Abstract
Riboswitch operation involves the complex interplay between the aptamer domain and the expression platform. During transcription, these two domains compete against each other for shared sequence. In this study, we explore the cooperative effects of ligand binding and Magnesium interactions in the SAM-I riboswitch in the context of aptamer collapse and anti-terminator formation. Overall, our studies show the apo-aptamer acts as (i) a pre-organized aptamer competent to bind ligand and undergo structural collapse and (ii) a conformation that is more accessible to anti-terminator formation. We show that both Mg2+ ions and SAM are required for a collapse transition to occur. We then use competition between the aptamer and expression platform for shared sequence to characterize the stability of the collapsed aptamer. We find that SAM and Mg2+ interactions in the aptamer are highly cooperative in maintaining switch polarity (i.e. aptamer ‘off-state’ versus anti-terminator ‘on-state’). We further show that the aptamer off-state is preferentially stabilized by Mg2+ and similar divalent ions. Furthermore, the functional switching assay was used to select for phosphorothioate interference, and identifies potential magnesium chelation sites while characterizing their coordinated role with SAM in aptamer stabilization. In addition, we find that Mg2+ interactions with the apo-aptamer are required for the full formation of the anti-terminator structure, and that higher concentrations of Mg2+ (>4 mM) shift the equilibrium toward the anti-terminator on-state even in the presence of SAM.
INTRODUCTION
Riboswitches represent a diverse group of functional RNAs that regulate protein expression. These RNAs are usually located in the 5′-untranslated region of the mRNAs they regulate. Here, they specifically bind small molecules and adjust expression to maintain homeostasis of various metabolites. This is accomplished by the adoption of one of two mutually exclusive folds. Depending on the riboswitch, the partitioning between these mutually exclusive secondary structures can modulate transcriptional termination (1–6), initiation of translation (7,8) or ribozyme self-cleavage (9). In the case of the SAM-I aptamer studied here, the binding of cognate ligand stabilizes the aptamer domain. The stabilized aptamer domain precludes the formation of a transcriptional anti-terminator and allows the formation of a rho-independent terminator helix in the expression platform. This is the off-state of the riboswitch (Figure 1a). In the absence of ligand, the nascent riboswitch transcript co-opts sequence from the aptamer and folds to the default anti-terminator helix. This is the on-state of the riboswitch. Although shifting the population of riboswitch RNA toward the aptamer, or off-state, is dependent on ligand concentration, the role divalent ions play in this process is not well understood (10).
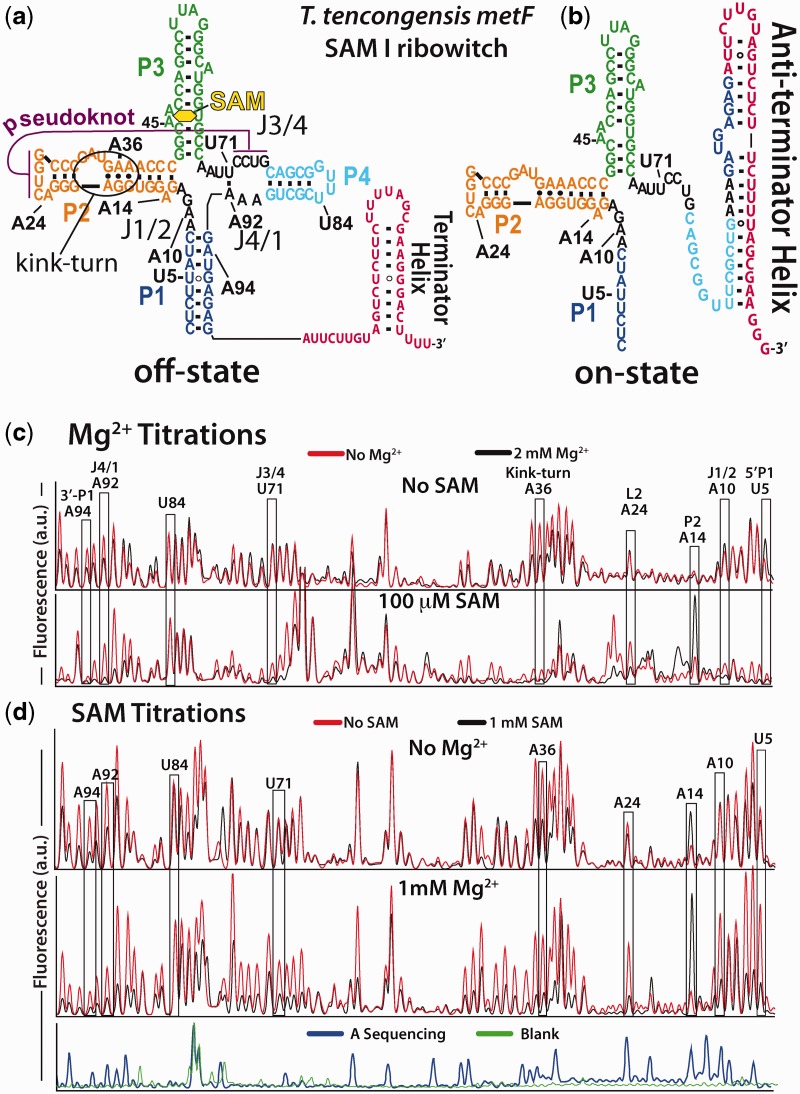
Figure 1.
SHAPE probing data for both the Mg2+ and SAM titrations. (a) The T. tengcongensis metF SAM I riboswitch in the SAM-bound off-state. This secondary structure is dominant in the presence of sufficient ligand. (b) In the absence of SAM interactions, the anti-terminator structure sequesters aptamer sequence to produce the riboswitch on-state. (c) SHAPE probing was performed on the aptamer domain. Full data sets are available in supplementary data (Supplementary Figures S1–S4). Example capillary electrophoresis traces from primer extension analysis of Mg2+ titrations in the absence (top) and presence (bottom) of 100 µM SAM. The overlaid traces are without Mg2+ (red) and 2 mM Mg2+ (black). Nucleotides from the eight regions analysed here are indicated. The reactivity of nucleotide U84 was invariant and was used to normalize the titration data. (d) SHAPE results for SAM titration without Mg2+ (top) and at 1 mM Mg2+ (bottom). Overlaid traces show no SAM (red), 1 mM SAM (black). Beneath is a primer extension control (green) and the dideoxy sequencing trace (blue) showing adenine positions.
Many riboswitch aptamers have been shown to undergo a large conformational rearrangement on ligand association. Oftentimes, this rearrangement consists of a collapse transition to the bound aptamer conformations (11–19). Hallmark features of this conversion in the SAM-I riboswitch are increased stability of helix joining regions in the core of the aptamer domain, stabilization of tertiary interactions and a decreased radius of gyration (11,15,20). The productive ligand interactions with the pre-collapse, apo-aptamer, result in the selection of the aptamer off-state over the expression platform on-state (Figure 1a). The nature of the apo-aptamer, the mechanism of the collapse transition, and the competition between the expression platform and the aptamer are of great importance in understanding riboswitch regulation (15,20).
In the well-studied group I ribozyme, the formation of tertiary contacts during equilibrium folding requires the presence of counterions (21–23,25,26). We show below that, in the SAM-I riboswitch, collapse also requires the presence of specifically bound divalent counterions. A key question that remains open is how do riboswitch aptamers coordinate the interaction of ligand and ions? Riboswitch operation involves a complex interplay between folding, ligand binding, transcription and Mg2+ interactions. In particular, ligand interactions must occur on a time scale that is comparable with folding and polymerization of the RNA. In addition, the ligand-free system must be sufficiently unstable to prevent Mg2+-induced stabilization of aptamer off-state, thus retaining the capacity to produce the expression platform derived on-state.
To understand how the interplay between Mg2+ and ligand determines the adopted functional state of a riboswitch, we used SHAPE probing to characterize the equilibrium folding properties of the SAM-I riboswitch aptamer domain. This study shows that both Mg2+ ions and SAM are required for collapse. Our results suggest that Mg2+ increases the affinity of the aptamer for SAM. Similarly, the presence of SAM may increase the affinity of Mg2+ for the aptamer. We find that these interactions help maintain switch polarity (i.e. ‘on’ versus ‘off’ state). Specifically, we have developed and used a functional assay that reports on the aptamer’s ability to control the switch state. We find that SAM interactions and Mg2+ binding sites in the aptamer are highly cooperative for maintaining the collapsed aptamer at the expense of the expression platform anti-terminator helix. The functional switching assay was then used as means of selection in a phosphorothioate interference assay. Functional selection identified the location of several important Mg2+ binding sites, and characterizes their coordinated role with SAM in aptamer stabilization.
