Abstract
Bacteriophage VP4 is a lytic phage of the Vibrio cholerae serogroup O1, and it is used in phage subtyping of V. cholerae biotype El Tor. Studies of phage infection mechanisms will promote the understanding of the basis of phage subtyping as well as the genetic differences between sensitive and resistant strains. In this study, we investigated the receptor that phage VP4 uses to bind to El Tor strains of V. cholerae and found that it infects strains through adsorbing the O antigen of V. cholerae O1. In some natural isolates that are resistant to VP4 infection, mutations were identified in the wb* cluster (O-antigen gene cluster), which is responsible for the biosynthesis of O antigen. Mutations in the manB, wbeE, and wbeU genes caused failure of adsorption of VP4 to these strains, whereas the observed amino acid residue mutations within wbeW and manC have no effect on VP4 infection. Additionally, although mutations in two resistant strains were found only in manB and wbeW, complementing both genes did not restore sensitivity to VP4 infection, suggesting that other resistance mechanisms may exist. Therefore, the mechanism of VP4 infection may provide a basis for subtyping the phage. Elaborate mutations of the O antigen may imbue V. cholerae strains with resistance to phage infection.
INTRODUCTION
Bacteriophages are found everywhere in the biosphere, and they outnumber prokaryotic cells by an estimated 10-fold (1). A specialized phage can infect only a narrow range of hosts, so phage typing schemes are used in epidemiological studies of many bacterial pathogens, such as Staphylococcus aureus (2), Salmonella enterica serovar Typhimurium (3), Vibrio cholerae (4, 5), and Mycobacterium tuberculosis (6–8). V. cholerae is the causative agent of the diarrheal disease cholera. Among the more than 208 O-antigen serogroups of V. cholerae, only the O1 and O139 serogroups have been found to cause epidemic cholera (9). The O1 serogroup is divided into the main serotypes Inaba and Ogawa and two distinct biotypes, designated classical and El Tor (10). Phage typing, as well as some other molecular subtyping methods, is also used for V. cholerae. A phage biotyping scheme has been used to subtype the Chinese O1 El Tor strains for 40 years (11), and it has played an important role in surveillance and source tracing of cholera. In the phage biotyping scheme, El Tor strains from patients in epidemics and from environmental samples during nonepidemic periods present different types and are therefore classified into “epidemic strains” and “nonepidemic strains” (11).
The study of phage typing mechanisms with various phage types may help to reveal genetic differences among the different bacterial strains, enhancing surveillance of the emergence of new phage-resistant clones in the environment and in epidemics (12, 13). In the natural environment, the phage-bacterium interaction promotes coevolution of both. The mutations in bacteria that lead to phage resistance allow these cells to dodge the phage attacks and have a survival advantage in their ecological niches. The mechanisms that bacteria use to avoid infection have hitherto been found in different steps of phage infection, i.e., adsorption resistance, prevention of host takeover, and abortive infection (14), which occur in the life cycles of phage adsorption, gene replication, and release, respectively (14, 15).
Adsorption to the host cell is the first step in the life cycle of phages. In this process, receptor-binding proteins (RBPs) (16) of the phage interact specifically with the receptors on the surface of host cells. Different bacterial surface components have been identified as the receptors for phages, including flagella (17), pili (18), lipopolysaccharides (LPS), capsular polysaccharides (19), teichoic acids (20), and outer member proteins such as OmpA (21), OmpC (22), LamB (23), FepA (24), and TonA (25). LPS are common phage receptors in Gram-negative bacteria; T phage, especially T3, T4, and T7, adsorb LPS on the surface of Shigella (26) and Escherichia (27, 28). Salmonella phages ϕX174 (29) and P22 (30), Klebsiella phage FC3-10 (31), and Yersinia enterocolitica phage ΦYeO3-12 (32) also use LPS as receptors. More recently, the Hep/Glc-Kdo/Ko region of Yersinia pestis and Yersinia pseudotuberculosis LPS have been identified as the receptor for phage ΦA1122 (33), the core oligosaccharide (OS) region of LPS was found to be necessary for binding of V. cholerae typing phage VP3 (34), and the O side chain was found to serve as the receptor for temperate phage CP-T1 (35). In addition, recent studies have reported some novel receptors. The phase-variable O-methyl phosphoramidate moiety, which is a common component of the diverse capsular polysaccharides of C. jejuni, has been identified as a receptor for phages (36). OmpLC was also identified as a receptor for phage infecting Edwardsiella ictaluri, and the extracellular loop 8 of OmpLC was predicted to be vital to the process of adsorption (37). Some phages use at least two receptors on the cell envelope (38, 39). Phage SPP1 of Bacillus subtilis interacts with the glucosylated wall teichoic acids and the membrane protein YueB (40).
