Abstract
Summary: Systems glycobiology studies the interaction of various pathways that regulate glycan biosynthesis and function. Software tools for the construction and analysis of such pathways are not yet available. We present GNAT, a platform-independent, user-extensible MATLAB-based toolbox that provides an integrated computational environment to construct, manipulate and simulate glycans and their networks. It enables integration of XML-based glycan structure data into SBML (Systems Biology Markup Language) files that describe glycosylation reaction networks. Curation and manipulation of networks is facilitated using class definitions and glycomics database query tools. High quality visualization of networks and their steady-state and dynamic simulation are also supported.
Availability: The software package including source code, help documentation and demonstrations are available at http://sourceforge.net/projects/gnatmatlab/files/.
Contact: neel@buffalo.edu or gangliu@buffalo.edu
1 INTRODUCTION
Glycosylation is an important post-translational modification that alters a majority of mammalian-secreted and cell surface proteins. Glycosylated proteins play structural and functional roles in diverse biological processes including inflammation, cancer and development. The recent development of advanced analytical tools has heralded the emergence of Glycomics, a field where entire glycomes of cell systems are characterized. These data are starting to be stored in glycosylation-specific databases like the GlycomeDB (Ranzinger et al., 2011) and the Consortium for Functional Glycomics (CFG) website (Raman et al., 2006). Bioinformatics and statistical approaches developed to query glycomics databases offer an avenue to study glycosylation (Konishi and Aoki-Kinoshita, 2012). Systems-based mathematical modelling represents an alternate methodology, particularly if the emphasis is on the quantitative analyses of biochemical pathways that contribute to a given experimentally measured glycan distribution (Krambeck et al., 2009; Liu et al., 2008). Software applications to construct and analyse glycosylation reaction networks do not exist.
This manuscript describes a new MATLAB-based toolbox called Glycosylation Network Analysis Toolbox (GNAT, pronounced næt). This package provides a streamlined approach for the construction, visualization and simulation of glycosylation reaction networks. It introduces a method to store glycan structure information in SBML format files (Hucka et al., 2003). It provides basic data structure manipulation tools to tailor glycans and associated networks. It enables an environment where data obtained by querying of glycomics databases can be applied to refine reaction network model structure. It provides an interactive GUI (Graphical User Interface) that allows linkage between individual glycans represented in glycosylation networks and corresponding data in glycomics repositories. Finally, the toolbox facilitates steady-state and dynamic simulation of glycosylation reaction networks.
2 FEATURES AND IMPLEMENTATION
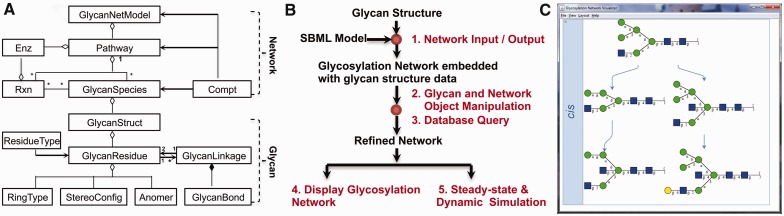
GNAT provides a user-friendly, user-extensible computational environment to handle glycan structures and glycosylation reaction networks. It is written in Java and MATLAB, and it uses previously developed libraries: SBMLToolbox (Keating et al., 2006), GlycanBuilder (Ceroni et al., 2007) and JGraph (http://www.jgraph.com/jgraph.html). GNAT includes new Java classes for network visualization, and MATLAB classes and functions for representation and manipulation of glycans and their networks (Fig. 1A). All functions and classes can be called from the MATLAB command line, and this facilitates user-extensibility and modularity. GNAT functions and classes enable five types of operations described below (numbered 1 to 5, Fig. 1B):
Fig. 1.
(A) UML class diagram (www.uml.org) defines the core classes of GNAT. Enz, Enzyme; Rxn, Reaction; Compt, Compartment. (B) Five operations, numbered 1 to 5, are enabled in the toolbox. (C) Reaction network model displayed in the Glycosylation Network Visualizer GUI. Glycans are represented in CFG nomenclature (http://www.functionalglycomics.org/static/consortium/Nomenclature.shtml)
2.1 Network input/output
GNAT provides a simple strategy to incorporate glycan structure information into SBML files. To this end, XML-format glycan structures are embedded in the annotation field of the SBML species element. SBML Toolbox and GlycanBuilder functions are used to import/export features related to the SBML files and Glycan structures, respectively. These operations primarily handle two of the classes shown in Figure 1B, GlycanStruct and GlycanNetModel. Here, GlycanStruct class uses the tree structure to store individual glycan information including residue and linkage data. GlycanNetModel class is constructed using graph structure. It describes the pairwise relationships between glycans. APIs from GlycanBuilder handle conversion between various glycan formats including GlycoCT, GlycoCT-condensed, GLYDE and LINUCS (Neelamegham and Liu, 2011).
