Abstract
All species of Chlamydia undergo a unique developmental cycle that transitions between extracellular and intracellular environments and requires the capacity to invade new cells for dissemination. A chlamydial protein called Tarp has been shown to nucleate actin in vitro and is implicated in bacterial entry into human cells. Colocalization studies of ectopically expressed enhanced green fluorescent protein (EGFP)-Tarp indicate that actin filament recruitment is restricted to the C-terminal half of the effector protein. Actin filaments are presumably associated with Tarp via an actin binding alpha helix that is also required for actin nucleation in vitro, but this has not been investigated. Tarp orthologs from C. pneumoniae, C. muridarum, and C. caviae harbor between 1 and 4 actin binding domains located in the C-terminal half of the protein, but C. trachomatis serovar L2 has only one characterized domain. In this work, we examined the effects of domain-specific mutations on actin filament colocalization with EGFP-Tarp. We now demonstrate that actin filament colocalization with Tarp is dependent on two novel F-actin binding domains that endow the Tarp effector with actin-bundling activity. Furthermore, Tarp-mediated actin bundling did not require actin nucleation, as the ability to bundle actin filaments was observed in mutant Tarp proteins deficient in actin nucleation. These data shed molecular insight on the complex cytoskeletal rearrangements required for C. trachomatis entry into host cells.
INTRODUCTION
The obligate intracellular bacterium Chlamydia trachomatis causes the most frequently reported sexually transmitted bacterial disease in the United States, with over 1 million cases reported to the Centers for Disease Control and Prevention annually (1). Worldwide, ocular infection with C. trachomatis (trachoma) is the leading cause of preventable blindness and is the subject of a global initiative led by the World Health Organization to eradicate trachoma by 2020 (2).
Species of Chlamydia utilize a unique developmental cycle in which bacteria transition from the infectious spore-like elementary body (EB) to the metabolically active reticulate body (RB) within the protective confines of a membrane-bound parasitophorous vacuole termed the inclusion (3). The invasive EB is formed in the middle to late stages of the intracellular development cycle as the RBs differentiate back to EBs and are packed with metabolites and proteins designed to facilitate extracellular survival and reinfection (4, 5). Additional infectious cycles arise from EBs that are released and disseminate from infected tissues (6).
C. trachomatis invasion is induced by cytoskeletal rearrangements initiated upon microbe contact with the host cell surface (7). Alterations of the host cytoskeleton are required for bacterial uptake, as drugs, such as cytochalasin D, that disrupt the cytoskeleton prevent C. trachomatis infections (7). A number of intracellular microorganisms harbor proteins that directly alter actin dynamics, which favor pathogen survival and propagation (8). These virulence factors can drive the formation of actin filaments and actin bundles or can lead to the disassembly of actin filaments. Cytoskeletal rearrangements initiated upon EB contact with the host cell surface may in part be triggered by the translocation of type III secreted effectors (9, 10). One of the effector proteins, called translocated actin recruiting protein (Tarp), is able to increase the rate of actin filament formation by directly nucleating actin (11). In addition, Tarp and the host cell Arp2-Arp3 actin-nucleating complex cooperate to increase the rate of actin filament formation, and both host- and bacterium-derived actin nucleators are implicated in C. trachomatis invasion (12–14).
Tarp contains a C-terminal actin binding and oligomerization domain required for actin nucleation and an N-terminal phosphorylation domain implicated in host cell signaling via association with host-derived proteins, such as phosphoinositide 3-kinase (PI3K) and Src homology 2 (SH2) domain-containing transforming protein 1 (SHC-1) (11, 15–17). Phosphorylated Tarp peptides have also been shown to immunoprecipitate a complex of proteins containing Sos1 and Vav2, two Rac guanine nucleotide exchange factors thought to participate in WAVE2 and Arp2-Arp3 complex recruitment (16). Colocalization studies of ectopically expressed enhanced green fluorescent protein (EGFP)-Tarp indicate that actin filament recruitment is restricted to the C-terminal half of the effector and is presumably associated with Tarp via the previously identified actin binding alpha helix required for actin nucleation in vitro (14, 15). Sequence and biochemical analyses of Tarp orthologs from C. pneumoniae, C. muridarum, C. caviae, and C. trachomatis serovars A, D, and L2 revealed the presence of between 1 and 4 actin binding sites (13). Although C. trachomatis Tarp (L2) appeared to harbor two putative actin binding domains (ABD), only one of the two alpha helices was found to associate with actin (13). In this work, we examined the effect of domain-specific mutations on actin filament colocalization with EGFP-Tarp. Here, we report that C. trachomatis L2 Tarp harbors two distinct filamentous-actin (F-actin) binding sites that allow the Tarp effector to bundle actin filaments. Furthermore, Tarp-mediated actin bundling did not require actin nucleation, as the ability to bundle actin filaments was observed in mutant Tarp proteins deficient in actin nucleation. These findings attribute a novel activity to the critical Tarp protein and provide molecular insight into the complex cytoskeletal rearrangements required for C. trachomatis entry into host cells.
MATERIALS AND METHODS
Cloning and protein expression.