The phage biotyping scheme includes five phages and some additional biochemical tests (11). VP4 is one of the five typing phages. In this study, we sought to investigate the receptor of VP4 and to understand the typing mechanism of VP4 from the perspective of receptor gene differences. The O side chain of LPS was identified as the VP4 receptor. In addition, some mutations in the wb* cluster (the O-antigen gene cluster that consists of open reading frames [ORFs] between VC0240 and VC0264 in El Tor strain N16961) of the natural strains confer resistance to VP4 infection.
MATERIALS AND METHODS
Bacterial strains, phage, plasmids, and culture conditions.
The phage, bacterial strains, and plasmids used in this work are described in Table 1. Phage VP4 was propagated on host strain 919c. The El Tor strain N16961 for which the whole genome has been sequenced (41), is sensitive to VP4. N16961-Sm, which is resistant to streptomycin (Sm) and sensitive to phage VP4, was selected by plating N16961 on Luria broth (LB) agar with 100 μg/ml of Sm. This strain was used in the conjugation test and was distinguished from Escherichia coli by its resistance to Sm. Unless otherwise stated, all strains were grown at 37°C in liquid or on solid (15 g/liter agar) LB medium, which could be supplemented with 100 μg/ml of kanamycin (Kan), 100 μg/ml of Sm, 10 μg/ml of chloramphenicol (Cm), or 100 μg/ml of ampicillin (Amp).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| N16961 | O1 El Tor, Inaba | Laboratory stock |
| N16961-Sm | Spontaneous mutant of N16961; Smr | This study |
| 919C | O1 El Tor | Laboratory stock |
| V07-16 | O1 El Tor, Inaba | Laboratory stock |
| 367 | O1 El Tor, Ogawa | Laboratory stock |
| V06-92 | O1 El Tor, Ogawa | Laboratory stock |
| 465 | O1 El Tor, Inaba | Laboratory stock |
| 470 | O1 El Tor, Ogawa | Laboratory stock |
| 417 | O1 El Tor, Inaba | Laboratory stock |
| 136 | O1 El Tor, Ogawa | Laboratory stock |
| 427-2A1 | N16961-Sm,vc0241::Tn, Smr Kanr VP4r | This study |
| 514-2G8 | N16961-Sm,vc0241::Tn, Smr Kanr VP4r | This study |
| 514-3F2 | N16961-Sm,vc0251::Tn, Smr Kanr VP4r | This study |
| 513-2E4 | N16961-Sm,vc0251::Tn, Smr Kanr VP4r | This study |
| 516-8B8 | N16961-Sm,vc0242::Tn, Smr Kanr VP4r | This study |
| 506-1C4 | N16961-Sm,vc0245::Tn, Smr Kanr VP4r | This study |
| 514-4C2 | N16961-Sm,vc0243::Tn, Smr Kanr VP4r | This study |
| 427-2H5 | N16961-Sm,vc0260::Tn, Smr Kanr VP4r | This study |
| 506-2B11 | N16961-Sm,vc2417::Tn, Smr Kanr VP4r | This study |
| N16961-Sm ΔmanB | manB deletion of N16961-Sm | This study |
| N16961-Sm ΔwbeU | wbeU deletion of N16961-Sm | This study |
| N16961-Sm ΔmanB/CmanB | manB complementation of N16961-Sm ΔmanB | This study |
| N16961-Sm ΔwbeU/CwbeU | wbeU complementation of N16961-Sm ΔwbeU | This study |
| 95001 CwbeE/CwbeU | wbeE and wbeU complementation of 95001 | This study |
| 367 CmanB | manB complementation of 367 | This study |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-TC::Mu Km λpir | 47 |
| Plasmids | ||
| pSC123 | Suicide plasmid carrying transposon; Kanr Cmr | 9 |
| pWM91 | Suicide plasmid; oriR oriT lacZ tetAR sacB | 40 |
| pWM91Δ0242 | pWM91 carrying upstream and downstream fragments flanking manB | This study |
| pWM91Δ2417 | pWM91 carrying upstream and downstream fragments flanking recJ | This study |
| pCVD442 | Suicide plasmid; R6K ori mob RP4 bla sacB | 13 |
| pCVD442-Δvc0260 | pCVD442 carrying upstream and downstream fragments flanking wbeU | This study |
| pBR322 | Cloning vector; ColE1 ori Ampr Tetr | TaKaRa |
| pBR322-c0260 | pBr322 carrying wbeU | |
| pACYC184 | Cloning vector; rep Cmr Tcr | 6 |
| pACYC184-c0242 | pACYC184 carrying manB | This study |
| pACYC184-c0244 | pACYC184 carrying wbeE | This study |
| pACYC184-c0261 | pACYC184 carrying wbeW | This study |
| pUC18 | Cloning vector; ori lacZ Ampr | TaKaRa |
| pUC18-c0260 | pUC18 carrying wbeU | This study |
Construction of transposon insertion library and selection of VP4-resistant mutants.
