Abstract
Originally identified as a response to starvation in yeast, autophagy is now understood to fulfill a variety of roles in higher eukaryotes, from the maintenance of cellular homeostasis to the cellular response to stress, starvation, and infection. Although genetics and biochemical studies in yeast have identified many components involved in autophagy, the findings that some of the essential components of the yeast pathway are missing in higher organisms underscore the need to study autophagy in more complex systems. This review focuses on the use of the fruitfly, Drosophila melanogaster as a model system for analysis of autophagy. Drosophila is an organism well-suited for genetic analysis and represents an intermediate between yeast and mammals with respect to conservation of the autophagy machinery. Furthermore, the complex biology and physiology of Drosophila presents an opportunity to model human diseases in a tissue specific and analogous context.
Keywords: Drosophila, Autophagy, Atg, Model system
The process of autophagy
There are three morphologically distinct forms of autophagy in mammalian cells: macroautophagy, microautophagy, and chaperone-mediated autophagy [13]. This review focuses on macroautophagy, an evolutionarily conserved mechanism for bulk degradation of organelles and long-lived proteins [61, 76]. Upon autophagy induction, a flat membrane cistern, called the isolation membrane or phagophore, envelops a portion of cytoplasm, eventually forming a closed double-membrane bound vesicle containing cytoplasm and organ-elles. The completed vesicle, called the autophagosome, fuses with the lysosome, where its inner membrane and contents are degraded by hydrolases. The resulting degradation products are transported back to cytoplasm where they can be reused for protein synthesis and adenosine triphosphate (ATP) production.
Studies in yeast have identified 33 ATG (autophagy-related) genes, involved in autophagy, many of which are conserved in higher organisms [55, 115]. However, several large gaps in our understanding of the process remain. In particular, it is not clear how the upstream signaling events known to trigger autophagy connect to the molecular machinery of autophagosome formation. Furthermore, although the core autophagy machinery appears to be conserved [110], there are significant differences between the genetics of autophagy in yeast and higher organisms. For example, the Atg1 kinase complex regulates autophagy in yeast, but several components of the complex are not conserved outside of fungi [79]. This suggests that there are a set of genes that have not yet been identified that are critical to the regulation of autophagy in mammals and other higher eukaryotes. These observations underscore the need to study autophagy in more complex genetically tractable systems.
Conservation of the core autophagy machinery in Drosophila
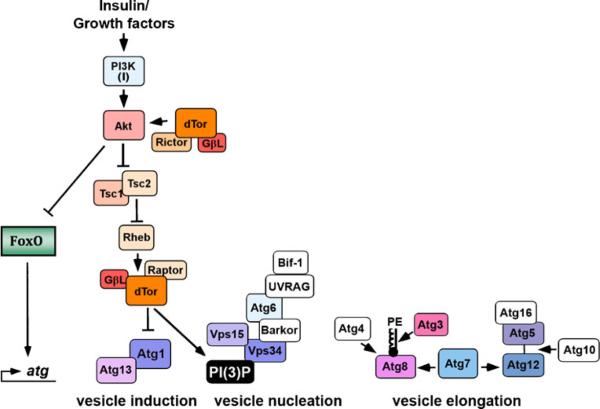
The molecular mechanism of autophagy can be divided into three major steps: (1) induction; (2) autophagosome nucleation; and (3) membrane expansion and completion (reviewed in Xie and Klionsky [110]). Figure 1 outlines the Drosophila proteins involved in these processes and the link between the Insulin/Tor pathway and the autophagy machinery. In all eukaryotes, autophagy is induced via the autophagy-related gene 1 (Atg1) complex. Autophagosomal membrane nucleation involves a complex containing Vps34 (the class III PI3K). Expansion of the autophagosome membrane requires two distinct sets of ubiquitin-like protein conjugation systems, Atg8 and Atg5-Atg12. Following autophagosome completion, the vesicles fuse with lysosomes, forming autolysosomes. Table 1 lists the core autophagy genes from Saccharomyces cerevisiae, humans, and their Drosophila orthologs.
Fig. 1.
Connecting the Insulin pathway to the Drosophila autophagy machinery. Both branches of the insulin pathway downstream of Akt regulate autophagy in Drosophila. Insulin signaling inhibits Tor, allowing for autophagic vesicle induction by a complex containing Atg1 and Atg13. The mechanism of autophagy induction by FOXO remains unknown, although it likely involves transcriptional regulation of autophagy-related (Atg) genes. Vesicle nucleation by the Class III PI3K/Vps34 complex is regulated, in part, by Tor/Atg1 activity. Elongation and completion of the autophagosome require the Atg8-PE and Atg12 ubiquitin-like conjugation systems. Many of the components of these complexes have been demonstrated to be active in Drosophila autophagy (colored boxes). However, the function of several conserved autophagy proteins (white boxes) in Drosophila remains undetermined
Table 1.
