Abstract
Template switching (TS) has been an inherent mechanism of reverse transcriptase, which has been exploited in several transcriptome analysis methods, such as CAGE, RNA-Seq and short RNA sequencing. TS is an attractive option, given the simplicity of the protocol, which does not require an adaptor mediated step and thus minimizes sample loss. As such, it has been used in several studies that deal with limited amounts of RNA, such as in single cell studies. Additionally, TS has also been used to introduce DNA barcodes or indexes into different samples, cells or molecules. This labeling allows one to pool several samples into one sequencing flow cell, increasing the data throughput of sequencing and takes advantage of the increasing throughput of current sequences. Here, we report TS artifacts that form owing to a process called strand invasion. Due to the way in which barcodes/indexes are introduced by TS, strand invasion becomes more problematic by introducing unsystematic biases. We describe a strategy that eliminates these artifacts in silico and propose an experimental solution that suppresses biases from TS.
INTRODUCTION
Reverse transcriptase (RT) has been widely used for the construction of cDNA libraries since its discovery (1,2) and has been subsequently used for gene expression studies. One intrinsic property of RT is that once it has reached the 5′ end of a RNA molecule, the 7-methylguanosine at the cap site is reverse transcribed to cytosine residues (3). This activity at the cap site has also been previously demonstrated on RNAs with an artificial adenosine cap, which was reverse-transcribed to thymidine (4). In addition to this mechanism, RT also exhibits terminal transferase activity that allows the addition of non-templated nucleotides (predominantly cytidines) once it reaches the 5′ end of a RNA molecule, especially in the presence of manganese (5). Combined, these two mechanisms form a cytosine overhang at the 3′ end of the cDNA after reverse transcription and serves as a useful marker for the 5′ site of the RNA. These properties have been taken advantage of in the construction of full-length cDNA libraries (6). More specifically, the library construction method uses oligonucleotides incorporating a stretch of consecutive ribo-guanosine nucleotides, r(G)3, at the 3′ end of the first strand cDNA that allows for the hybridization of the oligonucleotide with the cytosine overhang. Once hybridized, the RT then switches templates and starts polymerizing the oligonucleotide, thereby incorporating the oligonucleotide sequence with the cDNA sequence. This process is known as the template-switching (TS) mechanism.
Following original cDNA cloning protocols (6,7), several high-throughput transcriptome analyses protocols have incorporated the TS mechanism (8–12). The TS oligonucleotide used for the hybridization to the cytosine overhang is further used for incorporating priming sites for downstream steps in the respective protocols. Furthermore, in the experiments conducted by Plessy et al. (9) and Islam et al. (10), the TS oligonucleotide was used to incorporate DNA barcode sequences (also known as DNA indexes) into its cDNA libraries, allowing for pooled or multiplexed reactions. By including a set of known sequences (i.e. barcodes) directly upstream of the r(G)3 in the TS oligonucleotide, these sequences become identifiers for different samples. The pooling of several samples into a single sequencing reaction is a common strategy towards minimizing costs and labor (13) and increases the data throughput.
Given the constant increase of number of reads per sequencer run, techniques for multiplexing libraries are flourishing. For example, the current protocol of the HiSeq 2000 sequencer can produce up to 3 billion single reads that pass filtering on a single flow cell run (http://www.illumina.com/systems/hiseq_systems/hiseq_2000_1000/performance_specifications.ilmn). Methods that measure transcript expression levels by their 5′-end such as STRT (14), CAGE (15) or nanoCAGE (16) have a reduced complexity compared with RNA-Seq, and therefore take a particular advantage of multiplexing. In addition to TS, there are ligation- and polymerase chain reaction (PCR)-based methods that have been used for introducing barcodes into samples for multiplexed experiments. In single-read libraries using restriction enzymes to cleave sequence tags, the barcode is often added by ligation at the 5′ or 3′ end of the construct, like for CAGE (15), the cleaved version of nanoCAGE (9), SAGE protocols such as HT-SuperSAGE (17) or small RNA libraries (18). However, studies have demonstrated that ligation-based methods are heavily biased due to RNA ligases having sequence-specific biases (19,20). One strategy used for dealing with ligation-based biases has been to standardize the sequence at the end of the RNA adaptor that will be ligated (18). Another proposed strategy was to use a pool of RNA adaptors (20); however, Alon et al. (19) have further suggested that barcodes should be introduced via PCR-based methods, such as Illumina’s industry standard known as TruSeq. TruSeq uses 6-nt barcodes, which are detected as a separate step after sequencing the forward read or its mate pair. Read indexes are primed with a separate oligonucleotide, which gives a lot of flexibility in their placement in the 5′ and 3′ linkers. The designers of TruSeq protocols took this opportunity to place the index far from the reaction sites, usually in the tail of the primers. However, the indexes are introduced at a late step in the reaction, as there are no universal primers that would amplify the libraries and keep the indexes at the same time. As a consequence, it does not allow the pooling of the samples at early preparation steps, and for this reason, strategies where barcodes can be introduced as early as possible, such as via TS or ligation-based methods, are still preferred in situations that strongly benefit in terms of cost or logistics from early pooling. The question of which multiplexing approach to take is highly dependent on the nature of the research. For example, in a study by Kivioja et al. (21), they describe a method for introducing unique molecular identifiers via TS for quantifying transcript numbers. These identifiers are random bases in the TS oligonucleotides and function like random barcodes that index RNAs molecules instead of indexing samples. Double-stranded ligation and PCR are ruled out as alternatives for introducing indexes. In the case of ligation, it would be too difficult to produce the double-stranded adaptors because random sequences will not be reverse complementary. Indexing via PCR would be too late, as the purpose of these identifiers is to detect PCR duplicates. Lastly, Kivioja et al.(21) have envisioned that unique molecular identifiers can be combined with sample barcodes.
