-immunoprecipitation experiments of overexpressed GFP-TiPARP and AHR. TREx-FLP-IN 293 cells were transfected and treated with TCDD for 1.5 h. (D) Co-immunoprecipitation experiments of AHR and GFP-TiPARP truncations in TREx-FLP-IN 293 cells (upper panel). Co-immunoprecipitation experiments with AHR and GFP in TREx-FLP-IN 293 cells (lower panel). AHR (H-211) antibody was used for immunoprecipitation and GFP (JL-8) was used for immunoblotting. (E and F) Co-immunoprecipitation experiments of GFP-TiPARP and AHR truncations in TREx-FLP-IN 293 cells. GFP (632460) antibody was used for immunoprecipitation and AHR (N-19 or H-211) antibodies were used for immunoblotting.
We then tested whether overexpressed AHR co-localized with GFP-TiPARP in transfected HuH-7 cells treated with TCDD or DMSO for 1.5 h. In DMSO- and TCDD-treated cells, AHR was localized to both the cytoplasm and the nucleus, which was in agreement with previous reports for cells transiently transfected with AHR (18,33) (data not shown). In co-transfected cells, a fraction of the nuclear AHR was enriched in nuclear foci containing GFP-TiPARP (Figure 6B). Further analysis of 113 cells co-expressing transfected AHR and GFP-TiPARP revealed that AHR formed nuclear foci and co-localized with GFP-TiPARP in 75% of the cells. In the absence of transfected GFP-TiPARP, AHR formed nuclear foci in only 6% of 238 cells examined. This co-localization pattern was unaffected by TCDD treatment (data not shown).
Co-immunoprecipitation experiments were then performed using TREx FLP-IN 293 cells transfected with GFP-TiPARP, AHR and ARNT and treated with DMSO or TCDD for 1.5 h. Similar to the co-localization experiments, GFP-TiPARP and AHR co-immunoprecipitated in the presence or absence of ligand (Figure 6C). Deletion analysis revealed that AHR interacted with residues 275–448 of TiPARP (Figure 6D). Reciprocal co-immunoprecipitation experiments demonstrated that both the N-terminal (residue 1–425) and C-terminal transactivation domains (residue 464–848) of AHR co-immunoprecipitated with GFP-TiPARP (Figure 6E and F).
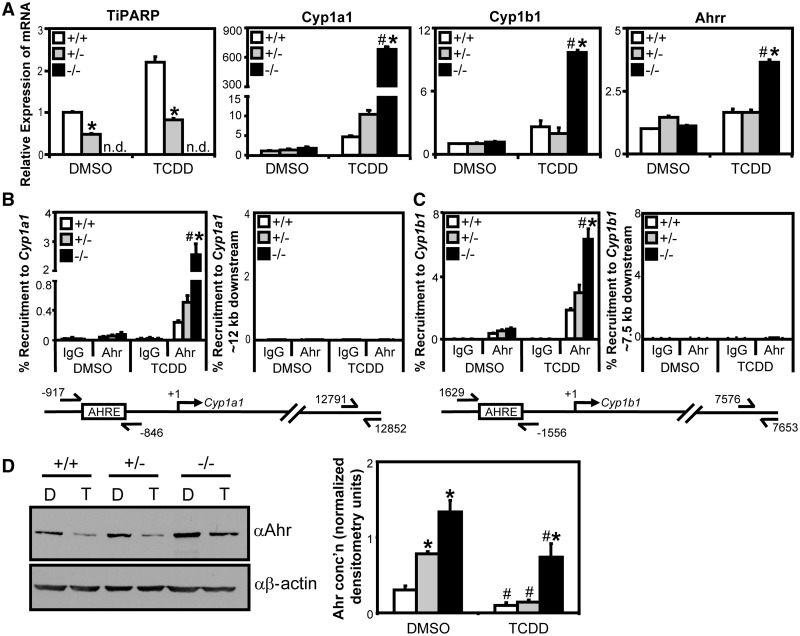
Tiparp knockout increased TCDD-dependent Ahr target transactivation and Ahr protein levels
To investigate the impact of TiPARP loss on AHR transactivation and protein expression, we created immortalized MEFs from wildtype, Tiparp+/− and Tiparp−/− siblings. As expected, Tiparp mRNA levels were induced in wildtype and Tiparp+/− cells after 24-h treatment with TCDD, but not in Tiparp−/− cells (Figure 7A). TCDD treatment of Tiparp−/− MEFs resulted in greater increases in Cyp1a1, Cyp1b1 and Ahrr mRNA levels compared with wildtype and Tiparp+/− cells (Figure 7A), while Gapdh mRNA levels were unaffected by TCDD treatment or by genotype (Supplementary Figure S10). Ectopic expression of Tiparp in Tiparp−/− MEFs reduced TCDD-dependent AHR transactivation compared with