Conservation of genes involved in autophagosome induction, nucleation, and expansion
| S. cerevisiae gene | D. melanogaster gene | Homo sapiens gene | References | |
|---|---|---|---|---|
| Induction | TOR | dTOR | mTOR | [23, 30, 41, 83, 94] |
| ATG1 | Atg1 | ULK1, ULK2 | [69, 93, 94, 111, 112] | |
| ATG13 | Atg13 | HARBI1 | [6, 7, 22] | |
| ATG17 | – | – | [42, 45] | |
| ATG29 | – | – | [50] | |
| ATG31 | – | – | [43] | |
| – | CG1347 | FIP200, RB1CC1 | [27] | |
| – | CG7053 | ATG101 | [31, 71] | |
| Nucleation | ATG6 | Atg6 | BECN1 | [39, 48, 51, 87, 94, 118] |
| VPS34 | Pi3K59F | PIK3C3 | [39, 51, 86] | |
| VPS15 | ird1 | PIK3R4 | [51, 65, 109] | |
| ATG14 | CG11877 | ATG14 (barkor) | [34, 48, 51, 101] | |
| – | CG6116 | UVRAG | [34, 63] | |
| – | endoB | SH3GLB1 | [102] | |
| – | buffy | BCL2 | [32, 85, 92] | |
| – | – | AMBRA1 | [20] | |
| Expansion | ATG3 | Atg3 | ATG3 | [33, 37, 106] |
| ATG4 | Atg4 | ATG4A,B,C,D | [29, 54, 95] | |
| ATG5 | Atg5 | ATG5 | [47, 75, 77, 91, 94] | |
| ATG7 | Atg7 | ATG7 | [38, 54, 75, 77, 104] | |
| ATG8 | Atg8a, Atg8b | LC3, GABARAP, GABARAPL2 | [3, 29, 39, 44, 91, 94, 105, 106] | |
| ATG10 | CG12821 | ATG10 | [75, 77] | |
| ATG12 | Atg12 | ATG12 | [75, 77, 94] | |
| ATG16 | CG31033 | ATG16L1, ATG16L2 | [59, 74] |
Atg1, the only serine/threonine kinase among the identified Atg proteins, is conserved in higher organisms and required for autophagy in Drosophila [70, 94] and mammals [69, 93, 94, 111, 112]. However, the Atg1 complex, comprised of several other Atg proteins [45], differs significantly both in its composition, and in its function, between yeast, fruitflies, and humans. In yeast, TOR regulates formation of an Atg1-Atg13-Atg17 complex [45] via phosphorylation of Atg13. Reduced TOR activity during starvation conditions causes Atg13 dephosphorylation, which increases its affinity for Atg1-Atg17, leading to autophagy induction [45]. Neither Drosophila nor humans possess clear orthologs for Atg17 or the Atg17-interacting proteins Atg29 and Atg31 [49, 50]. Rather, in Drosophila, Atg1 and Atg13 form a stable complex regardless of Tor activity. Likewise, the mammalian Atg1 ortholog, Unc-51-like kinase (Ulk1) interacts with Atg13 independent of nutrient conditions [30, 71]. In addition to Atg13, the mammalian Ulk1 complex also contains Atg101 and FIP200 [27, 31, 71], both of which are required for autophagosome formation. Drosophila orthologs of both Atg101 and FIP200 have been identified, but not yet tested. Importantly, although they contain many of the same core components, the Drosophila Atg1 and mammalian Ulk1 complexes do not function identically. For example, over-expression of Drosophila Atg1 induces autophagy, while overexpression of mammalian Ulk1 inhibits autophagy [6, 93]. It is not clear how this difference arises; speculation has focused on the influence of additional regulatory proteins, Atg13-Atg1 stoichiometry, and feedback from Atg1 to Tor. See Mizushima [8] and Chang and Neufeld [73] for more in-depth examinations of Atg1 regulation.
In yeast, Drosophila, and mammalian cells, following autophagy induction by the Atg1 complex, a phosphatidylinositol-3-phosphate (PI3P)-enriched structure appears at the site of autophagosome formation. PI3P is produced by phosphatidylinositol 3-kinase (PI3K), but its function at the nascent autophagosome is not yet clear. Possibly, PIP3 recruits additional factors, consistent with the observation that several yeast Atg proteins bind PIP3 and localize to the autophagosome in a PI3K-dependent manner [100]. However, of these proteins, only Atg18 has a Drosophila and mammalian ortholog, with only the former having been confirmed to be required for autophagy [3, 94].
The single yeast PI3K, Vps34, is required for a variety of membrane trafficking events, including autophagy [28]. Vps34, and the other components of the yeast PI3K complex, Vps15, Atg6, and Atg14, are conserved in both Drosophila and mammals, where they have an essential role in autophagosome formation [99]. It is important to note that unlike yeast, Drosophila and mammals possess three types of PI3K. The more familiar class I PI3K functions downstream of insulin signaling, and inhibits autophagy through activation of Tor [35]. As noted above, Vps34, the class III PI3K, activates autophagy through production of PIP3. Mammals have a further layer of complexity in that Vps34-Vps15-Atg6 is found associated with Atg14, Ambra1, UVRAG, or Rubicon, depending on the context [99]. Drosophila has orthologs of UVRAG, Rubicon and Atg14, however it is not yet clear how they function in the Vps34 complex.
Vesicle expansion is mediated by two ubiquitin-like groups, Atg5–Atg12–Atg16, and Atg8, both of which are highly conserved from yeast to mammals [84]. The Atg16 complex localizes to the autophagosome, and is required for membrane biogenesis [21, 59]. Atg12 is covalently attached to Atg5 in a ubiquitin conjugation-like process, involving Atg10 and Atg7, the latter homologous to the E1 ubiquitin-activating enzyme [75]. Atg10 functions like an E2 ubiquitin conjugating enzyme, although it is not homologous to those in the ubiquitin system [96]. Atg5-Atg12 is then noncovalently linked to Atg16, forming the completed complex [59]. Drosophila orthologs exist for each of these proteins, although neither Atg10 (CG12821) nor Atg16 (CG31033) have been shown to function in the autophagy pathway.
The second conjugation system involves the ubiquitin-like protein, Atg8, which is linked via an amide bond to the lipid phosphatidylethanolamine (PE) [33]. The conjugation process begins when the cysteine protease Atg4 cleaves Atg8, which is then bound by Atg7 [54]. Atg8 is then transferred to Atg3, an E2-like enzyme, which catalyzes the conjugation to PE. Humans possess four orthologs of yeast ATG8, MAP1LC3, GATE16, GABARAP, and ATG8L, each of which is conjugated to PE as occurs in yeast [29, 44, 105, 107]. Drosophila possesses two Atg8 genes (Atg8a and Atg8b), both of which localize to autophagosomes [39, 91, 94]. These proteins likely have some redundancy, as Atg8a loss of function alleles give a milder phenotype than would be expected for such an essential component [3, 93]. Orthologs of Atg3, Atg4, and Atg7 also exist in Drosophila, and have been shown to function in the autophagy pathway [38, 93, 94, 108].