One of the main advantages of using TS is the lack of purification and adaptor ligation steps, which eliminates ligation-introduced biases and also minimizes the loss of material. This has made TS highly suitable in studies working with a limited amount of RNA (9,10,12,22,23). Although TS is an inherent property of RTs, and is therefore only implemented in transcriptome studies, we may see an increase in the use of TS due to the growing interest in single cell transcriptomics (24). There are, however, intrinsic problems associated with the TS mechanism, such as the concatenation of TS oligonucleotides due to cycles of terminal transferase activity and TS oligonucleotide hybridization (25). Another issue that we address here is the interruption of first strand synthesis via strand invasion. Although TS is most efficient when RT has reached the end of the RNA template, the TS oligonucleotide may hybridize to the first strand cDNA due to sequence complementarity before the RT has finished polymerizing. This creates first strand cDNAs that are artificially shorter than the RNA due to the incomplete reverse transcription process. Furthermore, although this is usually a systematic bias, this becomes more problematic in protocols using varied TS oligonucleotides for barcoding purposes, as the strand invasion process is dependent on the oligonucleotide sequence. We study in detail the artifacts and biases created by strand invasion in a protocol using the TS mechanism and demonstrate how it is possible to remove such artifacts in silico. Lastly, we propose possible experimental strategies that may help reduce such artifacts and biases in protocols that use TS, and demonstrate it with the nanoCAGE protocol.
MATERIALS AND METHODS
NanoCAGE libraries were prepared from total RNA isolated from human whole blood samples (200 ng per sample) and rat whole body RNA (500 ng per sample) according to a previously published protocol (16), and sequenced using the Illumina GAIIx instrument on five (four for blood samples and one for rat samples) sequencing lanes. These quantities of starting material are well above the recommended quantity of 50 ng, and we therefore expected that the difference would not cause one set of samples to underperform compared with the other set. Blood samples were collected in PAXgene blood RNA tubes (PreAnalytix) following manufacturer’s instructions from seven donors (four male and three females) of the same ethnicity with an average age of 67 years and a standard deviation of 6.6 years and were labeled as 14–20P. Blood samples were collected following a fasting period and at the same hour of the day to help reduce variability. The rat whole body RNA were a generous donation from Dr. Alistair Forrest and are commercially available from BioChain (http://www.biocat.com/products/R4434567-1-BC).
We processed all five lanes of sequencing from the nanoCAGE libraries as follows. Using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), we extracted raw tags and distributed them into their respective samples based on their barcode sequence. Raw reads that did not match a barcode sequence were discarded mainly owing to poor or ambiguous base was calling. Barcode sequences (and the common spacer sequence in the rat libraries) and the leading guanosines were trimmed off from the sequenced read. Next, we filtered out artifactual reads using TagDust (26), a program that filters out reads resembling the primer, linker and adaptor sequences used during library construction, using a false discovery rate of 0.01. Lastly, reads mapping to the ribosomal sequences U13369.1, NR_003285.2, NR_003286.2 and NR_003287.2 with ≤2 mismatches were considered to be ribosomal sequences and removed (Supplementary Table S1). After all pre-processing stages, reads were mapped to the hg19 or rn4 genome depending on the sample using BWA (27) with a mismatch threshold of 2. Using SAMTools (28), we selected reads with a mapping quality (MAPQ) of 10 (90% accuracy) or better.
Identifying barcode-biased tags in the whole blood libraries
For the 21 human whole blood libraries, we used 14 barcodes, consisting of 12 unique barcode sequences (ACATAC, AGTACG, ATCACG, CACGAT, CGATAC, GAGACG, GCTATA, GCTCAG, GTAGTG, GTATAC, TATGTG and TCGACG) where the mean Hamming distance between all pairwise barcodes is 4.32. For each sample, three libraries were made using two barcodes, i.e. one technical replicate was made with one barcode sequence, and the other two technical replicates made with the other barcode sequence (Table 1).
Table 1.
A summary of the biological and technical replicates used in this study, along with the barcodes and the number of reads that were mapped at a MAPQ of ≥10
| Samples | Technical replicate | Barcodes | Number of reads mapping at q10 |
|---|---|---|---|
| Human | |||
| 14P | 1 | GCTATA | 597909 |
| 2 | CACGAT | 711936 | |
| 3 | CACGAT | 960204 | |
| 15P | 1 | GTAGTG | 445901 |
| 2 | CGATAC | 592336 | |
| 3 | CGATAC | 674823 | |
| 16P | 1 | TATGTG | 1040935 |
| 2 | GAGACG | 1163416 | |
| 3 | GAGACG | 756476 | |
| 17P | 1 | ACATAC | 722023 |
| 2 | GCTCAG | 538660 | |
| 3 | GCTCAG | 695706 | |
| 18P | 1 | ATCACG | 685146 |
| 2 | GTATAC | 889014 | |
| 3 | GTATAC | 884897 | |
| 19P | 1 | CACGAT | 371069 |
| 2 | TCGACG | 663186 | |
| 3 | TCGACG | 420775 | |
| 20P | 1 | CGATAC | 741908 |
| 2 | AGTACG | 1195816 | |
| 3 | AGTACG | 1431334 | |
| Rat | 1 | ACAGAT | 927429 |
| 2 | ATCGTG | 849609 | |
| 3 | CACGAT | 793598 | |
| 4 | CACTGA | 810155 | |
| 5 | CTGACG | 863029 | |
| 6 | GAGTGA | 895320 | |
| 7 | GTATAC | 1005221 | |
| 8 | TCGAGC | 823343 |
Next, using a present/absent criterion, we identified reads among technical replicates that were only present when using one barcode and absent when the other barcode is used. We used a threshold of ≥21 raw reads for our present criteria (Supplementary Table S4). Marioni et al. (29) reported that if technical replicates are sequenced, the read counts for a particular feature should vary according to the Poisson distribution. Thus, it is unlikely that our selected reads are a consequence of natural variation but rather are attained by the use of a different barcode sequence. Sequence logos (30) were created by extracting the nine nucleotides upstream of these mapped reads, and the sequence enrichment was calculated using unique upstream sequences.
Filtering strand invasion artifacts
From our selection of barcode-biased reads, we observed that the upstream region of these reads showed sequence complementary to the tail of the TS oligonucleotide (Figure 2), which is a consequence of strand invasion. This served as a marker for strand invasion artifacts, which was subsequently used as our strategy for their removal. Thus, once reads were mapped to a reference, the nine nucleotides immediately upstream were extracted, and using a global alignment approach (31), they were aligned to the last nine nucleotides of the TS oligonucleotide used for construction of that particular library. The edit distance was used as a metric for the alignment, and a single mismatch or gap constituted an edit distance of one. A perfect alignment would thus have zero edits. We observed the enrichment of at least two guanosine nucleotides directly upstream of where a strand invasion artifact mapped. Thus, we imposed this criterion to our filtering strategy; reads were only considered to be artifacts if two of the three nucleotides directly upstream were guanosines. Lastly, as an indication of the edit distance threshold to use for data filtering, we filtered libraries using edit distances of zero to five and measured the Spearman’s rank correlation coefficient between technical replicated libraries at each threshold. The filtering strategy was implemented using Perl, and an executable version of the script is available as supplementary data.