Plasmid pSC123 (42) was transferred by conjugation from E. coli donor SM10λpir (43) into N16961-Sm, and transconjugants were selected by streptomycin and kanamycin resistance (Smr and Kanr). The resulting strains contained a chromosomal insertion caused by the integration of the plasmid, which carries Cm and Kan resistance genes. Single colonies were picked and incubated in 96-well plates until the optical density at 600 nm (OD600) reached 0.1 to 0.2. The cultures were then mixed with a VP4 phage suspension (1 × 108 PFU/ml) at a ratio of 20:1 to 30:1 in new 96-well plates and incubated for 3 h. Cultures of strain N16961 with and without VP4 were used as negative and positive controls, respectively. The wells with an OD600 significantly higher than that of the negative control and nearly as high as that of the positive control were selected as candidates for phage-resistant mutants. These candidates were subsequently tested using a double-layer plaque assay (44).
Arbitrary PCR.
To identify the transposon insertion site, arbitrary PCR (45, 46) was performed with two rounds. In the first round, portions of the chromosomal DNA preparations were used as templates, and primers ARB-1, ARB-6, and 123-3 (Table 2) were used in amplification. PCR was performed under the following conditions: 95°C for 5 min; six cycles of 94°C for 30 s, 30°C for 30 s, and 72°C for 1 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and finally 72°C for 5 min. The second round of PCR was performed with a portion of the PCR product of round 1 as the template, and primers 123-4 and ARB-2 (Table 2) were used. PCR was performed under the following conditions: 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by 72°C for 5 min. Amplicons were sequenced using the primer 123-4.
Table 2.
Primers used in the study
| Use and primer name | Sequence | Restriction site |
|---|---|---|
| Mutagenesis of VC0260 of N16961-Sm | ||
| pCVD442Δ0260F1 | CGCGTCGACTTTTACTCTTTCCGGACCGTC | SalI |
| pCVD442Δ0260R2 | ATGTTTACCCCTATTTCGCCACAAAAATGA | |
| pCVD442Δ0260F3 | TCATTTTTGTGGCGAAATAGGGGTAAACAT | |
| pCVD442Δ0260R4 | GCGGCATGCTTAGCCTTAACCCCTTCG | SphI |
| InnerU-F | GATGGCTAGTGCCTTCCACATTCTC | |
| InnerU-R | CTTTATTCCCCGCAGTGTCCCCTTTA | |
| Mutagenesis of VC0242 of N16961-Sm | ||
| pWM91Δ02421F | CCGCTCGAGAATTGTTATATCTACGCCCC | XhoI |