In-frame amino-terminal glutathione S-transferase (GST) and carboxyl-terminal polyhistidine fusion proteins for full-length wild-type Tarp were generated by PCR by amplifying the corresponding coding regions from C. trachomatis serovar L2 LGV 434 genomic DNA (Qiagen genomic purification kit; Valencia, CA) as previously described (14). PCR was performed with custom synthesized oligonucleotide primers (Integrated DNA Technologies, Coralville, IA) engineered with SalI, SacI, or NotI linkers. PCR products were purified (Qiagen), digested with restriction enzymes (New England BioLabs, Beverly, MA), and subcloned into linearized pGEX-6P-1 to generate translational fusions with GST and polyhistidine. Tarp domain deletion mutants—phosphorylation domain deletion (ΔphoD; deletion of D125 to Y424), proline-rich-domain (PRD) deletion (ΔPRD; deletion of S625 to N650), actin binding domain deletion (ΔABD; deletion of amino acids [aa] 748 to 758), F-actin binding domain deletion 1 (ΔFAB1; deletion of L871 to L882), and F-actin binding domain deletion 2 (ΔFAB2; deletion of N942 to G967)—were generated by inverse PCR by amplifying the pGEX-6P-1 plasmids encoding the wild-type Tarp fusion protein. Multiple domain deletions in a single tarP gene (for example, ΔABD, ΔFAB1, and ΔFAB2) were generated sequentially by inverse PCR or by ligating individual deletion mutants together. The Tarp mutants described above were also cloned into pEGFP-C3 (BD Biosciences Clontech) to allow ectopic expression of EGFP-Tarp in HeLa cells. All pGEX-6P-1 plasmids were transformed into the BL21 strain of Escherichia coli (Novagen, Madison, WI). Protein expression and purification were performed according to the procedures outlined for Ni Sepharose 6 Fast Flow and glutathione Sepharose 4B in the Bulk GST Purification Module (GE Health Sciences, Piscataway, NY). In some experiments, the GST tag was removed prior to F-actin binding and bundling with PreScission Protease treatment according to the manufacturer's recommendation (GE Health Sciences).
GST fusion pulldown experiments.
GST fusion pulldown experiments were performed according to protocols previously described (11, 13). Briefly, HeLa 229 cells were suspended in 100 mM KCl, 10 mM HEPES (pH 7.7), 2 mM MgCl2, and 2 mM ATP (buffer A) and disrupted by sonication delivered in four consecutive bursts at 20-s intervals on setting 4 (ultrasonic sonicator processor XL equipped with a microtip; Misonix Incorporated, Farmingdale, NY). Insoluble material was removed by centrifugation (12,000 × g; 25 min; 4°C). Glutathione-Sepharose beads were incubated with 10 μg of GST fusion proteins or GST for 1 h at 4°C in PBS (GE Health Sciences). GST fusion protein-coated beads were washed twice with PBS and once with buffer A prior to the addition of approximately 100 μg of HeLa extract. The extracts and beads were incubated together for 2 h at 4°C and washed three times with fresh buffer A, and bound proteins were eluted using sample buffer.
SDS-PAGE and immunoblotting.
Proteins were separated on SDS-10% polyacrylamide gels and transferred to 0.45-μm pure nitrocellulose transfer and immobilization membranes (Schleicher & Schuell, Keene, NH). Immunoblotting employed peroxidase-conjugated secondary antibodies (Chemicon International, Temecula, CA) and Supersignal West Pico chemiluminescent substrate (Pierce, Rockford, IL). The anti-actin C4 monoclonal antibody was purchased from Chemicon International. The anti-phosphotyrosine 4G10 monoclonal antibody was purchased from Upstate (Millipore). Polyclonal rabbit antibodies directed toward C. trachomatis L2 LGV 434 Tarp (CT456) were developed at Rocky Mountain Laboratories as previously described (9). Peptide antibodies directed toward the Tarp actin binding domain and PRD were generated and purified by Sigma Genosys (Spring, TX) as previously described (13).
Transfection of HeLa cells and indirect immunofluorescence microscopy.
HeLa cells (2 × 105) were seeded in 6-well plates with coverslips and grown for 24 h in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). The cells were then transfected with transfection mixture containing 8 μl of Fugene HD (Promega) and 2.5 μg of the respective plasmid. After 24 h, the cells were fixed by adding 4% paraformaldehyde and incubating them at 4°C for 15 min. The cells were then treated with ice-cold 0.4% Triton X for 10 min, followed by blocking with 5% bovine serum albumin (BSA) for 45 min. To visualize tyrosine-phosphorylated protein, cells were first incubated with anti-phosphotyrosine primary antibody 4G10 (Upstate) at 1:1,000 dilution in 0.5% BSA at room temperature (RT) for 45 min, followed by incubation with anti-mouse secondary antibody conjugated to Alexa 350 (Invitrogen). To simultaneously visualize actin, phalloidin conjugated to Alexa 568 (Invitrogen) was added to the above-mentioned mixture containing secondary antibodies. Coverslips were rinsed and mounted in Prolong Gold antifade reagent (Invitrogen). The cells were examined with a Zeiss Axio Observer A1 microscope equipped with phase-contrast and epifluorescence optics. Images were obtained using an AxioCam MRm camera controlled by AxioVision 4.8.2 and further processed using Adobe Photoshop CS2.
F-actin binding and bundling.
Briefly, 5 μg of GST fusion proteins or control proteins (GST and α-actinin) was added to 40 μg of F actin (generated by adding 1/10 volume of polymerization buffer to globular actin [G actin] and incubating them at RT for 1 h) and allowed to incubate at RT for 30 min. F actin and the bound proteins were separated by differential sedimentation at 100,000 × g for 2 h at RT in a Beckman Optima TLX Ultracentrifuge using a TLA 55 or TLA 100.3 rotor (Beckman Coulter, Fullerton, CA). Proteins associated with the F-actin pellets were compared to unbound proteins that remained in the supernatant by resolving the proteins on 10% SDS-polyacrylamide gels, followed by Coomassie staining. Actin-bundling experiments were performed similarly to F-actin binding assays, except the actin bundles were isolated with a 15,000 × g spin.
Pyrene assay.