MATERIALS AND METHODS
RNA preparation
SAM-I aptamer sequence is derived from the Thermoanaerobacter tengcongensis Met F-Met H2 element. The aptamer sequence used includes sequence additions to the 5′ and 3′ (before and after P1 helix) to improve primer extension reads. The aptamer sequence with 5′- and 3′- extensions is: [5′-agc gac ugc acu uug acg cuc gac att a (begin aptamer) cu cuu auc aag aga ggu gga ggg acu ggc ccg aug aaa ccc ggc aac cag ccu uag ggc aug gug cca auu ccu gca gcg guu ucg cug aaa gau gag ag (end aptamer) a uuc uug ugg cau gcu c-3′]. Templates for RNA transcription were prepared as previously described (20). Briefly, PCR was used to prepare transcription templates using Ex-Taq polymerase (TaKaRa) for the amplification of long synthetic templates (ultramers, IDT). Following purification, the transcription templates were transcribed using Ampliscribe high-yield transcriptions kits (Epicentre). The RNA is precipitated by the addition of 1 volume 7 M ammonium acetate and centrifuged. Homogeneity of RNA is checked by PAGE (10% polyacrylamide, 7 M urea, 0.5× TBE). Phosphorothioate incorporated aptamers were prepared in a similar manner using the High yield transcription kit. The standard reaction was supplemented by the addition of α-phosphorothioate-NTP (Glen Research) at a 1:20 ratio to its parent NTP (0.375 mM:7.5 mM). After purification the level of incorporation was verified by 3′-fluorecent labeling, iodine cleavage and capillary electrophoresis (see below for details).
SHAPE probing
The reagent 1-methyl-7-nitroisatoic anhydride (1M7) was synthesized as described in (27). RNA samples were folded by heating to 95°C for 2 min in H2O followed by a 2-min incubation on ice. HMK buffer (50 mM HEPES pH 8.0, 100 mM KCl, with varying concentrations of MgCl2) was then added with the appropriate concentration of SAM (NEB). The RNA (final concentration, 0.5 µM) was equilibrated at 37°C for 10 min and cooled to 25°C. One-tenth volume of 1M7 (60 mM in DMSO) was added to the sample. The reaction was then incubated for 5 min at 25°C. RNA is precipitated by the addition of 3 volumes absolute ethanol, 1/10th volume 3 M sodium acetate and 25 µg glycogen (Ambion) followed by centrifugation. Aptamer RNA is then dissolved in 15 µl (1 µM RNA) primer extension mix containing 250 µM dNTPs, 3 pmoles of 5′-Alexa-488 labeled primer, in the supplied buffer and reverse transcriptase [200 U superscript III MMLV-RT (Invitrogen) or 10 U AMV-RT (Seikagaku)]. These reactions are incubated at 45°C for 1 h. Sequencing reactions are performed on unmodified RNA in the same manner, but the mix is supplemented to 100 µM with one of the four ddNTPs. Primer extension reactions are then loaded onto a P-6 micro-biospin column (Bio-Rad) and centrifuged to remove salt and nucleotides. The samples are then lyophilized and re-suspended in highly deionized (Hi-Di) formamide for analysis. Each sample is diluted 1:20 in Hi-Di formamide and heated to 95°C for 2 min. The samples are electrokinetically injected (30 s at 6 kV) onto an ABI Prism 3100 Avant quad-capillary instrument. A fluorescence electropherogram is then collected (∼60 m at 14 kV). The data are then integrated and aligned using an in-house software for the simultaneous fitting of multiple Gaussian peaks to the traces. Areas are then assigned to nucleotides based on dideoxy-sequencing data and normalized between runs based on the constant reactivity at nucleotide U84 in the loop of P4. Experiments were repeated a minimum of three times for each condition. [Mg2+]1/2 values were determined by fitting the data to a simple two-state binding model:
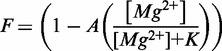 |
(1) |
where F is the fractional reactivity at a given residue as a function of Mg2+ concentration, A is the amplitude and K is the dissociation constant, or [Mg2+]1/2 value for the transition. Data were fit using Gnuplot’s non-linear least squares fitting algorithm. The unweighted non-linear least-squares regression was performed on representative data sets. The asymptotic standard fit errors for that parameter from the simultaneous fitting of amplitude and dissociation constant to the data set are reported.
Expression platform switching assay
The assay is based on our previously reported 2-aminopurine fluorescence-based switching system (20). Here, the folded aptamer domain sequence is challenged with an oligomer based on the anti-terminator sequence found in the expression platform of the T. tengcongensis met F-met H2 riboswitch. The oligomer has the potential to form the anti-terminator helix by base pairing with shared sequence in the aptamer domain. Within the aptamer, the shared sequence extends from the loop of P4 through the 3′ portion of P1 (see Figure 1a). The oligomer is a chimera of DNA and 2′-O-Methyl RNA with the following sequence: [5′-mGmAmA mUmCmU mCdTdC dAdTdC mUmUmU mCmAdG dCdGdA dA-3′]. Formation of the expression platform anti-terminator helix between the oligomer and the aptamer domain creates a hybrid DNA/RNA duplex. The duplex is a substrate for RNase H with the potential for two cleavage sites depending on the length of helix formed. The Mg2+ titration experiments were carried out in 1× HMK pH 7.5 and varying concentrations of MgCl2. As RNase H requires magnesium for activity, concentrations were kept above 50 µM. Aptamer RNA (0.5 µM) was folded as outlined earlier, after which RNase H (Ambion, 0.02 U/µl) was added with the chimera (1 µM). The reaction was allowed to equilibrate for 1 h at 37°C and then stopped by the addition of 3 volumes formamide with 2 mM EDTA pH 8.0 and heated to 90°C for 2 min. Then, 10 µl of the mix was then loaded on a 10% denaturing polyacrylamide gel and electrophoresed at 20 V/cm for 30 min to resolve the products. Gels were stained with ethidium bromide and scanned on a Hitachi FMBio III (532 nm excitation, 605 nm emission). Analysis was performed using Hitachi’s analysis software for lane trace integration. The data were then fit to the standard Hill equation using Gnuplot:
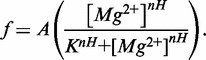 |
(2) |
Here, 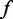 is the ratio of peak areas corresponding to
the fraction of full-length aptamer RNA over all aptamer RNA in that lane,
nH is the hill coefficient,
[Mg2+] is the concentration of magnesium,
A is the reaction amplitude and K the concentration at
which the reaction is at half maximum amplitude. After fitting, the amplitudes were
normalized to 0.5 for the ratio of cleaved to uncleaved (typically the ratio was within
10% of this value). The unweighted non-linear least-squares regression was
performed on representative data sets. Errors are the asymptotic standard fit errors
associated with that parameter during the simultaneous fitting of
nH and K.
is the ratio of peak areas corresponding to
the fraction of full-length aptamer RNA over all aptamer RNA in that lane,
nH is the hill coefficient,
[Mg2+] is the concentration of magnesium,
A is the reaction amplitude and K the concentration at
which the reaction is at half maximum amplitude. After fitting, the amplitudes were
normalized to 0.5 for the ratio of cleaved to uncleaved (typically the ratio was within
10% of this value). The unweighted non-linear least-squares regression was
performed on representative data sets. Errors are the asymptotic standard fit errors
associated with that parameter during the simultaneous fitting of
nH and K.
Phosphorothioate interference selection was achieved using the same RNase H cleavage protocol (above) with the following modification. Phosphorothioate incorporated aptamer RNA (see earlier in the text) was first 3′-end labeled using the Klenow fragment and Allyl-dUTP nucleotides. A DNA oligomer was used as a template to extend the 3′-end of the aptamer. A single adenosine was present in the template for the addition of allyl-dUTP (Ambion). The extension reaction mix included: aptamer RNA (1 µM), allyl-dUTP (50 µM), remaining dNTPs (100 µM) and Klenow fragment of DNA polymerase I from E. coli (NEB, 0.1 U/µl) in the supplied buffer. Reactions were incubated at 37°C for 2 h and then purified by HPLC (Dionex DNApac column), 0–40% buffer B (buffer A, 25 mM Tris–HCl pH 8.0, buffer B, 25 mM Tris–HCl pH 8.0 and 1 M NaClO4) in 45 min. After purification, the RNA was precipitated and labeled using amine reactive Alexa-488 SPD (Molecular Probes, ∼1 mM) in 100 mM NaBO4 pH 8.3. Reactions were incubated at RT for 6 h, precipitated (3 volumes ethanol, 300 mM NaOAc pH 6.5) and purified using the above HPLC gradient to separate labeled from unlabeled. The labeled RNA was then spiked into unlabeled phosporothioate incorporated RNA at a level sufficient for capillary electrophoresis analysis following selection. RNA (0.5 µM final concentration) was folded in HMK buffer (containing either 2 mM MgCl2 or 1 mM MgCl2 with 1 mM MnSO4) supplemented with 10 µM, 30 µM or 100 µM SAM as indicated. Selection was accomplished as described earlier. After equilibration with RNase H and the chimeric oligomer for 1 h at 37°C, the 3′-labeled RNA was desalted (micro-biospin P6 columns), lyophilized and resuspended in Hi-Di formamide. Phosphorothioate containing diester linkages were cleaved by the addition of 1/10th volume of 100 mM iodine in ethanol and heating to 95°C for 2 min. The samples were then analysed via capillary electrophoresis as outlined earlier.