Figure 2.
By preparing technical triplicates with different barcodes, we could identify reads present in a barcode specific manner, i.e. barcode-biased reads. The sequence logos were created using the sequence directly upstream of individually mapped barcode biased reads. The barcode sequence used to prepare each library and the three riboguanosines of the TS oligonucleotide are shown directly below the corresponding sequence logos. The enrichment profiles closely resemble the tailing sequence of the TS oligonucleotide used to construct that particular library.
Specificity and sensitivity
The specificity of a method relates to the ability of identifying negative results, assessed by the number of false positives. We created a negative set, i.e. putatively non-biased reads, by selecting for the least variable reads among technical triplicates. We first normalized reads by tags per million, and selected the top 20% of least variable reads among replicates. In contrast, the sensitivity refers to the ability of identifying positive results, assessed by the number of true positives. We created a positive set, i.e. strand invasion artifacts, in the same manner that we identified barcode-biased tags described above. With our negative and positive sets, we then applied our barcode filtering scheme described above with an edit distance of four. The specificity was calculated as the ‘number of true negatives/(number of true negatives + number of false positives)’, and the sensitivity was calculated as the ‘number of true positives/(number of true positives + number of false negatives)’.
Differential expression analysis
For the comparison of different libraries, we used a previously developed read/tag clustering method (32), as opposed to comparing individual reads. The clustering method aggregates reads that are mapped within a window of 20 nucleotides into single entity clusters; the expression of the cluster is the summation of all tags within the cluster. We conducted our differential expression analyses on tag clusters present among technical replicates using the edgeR_2.4.1 package (33) on R version 2.14.1. Within a technical triplicate set, technical replicates made with one barcode were tested against technical replicates made with the other barcode. For the comparison of the rat libraries, we arbitrarily tested the libraries made with the ACAGAT, ATCGTG, CACGAT and CACTGA barcodes against the libraries made with the CTGACG, GAGTGA, GTATAC and TCGAGC barcodes. We used an independent filtering criterion (34), selecting for tag clusters with ≥10 raw reads. The standard edgeR pipeline was carried out using a common dispersion approach (and tag-wise dispersion for the rat libraries) and the Benjamini and Hochberg’s (35) approach for controlling the false discovery rate. Tag clusters with an adjusted P-value of ≤0.01 were defined as differentially expressed.
RNA-Seq data sets
We processed two independently produced RNA-Seq data sets, made using two different protocols (10,36). Briefly, Islam et al. analysed the single cell transcriptomes of mouse embryonic fibroblasts and embryonic stem cells. The Islam et al. RNA-Seq libraries, which was made using TS and in a manner very similar to nanoCAGE, was downloaded directly from the author’s website and was processed in the same manner as our nanoCAGE libraries owing to the similarity between the protocols. Briefly, Guttman et al. produced RNA-Seq libraries from mouse embryonic stem cells, neuronal precursor cells and lung fibroblasts by mRNA fragmentation and random-primed reverse transcription. The Guttman et al. data set was downloaded from the DNA Data Bank of Japan under the accession number SRP002325, and the sequenced reads were mapped using TopHat (37) on the default settings. After all pre-processing steps, we compared the derived transcript structures between the fibroblasts libraries made by Islam et al. and by Guttman et al. In addition, we also compared different fibroblast libraries made with different barcodes in the Islam et al. data set.
RESULTS
Barcode specific reads in nanoCAGE libraries
Total RNA, isolated from whole blood samples derived from seven donors, was used to prepare 21 separate nanoCAGE libraries where each sample was made in triplicate (Table 1). Furthermore, libraries were prepared together to help limit batch effects. To study the effect of using different TS oligonucleotides and thus the barcode sequence, we prepared the same sample identically except for the TS oligonucleotides used; two barcodes were used per technical triplicate. As there are an odd number of replicates, two of the three replicates were prepared with one barcode and the remaining replicate prepared with the other barcode. NanoCAGE libraries were prepared following a previously published protocol (16). The 21 nanoCAGE libraries were then sequenced in multiplex using Illumina's GAIIx instrument on four sequencing lanes.
Sequenced reads in the nanoCAGE protocol represent the site at which TS occurred (Figure 1), which represents the 5′ end of a RNA molecule and thus the putative transcriptional starting site (TSS) (9). Hence, to identify artifacts, we could compare nanoCAGE reads that do not map to known promoters of transcripts, although these could represent previously uncharacterized transcripts. A more definitive approach not requiring transcript annotations is to search for intra sample differences, i.e. reads present only in one set of barcoded technical replicates. To correctly identify the corresponding transcript for a sequenced read, we selectively analysed 16 281 067 reads that could be mapped to the genome with 90% confidence (MAPQ of ≥10) (27). Finally, from this set, we identified 132 980 barcode specific reads, i.e. reads present only in one set of technical replicates using a particular barcode and not the other, where the variance is unlikely due to Poisson noise (see ‘Materials and Methods’ section).
Figure 1.