vector controls (Supplementary Figure S11). ChIP assays revealed significantly greater TCDD-induced Ahr recruitment to Cyp1a1 and Cyp1b1 in Tiparp−/− compared with wildtype or Tiparp+/− cells (Figure 7B and C). Because we observed increased AHR transactivation in Tiparp−/− compared with wildtype or Tiparp+/− MEFs, we examined the relative Ahr protein levels and ligand-induced Ahr degradation in the different MEF lines. Tiparp−/− cells had higher constitutive Ahr protein levels compared with wildtype and Tiparp+/− cells (Figure 7D). TCDD-induced degradation of Ahr was reduced in Tiparp−/− compared with wildtype and Tiparp+/− cells (Figure 7D). TCDD treatment for 24 h caused a 72% and 81% reduction in AHR protein levels in wildtype and Tiparp+/− cells, respectively, but only a 53% reduction in Tiparp−/− MEFs (Figure 7D). In contrast to increased Ahr protein levels, small but significant TCDD-dependent decreases in Ahr mRNA levels were detected in Tiparp−/− cells (Supplementary Figure S10).
Figure 7.
Tiparp knockout fibroblasts exhibited increased TCDD-induced Ahr transactivation. (A) wildtype (Tiparp+/+), Tiparp+/− and Tiparp−/− immortalized MEF lines were treated with 10 nM TCDD for 24 h and gene expression determined. Gene expression was normalized relative to DMSO-treated wildtype cells. Gene expression levels significantly (P < 0.05) different than treatment matched wildtype cells were denoted with an asterisk, and gene expression levels significantly (P < 0.05) different than treatment-matched Tiparp+/− (+/−) cells were denoted with a pound sign. (B and C) Tiparp−/− cells exhibit increased TCDD-induced Ahr recruitment to Cyp1a1 and Cyp1b1. (B) Ahr Recruitment to Cyp1a1 regulatory region and distal downstream control region. (C) Ahr recruitment to Cyp1b1 regulatory region and distal downstream control region. MEF lines were treated with 10 nM TCDD for 45 min and harvested for ChIP assays as described in ‘Materials and Methods’ section. Percent recruitment significantly greater (P < 0.05) than treatment-matched wildtype cells was denoted with an asterisk, and percent recruitment significantly greater (P < 0.05) than treatment-matched Tiparp+/− (+/−) cells was denoted with a pound sign. (D) Tiparp−/− cells have reduced TCDD-induced Ahr degradation. MEF lines following were treated with TCDD (T) or DMSO (D) for 24 h and Ahr protein expression was determined by western blot analysis. Left panel: representative western blot from three independent experiments. Right panel: quantification of Ahr protein levels, densitometry was performed using ImageJ analysis software (NIH). Asterisks denoted Ahr protein levels significantly (P < 0.05, two-tailed Student’s t-test) greater than treatment-matched wildtype cells. Pound sign denoted Ahr protein levels significantly (P < 0.05, two-tailed Student’s t-test) less than genotype-matched DMSO-treated cells.