Autophagy as an adaptive response to nutrient deprivation and cell stress
Like the ubiquitin-proteosomal degradation of short-lived proteins, autophagy is deployed at basal levels to recycle large protein complexes and damaged organelles. This nutrient recycling function is particularly important during periods of starvation. In nutrient rich conditions, yeast autophagy is almost undetectable. Within 30 min of nitrogen starvation, however, autophagosome formation is dramatically up-regulated [103]. Similarly, within one hour of starvation, autophagosomes appear in the Drosophila fat body, a nutrient storage organ analogous to the liver [94]. Likewise, in mice, autophagy increases in most organs under starvation conditions, with muscles showing a particularly clear response [78]. Interestingly, following birth, autophagy is up-regulated in various tissues in mouse neonates, apparently as an adaptive response to the severe starvation resulting from abrupt separation from the placenta [57]. Thus, the protective response to nutrient deprivation is a fundamental and well-conserved function of autophagy in eukaryotes.
In response to starvation, yeast cells regulate autophagy through the nutrient-sensing Tor pathway [35]. Higher eukaryotes also regulate autophagy through the insulin/class I phosphoinositide 3-kinase (PI3K) pathway upstream of Tor [12]. Recent studies have confirmed that both the Tor and Insulin pathways control autophagy in the Drosophila larval fat body (Fig. 1). Starvation, rapamycin treatment, or genetic inactivation of the Tor pathway induces a rapid autophagic response in fat body cells. [91, 93, 94]. Similarly, loss of PI3K or Insulin receptor function strongly induces autophagy in this tissue [91, 93, 94]. Thus, the signaling pathways controlling the autophagic response to starvation are conserved in Drosophila. Importantly, orthologs of several yeast ATG genes (Atg1, Atg5, Atg7, and Atg12) are required for the formation of autophagosomes in the fat body model, demonstrating that Drosophila autophagy utilizes conserved components of the ATG machinery.
Two recent studies demonstrate the conservation of Tor mediated autophagy regulation in Drosophila and mammalian cells, as well as the value of a complementary approach using both organisms. Kim et al. identified Rag GTPases as Tor activators in response to amino acid signals [52]. Using both Drosophila and mammalian cell culture they observed that reduced Rag gene expression suppressed the stimulatory effect of amino acids on Tor. In vivo analysis in the Drosophila fat body was then used to demonstrate a functional role for Rag in TOR-mediated regulation of autophagy and cell size regulation. The same group followed up these results with a similar analysis of Rab and Arf family GTPases, demonstrating that these also regulate Tor activity and autophagy, although unlike Rag this does not involve direct interation with Tor [62].
Modeling autophagy and disease in drosophila
In addition to autophagic recycling of essential nutrients and energy during periods of starvation, autophagy also protects the cell from injuries caused by oxidative stress, pathogenic infection, misfolded proteins, growth factor deprivation, and hypoxia. For example, mammalian cardiac muscles subjected to oxygen deprivation accumulate large numbers of autophagosomes, which appear to facilitate their survival [113]. This type of protective role has significant consequences for tumor cells, where autophagy may both promote survival [15] and inhibit growth [68]. Neurodegenerative diseases, such as Parkinson's disease, Alzheimer's disease, and Huntington's disease are accompanied by the accumulation of large mutant protein aggregates. Of all of the autophagy associated diseases in humans, those dealing with abnormal protein accumulations have the most highly developed model systems in Drosophila. The increased autophagosome formation observed in these diseases may play a protective role by degrading misfolded proteins [117]. Consistent with this notion, decreased neurodegeneration and huntingtin aggregate accumulation were reported in Drosophila following treatment with a rapamycin analog that induces autophagy [90]. Several recent reports have used Drosophila neurodegenerative disease models to explore the role of autophagy in Amyloid beta 42, polyQ repeat, and TDP-43 toxicity [26, 66, 88, 89]. Genetic analysis of the Drosophila ortholog of Alfy (autophagy-linked FYVE protein) using an eye-degeneration model of Huntington's disease has been essential in confirming the in vivo function of Alfy in clearance of ubiquitin-positive inclusions and suppression of neuronal degeneration [19, 97, 98].
Innate immunity and autophagy
The usefulness of Drosophila as a model system for immune system diseases is limited by the fact that insects lack the adaptive immune response typical of vertebrates. Thus, Drosophila studies cannot provide direct insight into the role of autophagy in T cell survival or the delivery of antigens to MHC class II compartments. However, in an evolutionarily conserved response to infection, autophagosomes can also directly envelop bacterial and viral pathogens. Several microbes, including group A streptococci and Mycobacterium tuberculosis are destroyed via the autophagy-lysosome system [25, 80]. Induction of autophagy has been shown to protect against sindbis virus in a mouse model of encephalitis [64]. Some pathogens, such as poliovirus and rhinoviruses, are able to avoid autophagic degradation, instead using the autophagic machinery to replicate [36].
The case of Listeria infection illuminates both the advantages and disadvantages of modeling bacterial autophagy in Drosophila. Listeria monocytogenes induces anti-bacterial autophagy in Drosophila cells in a manner dependant upon the peptidoglycan recognition protein LE [114]. In contrast, Listeria appears able to evade autophagy in mammalian macrophages, both in the cytoplasm via the actin-based motility protein ActA, and by Listeriolysin O, a pore-forming toxin that promotes the formation of non-degradative spacious Listeria-containing phagosomes [4, 72, 116]. These Listeria-containing phagosomes restrict bacterial replication and appear to form from abortive attempts at autophagosome/lysosome fusion. Genetic analysis of Listeria infection in Drosophila is therefore limited by the different response of autophagy machinery to the pathogen. Nonetheless, studying Listeria and autophagy in Drosophila cells is likely to reveal significant information related to the ability of Listeria to evade the full effects of autophagy.