| pWM91Δ02422R | TTAAATATCCAATTTAGTTAACTCTTTCAC | |
| pWM91Δ02423F | GTGAAAGAGTTAACTAAATTGGATATTTAA | |
| pWM91Δ02424R | AAGGAAGCGGCCGCATACGCATACATCTTTGCCACA | NotI |
| Mutagenesis of recJ of N16961-Sm | ||
| pWM91ΔrecJ1F | CCGCTCGAGCTAGGCATGAATAGCTACCGT | XhoI |
| pWM91ΔrecJ2R | TTGGCCTGTCACAGTCTGGAAGCGAAATGA | |
| pWM91ΔrecJ3F | TCATTTCGCTTCCAGACTGTGACAGGCCAA | |
| pWM91ΔrecJ4R | ATAGCGGCCGCGCCACTCAACGCCAAGA | NotI |
| Construction of complementary plasmids | ||
| pACYC118-E-F | ACATGCATGCCTAAAGATTACGTTAAGATGCA | SphI |
| pACYC118-E-R | ACGCGTCGACTCAAATACAGTTGAAATAATTT | SalI |
| pACYC184-W-F | CGCGGATCCTTTGTTGCCAGCACCCCA | BamHI |
| pACYC184-W-R | ACATGCATGCTTATGGCTTAACACGATCAC | SphI |
| pBR322-U-F | CGCAAGCTTTACACCAGACTACGCA | HindIII |
| pBR322-U-R | CGCGGATCCTCATTTTTGTGGCGA | BamHI |
| pBR322-B-F | ATAGGATCCGCTTCCGTGTTCTTA | BamHI |
| pBR322-B-R | CGCGTCGACTACTTCATACCCTTTTTC | SalI |
| pUC18-U-F | CGGGGTACCTACACCAGACTACGCA | KpnI |
| pUC18-U-R | CGCGGATCCTCATTTTTGTGGCGA | BamHI |
| Arbitrary PCR | ||
| ARB-1 | GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT | |
| ARB-6 | GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC | |
| ARB-2 | GGCCACGCGTCGACTAGTAC | |
| 123-3 | ACGCACTGAGAAGCCCTTAG | |
| 123-4 | GCCCGGGAATCATTTGAAGG | |
| Sequencing of wb∗ cluster | ||
| wbe-1F | ATAATTAAGATGGCTATAAACTACC | |
| wbe-1R | ATTGGGTTATGACTTGCGGTA | |
| wbe-2F | CAGATAAAACAGTTGTGGTAGGTG | |
| wbe-2R | GGGATTCGTGGTTAAAAAGTAT | |
| wbe-3F | ACTAAGTTTTATCAAGCCTCAA | |
| wbe-3R | GTTGCAAATTTCTTTTACTTGATCA | |
| wbe-4F | ATGGGTTGATGACCTTCTTAGA | |
| wbe-4R | AATAATCATCTACTCCCATTCTCCA | |
| wbe-5F | TTGGGAATGAAAATGCTTAAAGA | |
| wbe-5R | GCATCGATAATATCTTCAACAT | |
| wbe-6F | ATCTTATTGCAGGAATTCTTGA | |
| wbe-6R | CTTCAAGCTCTCCAATAACTCCA | |
| wbe-7F | ATGGCGTTAAAAGGTTTAGTAGC | |
| wbe-7R | ACATCATCAGGAGAAATACCCAAT | |
| wbe-8F | ATTTTTGGATTCACGTTTGTACTT | |
| wbe-8R | CACAGCGGTCCTCCTAGAGTTAC | |
| wbe-9F | CGACAGTGTACATGAAGATATCCA | |
| wbe-9R | CCTTTGACCGCCAATAAATAAT | |
| wbe-10F | GTGAACGCTCTTGCTACAGCA | |
| wbe-10R | ATCTTGAAAACGTTAAGCTAGTAG | |
| wbe-11F | CGCCATTGATGTTCACATAAGAT | |
| wbe-11R | AAATAGTTGCAGCACTTGGC | |
| wbe-12F | TTTTACGCCAGCTTCTATAGCT | |
| wbe-12R | CCGCCTCATATTTACTTAAGCC | |
| wbe-13F | GATGCCCTTAAAGCGTATCGTGA | |
| wbe-13R | CTTCCATGTTGTTTCTCCTGTTTTG |
Construction of mutants.