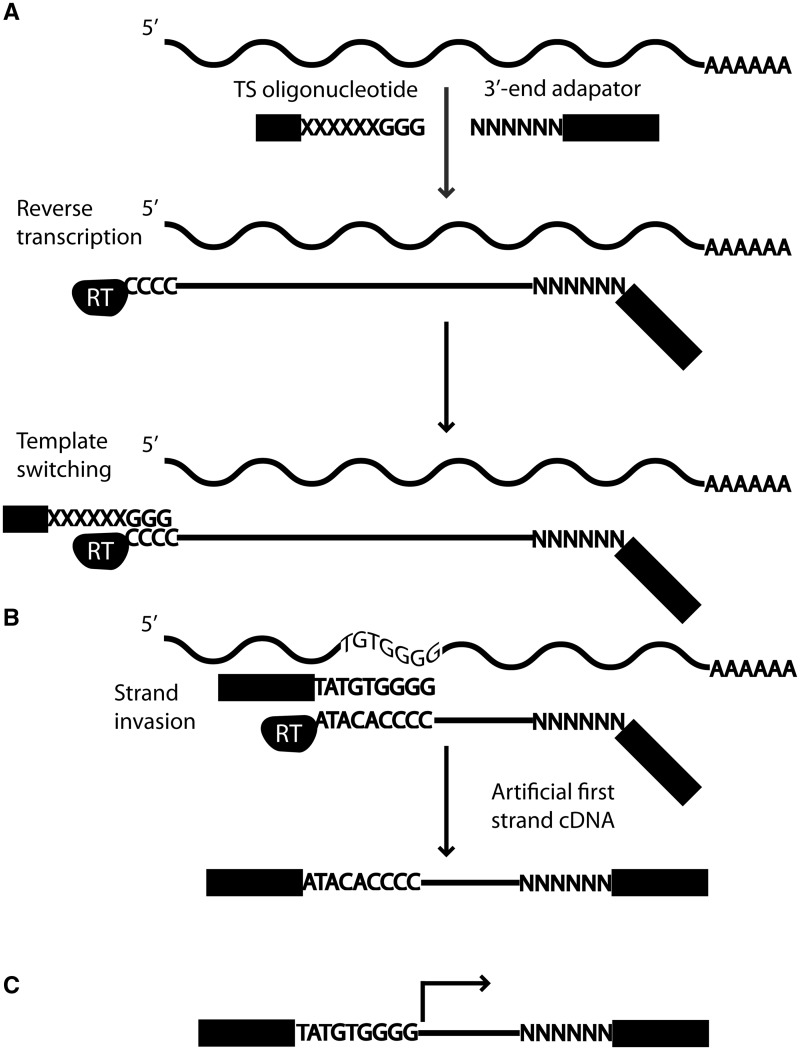
(A) The TS mechanism is used for first strand cDNA synthesis. First, an oligonucleotide hybridizes to the RNA molecule, and RT starts polymerizing. Once the RT reaches the 5′ end of the RNA, a cytosine overhang is formed. The TS oligonucleotide containing three riboguanosines hybridizes to the cytosine overhang and the RT switches template and polymerizes the TS oligonucleotide. (B) However, during RT synthesis, if the polymerized region has sequence complementary with the 3′ tail of the TS oligonucleotide, it may invade and hybridize with the first strand cDNA. RT then switches template and polymerizes the TS oligonucleotide. However, this strand invasion process has resulted in a cDNA that is shorter than the RNA. (C) With the nanoCAGE protocol, sequencing begins just upstream of the site of TS, which includes the barcode and the riboguanosine linker sequence. The barcode and linker sequences are trimmed off during processing steps, and the final read sequence is indicated by the black arrow.
From our barcode specific nanoCAGE reads, we analysed the region directly surrounding the reads. Interestingly, the upstream sequence of these barcode biased reads revealed an enrichment of nucleotides that resembled the 3′ end of the TS oligonucleotide used for that library (Figure 2). The sequence logos illustrate an enrichment of guanosines at positions −1 to −3, which corresponds to the r(G)3 tail of the TS oligonucleotide, whereby positions −4 to −9 show a varied enrichment of nucleotides that resemble the barcode used to produce the library, especially positions −4 to −6 (Figure 2). These results suggest the hybridization of the TS oligonucleotide to a complementary region on the first strand cDNA, i.e. strand invasion, and thus produces TS artifacts in a barcode dependent manner (Figure 1B). Although the r(G)3 tail of the TS oligonucleotide preferentially binds to the cytosine overhang created by the RT (9), the increase in sequence complementarity in the 3′ tail of the TS oligonucleotide may increase the hybridization of the TS oligonucleotide to the first strand cDNA (Figure 1B).
Filtering out strand invasion artifacts
Artifactual reads need to be removed before they are used for further downstream analyses (26). The TS mechanism is expected to occur at the cytosine overhang created by the RT (Figure 1A); thus, the sequence immediately upstream of a nanoCAGE tag should exhibit sequence complementarity only on a random basis, although this is largely dependent on the makeup of the genome. Under these assumptions, we devised a strategy for removing strand invasion artifacts by aligning the sequence immediately upstream of reads to the 3′ tail of the TS oligonucleotide. We chose to align the nine nucleotides directly upstream of a read (Figure 1C) to the last nine nucleotides of the TS oligonucleotide owing to the enrichment profiles previously observed (Figure 2).
Next, we analysed a range of sequence complementarity scores to determine the optimal threshold for classifying reads as artifacts. First, we carried out a global alignment (31) between the sequence upstream of a read and the TS oligonucleotide tail for all libraries. We directly used the edit distance of an alignment as a measurement of the sequence complementarity, where gaps and mismatches were individually constituted as one edit; a perfect alignment would thus have zero edits. In addition, we only classified reads as artifacts if two or more of the three nucleotides directly upstream were composed mainly of guanosines (see ‘Materials and Methods’ section). Lastly, we filtered out reads on a range of edit distances, from zero to five, and found that by removing such noise, we had technical replicates that correlated better with each other (Supplementary Table S2).
Effects of artifact filtering on library correlation
A common metric used to determine library similarity is by correlation. We assessed the correlation of technical replicates after our filtering strategy at various thresholds. Given the nature of CAGE reads and promoters, we first clustered reads into what are known as ‘tag clusters’ (32), enabling us to measure the correlation of libraries. Tag clusters are representative of putative promoter regions whereby the number of reads mapping to these regions represents the level of expression (see ‘Materials and Methods’ section). We calculated the Spearman’s rank correlation coefficient between all technical replicates, given the skewed expression rate of blood transcripts, i.e. the distribution of transcript expression is not linear; so we used a non-parametric measure of correlation. The distribution of transcripts in blood is largely skewed by the presence of globin transcripts, which resulted in the under sequencing of other transcripts. This under sampling resulted in an increase of noise, especially for transcripts that are lowly expressed, and subsequently lower correlations between replicates. The removal of globin transcripts would not significantly affect the Spearman’s rank correlation coefficient owing to the way the correlation is calculated. For technical replicates made with different barcodes, we observed a general increase in correlation between the libraries as we relaxed the similarity threshold, i.e. increasing the edit distance (Supplementary Table S2). The increase in correlation was the direct consequence of removing library specific reads, i.e. TS artifacts. The opposite effect, a decrease in correlation after read filtering, was observed when comparing technical replicates with the same barcode (Supplementary Table S2). The correlation of technical replicates was inflated owing to TS artifacts, and the removal of these reads decreased the correlation. For the comparison of libraries with different barcodes, the majority of correlations increased until an edit distance of four. This was due to the decrease in stringency, which resulted in real signal being removed by random chance that a loose alignment could be formed between the upstream sequence and the tail sequence of the TS oligonucleotide. Although the correlations between technical replicates are considered moderate, we demonstrated that we are able to identify TS artifacts, and the removal of these artifacts resulted in higher correlations between technical replicates.