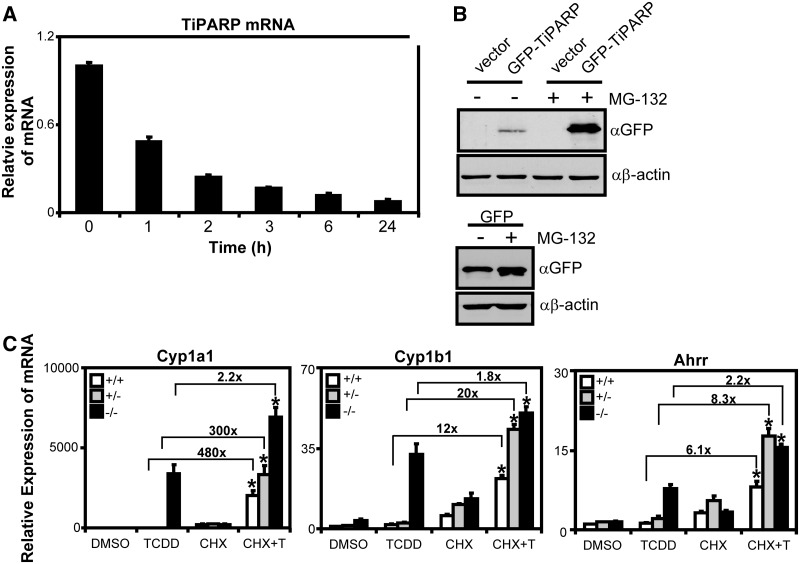
TiPARP is a labile factor that negatively regulates AHR transactivation
TCDD-induced proteolytic degradation of AHR has been proposed to be under the control of a labile factor, referred to as an AHR degradation promoting factor (ADPF) that when inhibited increases the expression of AHR target genes, such as CYP1A1 (34,35). In support of these findings, pre-treatment with CHX resulted in enhanced TCDD-dependent CYP1A1 mRNA levels (34,35). Because we observed that TiPARP was a negative regulator of AHR, we were interested to determine if TiPARP was a labile factor regulating AHR protein levels. To this end, we tested the stability of TiPARP mRNA by treating T-47D cells with actinomycin D and isolating RNA at the times indicated in Figure 8A. We observed a 50% loss in TiPARP mRNA after 1 h actinomycin D treatment (Figure 8A), which was in agreement with a previous report (30). Owing to the lack of a suitable antibody to detect endogenous TiPARP protein levels, we examined the stability of TiPARP by overexpressing GFP-tagged TiPARP in HuH-7 and treating with the 26S proteasome inhibitor, MG-132. After 6 h MG-132 treatment, we observed increased GFP-TiPARP expression compared with transiently transfected but untreated cells (Figure 8B).
Figure 8.
TiPARP is a labile negative regulator of AHR. (A) TiPARP mRNA was rapidly degraded after actinomycin D treatment. T-47D cells were treated with 1 µg/ml actinomycin D at the times indicated and Tiparp mRNA expression levels determined by qPCR. (B) GFP-TiPARP overexpression was increased after proteasome inhibition. HuH-7 cells were transfected with pEGFP-TiPARP, pcDNA (vector) and pEGFP for 24 h and treated with 25 µM MG-132 for 6 h, and GFP-TiPARP protein levels were determined by western blot using anti-GFP antibody. The data were from a representative western blot from three independent experiments. (C) Ahr target gene induction in MEF lines pre-treated with 10 µg/ml CHX. MEF cell lines were pre-treated with 10 µg/ml CHX for 1 h, then treated with TCDD for 6 h and gene expression was determined. Data were normalized to wildtype DMSO. Fold changes between TCDD alone and CHX + TCDD (CHX + T) were provided for each gene. Gene expression results were shown as means ± S.E.M for three independent experiments and significance analysed by one-way ANOVA and Tukey’s multiple comparisons test. Gene expression levels significantly different (P < 0.05) than TCDD alone within each genotype were denoted with an asterisk.
These data suggested that TiPARP was a potential candidate for the labile regulator of AHR protein levels and responsible for the increased expression of AHR target genes after pre-treatment with CHX (34,35). To test this notion, we pre-treated wildtype, Tiparp−/− and Tiparp+/− MEFs for 1 h with CHX before treatment with 6-h TCDD. Because we were interested in determining the effect of CHX on TCDD-dependent target gene expression, we also determined the fold increase of TCDD + CHX over TCDD alone for each genotype (Figure 8C). Cyp1a1 mRNA levels increased ∼480- and 300-fold following TCDD + CHX compared with TCDD alone in wildtype and Tiparp+/− MEFs, respectively. Cyp1b1 and Ahrr mRNA levels were also significantly increased following TCDD + CHX compared with TCDD alone, but the fold increases were an order of magnitude lower than those observed for Cyp1a1 (Figure 8C). For Tiparp−/− cells, treatment with CHX + TCDD resulted in significant but modest increases of approximately 2-fold for each of the AHR target genes examined compared to TCDD alone (Figure 8C). A similar pattern of Cyp1a1 and Cyp1b1 mRNA superinduction after TCDD treatment was observed after MG-132 pre-treatment, although the magnitude of induction was lower than that observed with CHX (Supplementary Figure S12). These data suggested TiPARP was an important factor regulating the CHX- and MG-132-dependent increases in ligand-activated AHR transactivation.
DISCUSSION
In this report, we identify TiPARP as a mART that ribosylates itself as well as core histones. We also provide evidence that TiPARP is a nuclear protein, a repressor of AHR transactivation and that its ability to repress AHR is dependent on its zinc-finger and catalytic domains. Our findings identify a new mechanism of negative feedback regulation in the AHR signalling pathway.