Pyrene actin polymerization assays were performed as previously described (11, 13, 14). Briefly, monomeric pyrene-labeled actin was prepared by diluting 100 μg of lyophilized pyrene actin (cytoskeleton) in 2 ml of 5 mM Tris (pH 8.0)-0.2 mM CaCl2-0.2 mM ATP (G buffer) and incubating for 1 h at RT, followed by 1 additional hour of incubation at 4°C. Monomeric pyrene actin was obtained by collecting the supernatant after a 2-h, 100,000 × g, 4°C spin in a Beckman Optima TLX Ultracentrifuge using a TLA 100.3 rotor (Beckman Coulter). Approximately 20 μg of pyrene-labeled actin was gently mixed with 5 μg of GST fusion proteins in a volume of 500 μl for 10 min before the addition of 1/20 volume of polymerization buffer (500 mM KCl, 20 mM MgCl2, 10 mM ATP). The reaction was monitored for 1 h with an LS 55 Luminescence spectrophotometer directed by FL WinLab software version 4.0 (Perkin-Elmer, Beaconsfield, Bucks, United Kingdom) with 2.5-nm bandwidth at 365-nm excitation wavelength and 2.5-nm bandwidth at 407-nm emission wavelength.
RESULTS
Mutant Tarp proteins exhibit unique actin binding and polymerization kinetics.
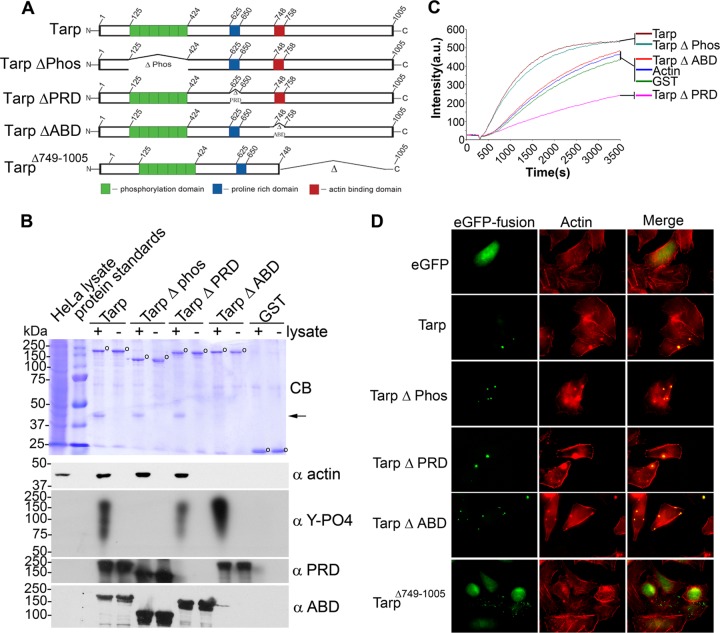
The actin-nucleating activity of Tarp results from distinct actin binding and proline-rich oligomerization domains in vitro (11). C. trachomatis L2 Tarp is a large, 1,005-aa protein, and studies to date have primarily focused on recombinant Tarp truncation mutants or Tarp peptides to identify the domains of the protein responsible for Tarp-mediated actin polymerization (9, 11, 13). To confirm that the previously identified domains were sufficient for actin binding and actin nucleation in the entire Tarp effector, we generated a series of GST-His and EGFP recombinant full-length L2 Tarp deletion mutants that are missing the phosphorylation, the actin binding, and/or the proline-rich oligomerization domain(s) to examine the contribution that each domain makes to actin kinetics biochemically and in HeLa cells (Fig. 1A). Wild-type Tarp and deletion mutant Tarp proteins with dual N-terminal GST and C-terminal polyhistidine affinity tags were purified and employed in actin binding and actin polymerization assays (Fig. 1B and C). All full-length Tarp mutants were able to associate with host cell actin in a GST pulldown assay, except for the ΔABD Tarp mutant harboring an 11-amino-acid deletion (aa 748 to 758) of the previously characterized actin binding domain (Fig. 1B) (11). The purified Tarp proteins also demonstrated distinct actin polymerization kinetics, as observed in in vitro pyrene actin polymerization assays (Fig. 1C). An increase in the rate of actin polymerization was observed in wild-type Tarp and the Tarp effector harboring a deletion in the phosphorylation domain compared to actin-only controls, which is in agreement with reports localizing the Tarp actin-nucleating activity to the C-terminal half of the protein (9, 11, 13, 14). Consistent with previous studies, a short PRD of 25 amino acids, implicated in Tarp oligomerization, was required for Tarp-mediated actin nucleation. Deletion of PRD resulted in the creation of an actin-sequestering protein (TarpΔPRD) that, in the pyrene actin polymerization assay, produced a curve that appeared below the actin-only and GST controls (Fig. 1C) (11). Surprisingly, when actin filament colocalization with EGFP-Tarp was examined in HeLa cells with Alexa-conjugated phalloidin, Tarp mutants lacking the actin binding alpha helix, amino acids 748 to 758 (TarpΔABD), known to be essential for Tarp-mediated actin nucleation (Fig. 1C) retained the ability to colocalize with actin filaments (Fig. 1D). A Tarp-EGFP fusion lacking amino acids 749 to 1005 (TarpΔ749-1005) did not colocalize with phalloidin and served as a negative control (Fig. 1D). EGFP-Tarp harboring mutations in both the phosphorylation and actin binding domains was also found to colocalize with actin filaments (data not shown). The actin pulldown and actin polymerization experiments differ from the actin filament colocalization experiments in that the first two experiments primarily use G actin, whereas the last primarily uses F actin. These data suggest that Tarp may harbor an as yet uncharacterized F-actin binding domain(s) distinct from the previously characterized G-actin binding domain that is essential for actin nucleation.
Fig 1.