Metal titrations
Titrations of various cation species were performed with a modified version of the expression platform switching assay. The activity of RNase H is dependent on Mg2+ ions. As such, we performed electrophoretic mobility shift assays to quantify the extent of anti-terminator formation. Here, the folded aptamer is challenged with a RNA oligo with the sequence: [5′-rGrArA rUrCrU rCrUrC rArUrC rUrUrU rCrArG rCrGrA rA-3′]. The aptamer is folded in the presence of 80 µM SAM and various concentrations of divalent ions; BaCl2, CaCl2, MgCl2, MnSO4 and SrCl2 (100 µM, 400 µM, 1 mM, 2 mM, 4 mM, 10 mM, 50 mM and 100 mM). A titration of KCl was also performed (200 mM, 400 mM, 800 mM, 1 M, 1.5 M and 2 M). After equilibration for 10 min at 37°C in the appropriate buffer, the aptamer was challenged with the oligomer for 1 h at 37°C (250 nM aptamer, 1 µM oligomer). The reaction was stopped by massive dilution into cold H2O (1:10 000) and analysed by Capillary electrophoresis. We found that dilution effectively stopped association of the RNA oligomer, and the distribution of products did not change when injected up to 24 h post-dilution (data not shown). A non-denaturing media for capillary electrophoresis was made by the polymerization of N,N-dimethylacrylamide (DMA, Sigma-Aldrich). A 5% (v/v) solution (10 ml) of DMA was made in 1× TBE and polymerized by the addition of 10 µl TEMED and 100 µl of ammonium persulfate. The reaction was stirred overnight at room temperature. The polymerized DMA was used on an Applied Biosystems 310 instrument with a 61 cm capillary. Thereafter, 20 µls of the diluted reaction was injected at 6 kV for 30 s and electrophoresed for 30 min at 12 kV. Sybr-Gold (Invitrogen) was added to the poly-dimethylacrylamide solution at a ratio of 1:1 × 106 to visualize the RNA in capillary by laser induced fluorescence. Data were processed as mentioned earlier (see SHAPE probing). After integration, the extent of switching is expressed as the ratio between the peak area of the free aptamer domain over the peak area of the aptamer-expression platform complex. Errors bars are the standard deviations calculated from three separate experiments.
RESULTS
SHAPE probing shows both Mg2+ and SAM are required to produce the collapsed state of the riboswitch aptamer domain
The SAM I aptamer domain undergoes a global structural collapse in response to ligand binding (11,15,16,20,28–31). The collapse involves the stabilization of joining regions between helical elements; J1/2, J3/4 and J4/1 (see Figure 1a), which results in decreased dynamics of P1. We previously demonstrated that the transition coincides with the formation of tertiary contacts in and around a pseudoknot interaction (20). We also found that these interactions were necessary to prevent formation of the anti-terminator on-state. That is, when the aptamer domain binds ligand, it adopts a more compact structure through the stabilization of tertiary interactions within and surrounding the pseudoknot. If this structure lacks one or more of these interactions, ligand affinity is decreased, and the bound form is unable to compete for shared sequence (Figure 1b). Here, we investigate the cumulative role of RNA-Mg2+ and RNA-SAM interactions in aptamer domain collapse.
To elucidate the interplay between ligand and Mg2+ interactions in facilitating structural collapse of the aptamer domain, we used Selective 2′-Hydroxyl Acylation analysed by Primer Extension (SHAPE) probing with 1M7. SHAPE probing reports on backbone mobility at each nucleotide (32), which provides insight into local and global structural dynamics of the aptamer domain. By following collapse as a function of Mg2+ and SAM concentration, we quantitatively describe their contributions to the collapsed aptamer structure. Nucleotide reactivities were quantified by integration of the corresponding capillary electrophoresis peaks and normalized. We outline eight regions in black boxes for easy comparison (Figure 1c and d). The regions and their representative nucleotides are the 3′ and 5′ strands of helix P1 (A94 and U5, respectively), the joining regions J1/2 and J4/1 flanking P1 (A10 and A92, respectively), the base of helix P2 (A14), the kink-turn motif in P2 (A36) and the pseudoknot motif joining the loop of P2 with the joining region J3/4 (A24 and U71, respectively). These nucleotides were selected based on their location in distinct secondary and tertiary structures in the aptamer, as well as their significant reactivity changes during the process of aptamer collapse (≥50%). All of these positions show reactivity changes that are consistent with other nucleotides in the region (see Supplementary Figures S1–S4 for complete data sets). The exception is A14, an isolated nucleotide that is flipped out of the P2 helix in the X-ray structure, which increases in reactivity during collapse.
We have performed four separate sets of titration experiments. Two Mg2+ titrations sets were performed, one with 100 µM SAM present and one without. Also, two SAM titration experiments were performed, one with 1 mM Mg2+ present and one without Mg2+. We first analyse the cooperative effects of SAM and Mg2+ by comparing the SHAPE reactivity profiles between the four titrations. The results are analysed by fitting a two-state binding isotherm for each titration. The analysis was performed using SHAPE reactivity data from a residue whose reactivity is altered significantly in all four titrations, mainly A92 in the J4/1 joining region. The Mg2+ and SAM titration data fit well to a standard two-state binding model yielding [Mg2+]1/2 and SAM KD values for the transitions within J4/1 (Figure 2a-d). The choice of residue does not alter these values. Analysis of reactivity changes at other positions in a given titration yielded similar results indicating the collapse process is concerted across the aptamer domain (data not shown). The comparison of SHAPE probing results for Mg2+ and SAM titrations demonstrates that SAM and Mg2+ interactions act cooperatively to induce collapse (Figure 2a-d). That is, the presence of Mg2+ shifts the SAM-binding isotherm and decreases the KD for SAM. Indeed, the ability of SAM or Mg2+ to alter the local backbone mobility is increased by the presence of the other.
Figure 2.
Plots of reactivity changes for the Mg2+ and SAM titration experiments at A92 with the best fit (red line) to a two-state binding isotherm. The [Mg2+]1/2 and KD values for SAM from the fits are shown. Fits were performed on representative data sets, and errors are the standard fit errors for that data set. (a) Mg2+ titration without SAM. (b) Mg2+ with SAM. (c) SAM titration without Mg2+. (d) SAM titration with 1 mM Mg2+.
For the Mg2+ titrations, the presence of 100 µM SAM decreased the concentration at which SHAPE reactivity is equal to one-half the reaction amplitude [denoted by (Mg2+)1/2] from ∼465 µM to ∼140 µM (Figure 2a and b). The SAM titration revealed a similar trend, albeit to a lesser extent, where the presence of 1 mM Mg2+ decreased the requirement for SAM from ∼28µM to ∼13 µM (Figure 2c and d). For SAM titration experiments in the presence of Mg2+, [Mg2+] = 1 mM was used to insure that the collapse transitions occurred over a similar SAM concentration range as those in the absence of Mg2+. Our earlier work demonstrated that the affinity of this aptamer for SAM was ∼36 nM at 2 mM Mg2+. This was also measured by SHAPE probing of the collapse process (20). This value is similar to the affinity measured for the Bacillus subtilis yit J SAM-I riboswitch at 2 mM Mg2+ (16,30).
SAM and Mg2+ cannot independently drive collapse
We next analyse the global reactivity profiles to delineate the different structural responses of the aptamer domain to SAM and Mg2+, either in isolation or together. Quantified SHAPE reactivity from representative titration data sets at the eight positions are displayed in Figure 3a. The quantified titration data from all eight regions show, as expected, that both SAM and Mg2+ together can induce a concerted structural response across the entire domain leading to collapse (Figure 3a, columns 2 and 4). Here, concerted reactivity changes are seen in all eight regions. On the other hand, titrations of SAM or Mg2+ alone cannot induce the reactivity profile associated with the collapsed aptamer. Comparison of reactivity changes across the domain reveals a subset of regions that become stabilized in isolation (Figure 3a, columns 1 and 3). When SAM was titrated without Mg2+, the titration proceeded to saturation. The resulting dynamical profile shows the important pseudoknot, and its associated tertiary interactions remain highly mobile (Figure 3a, column 3, A24 and U71). Similarly, when Mg2+ was titrated without the presence of SAM, the pseudoknot and related tertiary interactions also remained highly mobile even at 10 mM Mg2+ (Figure 3a, column 1, A24 and U71). These two observations show that (i) the cooperative interaction of SAM and Mg2+ is required to induce collapse; (ii) binding of SAM alone is uncoupled from collapse; and (iii) binding of Mg2+ alone is uncoupled from collapse.
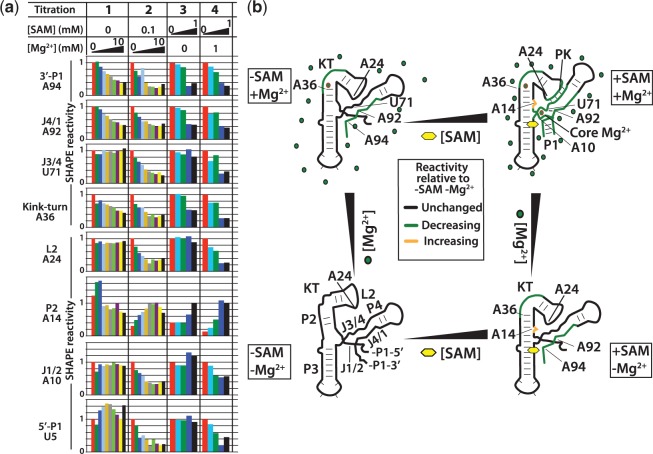
Figure 3.