Recent sequence and structural analyses reveal that the majority of PARPs may have mART rather than poly(ADP-ribose) polymer synthase activity (5). PARP-10 (ARTD10), PARP-14 (ARTD8) and PARP-15 (ARTD7) have recently been reported to exhibit mART rather than poly(ADP-ribose) polymer synthase activity (7,36). In support of these data, we show that TiPARP exhibits mART activity by targeting itself and core histones. We also show that isoleucine 631 (I631) of TiPARP is not required for its auto-ribosylation and the hetero-ribosylation of histones. The PARP-1 catalytic glutamate (E988), which is equivalent to I631 in TiPARP, is important for polymer elongation (26,37). Substitution of I631 to glutamate does not convert TiPARP catalytic activity to poly(ADP-ribose) polymer synthase activity. This is consistent with similar mutagenesis studies of PARP-10 showing that conversion of I987E reduces its mART activity rather than convert it to a poly(ADP-ribose) polymer synthase (7). Taken together, these data support that other structural features are important for poly(ADP-ribose) polymer formation (7).
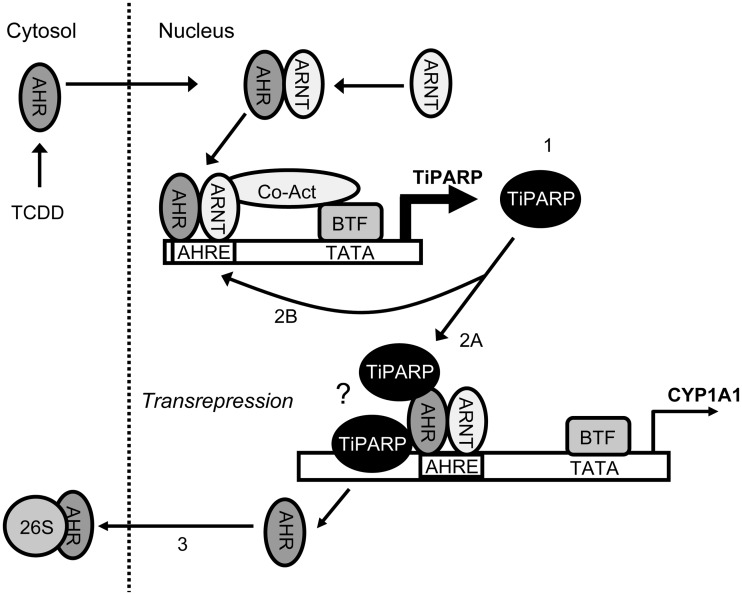
Our studies using human cell lines and MEFs isolated from wildtype and Tiparp−/− mice reveal that TiPARP modulates ligand-induced proteolytic degradation of AHR and functions as a repressor of AHR transactivation. In this model, TCDD-activated AHR induces TiPARP expression, TiPARP is recruited to AHR target genes either through direct DNA binding or tethering through AHR, increased levels of TiPARP enhance the proteolytic degradation of AHR, by either direct ribosylation or through the ribosylation of unidentified intermediary factor(s) to repress AHR transactivation (Figure 9).
Figure 9.
Proposed model of repression of AHR signalling by TiPARP. The AHR mediates the TCDD-induced expression of TiPARP (1). TiPARP is recruited to the AHR/ARNT-bound AHRE of targets to repress transcription (CYP1A1 in this example) (2A). TiPARP represses its own transcription and the regulation of other AHR target genes by binding to AHR/ARNT-bound to AHREs or potentially through direct DNA binding (indicated by the question mark), creating a negative feedback loop (2B) in which AHR dissociates from AHREs, translocates from the nucleus and is proteolytically degraded by the 26S proteasome (26S) (3). BTF, basal transcription factors; Co-act, co-activators.
The ability of TiPARP to repress AHR transactivation requires its zinc-finger and catalytic domains. Previous studies have reported that PARPs act as coregulators (co-activators or co-repressors) for a number of different transcriptional regulators, such as NF-κB, HES1, Oct-1, c-Myc, Sp1 and nuclear receptors, including peroxisome proliferator-activated receptor γ, and thyroid hormone receptor (31,38,39). In some cases, enzymatic activity is required, while in others it is not. PARP-1 alters chromatin structure but also directly binds chromatin to modulate gene expression (40–42). PARP-14 acts as a transcriptional switch for Stat6-dependent transcription whereby non-stimulated PARP-14 is present at Stat6 target promoters and acts as a transcriptional repressor (43). We also show that TiPARP is recruited to the CYP1A1 regulatory region and interacts with AHR, suggesting that TiPARP may directly target AHR. The TCDD-dependent recruitment of TiPARP at CYP1A1 also suggests the possibility that TiPARP recognizes a specific DNA sequence and that it might directly regulate the expression other AHR target genes or other transcription factors. It is not clear from our data if TiPARP binds directly to a specific recognition sequence in CYP1A1 or if it is recruited to CYP1A1 through its interactions with AHR. CCCH-type zinc fingers are associated with RNA binding capacity; however, these domains are also reported to bind to single-stranded DNA (44,45).