Tarp deletion mutants demonstrate distinct actin binding and actin polymerization kinetics. (A) Schematics of Tarp proteins indicating the locations of the actin binding domain (red box), the proline-rich domain (blue box), and the tyrosine-rich phosphorylation domain (green boxes). Δ indicates amino acids deleted in mutant Tarp proteins, and the numbers indicate amino acid positions encoded within the C. trachomatis tarP gene. Full-length and mutant Tarp proteins were tagged with N-terminal GST tags and C-terminal histidine tags to purify recombinant protein for biochemical assays or with EGFP for immunofluorescence assays. (B) Extracts from HeLa cells were incubated with GST or GST fusions to wild-type or mutant Tarp, and specifically bound proteins were resolved by SDS-PAGE and visualized by Coomassie blue staining (CB). GST alone and GST-Tarp fusion proteins are indicated by open circles, while the enriched actin protein is indicated by an arrow. Specifically, Tarp proteins harboring deletions in the phosphorylation domain (Δ phos), the proline-rich oligomerization domain (Δ PRD), and the actin binding domain (Δ ABD) were tested. The HeLa lysate shown in the first lane represents 1% of the material used in the + lysate pulldown lanes. Samples identical to those shown in the Coomassie-stained gel were subjected to immunoblotting with actin (α actin), phosphotyrosine (α Y-PO4), and peptide antisera specific for the proline-rich (α PRD) and actin binding (α ABD) Tarp domains. The molecular masses of protein standards are shown. (C) GST or GST-Tarp fusion proteins shown in panel B were incubated with 1 μM monomeric pyrene-labeled actin. A Tarp-mediated increase in actin polymerization after the addition of polymerization buffer at 300 s was measured as the arbitrary fluorescence intensity (arbitrary units [a.u.] over time) with excitation and emission wavelengths of 365 and 407 nm, respectively. GST and pyrene actin alone served as negative controls. (D) EGFP-Tarp mutants colocalize with filamentous actin in HeLa cells. Host cells expressing EGFP fusions of full-length Tarp (Tarp) or deletion mutants lacking the phosphorylation domain (Tarp Δ Phos), proline-rich domain (Tarp Δ PRD), or actin binding domain (Tarp Δ ABD) were fixed and stained with Alexa Fluor 568-conjugated phalloidin (Actin). Host cells expressing EGFP alone or an EGFP fusion with Tarp (1 to 748) were used as negative controls.
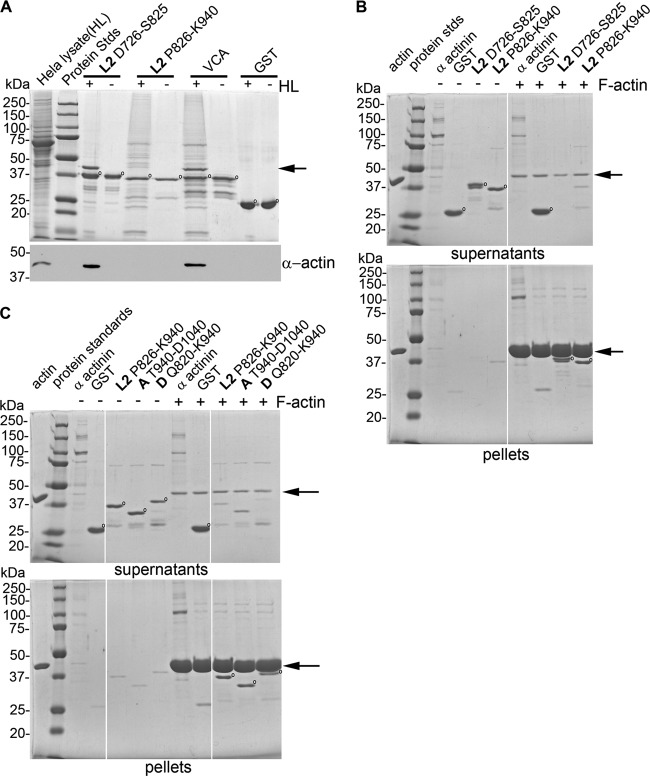
C. trachomatis L2 Tarp harbors a distinct F-actin binding domain. Tarp orthologs contain between one and four actin binding domains, according to GST pulldown assays performed with HeLa extracts (13). Previous reports have indicated that C. trachomatis L2 Tarp harbors one actin binding domain, which is consistent with the data presented in Fig. 1B (11, 13). Deletion of 11 amino acids contained within the L2 Tarp actin binding domain was sufficient to prevent actin binding in the GST pulldown assay with the mutant protein compared to the wild-type control. Interestingly, bioinformatics analysis of the entire L2 Tarp protein sequence identified a putative second actin binding domain with sequence similarity to the experimentally characterized L2 Tarp actin binding domain; however, this sequence lacked the ability to associate with host cell actin in a GST pulldown assay (13). This finding further supports the prediction that Tarp may harbor protein domains that differentiate between monomeric (globular) actin, found predominately in the HeLa-generated protein lysates, and filamentous actin, detected by fluorescent phalloidin in the transfected host cells. In order to examine whether the second actin binding-like domain found in the C. trachomatis L2 Tarp sequence (amino acids 871 to 883) was able to differentially associate with globular versus filamentous actin, GST-Tarp fusions to the domain were tested for their ability to associate with actin generated from HeLa lysates in a GST pulldown assay and an F-actin cosedimentation binding assay (Fig. 2). Similar to our previous findings, the 100-amino-acid peptide harboring the original actin binding domain was able to associate with actin generated from a HeLa lysate; however, the alternate putative actin binding sequence did not associate with actin generated from the same lysate (Fig. 2A) (11, 13). Interestingly, the two domains did cosediment with filamentous actin, indicating that the second domain preferentially associates with filamentous actin, while the original actin binding domain is able to associate with both monomeric and filamentous actin, as previously described (Fig. 2B) (11). Since the second actin binding sequence prefers F actin, we have called this site F-actin binding domain 1 (FAB1) to differentiate it from the originally characterized ABD, which associates with both G and F actin.