Effects of artifact filtering on differential expression detection
One of the core analyses conducted on transcriptome data is a statistical test that detects differential expression of transcripts. An observed difference is statistically significant only when the observed difference is greater than expected from random variation. Transcripts may be spuriously detected as differentially expressed owing to the introduction of experimental variations such as from using different barcodes. We tested this notion by conducting differential expression analyses using edgeR (33) on technical replicated libraries before artifact filtering, after filtering and after randomly removing reads (Figure 3). Given that our analyses were carried out on technical replicates, we would expect to find very few tag clusters that are detected as differentially expressed. However, a fraction of tag clusters were detected as differentially expressed between technical replicates before filtering (Supplementary Table S3). In all cases, the removal of strand invasion artifacts decreased the number of differentially expressed candidates (Supplementary Table S3). Using an edit distance of four for barcode filtering, on average, we observed a roughly 10-fold decrease in the number of differentially expressed candidates. In contrast, removing random reads resulted in a slight decrease of 1.2-fold in differentially expressed candidates.
Figure 3.
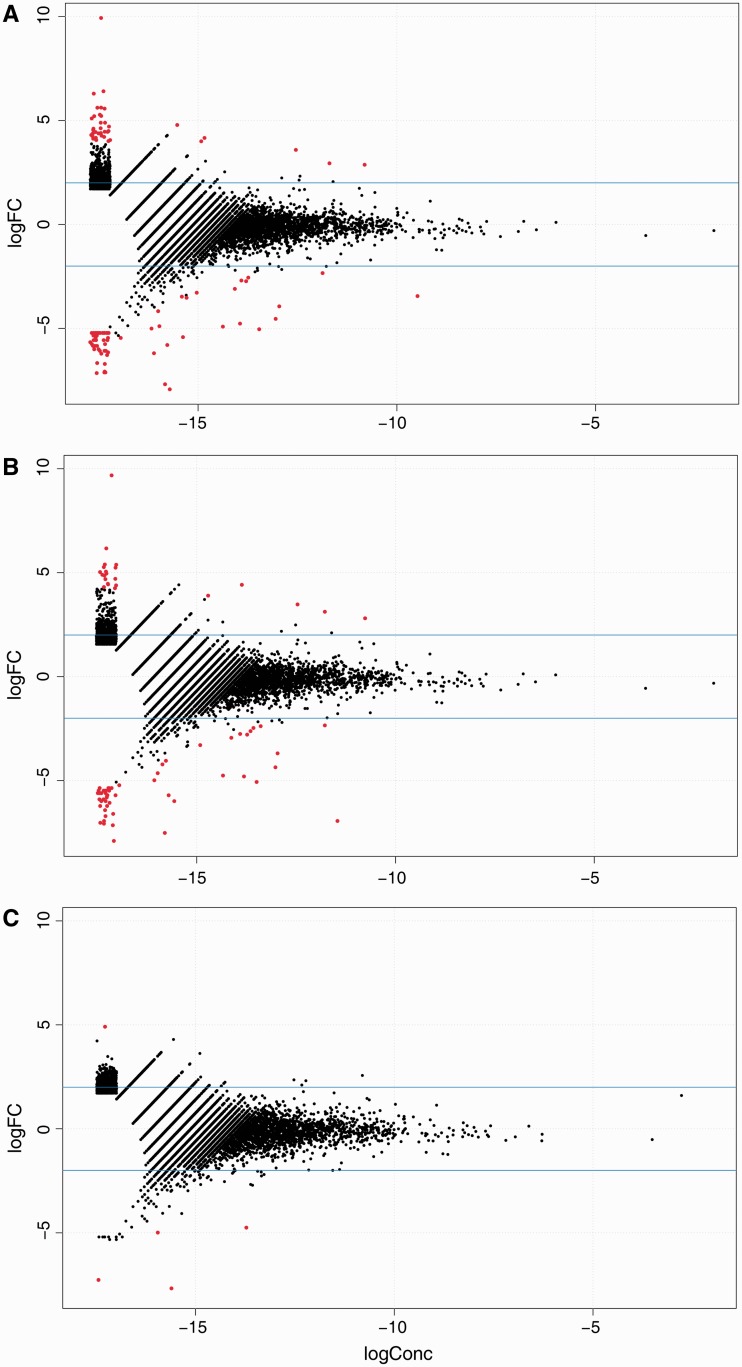
Differential expression analyses were carried out between technical replicates made from different barcode sequences using edgeR. The scatter plots show the log-fold change (y-axis) against the log concentration (x-axis) for tag clusters present among the 14P technical replicates. In red are tag clusters that were detected as differentially expressed at an adjusted P-value ≤ 0.01. Three separate analyses were carried out: test for differential expression (A) before strand invasion artifacts were filtered out, (B) when random reads were removed and (C) after strand invasion artifacts were filtered out. By removing artifacts, fewer tag clusters were detected as differentially expressed compared with no filtering and removing random reads.
Sensitivity and specificity of artifact filtering
We have experimentally prepared our libraries in such a way that we can identify TS artifacts. Using a set of nanoCAGE reads that were identified as TS artifacts (see ‘Materials and Methods’ section), i.e. true positives, we applied our filtering strategy to measure the sensitivity of the method. Reads not detected as artifacts in this set were considered as false negative. Using an edit distance metric of four, the average sensitivity was ∼94.3% across the entire data set; we could detect 125 357 of the 132 980 true positives (Supplementary Table S4).
The specificity of a method gives an estimate of the number of false positives. This measure is important for quantifying the potential amount of signal that is removed due to the random chance that the upstream region of a read resembles the 3′ tail of the TS oligonucleotide. To determine a true negative set, i.e. not barcode biased, we selected reads with the lowest amount of variance between the technical replicates (see ‘Materials and Methods’ section), and if any of these reads were filtered out, they were considered false positives. Of the subset of reads we considered to be a true negative set (n = 135 613), on average 6.7% +/− 2.1 of these reads were considered false positives (Supplementary Table S5). However, one should consider that even our true negative set may contain strand invasion artifacts, i.e. a false positive in the sense of being a true negative, and we only examined a small proportion of the total number of reads in a library; in reality, the false positive rate is likely to be much lower.