The ubiquitin-proteasome pathway plays an important role in TCDD-mediated degradation of AHR (34,46). PARP-1 plays a role in the reactive oxygen induced activation of the proteasome (47). Auto-modified PARP-1 interacts with and activates nuclear proteasome pathways (48). Inhibition of PARP-5a (ARTD5) and PARP-5b (ARTD6) activity prevents the degradation of axin through the ubiquitin-proteasome pathway and this increased half-life of axin promotes β-catenin degradation (49). Therefore, it is tempting to hypothesize that TiPARP activates components of the proteasome to enhance TCDD-mediated AHR degradation. It is also equally possible that TiPARP ADP-ribosylates AHR, targeting it for degradation.
The protein synthesis inhibitor CHX and 26S-proteasome inhibitor MG-132 inhibit TCDD-induced degradation of AHR, leading some investigators to hypothesize that AHR levels are regulated by a TCDD-inducible labile protein, termed ADPF, which modulates AHR transactivation by promoting its nuclear degradation (34,50). We demonstrate that Tiparp−/− MEFs are relatively insensitive to the increased AHR target gene expression after co-treatment with CHX + TCDD compared with TCDD alone. However, this lack of a further increase in AHR target gene expression in the Tiparp−/− MEFs may be due to the fact that these cells exhibit a highly active AHR signalling pathway that cannot be further increased compared with wildtype MEFs. We also show that GFP-TiPARP localizes to the nucleus and interacts with nuclear AHR and the GFP-TiPARP protein levels are stabilized by proteasome inhibition. TiPARP mRNA levels are also induced following protein synthesis and proteasomal inhibition, but this might be expected because TiPARP is an AHR target gene (30). We observe TiPARP knockdown or knockout increases AHR protein levels and AHR transactivation mimicking the AHR-dependent target gene superinduction observed after treatment with a protein synthesis inhibitor (34). Overall, our data support TiPARP as a potential candidate labile ADPF.
Much of our understanding of AHR repression comes from studies of AHRR, which is also part of negative feedback loop regulating AHR function (17,18,32). A comparison of what is known about the AHR repressive functions of both proteins reveals similarities but also some notable differences. Similarities between TiPARP- and AHRR-mediated inhibition of AHR signalling include the following: (i) both genes are induced by TCDD and regulated by AHR; (ii) AHRR and TiPARP localize to the nucleus; (iii) both interact with AHR; and (iv) increased ARNT expression fails to prevent AHRR- or TiPARP-dependent repression of AHR transactivation (17,18,32,51). In contrast, AHRR repression of AHR action does not involve increased AHR degradation, direct binding to AHREs or inhibition of the AHR-ARNT complex formation (32,52). This suggests that TiPARP and AHRR exhibit partially overlapping but also distinct mechanisms to repress AHR activity.
A recent study reported that the NAD+ precursor nicotinamide is an inhibitor of AHR target gene induction and a corrective agent for the TCDD-mediated suppression of avian hepatic glucose production and that these effects are mediated by TiPARP (19). TiPARP overexpression mimicked the suppressive effects of TCDD on gluconeogenesis and it was hypothesized the TCDD-mediated induction of TiPARP depleted hepatocytes of NAD+, inhibiting gluconeogenic genes and glucose output through the alteration of SIRT1 activity (19). In the present study, we observed that TiPARP represses rather than increases TCDD-dependent AHR activation. It is possible that the effects of TiPARP on gluconeogenic gene expression might be independent of its ability to feedback and repress AHR transactivation. Chicken (G. gallus) TiPARP (19) exhibited weak auto-ribosylation activity and did not inhibit AHR transactivation in transfected HuH7 cells, suggesting that TiPARP exhibits species- or model-specific differences. Our observation that TiPARP possesses mART rather than poly(ADP-ribose) polymer synthase activity suggests that either mART activity is sufficient to deplete cellular NAD+ levels (19) or that alterations in NAD+ levels occur through other downstream targets of TiPARP.