Fig 2.
Tarp harbors an F-actin binding domain that is separate from the actin nucleation domain. (A) Extracts from HeLa cells were incubated with GST alone (GST), GST fused to the Wiskott-Aldrich syndrome protein verprolin homology, cofilin homology, and acidic domain (VCA), or GST fusions to the 100-aa fragment of Tarp containing the Tarp ABD (L2 D726-S825) or the Tarp domain containing a similar sequence (L2 P826-K940). The bound proteins were resolved by SDS-PAGE and visualized by Coomassie blue staining. All GST fusion proteins are indicated by open circles, while the enriched actin protein is indicated by an arrow. The HeLa lysate shown in the first lane represents 1% of the material used in the + lysate pulldown lanes. Samples identical to those shown in the Coomassie-stained gel were subjected to immunoblotting with actin (α-actin). (B) GST-Tarp fusions were incubated with F actin and isolated by ultracentrifugation. Protein supernatants and pellets were resolved by SDS-PAGE and visualized by Coomassie blue staining. GST fusion proteins are indicated by open circles and differentially appear in the supernatant and/or pellet fraction following centrifugation. The actin protein is indicated by arrows. GST-L2 D726-S825 and L2 P826-K940 precipitated with F actin in the pellet fraction, as did the positive-control protein (α actinin). The GST control did not associate with the F-actin pellet and remained predominately in the supernatant. The actin shown in the first lane represents 2% of the material used in the + F actin lanes. The molecular masses of protein standards are shown. (C) Protein domains with sequence similarity to the C. trachomatis L2 Tarp FAB1 (L2 P826-K940) from C. trachomatis serovar A (A T940-D1040) and serovar D (D Q820-K940) were incubated with F actin and isolated by ultracentrifugation, as in panel B.
Previous studies examining the conserved domains within Tarp orthologs demonstrated that, like C. trachomatis L2 Tarp, C. trachomatis serovar A and D Tarps each harbored a domain that surprisingly did not associate with G actin in GST pulldown experiments (13). In order to determine if C. trachomatis serovar A and D Tarps harbored FAB1, GST-Tarp fusions harboring approximately 100 amino acids of each domain (C. trachomatis serovar A T940-D1040 and C. trachomatis serovar D Q820-K940) were tested in F-actin cosedimentation experiments (Fig. 2C). Similar to C. trachomatis L2 Tarp, the comparable protein domains within C. trachomatis serovar A and D Tarps were able to associate with filamentous actin (Fig. 2C).
Tarp harbors multiple F-actin binding sequences.
We identified a novel domain in the C-terminal region of the Tarp protein that specifically associated with F actin, now termed FAB1. This domain contains a 13-amino-acid peptide with similarity to the described ABD that is also predicted to form the alpha helix secondary structure required for actin binding. In order to demonstrate that ABD and FAB1 were the two protein domains responsible for colocalization with actin filaments, EGFP-Tarp harboring deletions of both the ABD and FAB1 were expressed in HeLa cells (Fig. 3). Surprisingly, extopically expressed EGFP-Tarp harboring deletions of both the ABD and FAB1 still colocalized with actin filaments, suggesting another actin binding domain was present in the C-terminal half of the Tarp open reading frame (Fig. 3). In order to identify the locations of additional actin binding sites, a series of C-terminal truncations were introduced into the EGFP-Tarp ABD and FAB1 double mutant (Fig. 3A). Truncation mutants that lacked amino acids 955 to 1005 and 943 to 1005 were no longer able to associate with actin filaments, suggesting an additional actin binding domain(s) may be located in this region of the protein (Fig. 3B). Further secondary-structure predictions of the C-terminal domain of Tarp revealed an additional alpha helix located between amino acids 942 and 967, although this peptide does not share sequence similarity with the ABD and FAB1 domains (data not shown). In order to determine if the ABD, FAB1, and/or the additional alpha helical domain was responsible for colocalization with filamentous actin in tissue culture cells, an EGFP fusion to Tarp lacking all three alpha helical sequences was tested for actin colocalization (Fig. 4). An EGFP-Tarp fusion harboring the single ABD deletion and an EGFP-Tarp fusion harboring a double mutant lacking the ABD and the FAB1 domain were also tested (Fig. 4A). As previously observed, the ΔABD mutant was able to colocalize with actin filaments, as was the double mutant harboring both ΔABD and ΔFAB1, supporting the possibility that the third alpha helix sequence is capable of promoting F-actin binding independently. Interestingly when all three sites were removed from Tarp, F-actin colocalization was no longer observed, suggesting that all three sites may serve to bind F actin (Fig. 4B). Thus, we have termed the last alpha helix F-actin binding domain 2 (FAB2). To confirm the EGFP colocalization results, GST fusions of Tarp (with the GST removed by PreScission protease treatment) harboring deletions in the ABD, FAB1, and FAB2 were used to biochemically examine cosedimentation of actin filaments with recombinant proteins. Consistent with the EGFP results, Tarp mutant proteins (with the GST removed) demonstrated an increased reduction in their ability to cosediment with preformed actin filaments as each domain of the protein was removed. This could be observed as both a reduction in the quantity of Tarp mutants fractioned to the pellet and an increase in the amounts of the proteins retained in the supernatant (Fig. 4C). Tarp proteins lacking all three actin binding sites showed the least F-actin binding (Fig. 4C).