Recent work by Cherry and colleagues [9, 95] has demonstrated the usefulness of Drosophila as a model for studying viral autophagy. This group showed that vesicular stomatitis virus (VSV) infects Drosophila cells both in culture, and in vivo, causing autophagy induction. Furthermore, RNAi-mediated knockdown of core autophagy genes greatly increased the virulence of VSV infection. Experimental manipulation of the insulin signaling pathway also impacted viral replication, consistent with the known influence of the PI3K/Tor pathway on autophagy. To date, this is the only work yet published that directly addresses viral autophagy in Drosophila. This model should be an attractive one for further research given the rapidity of Drosophila genetics and the fact that the evolutionary distance of host and virus allows for relatively straightforward identification of pathogen-autophagy interactions [16].
Tools and techniques
Autophagy detection
As is the case in human studies, autophagy in Drosophila is most commonly visualized by detection of the conserved lipid-conjugated ubiquitin-like protein Atg8, which localizes to autophagosomes in yeast, flies and mammals [75]. However, Drosophila Atg8 antibodies that are effective in immunocytochemistry are not readily available. Thus, several groups have constructed GFP-tagged versions of Atg8 that can be expressed using the tissue specific Gal4/UAS system, or under control of a heat shock promoter [39, 91, 94]. These transgenes have proved reliable indicators of autophagosome formation in a number of studies, but it is important to note that concerns have been raised about this reagent [58]. In brief, Kuma et al. found that in mammalian studies Atg8 tends to be incorporated into intracellular protein aggregates, independent of autophagy. Importantly, this association with aggregates includes endogenous Atg8 as well as ectopically expressed Atg8-GFP fusion protein. Thus, an Atg8 or Atg8-GFP-positive punctate dot can represent either an aggregate or a bone fide autophagosome. Given this issue, many Drosophila researchers supplement Atg8-based autophagosome detection with assays based on lysosomal markers Lyso Tracker Red and LAMP1 (lysosome associated membrane protein 1). Similarly, transcriptional upregulation of Atg genes has been used as a correlative measurement of autophagy activity [24, 40, 60]. Although it is non-quantitative, electron microscopy is the most conclusive demonstration of the presence of autophagosomes, and has been used in autophagy studies on both larval and adult Drosophila tissues.
Autophagosome detection approaches provide a snapshot of the autophagy activity of a cell but can sometimes be difficult to interpret. For example, an increase in autophagosome number could indicate either an increase in autophagy, or a functional decrease in autophagy due to failure of autophagosome-lysosme fusion. This becomes particularly important when determining the contribution of autophagy to disease pathogenesis. Thus, efforts are currently being made to measure autophagic flux, that is, the passage of organelles, cytoplasm, and other cargo through the autophagy-lysosmal degradation system. For example, a recent study used Western blots to examine the temporal profile of insoluble ubiquitinated aggregates isolated from Drosophila brain samples [14]. The researchers showed that as autophagic activity decreases in aging brains, ubiquitinated protein levels increase. Mutations in Atg8 exacerbated the build-up, while ectopic Atg8 expression prevented aggregate accumulation, demonstrating that the level of ubiquitinated proteins indicates changes in the rate of autophagic flux.
Autophagy mutants and RNAi
One great advantage of Drosophila as a model system is the extensive set of genetic tools readily available to the researcher. Loss-of-function alleles (point mutations, deletions, and transposon insertions) exist for nearly all of the core autophagy genes, and most of these have been tested for their function in the autophagy pathway in a variety of contexts. Two key exceptions are the Drosophila orthologs of Atg101 and FIP200, both newly identified components of the Atg1-Atg13 complex, for which there are no mutant alleles. Detailed information about all extant autophagy alleles and transgenic constructs can be found at Flybase, a comprehensive Drosophila genetics database.
Many of the tissues most affected by autophagy in Drosophila, such as fat, muscle, and neurons, are postmitotic, making clonal analysis difficult. Given the pleitropic function of autophagy genes, it is therefore not possible to clearly assay their tissue specific functions using classical alleles. Here, RNAi technology has been essential, as it allows for the spatial and temporal control of gene knockdown. In Caenorhabditis elegans, in vivo genome-wide RNAi screens of gene function are common practice based on feeding methods [46]. In Drosophila, as RNAi feeding is not possible, numerous efforts have been made to generate transgenes that express snapback/hairpin constructs. The most commonly used method relies on the Gal4/UAS system [5, 17, 82] to induce the specific expression of a doublestranded RNAi hairpin that triggers the posttranscriptional silencing of target genes. Based on the success of transgenic RNAi, large-scale efforts to generate genome-scale set of hairpin lines have been initiated [17]. In our lab we recently developed a system based on targeted integration of the hairpin constructs [81, 82]. Using this method, in collaboration with the Drosophila RNAi Screening Center, we are currently establishing a “second generation library”. Both groups have generated transgenic RNAi lines targeting most of the known core autophagy genes, which are available to the community through Flybase.
Genetic screens
The full potential of Drosophila genetics has not yet been realized in the field of autophagy. As noted above, the Drosophila tissues most often used as models for autophagy are polyploid and postmitotic, making the standard mosaic analyses much less efficient. Furthermore, it is a challenge to devise a screen for a cellular process like autophagy that has no easily scored morphology. RNAi-based analysis of autophagy in established Drosophila cell lines has shown promise, but the biological relevance of studies in such cell lines is questionable. Below, we discuss some of the approaches that have been applied to autophagy screening in Drosophila.