In summary, we show that TiPARP is a nuclear mART with both auto- and hetero-ribosylation activities. In addition, we provide evidence that TiPARP is a labile protein and transcriptional repressor of AHR involved in a new negative feedback loop regulating AHR function. Our study provides new insights into AHR signalling and further evidence for a role for PARP family members in the regulation of gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–12.
FUNDING
Canadian Institutes of Health Research operating grant [MOP-82715 to J.M.]; Cancer Research Society Inc. (to J.P.M.); Ontario Graduate Student Stipend Program (to L.M.); Canadian Institutes of Health Research New Investigator Award and Early Researcher Award from the Ontario Ministry of Innovation (to J.M.). Funding for open access charge: Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Patricia Harper (Hospital for Sick Children, Toronto, Canada) and Kevin Gardner (UT Southwestern Medical Center) for providing materials for this study, as well as all members of the Matthews’ laboratory for their help with the preparation of the manuscript.
REFERENCES
- 1.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 2.Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 3.Quenet D, El Ramy R, Schreiber V, Dantzer F. The role of poly(ADP-ribosyl)ation in epigenetic events. Int. J. Biochem. Cell Biol. 2009;41:60–65. doi: 10.1016/j.biocel.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 4.Hottiger MO. ADP-ribosylation of histones by ARTD1: an additional module of the histone code? FEBS Lett. 585:1595–1599. doi: 10.1016/j.febslet.2011.03.031. [DOI] [PubMed] [Google Scholar]
- 5.Otto H, Reche PA, Bazan F, Dittmar K, Haag F, Koch-Nolte F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs) BMC Genomics. 2005;6:139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Di Girolamo M, Dani N, Stilla A, Corda D. Physiological relevance of the endogenous mono(ADP-ribosyl)ation of cellular proteins. FEBS J. 2005;272:4565–4575. doi: 10.1111/j.1742-4658.2005.04876.x. [DOI] [PubMed] [Google Scholar]
- 9.Corda D, Di Girolamo M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003;22:1953–1958. doi: 10.1093/emboj/cdg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Salcedo JA, Gijon P, Nolan DP, Tebabi P, Pays E. A chromosomal SIR2 homologue with both histone NAD-dependent ADP-ribosyltransferase and deacetylase activities is involved in DNA repair in Trypanosoma brucei. EMBO J. 2003;22:5851–5862. doi: 10.1093/emboj/cdg553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aravind L. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 2001;26:273–275. doi: 10.1016/s0968-0004(01)01787-x. [DOI] [PubMed] [Google Scholar]
- 12.Ma Q, Baldwin KT, Renzelli AJ, McDaniel A, Dong L. TCDD-inducible poly(ADP-ribose) polymerase: a novel response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem. Biophys. Res. Commun. 2001;289:499–506. doi: 10.1006/bbrc.2001.5987. [DOI] [PubMed] [Google Scholar]
- 13.Van den Berg M, Birnbaum L, Bosveld AT, Brunstrom B, Cook P, Feeley M, Giesy JP, Hanberg A, Hasegawa R, Kennedy SW, et al. Toxic equivalency factors (TEFs) for PCBs, PCDDs, PCDFs for humans and wildlife. Environ. Health Perspect. 1998;106:775–792. doi: 10.1289/ehp.98106775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitlock JP., Jr Induction of cytochrome P4501A1. Annu. Rev. Pharmacol. Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- 15.Mimura J, Fujii-Kuriyama Y. Functional role of AhR in the expression of toxic effects by TCDD. Biochim. Biophys. Acta. 2003;1619:263–268. doi: 10.1016/s0304-4165(02)00485-3. [DOI] [PubMed] [Google Scholar]
- 16.Pollenz RS. The mechanism of AH receptor protein down-regulation (degradation) and its impact on AH receptor-mediated gene regulation. Chem. Biol. Interact. 2002;141:41–61. doi: 10.1016/s0009-2797(02)00065-0. [DOI] [PubMed] [Google Scholar]