Fig 3.
Tarp harbors a second site near its C terminus that distinctly binds F actin. (A) Schematics showing a series of truncated mutants of EGFP-Tarp ΔABD ΔFAB1 fusions having deletions in the C-terminal domain at increments of approximately 12 amino acids. The phosphorylation domain and proline-rich domain are represented by green and blue boxes, respectively. Δ indicates amino acids deleted in the mutant Tarp proteins, and the numbers indicate amino acid positions encoded within the C. trachomatis tarP gene. (B) EGFP-ΔABD ΔFAB1 Tarp mutants, as depicted in panel A, were ectopically expressed in HeLa cells and tested for the ability to form actin aggregates that colocalize with Alexa Fluor 568-conjugated phalloidin (Actin). EGFP-ΔABD ΔFAB1 Tarp mutants harboring C-terminal deletions between amino acids 966 and 1005 retained the ability to colocalize with phalloidin-stained actin, whereas EGFP-ΔABD ΔFAB1 Tarp mutants harboring larger C-terminal deletions that went beyond amino acid 966 of the Tarp protein did not colocalize with phalloidin-stained actin.
Fig 4.
Tarp's ability to bind filamentous actin is dependent on two F-actin binding domains and one F- and G-actin binding domain. Tarp deletion mutants expressed as either GST or EGFP fusions were examined for the ability to associate with filamentous actin. (A) Schematic of the GST- or EGFP-Tarp fusion proteins, indicating the locations of the actin binding domain (red box), the proline-rich domain (blue box), the tyrosine-rich phosphorylation domain (green boxes), and the newly characterized F-actin binding domains (yellow and purple boxes). Δ indicates amino acids deleted in the mutant Tarp proteins, and the numbers indicate amino acid positions encoded within the C. trachomatis tarP gene. (B) Tarp mutants ectopically expressed as enhanced green fluorescent protein fusions were examined for the ability to localize with actin filaments. Host cells expressing EGFP alone, EGFP fusions of full-length Tarp (Tarp), or single-deletion mutants lacking the actin binding domain (Tarp Δ ABD), a double-deletion mutant lacking the actin binding domain and first F-actin binding domain (Tarp Δ ABD Δ FAB1), or a triple-deletion mutant lacking the actin binding domain and both F-actin binding domains (Tarp Δ ABD Δ FAB1-2) were fixed and stained with Alexa Fluor 568-conjugated phalloidin (Actin). (C) GST-Tarp fusions (with the GST removed by PreScission Protease treatment) were incubated with F actin and isolated by ultracentrifugation. Protein supernatants and pellets were resolved by SDS-PAGE and visualized by Coomassie blue staining. GST alone is indicated by open circles, while the Tarp proteins with GST removed are indicated by asterisks. GST and Tarp differentially appear in the supernatant and/or pellet fraction following centrifugation. The actin protein is indicated by arrows. Tarp precipitated with F actin in the pellet fraction, as did the positive-control protein (α-actinin). Recombinant Tarp mutants harboring deletions in the ABD, FAB1, and FAB2 domains precipitated with F actin to a lesser extent than Tarp. The GST control did not associate with the F-actin pellet and remained predominately in the supernatant. The actin shown in the first lane represents 2% of the material used in the + F actin lanes. The molecular masses of protein standards are shown.
Tarp bundles actin filaments.
Tarp has previously been shown to function as an actin nucleator (11). The actin-nucleating activity was localized to a 200-amino-acid region of the Tarp protein sequence that was found to contain a proline-rich region responsible for protein oligomerization and a solitary actin binding domain (11). This actin binding domain was able to associate with monomeric and filamentous actin (11). In light of the identification of two additional F-actin binding domains, we sought to examine whether the Tarp protein was capable of bundling actin filaments (Fig. 5). Actin bundles sediment at a higher rate than actin filaments and monomeric actin. Therefore, proteins capable of bundling actin filaments will appear in the pellet upon low-speed centrifugation. Interestingly, Tarp functioned to bundle actin filaments (Fig. 5A); however, actin bundling was not dependent on Tarp-mediated actin nucleation, as the Tarp ΔPRD mutant, which fails to nucleate actin in vitro (Fig. 1), retained actin-bundling activity (Fig. 5B). The Tarp triple mutant lacking the ABD, FAB1, and FAB2 alpha helices was unable to bundle actin filaments, which is consistent with both the EGFP colocalization and F-actin cosedimentation results (Fig. 5A).
Fig 5.

Tarp bundles actin filaments. (A) Purified recombinant Tarp (with the GST removed by PreScission Protease treatment) were incubated with filamentous actin (F actin) and isolated by low-speed centrifugation. Protein supernatants (S) and pellets (P) were resolved by SDS-PAGE and visualized by Coomassie blue staining. GST alone is indicated by open circles, and recombinant Tarp proteins with the GST removed are indicated by asterisks. GST and Tarp differentially appear in the supernatant and/or pellet fraction following centrifugation. The actin protein is indicated by arrows. Tarp associated with actin filaments to form actin bundles, which appeared in the pellet fraction, as did the positive actin-bundling control protein (α-actinin). Neither the Tarp mutant, harboring three actin binding domain deletions, nor the GST control associate with actin and remained predominately in the F-actin supernatant. The molecular masses of protein standards are shown. (B) An experiment similar to that in panel A was performed using a Tarp mutant harboring a proline-rich domain deletion (Tarp Δ PRD). Purified Tarp Δ PRD (with GST removed) retained the ability to cosediment with bundled F actin.