Degree of bias from different barcode sequences
Strand invasion occurs during first strand cDNA synthesis, and successful hybridization depends on the degree of sequence complementarity between the cDNA and the 3′ tail of the TS oligonucleotide. Therefore, the number of TS artifacts becomes a function of the number of RNA molecules that contains sequence complementarity to the TS oligonucleotide. Barcode sequences that occur more prominently among RNA molecules would result in a higher number of TS artifacts. To test this hypothesis, we scanned the genome in a sliding window manner. Given that the last six nucleotides of the TS oligonucleotide are the most important for strand invasion (Figure 2), we tallied the number of all possible 6-mers that end in GGG (total of 64 6-mer combinations) across the human genome (hg19) on both strands. For the sake of simplicity and owing to a lack of a complete transcriptome, we chose to tally this number across the genome as opposed to a defined transcriptome. We previously defined a set of TS artifacts by examining technical replicates made with different barcodes and used this number as an estimate of the number of TS artifacts. We then calculated the Spearman’s rank correlation coefficient between the number of TS artifacts and the tallied number for the 6-mer that corresponded to the sequence at the end of the TS oligonucleotide. As expected, a positive correlation (Spearman’s rho ∼ 0.67) was observed (Supplementary Table S6), which supported the hypothesis that choosing a barcode sequence, which is present more often in the genome, leads to a larger number of strand invasion artifacts.
An experimental strategy for suppressing artifacts and barcode bias
Our results have suggested that first strand cDNAs with regions of sequence complementarity to the last six nucleotides of the TS oligonucleotide are potential sites for strand invasion. Given this information, intuitively it is expected that if the last six nucleotides of the TS oligonucleotide occur more frequently amongst RNA molecules, there would be a higher number of TS artifacts. We established this hypothesis that barcode sequences that are present more frequently in the genome have more strand invasion artifacts (Supplementary Table S6). So to suppress the number of artifacts, one should select barcodes less frequent in the genome. We have also observed that barcodes that end with a guanosine, thus creating a sequence tail of four guanosines in the TS oligonucleotide, have much higher number of TS artifacts (Supplementary Table S6). Furthermore, at transcriptional starting sites, libraries made with a barcode ending with a guanosine have much higher counts of certain transcripts compared with barcodes that do not end with a guanosine (Figure 4); this type of bias cannot be mitigated by our artifact filterer. To suppress this barcoding bias, it is necessary that the sequence directly upstream of the riboguanosines is standardized, a strategy similar to standardizing the adaptor sequence in ligation-based barcoding (18). Additionally, our strategy for the choice of a standard spacer is one that occurs less frequently in the genome. Thus, we can potentially suppress the extent of strand invasion and systematically remove the barcode bias effect.
Figure 4.
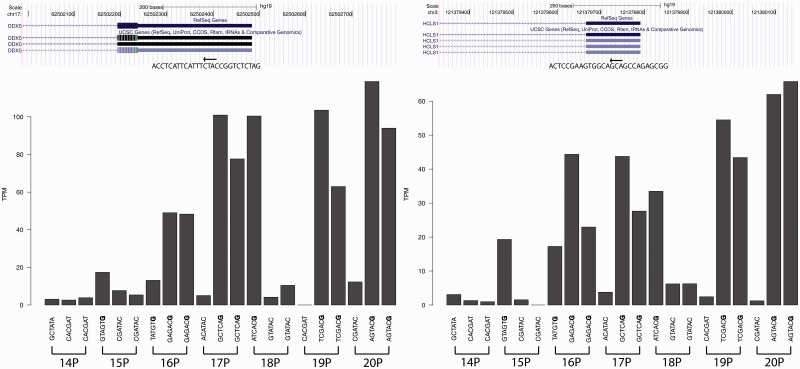
Barcode bias in nanoCAGE technical replicated libraries. The choice of barcode sequence can affect the read count pertaining to the transcriptional starting site. Here, we show two examples for the genes DDX5 and HCLS1, where the read count fluctuates according to the barcode sequence and not by the sequencing depth or by the library. Libraries made with barcodes ending with a guanosine (shown in bold in the bar plot) have a much higher tags per million (TPM) count than barcodes that end with other nucleotides.
To test this strategy, we redesigned the TS oligonucleotide to include a 6-nucleotide spacer (GCTATA) directly upstream of the riboguanosines. We produced eight nanoCAGE libraries using eight barcodes from rat whole body RNA, i.e. technical replicates, and sequenced them on one lane on the Illumina GAIIx platform. We processed these libraries in the same manner as our blood nanoCAGE libraries and obtained around 8 million reads in total after processing (Supplementary Table S7). Next, using the tag-clustering method previously described, we aggregated our reads and measured the pairwise correlations of each library; the average Spearman’s rank correlation coefficients was ∼0.75 (Supplementary Table S7), a vast improvement to the blood nanoCAGE libraries. To investigate how much of the variance in the data is explained by sequencing noise, for each tags per million -normalized tag cluster, we calculated the mean and the exact 95% confidence intervals (CIs) for the mean assuming a Poisson distribution. For each tag cluster and the respective library expression, we tallied the number of times an expression value was inside the CIs; approximately 92% of the total expression values fall inside the 95% CIs. The nanoCAGE protocol is designed to work with few nanograms of total RNAs and require a relatively large number of semi-suppressive PCR cycle, which in addition to the Poisson noise, may account for points that fall outside of the 95% CIs. Semi-suppressive PCR allows the use of random primers, which can capture non-coding RNAs, but, however, shows suboptimal yields at each PCR cycles. Additionally, a second PCR reaction is needed to add sequencing adapters after the semi-suppressive PCR. In summary, although nanoCAGE can also identify non-polyA RNAs from low starting material (9), it requires two PCR cycles, which may be a source of noise.
When we applied the filtering strategy on the libraries made with the common spacer, we found that on average 4.5% +/− standard deviation of 0.12 (Supplementary table S7) of the total reads were detected as putative TS artifacts compared with an average of 11.1% +/− standard deviation of 6.65 (Supplementary Table S1) for the libraries made without the common spacer. By using a common spacer, all libraries had roughly the same number of putative artifacts, i.e. very low standard deviation, which is also lower than the number of putative artifacts detected in most libraries made without the common spacer. Although the older data set detected an average of ∼11%, the number of artifacts is highly dependent on the barcode (Supplementary Table S1), which is the reason for a much higher strand deviation. For example, by using the GCTCAG barcode without a spacer, up to ∼25% of the reads were detected as artifacts. By using a common spacer, the biases will affect the same transcripts in the same way in different samples. This is particularly important when conducting differential expression analyses and because of this, it is not necessary to filter out putative artifacts. To test this, we performed a differential expression analysis on the common spacer libraries, and indeed none of the tag clusters were detected as significantly differentially expressed.