SAM-Mg2+ landscape for aptamer collapse: quantified SHAPE probing results following the aptamer collapse process for SAM and Mg2+ titrations. (a) Peak integration results for four representative titration experiments at the eight regions analysed. Peaks are integrated and normalized to the reactivity at position U84. The reactivity at the lowest titrated concentration is set to 1. The exception is A14 in helix P2 where the collapsed aptamer conformation produces increased reactivity consistent with the observed conformation in the X-ray structure. Here, the reactivity at 2 mM Mg2+ and 1 mM SAM for the Mg2+ and SAM titrations, respectively, is set to 1. For the Mg2+ titrations, the concentrations are 0, 100 µM, 200 µM, 300 µM, 400 µM, 800 µM, 1 mM, 2 mM, 4 mM and 10 mM. SAM titration concentrations are 0, 1 µM, 10 µM, 100 µM and 1 mM. (b) Schematic representation of the collapse process observed in (a). Green, regions that decrease in mobility with increasing SAM or Mg2+ relative to no SAM and no Mg2+. Black, regions that are unchanged in mobility with increasing SAM or Mg2+ relative to no SAM and no Mg2+. Yellow, regions that increase in mobility with increasing SAM or Mg2+ relative to no SAM and no Mg2+. The final collapsed structure is only observed with both SAM and Mg2+ interactions.
For the Mg2+ titration without SAM, the 3′ strand of P1, the adjacent J4/1 region and the kink-turn show reductions in SHAPE reactivity, indicating stabilization as a function of Mg2+ concentration (Figure 3, column 1).
Similarly, in the SAM titration without Mg2+, SAM altered the reactivity of only a subset of the regions surveyed. The pattern was similar to that found in the Mg2+ titration without SAM. This indicates that pre-organization of the aptamer by Mg2+ populates an apo-aptamer ensemble that is also accessible through SAM interactions alone (Figure 3b). The 3′ strand of P1, J4/1 and the kink-turn had reduced mobility. Neither of the pseudoknot partners, J3/4 and L2, displayed decreased reactivity. Unlike the Mg2+ titration, nucleotide A14 at the base of P2, can be seen to increase in reactivity. In the absence of SAM, this nucleotide has very low reactivity irrespective of Mg2+ concentration. This nucleotide adopts a hyper-reactive conformation during collapse, showing that helix P2 is induced to form a collapsed-like conformation at the base of P2 by SAM interactions alone, without the adoption of a stable pseudoknot interaction from its loop to J3/4. This shows that, although both SAM and Mg2+ can independently induce a similar apo-aptamer conformation or ensemble of conformations, the mechanisms of induction are distinct. Although these aptamer regions respond to SAM alone, others are dependent on the presence of Mg2+ to adopt the ‘collapsed’ reactivity profile. Just as with the Mg2+ titration without SAM, there is a lack of stabilization at the top of P1 near J1/2. Indeed, both the pseudoknot and J1/2 require the presence of SAM and Mg2+ for stabilization.
Expression platform experiments show that SAM and Mg2+ are highly cooperative in maintaining the riboswitch off-state
The aforementioned results show that aptamer collapse is dependent on the presence of both ligand and Mg2+. Here, we use expression platform switching experiments (20) to examine the cooperativity between SAM and Mg2+ in the functional context of changes in secondary structure. We find that SAM effects and Mg2+ effects are highly cooperative in preserving the aptamer off-state, as the aptamer competes with the expression platform for shared sequence.
We use an assay that allows us to monitor the ability of aptamer RNA to compete for shared sequence under a variety of conditions. This assay is analogous to our previous fluorescence-based assay, which used a 2-aminopurine label in the expression platform sequence that base pairs with aptamer domain sequence to monitor this change in secondary structure (20). This assay relies on RNase H digestion of RNA in hybrid DNA/RNA helices to report anti-terminator helix formation (Figure 4a). In this experiment, the oligomer (or anti-terminator strand) is an RNA/DNA chimera of 2′-O-methyl RNA and DNA residues. Formation of the anti-terminator helix creates sites for RNase H cleavage. After folding, the aptamer is challenged with the anti-terminator strand in the presence of RNase H and allowed to equilibrate for 1 h. The degree of cleavage at equilibrium is a direct measure of the thermodynamic stability of the aptamer domain relative to that of the anti-terminator helix.
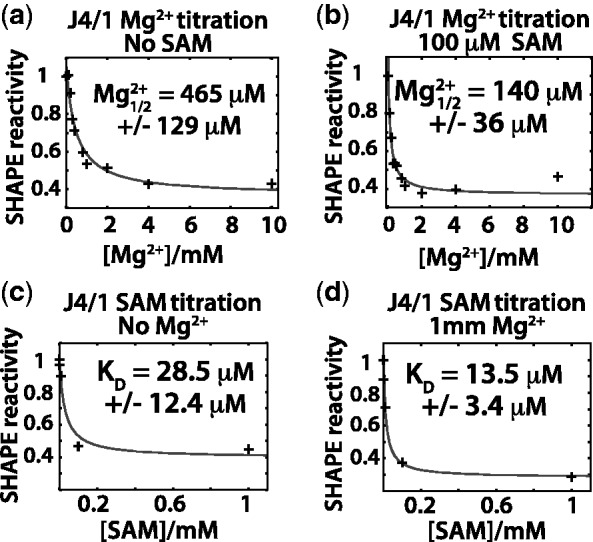
Figure 4.
Switching assay results. (a) Aptamer domain RNA is folded and challenged with a chimeric RNA/DNA oligomer based on the native expression platform sequence. Instability in the aptamer domain allows the expression platform sequence to compete for shared sequence in the aptamer domain. Formation of the anti-terminator helix produces a substrate RNA–DNA duplex for RNase H resulting in cleavage of the aptamer domain (right). (b) Example denaturing PAGE gels used to analyse the Mg2+ titrations at various concentrations of SAM. Mg2+ concentrations were chosen for each SAM concentration to best resolve the transition from destabilized (cleaved) to stable (uncleaved) aptamer. (c) After quantification of the bands representing the cleaved (sum of both cleavage products) and uncleaved fractions, the data [(fluorescence uncleaved aptamer)/(fluorescence cleaved + uncleaved aptamer)] were plotted versus [Mg2+] and fit to the Hill equation (methods, equation 2). Fits yielded the [Mg2+]1/2 (the concentration at which the transition was 50% complete) and Hill coefficients (nH) for the transitions at each concentration of SAM. Fits to the Hill model were performed on representative data sets, and errors represent the standard errors for the fitting of that parameter. Hill coefficients were not determined for the experiments with 200 µM SAM. High fit errors were caused by too few data points representing fully cleaved aptamer at low Mg2+ concentrations (standard errors exceeded 100%).
The interplay between SAM and Mg2+ was analysed by performing Mg2+ titrations at increasing concentrations of SAM. The amount of switching was determined by denaturing polyacrylamide gel electrophoresis (Figure 4b), after which, bands were quantified, and the fraction of uncleaved aptamer domain was plotted versus the concentration of Mg2+ (Figure 4c). The data for each concentration of SAM were then fit to the Hill equation. The fits yielded [Mg2+]1/2 values at each SAM concentration and the Hill coefficient, nH (Equation 2), expressing the degree cooperativity. The Hill coefficient in this context describes the cooperativity of the transition from destabilized, ‘switchable’ aptamer to stable aptamer. As expected from ligand affinity measurements, the amount of Mg2+ required to stabilize the domain was inversely related to the concentration of SAM. The level of cooperativity, as reflected by the Hill coefficient, was high at low concentrations of SAM. There was also an apparent decrease in the Hill coefficient as SAM concentrations increased. This likely reflects the cooperative interaction of one or more Mg2+ and SAM at lower SAM concentrations, which decreases as the SAM concentration becomes saturating even in the absence of Mg2+.
Mg2+ pre-organization of the apo-aptamer enhances anti-terminator formation
In the previous experiment, we used RNase H cleavage to follow the transition in the aptamer, as Mg2+ increases in the presence of SAM. There, the aptamer population shifts from susceptible to anti-terminator formation (cleaved by RNase H) to resistant to anti-terminator formation (uncleaved). The chimeric anti-terminator strand contains two potential RNase H cleavage sites (Figure 5a). This allowed us to also follow the extent of anti-terminator helix formation as a function of Mg2+ in the absence of SAM. The first cleavage site becomes available through partial formation of the anti-terminator by sequestering aptamer sequence found in P1. The second site is only available in the full anti-terminator helix (Figure 5b). Unlike the earlier experiment, here we are investigating the dependence of the extent of anti-terminator formation on Mg2+. The goal is to probe the contribution of aptamer pre-organization in promoting the adoption of anti-terminator on-state. We quantify the RNase H cleavage at the two available sites and follow the transition from aptamer that is only partially accessible to anti-terminator (site 1 cleavage) to aptamer that is fully accessible to anti-terminator (site 2 cleavage). The experiments were performed under equilibrium conditions at increasing [Mg2+] where all of the aptamer domain RNA is cleaved (Figure 5b). The data are expressed as the ratio of (second site cleavage)/(cleavage at both sites), which was then fit to the Hill equation.
Figure 5.