Simonsen et al. [98] performed a classic genetic modifier screen, taking advantage of a dominant eye phenotype associated with ectopic expression of the gene blue cheese (bchs). Bchs and its mammalian ortholog Alfy (autophagy-linked FYVE protein) are essential for the autophagic clearance of protein aggregates through an unknown mechanism [19]. Using deficiency stocks and select mutant alleles, the researchers found that mutations in lysosmal trafficking genes, as well as mutations in the SUMO and ubiquitin signaling pathways modified the dominant Bchs eye phenotype. Importantly, Atg1, Atg6, and Atg18 were identified as enhancers of the phenotype, suggesting that this type of approach could successfully identify genes involved in the autophagy pathway. In an attempt to directly screen autophagy regulators, Arsham and Neufeld [1] combined standard mosaic analysis with live-cell imaging of LysoTracker Red and fixed-cell imaging of autophagy-specific fluorescent protein markers. The FLPFRT recombination system was used to generate homozygous mutant clones for each of 383 lethal transposon insertions on chromosome 2 L. Mosaic fat bodies from the larvae were then dissected, and immediately stained with LysoTracker Red to compare the number of acidic lysosomes in the mutant cells versus surrounding wild-type tissue. Using this approach the reearchers identified 79 transposon insertions that caused an increase in lysosmal activity. The insertions were enriched in genes involved in protein synthesis, folding, transport, and degradation, and mitochondrial function and morphology.
Given the difficulty of performing large-scale mutagenesis screens for autophagy in Drosophila, several labs have turned to transcriptional and proteomic analysis to identify new factors involved in the pathway. Two studies published at the same time [24, 60] involved genome-wide analyses of transcripts from salivary glands undergoing autophagic cell death. Using microarrays, Lee et al. found several fly ATG genes (Atg2, Atg4, Atg5, and Atg7) that were transcriptionally up-regulated. A later report from the same lab demonstrated that dynein light chain 1, a gene that was previously detected in their microarray study, is required for autophagy induction during during salivary gland cell death [2]. Gorski et al. performed serial analysis of gene expression (SAGE) on salivary glands undergoing autophagic cell death. In addition to previously known autophagy and cell death genes, this group identified more than 732 differentially expressed genes with unknown functions. Another more recent study used microarrays to analyze autophagy induction in the larval fat body [40]. This group found that FK506-binding protein of 39 kDa (FKBP39) was down-regulated during fat body autophagy, and showed that it functions in vivo as an inhibitor of autophagy, likely through modulation of the transcription factor Foxo.
High-throughput proteomic techniques have also been used to identify proteins expressed during salivary gland and fat body autophagy in Drosophila [56, 67]. Martin et al. used a shotgun proteomics approach to identify proteins that are expressed during autophagic programmed cell death of the larval salivary glands. There was significant overlap between the resulting protein set and the previous microarray and SAGE studies. However, they also identified proteins not previously known to be expressed in dying salivary glands, including warts, a serine/threonine kinase in the Hippo signaling pathway. Subsequent work has shown that Warts is required for salivary gland programmed cell death via regulation of autophagy [18]. Kohler et al. used Isotope coded affinity tag and mass spectrometry to use identify components of starvation-induced autophagic responses in the Drosophila fat body. By comparing the proteins from starved versus wild-type fat body, they found 110 proteins that were differentially regulated. Among these, the lipid desaturase Desat1 was up-regulated in the starved sample, and was found to be required for starvation-induced autophagy and to localize to Atg5-and Atg8-positive structures.
Drosophila has proven an excellent system for systematic genome-wide cell-based RNAi high-throughput screens (RNAi HTS). Existing Drosophila cell culture lines (S2, Kc, Cl8, and BG3) rapidly take up long dsRNAs added to the medium, causing efficient target knockdown [11]. Furthermore, comprehensive dsRNA libraries allow for large-scale screens to systematically interrogate the function of all genes predicted from genomic sequencing. Thus, it is somewhat surprising that this system has not yielded a published report of a genome-wide autophagy screen. Nonetheless it is clear that Drosophila cell culture can be used to model the role of autophagy in immunity, cell death and starvation [32, 53, 95, 114]. In one moderately scaled screen, Chittaranjan et al. assayed 460 genes previously identified by expression studies of autophagic cell death in the salivary gland [10]. The authors induced autophagic cell death in a tumorous Drosophila hemocyte cell line by application of the hormone, ecdysone, which promotes metamorphosis and autophagic cell death in vivo. Screening for dsRNAs that significantly increased or decreased cell survival, they identified 25 genes for further analysis. Knockdown of Atg2, Atg3, Atg5, Atg6, Atg7, Atg8a, Atg8b caused decreased viability irrespective of ecdysone application.
Conclusions
There is an impressive repertoire of tools available to Drosophila researchers for studying autophagy. We have discussed how classical genetic approaches, RNAi both in vivo and in culture, transcription analyses, and a growing number of proteomic techniques have been applied to autophagy in a range of contexts. It is remarkable how conserved the autophagy machinery is between fruitfly and human, and cross-species studies have proven the value of the Drosophila model system in uncovering new autophagy pathway components and interactions. Given the establishment of cultured cell models of bacterial autophagy, viral autophagy and autophagic cell death, the next few years are likely to see a greater number RNAi screens focused on these phenomena. If previous experience is a guide, such research will provide insights into the molecular mechanisms underlying autophagy in both Drosophila biology and human disease.
Acknowledgments
This work was supported by the National Institutes of Health (R01 AR057352) and the Howard Hughes Medical Institute.