DISCUSSION
The Tarp effector is a multifunctional protein that primes the host cell for bacterial entry and residence. We now demonstrate that, in addition to the previously characterized G-actin binding/nucleating domain (11), the Tarp protein harbors two distinct F-actin binding/bundling domains (FAB1 and -2). All three domains are similar in that they mediate a direct link to the host cytoskeleton, yet biochemically, they are discrete sites that specifically associate with globular or filamentous actin. A comparison of Tarp orthologs indicates that the FAB1 and FAB2 sites are conserved among serovars of C. trachomatis and may also be present in C. caviae and C. muridarum. F-actin-specific binding sites might also be found in other species, such as C. pneumoniae, but will have to be located biochemically, as sequences of some Tarp orthologs are more divergent, making domain comparisons more difficult to predict.
EB attachment to the surface of an epithelial cell ultimately results in the formation of an actin-rich pedestal at the site of contact and is associated with bacterial invasion (7). Actin filament-destabilizing drugs, such as cytochalasin D, inhibit the formation of these projections and subsequent uptake of C. trachomatis (7). The arrangement of the actin filaments within the pedestal is unknown, but presumably, the actin filaments form polarized actin bundles (actin filaments sharing the same orientation with respect to their barbed [+] and pointed [−] ends) similar to those characterized in microvilli and filopodia (18). Actin-bundling proteins, such as fascin 1, colocalize with filopodia on the leading edge of the growth cones of developing nerve cells and are implicated in the formation of actin bundles (19). Similarly, Tarp may play a role in the formation of actin bundles located directly beneath the invading microbe. C. trachomatis entry into host cells in vitro is temperature dependent and involves the recruitment of actin to the site of EB attachment (7). Once internalized, the recruited actin quickly disseminates. The molecular details of actin disassembly are not well defined, but the process may involve the translocation of the chlamydial effector CT694 (10). CT694 associates with the human AHNAK protein, and ectopic expression of CT694 in HeLa cells is associated with a reduction in stress fibers (10). It is interesting to speculate that actin depolymerization may drive EB entry, and the recent examination of chlamydial invasion in the presence of actin filament-stabilizing drugs, such as jasplakinolide (Jas), supports this hypothesis, as Jas was found to inhibit EB entry (14, 20). However, the effects of Jas on actin filaments in vivo is controversial, as changes in cell morphology that are consistent with a reduction in filamentous actin are observed in some Jas-treated cells (21).
Similar to Tarp, the Salmonella enterica serovar Typhimurium SipC effector is able to nucleate actin and to bundle actin filaments (22). Recently, mutant bacteria lacking the C-terminal region of SipC responsible for F-actin binding and bundling were found to be less invasive than wild-type Salmonella, suggesting that the bundling activity of SipC plays a role in pathogen entry into HeLa cells (23). Whether Tarp's ability to bundle actin filaments also contributes to pathogen entry is unknown but worthy of investigation, as new molecular tools continue to be developed to examine the genetic requirements of C. trachomatis pathogenicity (24, 25).
Actin bundles are tightly controlled by a variety of actin binding proteins (ABPs) that drive specific cytoskeletal processes and result in actin assemblies of defined thickness, length, and organization. The architecture of Tarp-mediated actin bundles has not been investigated, and it will be intriguing to compare it to the structure of actin bundles formed in the presence of other bacterial effectors, such as SipC. It is possible that evolutionarily divergent bacterial species employ unique effector proteins to create similar actin bundles for the same purpose of host cell invasion.
The phosphorylation of bacterial effector proteins by host cell kinases is implicated in the pathogenesis of multiple microorganisms (26). C. trachomatis Tarp orthologs are phosphorylated in the mammalian host by a variety of tyrosine kinases, including Src family kinases, Abl, and Syk (27, 28). SH2 domains found in the host proteome specifically associate with phosphotyrosine-containing polypeptides and are potential Tarp-interacting proteins (29). Recent studies have revealed the ability of phosphorylated Tarp to interact with SHC-1 and PI3K (16, 17). Tarp binding to SHC-1 is believed to alter host cell signal transduction cascades that favor host cell survival during early chlamydial development (17). The significance of Tarp's association with PI3K still requires further investigation, although C. trachomatis infection of host cells has been found to lead to PI3K activation, and this activation is thought to contribute to host cell survival (30). We have previously reported that Tarp phosphorylation in vitro does not alter actin polymerization kinetics (14). The data presented in this report further corroborate the model in which Tarp appears to be divided into two separate functional parts that reside within each half of the Tarp polypeptide. Tarp phosphorylation and subsequent association with host cell signaling and adapter proteins is restricted to the amino-terminal half of the protein, whereas Tarp-mediated actin nucleation and actin filament binding is restricted to the carboxyl-terminal half of the Tarp protein. The report further defines the biochemical bridge between the Tarp effector and the cytoskeleton of the host cell by examining the two newly characterized FAB domains. These findings attribute a novel activity to the critical Tarp protein and provide molecular insight into the complex cytoskeletal rearrangements required for C. trachomatis entry into host cells.
ACKNOWLEDGMENTS
We thank Mollie Jewett and Ted Hackstadt for careful review of the manuscript and acknowledge the technical assistance of Talia Chavez.
This work was supported by the NIAID, NIH, K award 5K22AI81729-2 and a University of Central Florida grant to T.J.J.