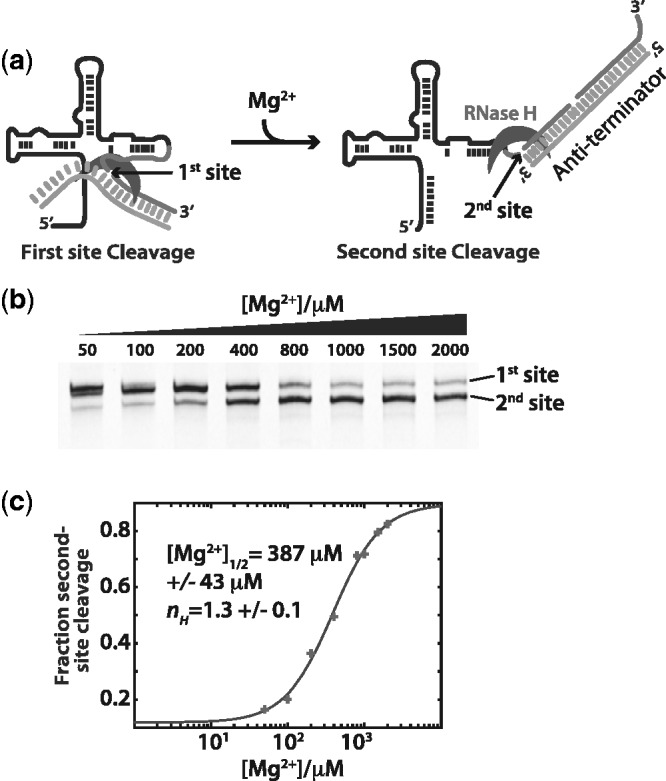
The extent of anti-terminator strand invasion is influenced by Mg2+ concentration. (a) The RNase H cleavage assay (Figure 4) uses a RNA/DNA chimera as an analog of the expression platform sequence. Partial formation of the anti-terminator helix creates a single site for RNase H cleavage. Full association creates a second site. (b) Denaturing poly-acrylamide gels showing the cleavage site selection on the aptamer domain. As Mg2+ concentrations increase, the ability of the expression platform to fully form and become a substrate for RNase H increases. (c) Plot of fractional peak areas for the second site cleavage product [(area second site)/(area both sites)]. The curve represents the best fit of the data to the Hill equation (equation 2, methods). Errors are the standard fit errors for that parameter.
We find that as Mg2+ increases, the second site becomes increasingly available (Figure 5c). Although the first cleavage site is dominant at low [Mg2+], the second site becomes available only at higher [Mg2+]. The transition from site 1 to site 2 occurs over the same Mg2+ range ([Mg2+]1/2 = 387 µM ± 43 µM, nH = 1.3) as the conformational rearrangements found with SHAPE probing in the absence of ligand (Figures 2 and 3). These results suggest that Mg2+ pre-organization of the apo-aptamer structure facilitates both collapse in the aptamer domain (off-state) through the cooperative interaction of Mg2+ and ligand, as well as the formation of the full anti-terminator helix (on-state) in the absence of ligand.
Mg2+ is more active in stabilizing the aptamer than alternative divalent ions
We next assayed the capacity of divalent cations; Mn2+, Sr2+, Ca2+, Ba2+ and monovalent cation K+ to stabilize the aptamer off-state. If the stability of the aptamer is owing to specifically bound Mg2+ ion(s), then there should be a differential in the capacity of ions to occupy the site(s) based on their coordination properties. We expect the best-performing ions to possess similar ionic radii and coordination numbers. We tested the capacity of these cations to replace Mg2+ in stabilizing the SAM-bound off-state using a modified version of the RNase-H assay (above). As Mg2+ is required for RNase H catalytic activity (33), we followed the association of a RNA anti-terminator oligo with the aptamer using an electrophoretic mobility shift assay. Here, the aptamer and anti-terminator oligo are equilibrated for 1 h with increasing concentrations of cations. The reactions were stopped by 10 000-fold dilution into cold H2O. Reactions were then separated by non-denaturing poly-dimethylacrylamide capillary electrophoresis. The anti-terminator associated on-state, and free off-state forms of the aptamer were resolved by the inclusion of the intercalating fluorescent dye, Sybr-gold, in the separation matrix. The peaks for both free and anti-terminator forms were integrated, and the results plotted as the ratio of areas (area free aptamer/area aptamer:anti-terminator complex) (Figure 6a).
Figure 6.
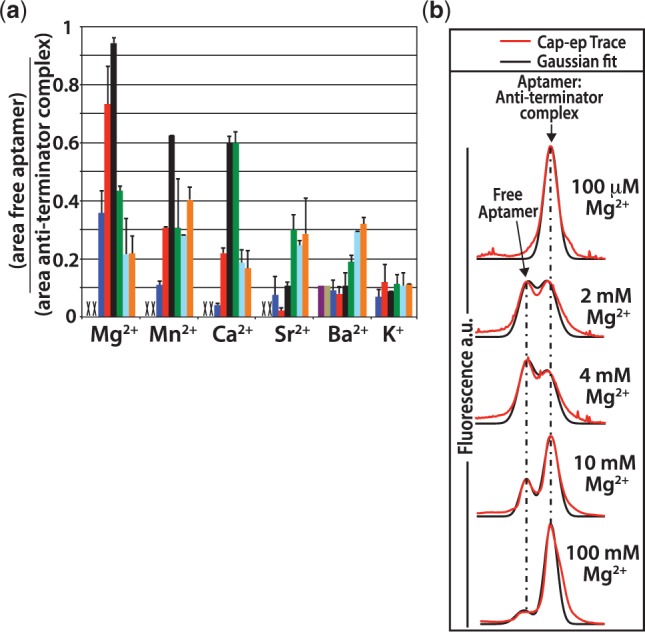
Titration results of various cations. An electrophoretic mobility shift assay was used to follow the extent of association of the aptamer domain with an RNA oligomeric analog of the expression platform anti-terminator sequence. After equilibration with the RNA anti-terminator oligo, the samples were diluted 10 000-fold into cold H2O and analysed by capillary electrophoresis. (a) Titrations were performed for divalent metals Mg2+, Mn2+, Ca2+, Sr2+ and Ba2+, as well as the monovalent ion K+. The ratio of the peak area from the free aptamer over the peak area for the aptamer-anti-terminator complex is plotted versus the ion concentration. The concentrations of divalent ions were as follows: 100 µM (purple), 400 µM (tan), 1 mM (blue), 2 mM (red), 4 mM (black), 10 mM (green), 50 mM (cyan) and 100 mM (orange). The concentrations for K+ were as follows: 200 mM (blue), 400 mM (red), 800 mM (black), 1 M (green), 1.5 M (cyan) and 2 M (orange). Error bars are the standard deviations for three separate experiments. Concentrations where there was no detectable free aptamer remaining are marked with an ‘X’. (b) Example capillary electrophoresis traces from the titration of Mg2+ showing the shift toward stabilized free aptamer as Mg2+ concentrations increase. This is followed by a shift in equilibrium toward the anti-terminator complex at Mg2+ concentrations >4 mM.
The results from this experiment show a clear preference for Mg2+ ions in the ligand-mediated stabilization of the aptamer domain off-state. The other divalent cations show varying capacity to replace Mg2+. The ability of the other cations to replace Mg2+ correlated well with their similarity to Mg2+. For cation interactions with anions of period 2, such as oxygen, the progression of most similar to most dissimilar to Mg2+ from the standpoint of ionic radii and predicted coordination number is as follows: Mn2+ > Ca2+ > Sr2+ > Ba2+ (34). Our results show the same progression in the stabilization of the aptamer off-state at 80 µM SAM. Both Mn2+ and Ca2+ were capable of stabilizing the aptamer domain, whereas Sr2+ and Ba2+ were much less effective. The titration of the monovalent ion, K+, yielded only a small fraction of free aptamer at equilibrium with no indication of increasing stabilization as K+ concentrations increased to 2 M. Taken together, these results indicate that site-specific cation interactions are likely required for the ligand-mediated stabilization of the SAM-I aptamer.
Interestingly, we also observe that as the concentration of Mg2+ ions is increased above 4 mM, the equilibrium was shifted toward the on-state, aptamer:anti-terminator oligo complex (Figure 6b). The same trend was observed for Mn2+ and Ca2+ ions. As such, divalent ions appear to stabilize the off-state in the physiological range and stabilize the on-state at higher concentrations.
Specific Mg2+ chelation sites in key regions of the aptamer domain are required to prevent the transition to the expression platform
We performed phosphorothioate interference mapping experiments to isolate the location of site-specifically bound Mg2+ ions that act cooperatively with SAM. We again use RNase H cleavage of the expression platform helix (see earlier in the text). Here, we use the cleavage to select against specific phosphorothioate substitutions that interfere with the stability of the aptamer off-state. Because phosphorothioates often reduce the affinity of a Mg2+ binding site, our assay can localize Mg2+ ions important for aptamer stability. More specifically, as Mg2+ is a hard Lewis acid, it prefers to be chelated by hard, electronegative Lewis bases. As such, it prefers interactions with oxygen or nitrogen ligands versus the softer sulfur atom found in a phosphorothioate diester linkage (35–37).
In these experiments, the SAM-I aptamer RNAs are incorporated to a level of ∼5% with different α-phosphorothioate-NTPs during transcription (38). The RNA is then fluorescently labeled on the 3′ terminus and purified. After folding in a buffer containing 2 mM Mg2+ and different concentrations of SAM, the RNA is allowed to equilibrate with anti-terminator strand as described previously (Figure 4). RNase H cleavage of aptamers in the anti-terminator helix, effectively removes them from the pool by cleaving the 3′-fluorescent label aptamers RNA in the anti-terminator on-state. A rescue experiment was also performed to verify the effect is owing to loss of a specific Mg2+ interaction. The samples are rescued using a buffer with [Mg2+] = 1 mM, [Mn2+] = 1 mM and the lowest concentration of SAM (10 µM). As manganese is a softer Lewis acid than magnesium, it has the potential to form a more stable inner-shell contact with the phosphorothioate sulfur. Analysis is performed by capillary electrophoresis following cleavage of the phosphorothioate linkages with molecular iodine (Figure 7).