DISCUSSION
The TS mechanism has been exploited in full-length cDNA library construction owing to its technical simplicity (6), in transcriptome analyses due to its ability to mark the 5′ end of a RNA molecule (9) and its flexibility in incorporating DNA barcodes for multiplexing (10) and for incorporating DNA fingerprints for quantifying the absolute number of molecules (21). Owing to the elimination of adaptor mediated steps, RNA material can be conserved, making TS an attractive choice when working with limited amounts of sample, such as with single cell type analyses (10,12). Furthermore, the decreasing costs of DNA sequencing are driven by an increase of throughput in a constant number of sequencing lanes, which makes multiplex sequencing determinant for cost and time efficiency. Although TS has been used to incorporate barcodes during reverse transcription for multiplexing, one particular drawback of this approach is that the barcode sequences may skew the following reactions, in particular PCR, in favor of one sample. For this reason, strategies where the barcode is added at a last step are sometimes preferred, for instance in the protocols of Illumina’s TruSeq product line. Nevertheless, this has the drawback that samples cannot be pooled as early as with TS, which increase the work load and cost of the experiment. In addition, because these two methods of barcoding are directed at different parts of the library constructs, they can be used together to implement combinatorial multiplexing. By combining two barcodes together, the index diversity is greatly increased, and this allows the unique labeling of all transcripts in a sample (38). This approach takes advantage of the very high throughput of current sequencers, which can also be applied to labeling several thousands of low complexity single cell libraries.
Here, we have characterized and investigated a source of bias that is inherent to TS: the production of spurious, sequence-specific reads owing to strand invasion and different hybridization rates as a consequence of choosing different barcodes. We have shown that the extent of strand invasion depends highly on the sequence of the TS oligonucleotide, especially the last six nucleotides. All oligonucleotides in the reverse transcription reactions can interrupt the first-strand cDNA synthesis by strand invasion, and we have previously observed oligo-dT primers being template switched at the 5′ of T-rich regions of the mRNAs (data not shown). This strand invasion becomes more problematic when different sets of TS oligonucleotides are used to barcode specific samples, as this subjects the samples to different degrees of bias. In these multiplexed libraries, the strand invasion artifacts will produce sample-specific signals, which will can wrongly suggest correlations or in contrary mask the similarity between related samples. For example, samples using two barcodes that end in the same six nucleotides may artificially cluster together irrespective of the sample condition. Even in non-multiplexed libraries, strand invasion produce shortened cDNAs that can systematically bias expression levels and create artifacts that do not reflect the transcriptome. We compared two different RNA-Seq protocols, one using TS (and with the same multiplexing strategy as nanoCAGE) (10) and another by conventional RNA fragmentation on mouse fibroblasts and observed different coverage patterns (Supplementary Figure S1). The transcript profile observed in the TS RNA-Seq protocol is likely a consequence of strand invasion. Moreover, we have also observed different transcript profiles in biologically replicated samples that were made from different barcodes (Supplementary Figure S2). As we have demonstrated in our work, it is crucial to control strand invasion products especially with respect to introducing barcodes by TS.
It is possible to identify strand invasion products in silico and consequently have them removed. We have shown that by analysing the sequence upstream of where a sequenced read maps, artifactual reads could be identified with high specificity and sensitivity. Importantly, by removing such noise, replicated libraries made using different barcodes correlated better to each other. By performing a differential expression analysis on the filtered data sets, on average, a 10-fold decrease in the number of tag clusters called as differentially expressed was observed. The removal of strand invasion artifacts, which contribute to an increased variance among samples, is crucial in differential expression analyses using digital gene expression data such as CAGE and RNA-Seq. However, it is ideal to design an experimental protocol that limits as much as possible biases that are a consequence of the barcoding strategy (19). We proposed a strategy, which we tested in the nanoCAGE method, by updating the sequence of the TS oligonucleotide by inserting a 6-nucleotide long standard spacer between the barcode and the ribo-guanosines. In addition, we chose a spacer sequence that had less potential for strand invasion. The main purpose of the common spacer is to ensure that any TS bias systematically affects all libraries in the same manner. We confirmed this by conducting a differential expression analysis on the libraries made with the common spacer, and indeed no tag clusters were detected as significantly differentially expressed (Supplementary Table S7). A potential downside to the common spacer approach is the addition of six more nucleotides to a sequenced read. However, when sequenced on a HiSeq instrument with the standard read length of 50 nucleotide, the resulting libraries can be aligned accurately with standard tools such as BWA (27), as 35 informative bases are remaining after removing the barcode, the spacer and the linker. Alternative strategies could be conceived, and the spacer could be extended or replaced by a random sequence (21,39).
We have described in this article an inherent problem that exists with the TS mechanism, which we could suppress by combining experimental and computational strategies; however, TS artifacts cannot be entirely abolished. What distinguishes the artifacts from bona fide full-length cDNAs is the presence of the remaining 5′ part of the mRNA as a possibly long tail in the mRNA/cDNA/oligonucleotide triplex. By using an experimental protocol called CAP Trapper (40), which is used in CAGE protocols, it is possible to identify this triplex due to the presence of a 7-methylguanosine cap, therefore accurately identifying transcriptional starting sites as opposed to strand invasion products. This concept of combining TS and CAP Trapper has been shown to produce multiplexed libraries that capture promoters with high fidelity (41). However, as this methodology requires additional preparation steps and is not favored in most TS protocols, where the starting material is limited such as in single cell analyses. Despite the remaining artifacts, our proposed strategy allows one to directly compare different samples, such as between normal and diseased samples. Given that TS is garnering interest again, as seen by the number of recent publications that have used TS, it is important that investigators become aware of TS artifacts. It is clear that more investigations are needed to fully understand the TS mechanism, especially with respect to the types of biases that could potentially be introduced.
DATA DEPOSITION
Sequence data have been deposited in the DNA Data Bank of Japan under accession code DRA000552.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–7 and Supplementary Figures 1 and 2.