Footnotes
Published ahead of print 30 November 2012
REFERENCES
- 1. CDC 2011. CDC Grand Rounds. Chlamydia prevention: challenges and strategies for reducing disease burden and sequelae. MMWR Morb. Mortal. Wkly. Rep. 60:370–373 [PubMed] [Google Scholar]
- 2. Wright HR, Turner A, Taylor HR. 2008. Trachoma. Lancet 371:1945–1954 [DOI] [PubMed] [Google Scholar]
- 3. Moulder JW. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55:143–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Betts-Hampikian HJ, Fields KA. 2010. The chlamydial type III secretion mechanism: revealing cracks in a tough nut. Front. Microbiol. 1:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saka HA, Thompson JW, Chen YS, Kumar Y, Dubois LG, Moseley MA, Valdivia RH. 2011. Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms. Mol. Microbiol. 82:1185–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hybiske K, Stephens RS. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. U. S. A. 104:11430–11435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carabeo RA, Grieshaber SS, Fischer E, Hackstadt T. 2002. Chlamydia trachomatis induces remodeling of the actin cytoskeleton during attachment and entry into HeLa cells. Infect. Immun. 70:3793–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haglund CM, Welch MD. 2011. Pathogens and polymers: microbe-host interactions illuminate the cytoskeleton. J. Cell Biol. 195:7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. U. S. A. 101:10166–10171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hower S, Wolf K, Fields KA. 2009. Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol. Microbiol. 72:1423–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jewett TJ, Fischer ER, Mead DJ, Hackstadt T. 2006. Chlamydial TARP is a bacterial nucleator of actin. Proc. Natl. Acad. Sci. U. S. A. 103:15599–15604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carabeo RA, Dooley CA, Grieshaber SS, Hackstadt T. 2007. Rac interacts with Abi-1 and WAVE2 to promote an Arp2/3-dependent actin recruitment during chlamydial invasion. Cell Microbiol. 9:2278–2288 [DOI] [PubMed] [Google Scholar]
- 13. Jewett TJ, Miller NJ, Dooley CA, Hackstadt T. 2010. The conserved Tarp actin binding domain is important for chlamydial invasion. PLoS Pathog. 6:e1000997 doi:10.1371/journal.ppat.1000997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiwani S, Ohr RJ, Fischer ER, Hackstadt T, Alvarado S, Romero A, Jewett TJ. 2012. Chlamydia trachomatis Tarp cooperates with the Arp2/3 complex to increase the rate of actin polymerization. Biochem. Biophys. Res. Commun. 420:816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clifton DR, Dooley CA, Grieshaber SS, Carabeo RA, Fields KA, Hackstadt T. 2005. Tyrosine phosphorylation of the chlamydial effector protein Tarp is species specific and not required for recruitment of actin. Infect. Immun. 73:3860–3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lane BJ, Mutchler C, Al Khodor S, Grieshaber SS, Carabeo RA. 2008. Chlamydial entry involves TARP binding of guanine nucleotide exchange factors. PLoS Pathog. 4:e1000014 doi:10.1371/journal.ppat.1000014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mehlitz A, Banhart S, Maurer AP, Kaushansky A, Gordus AG, Zielecki J, Macbeath G, Meyer TF. 2010. Tarp regulates early Chlamydia-induced host cell survival through interactions with the human adaptor protein SHC1. J. Cell Biol. 190:143–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brown JW, McKnight CJ. 2010. Molecular model of the microvillar cytoskeleton and organization of the brush border. PLoS One 5:e9406 doi:10.1371/journal.pone.0009406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Edwards RA, Bryan J. 1995. Fascins, a family of actin bundling proteins. Cell Motil. Cytoskeleton 32:1–9 [DOI] [PubMed] [Google Scholar]
- 20. Carabeo R. 2011. Bacterial subversion of host actin dynamics at the plasma membrane. Cell Microbiol. 13:1460–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bubb MR, Spector I, Beyer BB, Fosen KM. 2000. Effects of jasplakinolide on the kinetics of actin polymerization. An explanation for certain in vivo observations. J. Biol. Chem. 275:5163–5170 [DOI] [PubMed] [Google Scholar]
- 22. Hayward RD, Koronakis V. 1999. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 18:4926–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Myeni SK, Zhou D. 2010. The C terminus of SipC binds and bundles F-actin to promote Salmonella invasion. J. Biol. Chem. 285:13357–13363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kari L, Goheen MM, Randall LB, Taylor LD, Carlson JH, Whitmire WM, Virok D, Rajaram K, Endresz V, McClarty G, Nelson DE, Caldwell HD. 2011. Generation of targeted Chlamydia trachomatis null mutants. Proc. Natl. Acad. Sci. U. S. A. 108:7189–7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. 2011. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 7:e1002258 doi:10.1371/journal.ppat.1002258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Selbach M, Paul FE, Brandt S, Guye P, Daumke O, Backert S, Dehio C, Mann M. 2009. Host cell interactome of tyrosine-phosphorylated bacterial proteins. Cell Host Microbe 5:397–403 [DOI] [PubMed] [Google Scholar]
- 27. Jewett TJ, Dooley CA, Mead DJ, Hackstadt T. 2008. Chlamydia trachomatis Tarp is phosphorylated by src family tyrosine kinases. Biochem. Biophys. Res. Commun. 371:339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mehlitz A, Banhart S, Hess S, Selbach M, Meyer TF. 2008. Complex kinase requirements for Chlamydia trachomatis Tarp phosphorylation. FEMS Microbiol. Lett. 289:233–240 [DOI] [PubMed] [Google Scholar]
- 29. Koch CA, Anderson D, Moran MF, Ellis C, Pawson T. 1991. SH2 and SH3 domains: elements that control interactions of cytoplasmic signaling proteins. Science 252:668–674 [DOI] [PubMed] [Google Scholar]
- 30. Verbeke P, Welter-Stahl L, Ying S, Hansen J, Hacker G, Darville T, Ojcius DM. 2006. Recruitment of BAD by the Chlamydia trachomatis vacuole correlates with host-cell survival. PLoS Pathog. 2:e45 doi:10.1371/journal.ppat.0020045 [DOI] [PMC free article] [PubMed] [Google Scholar]