Figure 7.
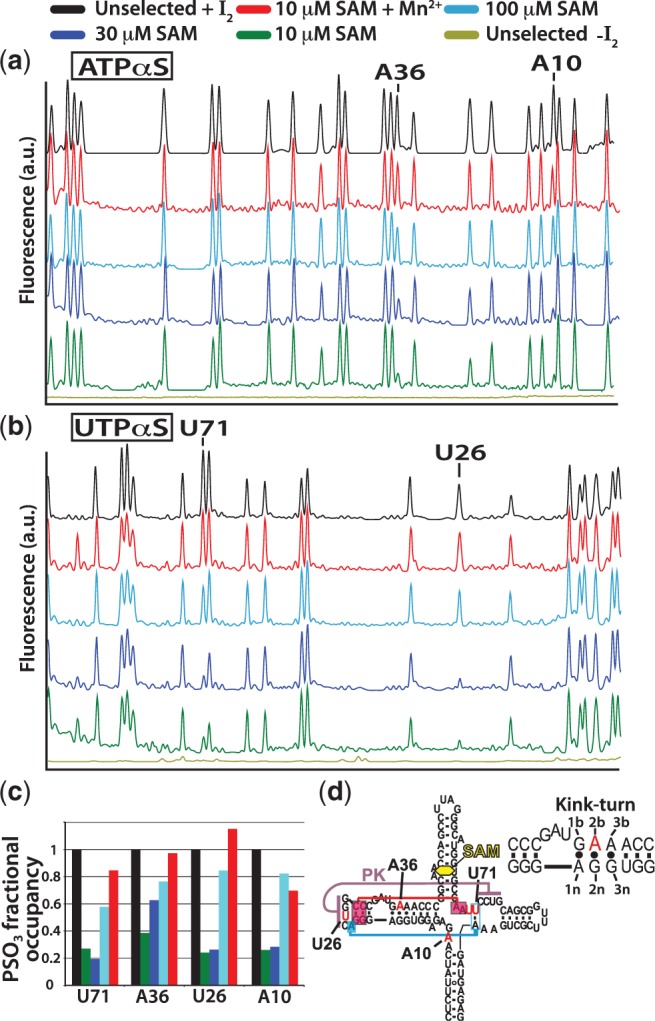
The expression platform switching assay was used as a selection screen in a phosphorothioate interference assay. A schematic detailing the selection methodology is available in supplementary data (Supplementary Figure S5). Selection was performed using RNase H to cleave destabilized aptamers (see Figure 4). Aptamer RNA is randomly incorporated to ∼5% with one of the four α-phosphorothioate-rNTPs. The RNA is 3′-end labeled with the Alexa-488 fluorophore. RNase H cleavage removes the label from aptamers unfit to compete for shared sequence. Populations of each phosphorothiate position are resolved by phosphorothioate cleavage with iodine after selection and before capillary electrophoresis. (a) Capillary electrophoresis traces of selected and unselected RNA incorporated with ATPαS. Experiments were performed at various concentrations of SAM; black (unselected control RNA), green (10 µM SAM), blue (30 µM SAM), cyan (100 µM SAM), red (rescue at 10 µM SAM with 1 mM Mn2+) and brown (unselected control without iodine cleavage). Positions showing phophorothioate interference are indicated. As SAM concentrations increase, the population of phosphorothioate at that position returns to normal. (b) Electropherograms for UTPαS interference assay (colors the same as in a). (c) Traces are integrated and the areas normalized to peaks that display no selection. Bar graph color-code is the same as that for the cap-EP traces above. (d) Secondary structure plot showing the positions of interference with an inset showing the kink-turn element with residue numbering. Red and blue boxed nucleotides show important tertiary interaction (base-triple contacts) proximal to the central Mg2+-binding site formed by A10 and U71.
The results from this assay show 3 Mg2+ binding sites that are cooperative with SAM in preventing association of the anti-terminator strand (Figure 7). The sites are located at A and U residues. There was no interference found from phosphorothioate incorporation at G or C positions (see Supplementary Figure S6). There was strong interference at the A10/U71 Mg2+ binding site (Figure 8). Between 10 µM and 30 µM SAM, phosphorothioates at both A10 and U71 were almost completely depopulated (Figure 7c). The interference was relieved by increasing the concentration of SAM to the concentration found to be saturating in the absence of Mg2+ (i.e., 100 µM) (Figure 3). These sites of interference were also rescued by the addition of manganese containing buffer at the lowest concentration of SAM, 10 µM (Figure 7c). In our previously published Nucleotide Analog Interference Mapping study of collapse, the A10 position was also the only adenosine found to be a site of phosphorothioate interference (20). This demonstrates the importance of this particular Mg2+ interaction for aptamer stability and reinforces the cooperative nature of Mg2+ and SAM interactions in the core. Both A10 and U71 can be seen in the X-ray structure (39) to interact with the magnesium via their pro-Rp non-bridging phosphate oxygens, which are also the phosporothioate diastereomer produced by T7 transcription (40) (Figure 8). A phosphorothioate at U70 also displayed a moderate amount of interference, which may be owing to its proximity to the Mg2+ chelated by U71 and A10. Two more sites of strong interference were also identified that displayed similar response to SAM and rescue by Mn2+. The second site of interference is at U26, which participates in the pseudoknot interaction from the loop of P2. This site is occupied by an iridium complex in the SAM I X-ray structure from the Batey lab (Figure 8). The third site is at A36 in the kink-turn position [‘2b’ in the nomenclature of Lilley et al. (41), Figure 7d]. The location of this Mg2+ binding site can again be seen in the X-ray structure to be occupied by an iridium complex (Figure 8) (39). At A36, both the pro-Rp and pro-Sp oxygens are in a position to interact. The tight architecture of the kink-turn also allows for guanosine base functional groups at the 2 n and/or 3 n positions to chelate a bound metal. Unlike the other sites, interference at A36 was not as severe at 10 µM SAM. There was also significant relief of interference at 30 µM SAM. This suggests two possibilities: (i) Mg2+ binding at this site is not severely perturbed by the inclusion of a pro-Rp phosphorothioate or (ii) disruption by the phosphorothioate is more easily compensated for by other stabilizing interactions. Overall, our results confirm that specific Mg2+ interactions act in a cooperative manner with SAM to maintain the off-state.
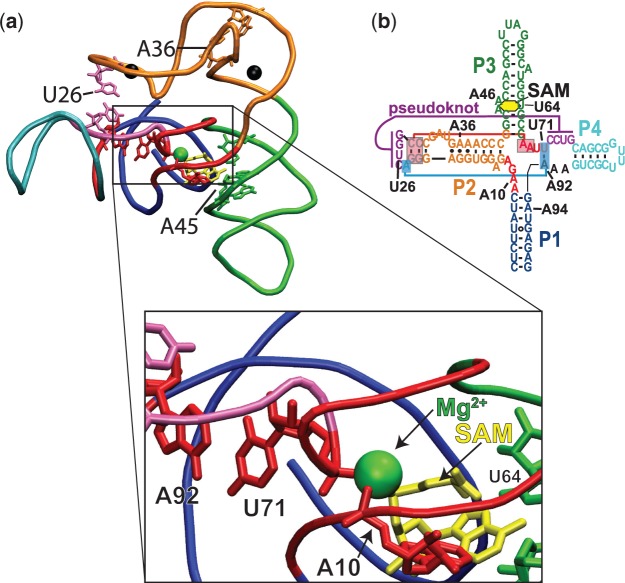
Figure 8.
Structure of the T. tengcongensis aptamer domain. (a) Structural model based on the original T. tengcongensis SAM I aptamer structure of Montange and Batey (39), but using the full native sequence (20). Helical elements in the structure are color coded to the secondary structure (b). Nucleotides of interest are shown in red. The bound SAM molecule (yellow) can be seen in the enlarged view to interact with J1/2 nucleotide A10 that also acts as a ligand coordinating a bound Mg2+ ion (green), bridging J1/2 and J3/4 at U71. Other positions where Mg2+ is found to interact by phosphorothioate interference are A36 and U26. In the X-ray structure, these positions are occupied by iridium complexes, which are indicated by black spheres. Data show these three sites are involved in the chelation of Mg2+ ions. Important secondary and tertiary interactions that are ligand dependent are shown including the U71:A92 base pair that joins J4/1 with J3/4 and the pseudoknot interaction (magenta). (b) Secondary structure of the full T. tengcongensis riboswitch element noting positions of interest including three ligand-dependent base-triple interactions (blue and red boxes).
DISCUSSION
The collapse transition in the aptamer is dependent on the cooperative interaction of SAM, Mg2+ and intramolecular contacts
The results presented here show that the adoption of the collapsed off-state requires the interactions of both SAM and Mg2+. There are many examples of RNAs that adopt their native fold in the presence of high concentrations of monovalent ions alone (23,42,43). There are also well-studied RNA structures that can only adopt their native fold in the presence of divalent cations, some of which, require a site-specifically bound Mg2+ ion (25,44,45).