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Arsham AM, Neufeld TP. A genetic screen in Drosophila reveals novel cytoprotective functions of the autophagy-lysosome pathway. PLoS ONE. 2009;4(6):e6068. doi: 10.1371/journal.pone.0006068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batlevi Y, Martin DN, Pandey UB, Simon CR, Powers CM, Taylor JP, Baehrecke EH. Dynein light chain 1 is required for autophagy, protein clearance, and cell death in Drosophila. Proc Natl Acad Sci USA. 2010;107(2):742–747. doi: 10.1073/pnas.0907967107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131(6):1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. Listeriolysin o allows listeria monocytogenes replication in macrophage vacuoles. Nature. 2008;451(7176):350–354. doi: 10.1038/nature06479. [DOI] [PubMed] [Google Scholar]
- 5.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 6.Chan EY, Kir S, Tooze SA. SiRNA screening of the kinome identifies ulk1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282(35):25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 7.Chang YY, Neufeld TP. An atg1/atg13 complex with multiple roles in tor-mediated autophagy regulation. Mol Biol Cell. 2009;20(7):2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang YY, Neufeld TP. Autophagy takes flight in Drosophila. FEBS Lett. 2010;584(7):1342–1349. doi: 10.1016/j.febslet.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherry S. Vsv infection is sensed by Drosophila, attenuates nutrient signaling, and thereby activates antiviral autophagy. Autophagy. 2009;5(7):1062–1063. doi: 10.4161/auto.5.7.9730. [DOI] [PubMed] [Google Scholar]
- 10.Chittaranjan S, McConechy M, Hou YC, Freeman JD, Devorkin L, Gorski SM. Steroid hormone control of cell death and cell survival: Molecular insights using RNAi. PLoS Genet. 2009;5(2):e1000379. doi: 10.1371/journal.pgen.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci USA. 2000;97(12):6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corradetti MN, Guan KL. Upstream of the mammalian target of rapamycin: do all roads pass through mtor? Oncogene. 2006;25(48):6347–6360. doi: 10.1038/sj.onc.1209885. [DOI] [PubMed] [Google Scholar]
- 13.Cuervo AM. Autophagy: many paths to the same end. Mol Cell Biochem. 2004;263(1–2):55–72. doi: 10.1023/B:MCBI.0000041848.57020.57. [DOI] [PubMed] [Google Scholar]
- 14.Cumming RC, Simonsen A, Finley KD. Quantitative analysis of autophagic activity in Drosophila neural tissues by measuring the turnover rates of pathway substrates. Methods Enzymol. 2008;451:639–651. doi: 10.1016/S0076-6879(08)03235-7. [DOI] [PubMed] [Google Scholar]
- 15.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10(1):51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5(6):527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448(7150):151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 18.Dutta S, Baehrecke EH. Warts is required for pi3k-regulated growth arrest, autophagy, and autophagic cell death in Drosophila. Curr Biol. 2008;18(19):1466–1475. doi: 10.1016/j.cub.2008.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, Krainc D, Brech A, Stenmark H, Simonsen A, Yamamoto A. The selective macroautophagic degradation of aggregated proteins requires the pi3p-binding protein alfy. Mol Cell. 2010;38(2):265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447(7148):1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 21.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The atg16l complex specifies the site of lc3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19(5):2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of apg13 gene involved in autophagy in yeast, saccharomyces cerevisiae. Gene. 1997;192(2):207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 23.Ganley IG, du Lam H, Wang J, Ding X, Chen S, Jiang X. Ulk1.Atg13.Fip200 complex mediates mtor signaling and is essential for autophagy. J Biol Chem. 2009;284(18):12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorski SM, Chittaranjan S, Pleasance ED, Freeman JD, Anderson CL, Varhol RJ, Coughlin SM, Zuyderduyn SD, Jones SJ, Marra MA. A sage approach to discovery of genes involved in autophagic cell death. Curr Biol. 2003;13(4):358–363. doi: 10.1016/s0960-9822(03)00082-4. [DOI] [PubMed] [Google Scholar]
- 25.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119(6):753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 26.Hanson KA, Kim SH, Wassarman DA, Tibbetts RS. Ubiquilin modifies tdp-43 toxicity in a Drosophila model of amyotrophic lateral sclerosis (als) J Biol Chem. 2010;285(15):11068–11072. doi: 10.1074/jbc.C109.078527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. Fip200, a ulk-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181(3):497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, apg4b, is specific for the autophagy-related ubiquitin-like proteins gate-16, map1-lc3, gabarap, and apg8l. J Biol Chem. 2003;278(51):51841–51850. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- 30.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mtorc1 association with the ulk1-atg13-fip200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with atg13. Autophagy. 2009;5(7):973–979. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- 32.Hou YC, Chittaranjan S, Barbosa SG, McCall K, Gorski SM. Effector caspase dcp-1 and iap protein bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol. 2008;182(6):1127–1139. doi: 10.1083/jcb.200712091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408(6811):488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 34.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian atg14 and uvrag. Mol Biol Cell. 2008;19(12):5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacinto E, Hall MN. Tor signalling in bugs, brain and brawn. Nat Rev Mol Cell Biol. 2003;4(2):117–126. doi: 10.1038/nrm1018. [DOI] [PubMed] [Google Scholar]
- 36.Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3(5):e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juhasz G, Csikos G, Sinka R, Erdelyi M, Sass M. The Drosophila homolog of aut1 is essential for autophagy and development. FEBS Lett. 2003;543(1–3):154–158. doi: 10.1016/s0014-5793(03)00431-9. [DOI] [PubMed] [Google Scholar]
- 38.Juhasz G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21(23):3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Juhasz G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class iii pi(3)k vps34 promotes autophagy and endocytosis but not tor signaling in Drosophila. J Cell Biol. 2008;181(4):655–666. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Juhasz G, Puskas LG, Komonyi O, Erdi B, Maroy P, Neufeld TP, Sass M. Gene expression profiling identifies fkbp39 as an inhibitor of autophagy in larval Drosophila fat body. Cell Death Differ. 2007;14(6):1181–1190. doi: 10.1038/sj.cdd.4402123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTor regulation of autophagy. FEBS Lett. 2010;584(7):1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y. Atg17 functions in cooperation with atg1 and atg13 in yeast autophagy. Mol Biol Cell. 2005;16(5):2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kabeya Y, Kawamata T, Suzuki K, Ohsumi Y. Cis1/atg31 is required for autophagosome formation in saccharomyces cerevisiae. Biochem Biophys Res Commun. 2007;356(2):405–410. doi: 10.1016/j.bbrc.2007.02.150. [DOI] [PubMed] [Google Scholar]