FUNDING
The European Union Seventh Framework Programme under grant agreement [FP7-People-ITN-2008-238055] (‘BrainTrain’ project) (to P.C.); the Research Grant for RIKEN Omics Science Center from Ministry of Education, Culture, Sports, Science and Technology; the Grant-in-Aids for Scientific Research (A) No. 20241047 for nanoCAGE (to P.C.); the 7th Framework Programme Dopaminet Project from the EU (to P.C.); the Telethon grant [GGP10224] (to S.G.). Funding for open access charge: the European Union Seventh Framework Programme under grant agreement [FP7-People-ITN-2008-238055] (‘BrainTrain’ project) (to P.C.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Michiel de Hoon for assistance with the statistical analyses and Dr. Alistair Forrest for the rat samples.
REFERENCES
- 1.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 2.Temin HM, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 3.Hirzmann J, Luo D, Hahnen J, Hobom G. Determination of messenger RNA 5′-ends by reverse transcription of the cap structure. Nucleic Acids Res. 1993;21:3597–3598. doi: 10.1093/nar/21.15.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohtake H, Ohtoko K, Ishimaru Y, Kato S. Determination of the capped site sequence of mRNA based on the detection of cap-dependent nucleotide addition using an anchor ligation method. DNA Res. 2004;11:305–309. doi: 10.1093/dnares/11.4.305. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt WM, Mueller MW. CapSelect: a highly sensitive method for 5′ CAP-dependent enrichment of full-length cDNA in PCR-mediated analysis of mRNAs. Nucleic Acids Res. 1999;27:e31. doi: 10.1093/nar/27.21.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques. 2001;30:892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]
- 7.Matz M, Shagin D, Bogdanova E, Britanova O, Lukyanov S, Diatchenko L, Chenchik A. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 1999;27:1558–1560. doi: 10.1093/nar/27.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 9.Plessy C, Bertin N, Takahashi H, Simone R, Salimullah M, Lassmann T, Vitezic M, Severin J, Olivarius S, Lazarevic D, et al. Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nat. Methods. 2010;7:528–534. doi: 10.1038/nmeth.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, Linnarsson S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko JH, Lee Y. RNA-conjugated template-switching RT-PCR method for generating an Escherichia coli cDNA library for small RNAs. J. Microbiol. Methods. 2006;64:297–304. doi: 10.1016/j.mimet.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 12.Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeda N, Nishiyori H, Nakamura M, Kawazu C, Murata M, Sano H, Hayashida K, Fukuda S, Tagami M, Hasegawa A, et al. Development of a DNA barcode tagging method for monitoring dynamic changes in gene expression by using an ultra high-throughput sequencer. Biotechniques. 2008;45:95–97. doi: 10.2144/000112814. [DOI] [PubMed] [Google Scholar]
- 14.Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, Linnarsson S. Highly multiplexed and strand-specific single-cell RNA 5′ end sequencing. Nat. Protoc. 2012;7:813–828. doi: 10.1038/nprot.2012.022. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi H, Lassmann T, Murata M, Carninci P. 5′ end-centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat. Protoc. 2012;7:542–561. doi: 10.1038/nprot.2012.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salimullah M, Sakai M, Plessy C, Carninci P. NanoCAGE: a high-resolution technique to discover and interrogate cell transcriptomes. Cold Spring Harb. Protoc. 2011;2011 doi: 10.1101/pdb.prot5559. pdb prot5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumura H, Yoshida K, Luo S, Kimura E, Fujibe T, Albertyn Z, Barrero RA, Kruger DH, Kahl G, Schroth GP, et al. High-throughput SuperSAGE for digital gene expression analysis of multiple samples using next generation sequencing. PLoS One. 2010;5:e12010. doi: 10.1371/journal.pone.0012010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawano M, Kawazu C, Lizio M, Kawaji H, Carninci P, Suzuki H, Hayashizaki Y. Reduction of non-insert sequence reads by dimer eliminator LNA oligonucleotide for small RNA deep sequencing. Biotechniques. 2010;49:751–755. doi: 10.2144/000113516. [DOI] [PubMed] [Google Scholar]
- 19.Alon S, Vigneault F, Eminaga S, Christodoulou DC, Seidman JG, Church GM, Eisenberg E. Barcoding bias in high-throughput multiplex sequencing of miRNA. Genome Res. 2011;21:1506–1511. doi: 10.1101/gr.121715.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jayaprakash AD, Jabado O, Brown BD, Sachidanandam R. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 2011;39:e141. doi: 10.1093/nar/gkr693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kivioja T, Vaharautio A, Karlsson K, Bonke M, Enge M, Linnarsson S, Taipale J. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods. 2011;9:72–74. doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
- 22.Goetz JJ, Trimarchi JM. Transcriptome sequencing of single cells with Smart-Seq. Nat. Biotechnol. 2012;30:763–765. doi: 10.1038/nbt.2325. [DOI] [PubMed] [Google Scholar]
- 23.Fan JB, Chen J, April CS, Fisher JS, Klotzle B, Bibikova M, Kaper F, Ronaghi M, Linnarsson S, Ota T, et al. Highly parallel genome-wide expression analysis of single mammalian cells. PLoS One. 2012;7:e30794. doi: 10.1371/journal.pone.0030794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, Bodovitz S. Single cell analysis: the new frontier in ‘omics’. Trends Biotechnol. 2010;28:281–290. doi: 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapteyn J, He R, McDowell ET, Gang DR. Incorporation of non-natural nucleotides into template-switching oligonucleotides reduces background and improves cDNA synthesis from very small RNA samples. BMC Genomics. 2010;11:413. doi: 10.1186/1471-2164-11-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lassmann T, Hayashizaki Y, Daub CO. TagDust—a program to eliminate artifacts from next generation sequencing data. Bioinformatics. 2009;25:2839–2840. doi: 10.1093/bioinformatics/btp527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 32.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 33.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. Proc. Natl Acad. Sci. USA. 2010;107:9546–9551. doi: 10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 36.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiroguchi K, Jia TZ, Sims PA, Xie XS. Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc. Natl Acad. Sci. USA. 2012;109:1347–1352. doi: 10.1073/pnas.1118018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carninci P, Kvam C, Kitamura A, Ohsumi T, Okazaki Y, Itoh M, Kamiya M, Shibata K, Sasaki N, Izawa M, et al. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics. 1996;37:327–336. doi: 10.1006/geno.1996.0567. [DOI] [PubMed] [Google Scholar]
- 41.Batut PJ, Dobin A, Plessy C, Carninci P, Gingeras TR. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 2012;23:169–180. doi: 10.1101/gr.139618.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.