- 17.Mimura J, Ema M, Sogawa K, Fujii-Kuriyama Y. Identification of a novel mechanism of regulation of Ah (dioxin) receptor function. Genes Dev. 1999;13:20–25. doi: 10.1101/gad.13.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karchner SI, Jenny MJ, Tarrant AM, Evans BR, Kang HJ, Bae I, Sherr DH, Hahn ME. The active form of human aryl hydrocarbon receptor (AHR) repressor lacks exon 8, and its Pro 185 and Ala 185 variants repress both AHR and hypoxia-inducible factor. Mol. Cell Biol. 2009;29:3465–3477. doi: 10.1128/MCB.00206-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diani-Moore S, Ram P, Li X, Mondal P, Youn DY, Sauve AA, Rifkind AB. Identification of the aryl hydrocarbon receptor target gene TiPARP as a mediator of suppression of hepatic gluconeogenesis by 2,3,7,8-tetrachlorodibenzo-p-dioxin and of nicotinamide as a corrective agent for this effect. J. Biol. Chem. 2010;285:38801–38810. doi: 10.1074/jbc.M110.131573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmahl J, Raymond CS, Soriano P. PDGF signaling specificity is mediated through multiple immediate early genes. Nat. Genet. 2007;39:52–60. doi: 10.1038/ng1922. [DOI] [PubMed] [Google Scholar]
- 21.Tamblyn L, Li E, Sarras H, Srikanth P, Hande MP, McPherson JP. A role for Mus81 in the repair of chromium-induced DNA damage. Mutat. Res. 2009;660:57–65. doi: 10.1016/j.mrfmmm.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 22.Macpherson L, Matthews J. Inhibition of aryl hydrocarbon receptor-dependent transcription by resveratrol or kaempferol is independent of estrogen receptor alpha expression in human breast cancer cells. Cancer Lett. 299:119–129. doi: 10.1016/j.canlet.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed S, Valen E, Sandelin A, Matthews J. Dioxin increases the interaction between aryl hydrocarbon receptor and estrogen receptor alpha at human promoters. Toxicol. Sci. 2009;111:254–266. doi: 10.1093/toxsci/kfp144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthews J, Wihlen B, Thomsen J, Gustafsson JA. Aryl hydrocarbon receptor-mediated transcription: ligand-dependent recruitment of estrogen receptor alpha to 2,3,7,8-tetrachlorodibenzo-p-dioxin-responsive promoters. Mol. Cell Biol. 2005;25:5317–5328. doi: 10.1128/MCB.25.13.5317-5328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holbourn KP, Shone CC, Acharya KR. A family of killer toxins. Exploring the mechanism of ADP-ribosylating toxins. FEBS J. 2006;273:4579–4593. doi: 10.1111/j.1742-4658.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- 26.Marsischky GT, Wilson BA, Collier RJ. Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation. Evidence for active site similarities to the ADP-ribosylating toxins. J. Biol. Chem. 1995;270:3247–3254. doi: 10.1074/jbc.270.7.3247. [DOI] [PubMed] [Google Scholar]
- 27.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 28.Kelly SM, Pabit SA, Kitchen CM, Guo P, Marfatia KA, Murphy TJ, Corbett AH, Berland KM. Recognition of polyadenosine RNA by zinc finger proteins. Proc. Natl Acad. Sci. USA. 2007;104:12306–12311. doi: 10.1073/pnas.0701244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messner S, Altmeyer M, Zhao H, Pozivil A, Roschitzki B, Gehrig P, Rutishauser D, Huang D, Caflisch A, Hottiger MO. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 38:6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Q. Induction and superinduction of 2,3,7,8-tetrachlorodibenzo-rho-dioxin-inducible poly(ADP-ribose) polymerase: role of the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator transcription activation domains and a labile transcription repressor. Arch. Biochem. Biophys. 2002;404:309–316. doi: 10.1016/s0003-9861(02)00339-9. [DOI] [PubMed] [Google Scholar]
- 31.Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans BR, Karchner SI, Allan LL, Pollenz RS, Tanguay RL, Jenny MJ, Sherr DH, Hahn ME. Repression of aryl hydrocarbon receptor (AHR) signaling by AHR repressor: role of DNA binding and competition for AHR nuclear translocator. Mol. Pharmacol. 2008;73:387–398. doi: 10.1124/mol.107.040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrulis JR, Hord NG, Perdew GH. Subcellular localization of the aryl hydrocarbon receptor is modulated by the immunophilin homolog hepatitis B virus X-associated protein 2. J. Biol. Chem. 2000;275:37448–37453. doi: 10.1074/jbc.M006873200. [DOI] [PubMed] [Google Scholar]