- 44.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. Lc3, gabarap and gate16 localize to autophagosomal membrane depending on form-ii formation. J Cell Sci. 2004;117(Pt 13):2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 45.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an apg1 protein kinase complex. J Cell Biol. 2000;150(6):1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421(6920):231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 47.Kametaka S, Matsuura A, Wada Y, Ohsumi Y. Structural and functional analyses of apg5, a gene involved in autophagy in yeast. Gene. 1996;178(1–2):139–143. doi: 10.1016/0378-1119(96)00354-x. [DOI] [PubMed] [Google Scholar]
- 48.Kametaka S, Okano T, Ohsumi M, Ohsumi Y. Apg14p and apg6/vps30p form a protein complex essential for autophagy in the yeast, saccharomyces cerevisiae. J Biol Chem. 1998;273(35):22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- 49.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19(5):2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawamata T, Kamada Y, Suzuki K, Kuboshima N, Akimatsu H, Ota S, Ohsumi M, Ohsumi Y. Characterization of a novel autophagy-specific gene, atg29. Biochem Biophys Res Commun. 2005;338(4):1884–1889. doi: 10.1016/j.bbrc.2005.10.163. [DOI] [PubMed] [Google Scholar]
- 51.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase y sorting in saccharomyces cerevisiae. J Cell Biol. 2001;152(3):519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of torc1 by rag gtpases in nutrient response. Nat Cell Biol. 2008;10(8):935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim YI, Ryu T, Lee J, Heo YS, Ahnn J, Lee SJ, Yoo O. A genetic screen for modifiers of Drosophila caspase dcp-1 reveals caspase involvement in autophagy and novel caspase-related genes. BMC Cell Biol. 2010;11:9. doi: 10.1186/1471-2121-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of apg8/aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151(2):263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klionsky DJ, Cregg JM, Dunn WA, Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5(4):539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 56.Kohler K, Brunner E, Guan XL, Boucke K, Greber UF, Mohanty S, Barth JM, Wenk MR, Hafen E. A combined proteomic and genetic analysis identifies a role for the lipid desaturase desat1 in starvation-induced autophagy in Drosophila. Autophagy. 2009;5(7):980–990. doi: 10.4161/auto.5.7.9325. [DOI] [PubMed] [Google Scholar]
- 57.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 58.Kuma A, Matsui M, Mizushima N. Lc3, an autophago-some marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of lc3 localization. Autophagy. 2007;3(4):323–328. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 59.Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kda apg12-apg5.Apg16 multimeric complex, mediated by apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277(21):18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- 60.Lee CY, Clough EA, Yellon P, Teslovich TM, Stephan DA, Baehrecke EH. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr Biol. 2003;13(4):350–357. doi: 10.1016/s0960-9822(03)00085-x. [DOI] [PubMed] [Google Scholar]
- 61.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6(4):463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 62.Li L, Kim E, Yuan H, Inoki K, Goraksha-Hicks P, Schiesher RL, Neufeld TP, Guan KL. Regulation of mtorc1 by the rab and arf gtpases. J Biol Chem. 2010;285(26):19705–19709. doi: 10.1074/jbc.C110.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel beclin1-binding protein uvrag. Nat Cell Biol. 2006;8(7):688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 64.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal sindbis virus encephalitis by beclin, a novel bcl-2-interacting protein. J Virol. 1998;72(11):8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lindmo K, Brech A, Finley KD, Gaumer S, Contamine D, Rusten TE, Stenmark H. The pi 3-kinase regulator vps15 is required for autophagic clearance of protein aggregates. Autophagy. 2008;4(4):500–506. doi: 10.4161/auto.5829. [DOI] [PubMed] [Google Scholar]
- 66.Ling D, Song HJ, Garza D, Neufeld TP, Salvaterra PM. Abeta42-induced neurodegeneration via an age-dependent autophagic-lysosomal injury in Drosophila. PLoS ONE. 2009;4(1):e4201. doi: 10.1371/journal.pone.0004201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin DN, Balgley B, Dutta S, Chen J, Rudnick P, Cranford J, Kantartzis S, DeVoe DL, Lee C, Baehrecke EH. Proteomic analysis of steroid-triggered autophagic programmed cell death during Drosophila development. Cell Death Differ. 2007;14(5):916–923. doi: 10.1038/sj.cdd.4402098. [DOI] [PubMed] [Google Scholar]
- 68.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21(11):1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in saccharomyces cerevisiae. Gene. 1997;192(2):245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 70.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301(5638):1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 71.Mercer CA, Kaliappan A, Dennis PB. A novel, human atg13 binding protein, atg101, interacts with ulk1 and is essential for macroautophagy. Autophagy. 2009;5(5):649–662. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- 72.Meyer-Morse N, Robbins JR, Rae CS, Mochegova SN, Swanson MS, Zhao Z, Virgin HW, Portnoy D. Listeriolysin o is necessary and sufficient to induce autophagy during listeria monocytogenes infection. PLoS One. 2010;5(1):e8610. doi: 10.1371/journal.pone.0008610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mizushima N. The role of the atg1/ulk1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22(2):132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 74.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse apg16l, a novel wd-repeat protein, targets to the autophagic isolation membrane with the apg12-apg5 conjugate. J Cell Sci. 2003;116(Pt 9):1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 75.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395(6700):395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 76.Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27(6):421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- 77.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast apg12p conjugation system essential for autophagy. J Biol Chem. 1998;273(51):33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- 78.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nair U, Klionsky DJ. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem. 2005;280(51):41785–41788. doi: 10.1074/jbc.R500016200. [DOI] [PubMed] [Google Scholar]