In the well-studied group I ribozyme, the complexity of the collapse process is directly related to the cooperativity and overall stability of multiple weak tertiary interactions found in the native state (21). As a result, these RNA molecules undergo cooperative collapse transitions (21,23). For the group I ribozyme from Azoarcus, the greater stability of its tertiary interactions allows the formation of near-native conformations in the presence of monovalent ions alone. However, a fully organized catalytic core structure requires the specific interaction of Mg2+ ions (25). Together, these facts result in a Mg2+-induced structural collapse that approaches a cooperative two-state process, but where destabilization of one tertiary interaction decreases the stability of all tertiary interactions and also decreases the cooperativity of the overall transition (46). Similarly, in our previous study of the T. tengcongensis SAM-I aptamer, we demonstrated that the ligand-induced stabilization of key tertiary interactions within the aptamer resulted in the ability of the domain to collapse and compete for shared sequence. Destabilizing any one of these interactions through mutation decreased the affinity of the domain for ligand and allowed the transition to the anti-terminator on-state. Unlike folding transitions in the group I ribozymes in response to Mg2+, the transition of the SAM-I aptamer toward collapse requires both divalent ions and the ligand-dependent organization of core structures.
We have shown here that the resulting structures from SAM titration (no Mg2+) and Mg2+ titration (no SAM) are similar but distinct from the collapsed aptamer (with SAM and Mg2+) (Figure 3). A recent study of the SAM-I aptamer using FRET labels in the loop regions of aptamer helices also found that the Mg2+-induced structure was different than the ligand-bound collapsed form (18). Here, we show using SHAPE analysis of the collapse process, that the collapsed state is the product of the interactions of both SAM and Mg2+. The collapse process appears to follow a simple two-state binding isotherm. Although the affinity of the aptamer for SAM is increased by increasing [Mg2+] and is therefore cooperative with Mg2+ in driving collapse, the collapse process has an apparent Hill coefficient of ∼1. An earlier study by Heppell et al. (16) also examined the collapse process as a function of Mg2+ in the presence of SAM. They used the intrinsic nucleic acid fluorophore, 2-aminopurine at the functionally important A92 residue (A92 makes a ligand-dependent base pair with U71) and found the Hill coefficient for the Mg2+-dependent transition in the presence of SAM was ∼1. A more recent study by Heppell et al. (28), found rearrangements between helices 1 and 3, measured by FRET as a function [Mg2+] were cooperative (nH∼ 4). However, other rearrangements, measured between other helices, were apparently not cooperative. This indicates that global collapse and stabilization in the SAM-I riboswitch aptamer is a complex multi-state process. These results also suggest that some methods used to follow aptamer collapse can be insensitive to the cooperative nature of functionally important local rearrangements. To develop a clear mechanistic model of the process, we must deconvolute the many cumulative interactions that contribute to the functionally relevant states. Our results, assaying the transition of the aptamer from instability to stability in the off-state, identified the cooperative interaction of Mg2+ and ligand. We also show an absolute requirement for ligand in both collapse and function. As a result, it seems that despite the structural hallmarks of collapse that have been observed in earlier studies on the SAM-I aptamer at high Mg2+ concentrations using chemical probing and SAXS analysis without SAM (11,15), the aptamer domain is not competent to resist refolding to the alternative on-state. These earlier studies support a proposed mechanism of conformational selection. Although the results in this study are not inconsistent with conformational selection, they do add constraints to the mechanism. Mainly, although the apo-aptamer is capable of adopting global conformations with similar SAXS profiles to those of the ligand bound state, there are ligand-dependent local rearrangements necessary for functional stability. As such, fluctuations in the apo-aptamer toward the collapsed state can be captured by ligand, but capture may be followed by further important rearrangements that are not accessible in its absence. Further work will be required to correlate the interdependence, cooperativity and nature of specific local interactions to structure and function and derive a clear mechanistic model for ligand-mediated riboswitch regulation. Specifically, a clear understanding of the mechanism of coupling between SAM and Mg2+ interactions in the aptamer will be required.
Studies of the collapse process for the group I ribozymes have shown that increasing charge density of diffuse solvated counterions decreases the concentration requirements for collapse to occur (21). Here, we find that in addition to loosely associated diffuse ions, the SAM-I aptamer requires site-specific metal-RNA interactions to maintain the off-state. This result verifies a prediction made in a recent computational study by our group (47), that a core Mg2+ ion (Figure 8) is chelated by the aptamer RNA at positions A10 and U71. Studies by Lipfert et al. (18) have previously characterized the Mg2+ dependence of collapse in the Vibrio cholerae tandem glycine aptamer. They find that the adoption of the ligand-bound off-state is the result of the cooperative interaction between ligand and Mg2+, requiring the specific binding of Mg2+. They also found that in addition to Mg2+, Ca2+ and Mn2+ were also capable of stimulating glycine binding. We find here that the functional stabilization of the aptamer is also supported by these three ions at concentrations between 1 mM and 10 mM, but not by Sr2+, Ba2+ or even 2M K+. Furthermore, the phosphorothioate interference assay provides evidence for multiple interaction sites that are cooperative with SAM for the stabilization of the aptamer domain. One determination that remains to be made concerns the order of events surrounding the cooperative effects of ligand and Mg2+. The Lafontaine group has shown that Mg2+ pre-organizes the aptamer RNA and is required for aptamer collapse. They have also revealed some of the details of the structural rearrangements using smFRET, bulk FRET and 2-aminopurine fluorescence studies (16,28). In their proposed folding model, the Mg2+ pre-organized apo-aptamer is fully associated with Mg2+. We propose an important addition to their model, where one or more specific Mg2+ interactions are interdependent with ligand interactions (Figure 9). Here, Mg2+-binding events are stabilized by ligand interactions and aid in promoting collapse. During the folding of the nascent polymerizing RNA, an ensemble of folding intermediates (If) are pre-organized by Mg2+ to form the decisive-switching intermediate (Isw). The final transition requires the stabilization of one or more specific Mg2+-binding site(s). The core Mg2+ chelated by nucleotides A10 in J1/2 would be an obvious candidate (Figure 8). The nucleotides in J1/2 make direct contact with both Mg2+ and SAM. They also tie pseudoknot stabilization to ligand interactions through U71 interactions in J3/4 with the same Mg2+. This is supported by SHAPE analysis, where we find a lack of Mg2+ stabilization of J1/2 and J3/4 in the absence of ligand (Figure 3). It is also supported by the cooperative functional transitions observed in our switching assay (Figure 4). We found that Mg2+ pre-organization observed with SHAPE probing occurred with a [Mg2+]1/2 ∼500 µM (Figure 2), which was nearly identical to that observed in the smFRET analysis of Mg2+ pre-organization (between P1 and P3) by Heppell and Lafontaine (28). However, the cooperative transitions to stable aptamer occur from ∼500 µM to ∼2.5 mM, depending on SAM concentration. If Mg2+ pre-organization includes the full complement of associated ions then the cooperative collapse transition should be in a Mg2+ concentration range <500 µM. Instead, there seems to be a separate Mg2+-dependent transition that is cooperative with SAM interactions. This is the transition from Isw to the native-collapsed aptamer (Napt) (Figure 9). This mechanistic proposal was also suggested for the collapse transition in the VCI-II glycine tandem aptamer (18). Further detailed structural analysis will be required to confirm this hypothesis.
Figure 9.
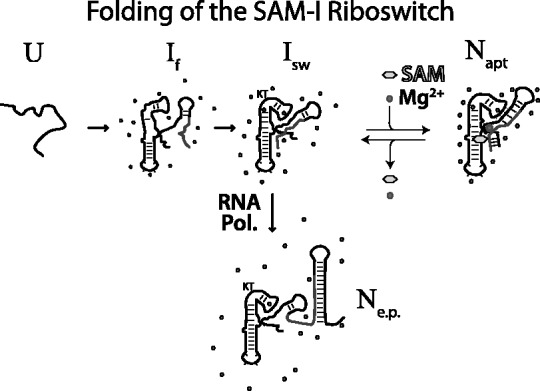
A proposed folding scheme for the SAM-I riboswitch RNA. Here, the nascent polymerizing unfolded RNA (U) rapidly folds to an ensemble of folding intermediates (If). Interaction with Mg2+ ions pre-organizes the folding intermediates to form the ligand-binding competent aptamer (Isw). Binding of ligand allows the RNA to access the native aptamer state (Napt) by stabilizing the site-specific binding of ions. An alternative Native state is the on-state (Ne.p.) that contains many of the pre-organized features found in (Isw) and those formed by the polymerized expression platform sequence.
Functional instability: aptamer pre-organization may function to promote the transition to the ‘ON’ state
Here, we have shown that the apo-aptamer may serve two roles necessary for function: (i) as a pre-organized aptamer competent to bind ligand and undergo structural collapse and (ii) as a structure that is more accessible to anti-terminator formation. The latter is apparent in the analysis of the extent of anti-terminator formation in the expression platform switching study (Figure 5). The formation of the apo-aptamer conformations as Mg2+ concentrations increase (in the absence of SAM, Figure 2) coincides with the increased potential for the adoption of the full anti-terminator helix. The apo-aptamer is inherently unstable, with respect to tertiary interactions. Yet, the magnitude of fluctuations does not increase significantly as a function of Mg2+ concentration. As such, the increased potential for the formation of the anti-terminator may be related to the pre-organization of the apo-aptamer rather than its instability. We also observe a shift in equilibrium toward the anti-terminator helix as divalent ion (Mg2+, Mn2+ and Ca2+) concentrations increase in the presence of ligand (Figure 6). This may be evidence that Mg2+ can prevent productive SAM interactions. Alternatively, this may result from competing Mg2+ interaction sites whose affinities are modulated by ligand interactions. Here, in the absence of ligand, the interaction of a divalent ion shifts the equilibrium toward anti-terminator helix formation. This would impact function by increasing the kinetics of partitioning the nascent riboswitch RNA to the anti-terminator on-state (Ne.p., Figure 9). Transcriptionally regulating riboswitches are subject to control governed by the kinetic parameters of polymerization, folding and ligand binding (48,49). As such, an enhancement of the transition from apo-aptamer to on-state could have large functional relevance, allowing the proper formation of the anti-terminator before the polymerization of the terminator sequence. Overall, our results add more insight into the complex folding landscape of the SAM-I riboswitch including the expression platform.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6.