2.2 Glycan and network object manipulation
Construction and manipulation of glycan and network objects is enabled by defining various classes. These can be categorized into two groups depending on whether they handle ‘Glycans’ (bottom half, Fig. 1A) or ‘Networks’ (top half, Fig. 1A). Whereas the former group deals with the properties of individual glycans that are described by GlycanLinkage, GlycanResidue, etc., the latter includes classes that describe network components like GlycanSpecies and Rxn (Reaction). Manipulation of individual class properties using methods allows refinement of individual glycans and glycosylation network models. The GettingStarted.pdf document (provided with the GNAT package at the SourceForge website) and the MATLAB ‘help classname’ command exhaustively describe all properties and methods related to the individual classes.
2.3 Database query
Refinement of reaction network models is possible using knowledge stored in glycomics databases. GNAT enables this by providing facilities for querying databases. Such queries to the GlycomeDB or CFG databases return information regarding glycan family, species where specific glycans have been detected, etc. Most of the functions query databases using their respective glycan ID, and this is enabled after basic GNAT installation. Advanced database commands that enable XML-structure-based querying of GlycomeDB requires a PostgreSQL server.
2.4 Display glycosylation network
A user-friendly GUI called Glycosylation Network Visualizer (GNV, Fig. 1C) within GNAT provides high-quality visualization of glycosylation pathways. This environment enables visualization of multi-compartment glycosylation networks. Editing of network layout (Circle, Hierarchical, Compact Tree), scaling of graphics, mouse wheel-based zoom-in/out functions and exporting of graphics in PNG or GIF formats are also possible. Display options can be specified by command line operations. Linkage between individual glycans in the GNV window and the GlycomeDB database is enabled using the database connectivity functions. This module uses APIs from GlycanBuilder and JGraph.
2.5 Steady-state and dynamic simulation
Computation of network dynamic states is implemented using MATLAB’s built-in Ordinary Differential Equation (ODE) solvers. Steady-state solutions are calculated using the MATLAB Optimization toolbox function ‘fsolve’. Additional analysis of results is feasible using a range of functions available in MATLAB.
3 INSTALLATION AND AVAILABILITY
GNAT is a platform-independent MATLAB toolbox that has been tested in Windows, Linux and Mac OS. It uses MATLAB Release 14.05 (R2011b) or later versions. No compilation is required. Basic toolbox installation, which enables a majority of GNAT functions, consists of two steps: (i) installing compressed files, and (ii) editing java library path. Advanced database functionality described in Section 2.3 requires the MATLAB Database Toolbox and the GlycomeDB PostgreSQL server.
The package includes instructions for installation, usage and examples. Two detailed case studies that focus on database connectivity and simulation of glycosylation pathways are provided in the GettingStarted.pdf document. The latter describes the construction, visualization and simulation of the N-linked glycosylation reaction network described by (Umana and Bailey, 1997).
4 SUMMARY
GNAT is the first toolbox for glycosylation reaction network analysis. It can facilitate research in the emerging field of Systems Glycobiology (Neelamegham and Liu, 2011).
Funding: Supported by the National Institutes of Health (HL103411 and Program of Excellence in Glycosciences HL107146).
Conflict of Interest: none declared.
REFERENCES
- Ceroni A, et al. The GlycanBuilder: a fast, intuitive and flexible software tool for building and displaying glycan structures. Source Code Biol. Med. 2007;2:3. doi: 10.1186/1751-0473-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucka M, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Keating SM, et al. SBMLToolbox: an SBML toolbox for MATLAB users. Bioinformatics. 2006;22:1275–1277. doi: 10.1093/bioinformatics/btl111. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Aoki-Kinoshita KF. The GlycomeAtlas tool for visualizing and querying glycome data. Bioinformatics. 2012;28:2849–2850. doi: 10.1093/bioinformatics/bts516. [DOI] [PubMed] [Google Scholar]
- Krambeck FJ, et al. A mathematical model to derive N-glycan structures and cellular enzyme activities from mass spectrometric data. Glycobiology. 2009;19:1163–1175. doi: 10.1093/glycob/cwp081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, et al. Systems-level modeling of cellular glycosylation reaction networks: O-linked glycan formation on natural selectin ligands. Bioinformatics. 2008;24:2740–2747. doi: 10.1093/bioinformatics/btn515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelamegham S, Liu G. Systems glycobiology: biochemical reaction networks regulating glycan structure and function. Glycobiology. 2011;21:1541–1553. doi: 10.1093/glycob/cwr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman R, et al. Advancing glycomics: implementation strategies at the consortium for functional glycomics. Glycobiology. 2006;16:82R–90R. doi: 10.1093/glycob/cwj080. [DOI] [PubMed] [Google Scholar]
- Ranzinger R, et al. GlycomeDB—a unified database for carbohydrate structures. Nucleic Acids Res. 2011;39:D373–D376. doi: 10.1093/nar/gkq1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umana P, Bailey JE. A mathematical model of N-linked glycoform biosynthesis. Biotechnol. Bioeng. 1997;55:890–908. doi: 10.1002/(SICI)1097-0290(19970920)55:6<890::AID-BIT7>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]