- 80.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T. Autophagy defends cells against invading group a streptococcus. Science. 2004;306(5698):1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 81.Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer B, Markstein M, Wang H, Villalta C, Laverty T, Perkins L, Perrimon N. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009;182(4):1089–1100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ni JQ, Markstein M, Binari R, Pfeiffer B, Liu LP, Villalta C, Booker M, Perkins L, Perrimon N. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat Methods. 2008;5(1):49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273(7):3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 84.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2(3):211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 85.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 86.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in ht-29 cells. J Biol Chem. 2000;275(2):992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 87.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112(12):1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ravikumar B, Acevedo-Arozena A, Imarisio S, Berger Z, Vacher C, O'Kane CJ, Brown SD, Rubinsztein DC. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet. 2005;37(7):771–776. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- 89.Ravikumar B, Imarisio S, Sarkar S, O'Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci. 2008;121(Pt 10):1649–1660. doi: 10.1242/jcs.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O'Kane CJ, Rubinsztein DC. Inhibition of mtor induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of huntington disease. Nat Genet. 2004;36(6):585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 91.Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the pi3k pathway. Dev Cell. 2004;7(2):179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 92.Saeki K, Yuo A, Okuma E, Yazaki Y, Susin SA, Kroemer G, Takaku F. Bcl-2 down-regulation causes autophagy in a caspase-independent manner in human leukemic hl60 cells. Cell Death Differ. 2000;7(12):1263–1269. doi: 10.1038/sj.cdd.4400759. [DOI] [PubMed] [Google Scholar]
- 93.Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17(1):1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7(2):167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 95.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30(4):588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18(19):5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4(2):176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 98.Simonsen A, Cumming RC, Finley KD. Linking lysosomal trafficking defects with changes in aging and stress response in Drosophila. Autophagy. 2007;3(5):499–501. doi: 10.4161/auto.4604. [DOI] [PubMed] [Google Scholar]
- 99.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III pi3-kinase complexes. J Cell Biol. 2009;186(6):773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stromhaug PE, Reggiori F, Guan J, Wang CW, Klionsky DJ. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of atg8 during uptake of aminopeptidase i by selective autophagy. Mol Biol Cell. 2004;15(8):3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of barkor as a mammalian autophagy-specific factor for beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 2008;105(49):19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, Pledger WJ, Wang HG. Bif-1 interacts with beclin 1 through uvrag and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9(10):1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119(2):301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E. Apg7p/cvt2p: a novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10(5):1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tanida I, Sou YS, Minematsu-Ikeguchi N, Ueno T, Kominami E. Atg8l/apg8l is the fourth mammalian modifier of mammalian atg8 conjugation mediated by human atg4b, atg7 and atg3. FEBS J. 2006;273(11):2553–2562. doi: 10.1111/j.1742-4658.2006.05260.x. [DOI] [PubMed] [Google Scholar]
- 106.Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human apg3p/aut1p homologue is an authentic e2 enzyme for multiple substrates, gate-16, gabarap, and map-lc3, and facilitates the conjugation of hapg12p to hapg5p. J Biol Chem. 2002;277(16):13739–13744. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- 107.Tanida I, Tanida-Miyake E, Nishitani T, Komatsu M, Yamazaki H, Ueno T, Kominami E. Murine apg12p has a substrate preference for murine apg7p over three apg8p homologs. Biochem Biophys Res Commun. 2002;292(1):256–262. doi: 10.1006/bbrc.2002.6645. [DOI] [PubMed] [Google Scholar]
- 108.Thumm M, Kadowaki T. The loss of Drosophila apg4/aut2 function modifies the phenotypes of cut and notch signaling pathway mutants. Mol Genet Genomics. 2001;266(4):657–663. doi: 10.1007/s004380100585. [DOI] [PubMed] [Google Scholar]
- 109.Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 110.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 111.Yan J, Kuroyanagi H, Kuroiwa A, Matsuda Y, Tokumitsu H, Tomoda T, Shirasawa T, Muramatsu M. Identification of mouse ulk1, a novel protein kinase structurally related to C. Elegans unc-51. Biochem Biophys Res Commun. 1998;246(1):222–227. doi: 10.1006/bbrc.1998.8546. [DOI] [PubMed] [Google Scholar]
- 112.Yan J, Kuroyanagi H, Tomemori T, Okazaki N, Asato K, Matsuda Y, Suzuki Y, Ohshima Y, Mitani S, Masuho Y, Shirasawa T, Muramatsu M. Mouse ulk2, a novel member of the unc-51-like protein kinases: unique features of functional domains. Oncogene. 1999;18(43):5850–5859. doi: 10.1038/sj.onc.1202988. [DOI] [PubMed] [Google Scholar]
- 113.Yan L, Sadoshima J, Vatner DE, Vatner SF. Autophagy: a novel protective mechanism in chronic ischemia. Cell Cycle. 2006;5(11):1175–1177. doi: 10.4161/cc.5.11.2787. [DOI] [PubMed] [Google Scholar]
- 114.Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Takada H, Goldman WE, Fukase K, Silverman N, Yoshimori T, Kurata S. Autophagic control of listeria through intracellular innate immune recognition in Drosophila. Nat Immunol. 2008;9(8):908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, Kakizuka A, Sztul E, Chakraborty T, Sasakawa C. Listeria monocytogenes acta-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11(10):1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 117.Yuan J, Lipinski M, Degterev A. Diversity in the mechanisms of neuronal cell death. Neuron. 2003;40(2):401–413. doi: 10.1016/s0896-6273(03)00601-9. [DOI] [PubMed] [Google Scholar]
- 118.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100(25):15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]