FUNDING
Funding for open access charge: Los Alamos National Laboratory, Laboratory Directed Research and Development-Exploratory Research [#20090163ER to K.Y.S.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Cliff Unkefer and staff at the National Stable Isotope Resource for lab space. We would also like to thank Dr. Paul Whitford for his assistance with this manuscript.
REFERENCES
- 1.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 2.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc. Natl Acad. Sci. USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 4.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 5.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nahvi A, Barrick JE, Breaker RR. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004;32:143–150. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 9.Gong B, Klein DJ, Ferre-D'Amare AR, Carey PR. The glmS ribozyme tunes the catalytically critical pK(a) of its coenzyme glucosamine-6-phosphate. J. Am. Chem. Soc. 133:14188–14191. doi: 10.1021/ja205185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu VB, Bai Y, Lipfert J, Herschlag D, Doniach S. A repulsive field: advances in the electrostatics of the ion atmosphere. Curr. Opin. Chem. Biol. 2008;12:619–625. doi: 10.1016/j.cbpa.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baird NJ, Ferre-D’Amare AR. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA. 16:598–609. doi: 10.1261/rna.1852310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon M, Strobel SA. Chemical basis of glycine riboswitch cooperativity. RNA. 2008;14:25–34. doi: 10.1261/rna.771608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen B, Zuo X, Wang YX, Dayie TK. Multiple conformations of SAM-II riboswitch detected with SAXS and NMR spectroscopy. Nucleic Acids Res. 2012;40:3117–3130. doi: 10.1093/nar/gkr1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulshina N, Edwards TE, Ferre-D’Amare AR. Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch. RNA. 16:186–196. doi: 10.1261/rna.1847310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heppell B, Lafontaine DA. Folding of the SAM aptamer is determined by the formation of a K-turn-dependent pseudoknot. Biochemistry. 2008;47:1490–1499. doi: 10.1021/bi701164y. [DOI] [PubMed] [Google Scholar]
- 17.Lipfert J, Das R, Chu VB, Kudaravalli M, Boyd N, Herschlag D, Doniach S. Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae. J. Mol. Biol. 2007;365:1393–1406. doi: 10.1016/j.jmb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipfert J, Sim AY, Herschlag D, Doniach S. Dissecting electrostatic screening, specific ion binding, and ligand binding in an energetic model for glycine riboswitch folding. RNA. 16:708–719. doi: 10.1261/rna.1985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ali M, Lipfert J, Seifert S, Herschlag D, Doniach S. The ligand-free state of the TPP riboswitch: a partially folded RNA structure. J. Mol. Biol. 2010;396:153–165. doi: 10.1016/j.jmb.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hennelly SP, Sanbonmatsu KY. Tertiary contacts control switching of the SAM-I riboswitch. Nucleic Acids Res. 39:2416–2431. doi: 10.1093/nar/gkq1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moghaddam S, Caliskan G, Chauhan S, Hyeon C, Briber RM, Thirumalai D, Woodson SA. Metal ion dependence of cooperative collapse transitions in RNA. J. Mol. Biol. 2009;393:753–764. doi: 10.1016/j.jmb.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heilman-Miller SL, Pan J, Thirumalai D, Woodson SA. Role of counterion condensation in folding of the Tetrahymena ribozyme: part II. Counterion-dependence of folding kinetics. J. Mol. Biol. 2001;309:57–68. doi: 10.1006/jmbi.2001.4660. [DOI] [PubMed] [Google Scholar]
- 23.Heilman-Miller SL, Thirumalai D, Woodson SA. Role of counterion condensation in folding of the Tetrahymena ribozyme: part I. Equilibrium stabilization by cations. J. Mol. Biol. 2001;306:1157–1166. doi: 10.1006/jmbi.2001.4437. [DOI] [PubMed] [Google Scholar]
- 24.Lipfert J, Herschlag D, Doniach S. Riboswitch conformations revealed by small-angle X-ray scattering. Methods Mol. Biol. 2009;540:141–159. doi: 10.1007/978-1-59745-558-9_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rangan P, Woodson SA. Structural requirement for Mg2+ binding in the group I intron core. J. Mol. Biol. 2003;329:229–238. doi: 10.1016/s0022-2836(03)00430-3. [DOI] [PubMed] [Google Scholar]
- 26.Woodson SA. Compact intermediates in RNA folding. Annu. Rev. Biophys. 39:61–77. doi: 10.1146/annurev.biophys.093008.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 28.Heppell B, Blouin S, Dussault AM, Mulhbacher J, Ennifar E, Penedo JC, Lafontaine DA. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat. Chem. Biol. 7:384–392. doi: 10.1038/nchembio.563. [DOI] [PubMed] [Google Scholar]
- 29.McDaniel BA, Grundy FJ, Henkin TM. A tertiary structural element in S box leader RNAs is required for S-adenosylmethionine-directed transcription termination. Mol. Microbiol. 2005;57:1008–1021. doi: 10.1111/j.1365-2958.2005.04740.x. [DOI] [PubMed] [Google Scholar]
- 30.Tomsic J, McDaniel BA, Grundy FJ, Henkin TM. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: s-box elements in bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro. J. Bacteriol. 2008;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl Acad. Sci. USA. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J. Am. Chem. Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Black CB, Cowan JA. Magnesium Activation of Ribonuclease H. Evidence for One Catalytic Metal Ion? Inorg. Chem. 1994;33:5805–5808. [Google Scholar]
- 34.Brown ID. What factors determine cation coordination number? Acta Crystallogr. 1988;B44:545–553. [Google Scholar]
- 35.Christian EL, Yarus M. Metal coordination sites that contribute to structure and catalysis in the group I intron from Tetrahymena. Biochemistry. 1993;32:4475–4480. doi: 10.1021/bi00068a001. [DOI] [PubMed] [Google Scholar]
- 36.Christian EL, Yarus M. Analysis of the role of phosphate oxygens in the group I intron from Tetrahymena. J. Mol. Biol. 1992;228:743–758. doi: 10.1016/0022-2836(92)90861-d. [DOI] [PubMed] [Google Scholar]
- 37.Strauss-Soukup JK, Strobel SA. A chemical phylogeny of group I introns based upon interference mapping of a bacterial ribozyme. J. Mol. Biol. 2000;302:339–358. doi: 10.1006/jmbi.2000.4056. [DOI] [PubMed] [Google Scholar]
- 38.Ryder SP, Strobel SA. Nucleotide analog interference mapping. Methods. 1999;18:38–50. doi: 10.1006/meth.1999.0755. [DOI] [PubMed] [Google Scholar]
- 39.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 40.Griffiths AD, Potter BV, Eperon IC. Stereospecificity of nucleases towards phosphorothioate-substituted RNA: stereochemistry of transcription by T7 RNA polymerase. Nucleic Acids Res. 1987;15:4145–4162. doi: 10.1093/nar/15.10.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner B, Lilley DM. The importance of G.A hydrogen bonding in the metal ion- and protein-induced folding of a kink turn RNA. J. Mol. Biol. 2008;381:431–442. doi: 10.1016/j.jmb.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 42.Grilley D, Misra V, Caliskan G, Draper DE. Importance of partially unfolded conformations for Mg(2+)-induced folding of RNA tertiary structure: structural models and free energies of Mg2+ interactions. Biochemistry. 2007;46:10266–10278. doi: 10.1021/bi062284r. [DOI] [PubMed] [Google Scholar]
- 43.Lambert D, Leipply D, Shiman R, Draper DE. The influence of monovalent cation size on the stability of RNA tertiary structures. J. Mol. Biol. 2009;390:791–804. doi: 10.1016/j.jmb.2009.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grilley D, Soto AM, Draper DE. Mg2+-RNA interaction free energies and their relationship to the folding of RNA tertiary structures. Proc. Natl Acad. Sci. USA. 2006;103:14003–14008. doi: 10.1073/pnas.0606409103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leipply D, Draper DE. Evidence for a thermodynamically distinct Mg2+ ion associated with formation of an RNA tertiary structure. J. Am. Chem. Soc. 133:13397–13405. doi: 10.1021/ja2020923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Behrouzi R, Roh JH, Kilburn D, Briber RM, Woodson SA. Cooperative tertiary interaction network guides RNA folding. Cell. 2012;149:348–357. doi: 10.1016/j.cell.2012.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayes RL, Noel JK, Mohanty U, Whitford PC, Hennelly SP, Onuchic JN, Sanbonmatsu KY. Magnesium fluctuations modulate RNA dynamics in the SAM-I riboswitch. J. Am. Chem. Soc. 2012;134:12043–12053. doi: 10.1021/ja301454u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 49.Lemay JF, Desnoyers G, Blouin S, Heppell B, Bastet L, St-Pierre P, Masse E, Lafontaine DA. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 2011;7:e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.