- 34.Ma Q, Baldwin KT. 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced degradation of aryl hydrocarbon receptor (AhR) by the ubiquitin-proteasome pathway. Role of the transcription activaton and DNA binding of AhR. J. Biol. Chem. 2000;275:8432–8438. doi: 10.1074/jbc.275.12.8432. [DOI] [PubMed] [Google Scholar]
- 35.Ma Q. Aryl hydrocarbon receptor degradation-promoting factor (ADPF) and the control of the xenobiotic response. Mol. Interv. 2007;7:133–137. doi: 10.1124/mi.7.3.4. [DOI] [PubMed] [Google Scholar]
- 36.Aguiar RC, Takeyama K, He C, Kreinbrink K, Shipp MA. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 2005;280:33756–33765. doi: 10.1074/jbc.M505408200. [DOI] [PubMed] [Google Scholar]
- 37.Ruf A, Rolli V, de Murcia G, Schulz GE. The mechanism of the elongation and branching reaction of poly(ADP-ribose) polymerase as derived from crystal structures and mutagenesis. J. Mol. Biol. 1998;278:57–65. doi: 10.1006/jmbi.1998.1673. [DOI] [PubMed] [Google Scholar]
- 38.Bai P, Houten SM, Huber A, Schreiber V, Watanabe M, Kiss B, de Murcia G, Auwerx J, Menissier-de Murcia J. Poly(ADP-ribose) polymerase-2 [corrected] controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/peroxisome proliferator-activated receptor-gamma [corrected] heterodimer. J. Biol. Chem. 2007;282:37738–37746. doi: 10.1074/jbc.M701021200. [DOI] [PubMed] [Google Scholar]
- 39.Miyamoto T, Kakizawa T, Hashizume K. Inhibition of nuclear receptor signalling by poly(ADP-ribose) polymerase. Mol. Cell Biol. 1999;19:2644–2649. doi: 10.1128/mcb.19.4.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 41.Madison DL, Lundblad JR. C-terminal binding protein and poly(ADP)ribose polymerase 1 contribute to repression of the p21(waf1/cip1) promoter. Oncogene. 29:6027–6039. doi: 10.1038/onc.2010.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Mehrotra P, Riley JP, Patel R, Li F, Voss L, Goenka S. PARP-14 functions as a transcriptional switch for Stat6-dependent gene activation. J. Biol. Chem. 286:1767–1776. doi: 10.1074/jbc.M110.157768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bai C, Tolias PP. Cleavage of RNA hairpins mediated by a developmentally regulated CCCH zinc finger protein. Mol. Cell Biol. 1996;16:6661–6667. doi: 10.1128/mcb.16.12.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collart C, Remacle JE, Barabino S, van Grunsven LA, Nelles L, Schellens A, Van de Putte T, Pype S, Huylebroeck D, Verschueren K. Smicl is a novel Smad interacting protein and cleavage and polyadenylation specificity factor associated protein. Genes Cells. 2005;10:897–906. doi: 10.1111/j.1365-2443.2005.00887.x. [DOI] [PubMed] [Google Scholar]
- 46.Pollenz RS. Specific blockage of ligand-induced degradation of the Ah receptor by proteasome but not calpain inhibitors in cell culture lines from different species. Biochem. Pharmacol. 2007;74:131–143. doi: 10.1016/j.bcp.2007.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ullrich O, Reinheckel T, Sitte N, Grune T. Degradation of hypochlorite-damaged glucose-6-phosphate dehydrogenase by the 20S proteasome. Free Radic. Biol. Med. 1999;27:487–492. doi: 10.1016/s0891-5849(99)00060-x. [DOI] [PubMed] [Google Scholar]
- 48.Mayer-Kuckuk P, Ullrich O, Ziegler M, Grune T, Schweiger M. Functional interaction of poly(ADP-ribose) with the 20S proteasome in vitro. Biochem. Biophys. Res. Commun. 1999;259:576–581. doi: 10.1006/bbrc.1999.0824. [DOI] [PubMed] [Google Scholar]
- 49.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 50.Ma Q, Baldwin KT. A cycloheximide-sensitive factor regulates TCDD-induced degradation of the aryl hydrocarbon receptor. Chemosphere. 2002;46:1491–1500. doi: 10.1016/s0045-6535(01)00270-3. [DOI] [PubMed] [Google Scholar]
- 51.Lo R, Celius T, Forgacs AL, Dere E, MacPherson L, Harper P, Zacharewski T, Matthews J. Identification of aryl hydrocarbon receptor binding targets in mouse hepatic tissue treated with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Appl. Pharmacol. 257:38–47. doi: 10.1016/j.taap.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 52.Hahn ME, Allan LL, Sherr DH. Regulation of constitutive and inducible AHR signaling: complex interactions involving the AHR repressor. Biochem. Pharmacol. 2009;77:485–497. doi: 10.1016/j.bcp.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.