An in-frame wbeU deletion mutant of V. cholerae N16961-Sm was constructed by homologous recombination using the suicide plasmid pCVD442 (47). The deletion fragment inserted into pCVD442 was produced using overlap extension PCR. The ca. 700-bp regions upstream and downstream of the wbeU gene were amplified by PCR from N16961-Sm chromosomal DNA using primers pCVD442Δ0260F1/pCVD442Δ0260R2 and primers pCVD442Δ0260F3/pCVD442Δ0260R4 (Table 2), respectively. The two amplicons overlapped by 30 bp. The overlapping fragments were purified with the QIAquick gel extraction kit (catalog no. 28706; Qiagen, Germany) and mixed as templates to generate heteroduplexes that were extended by PCR (with primers pCVD442Δ0260F1 and pCVD442Δ0260R4). The fragment was then cloned into pCVD442 to generate plasmid pCVD442-Δ0260, which was conjugally transferred into N16961-Sm from the donor strain E. coli SM10λpir. Transconjugants were selected on LB agar with Cm and Amp. Clones were streaked onto LB agar with 10% sucrose and without NaCl at 22°C. Colonies from the sucrose selection medium were then streaked on LB agar plates with Amp. Failure to grow indicated that the plasmid was absent and a double crossover had occurred. Clones were screened by PCR with special primers innerU-F and innerU-R (Table 2). PCR-negative clones were amplified with primers pCVD442Δ0260F1/pCVD442Δ0260R4 (Table 2), producing amplicons of 1.4 kb, which are shorter than the approximately 3.3 kb for the wild type N16961-Sm. Sequencing confirmed the wbeU deletion mutants.
The suicide plasmid pWM91 (40) was used to construct a manB deletion mutant of N16961-Sm in a similar way. Primer pairs pWM91Δ02421F/pWM91Δ02422R and pWM91Δ02423F/pWM91Δ02424R (Table 2) were used to amplify the 5′ and 3′ flanking sequences of manB, respectively. Primers pWM91Δ02421F and pWM91Δ02424R (Table 2) were used to amplify the heteroduplexes produced by mixing the two PCR products. The resulting fragment was subsequently cloned into pWM91 to construct the plasmid pWM91Δ0242. The construction of the recJ gene deletion mutation with pWM91 was performed using a similar protocol.
Complementation of mutations.
Primers pBR322-U-F and pBR322-U-R, incorporating HindIII and BamHI restriction sites (Table 2), amplified a 2,666-bp region of the N16961 genome that included an 800-bp upstream sequence in addition to the ORF of wbeU. The amplicon was digested with BamHI and HindIII (New England BioLabs) and ligated into the BamHI- and HindIII-digested plasmid pBR322 (D3050; TaKaRa, Japan), producing plasmid pBR322-c0260, which was electroporated into the wbeU gene deletion mutant strain. The manB gene and the 800-bp upstream region were amplified with primers pBR322-B-F and pBR322-B-R and cloned into pBR322. This process created pBR322-c0251, which was electroporated into the manB gene deletion mutant and O1 El Tor strains (strain 367, strain v07-16, and strain v06-92). Using the same technique, we used pACYC184 (48) to construct plasmids pACYC184-c0244 and pACYC184-c0261 with primers pACYC184-E-F/pACYC184-E-R and pACYC184-W-F/pACYC184-W-R, respectively (Table 2). In addition, we constructed plasmid pUC18-c0260 from pUC18 using primers pUC18-U-F/pUC18-U-R (Table 2). The plasmids pACYC184-c0244 and pUC18-c0260 were transformed into strain 95001, and pACYC184-c0261 and pBR322-c0251 were transformed into strains V07-16 and V06-92 via electroporation; transformants were selected by Cm and Amp resistance. All the recombinant clones described above were confirmed by PCR and sequenced to ensure that they contained the correct insert sequence. The VP4 sensitivity of the complemented mutants was examined by a double-layer plaque assay and compared with that of its respective mutant strain containing the empty vector.
Phage adsorption assay.
Five hundred microliters of VP4 (approximately 107 PFU/ml) was mixed with 500 μl of bacterial culture (108 CFU/ml), incubated at 37°C for 10 min, and centrifuged at 10,000 × g for 3 min. The phage titer remaining in the supernatant was determined by a double-layer plaque assay. LB broth was mixed with the phage without the bacteria and used as a control. Each assay was performed in duplicate and repeated three times.
Adsorption of SYBR gold-stained phage VP4 on the surface of bacteria.
A phage lysate with a titer of at least 108 PFU/ml was mixed at 50,000:1 (vol/vol) with a SYBR gold nucleic acid gel stain stock solution (S11494; Invitrogen) and incubated for 30 min in the dark at room temperature. The mixture was filtered through 0.02-μm-pore-size filters (6809-5002; Waterman, Germany). The filters were washed with phosphate-buffered saline (PBS) three times and then backwashed with 1 ml of PBS to gather labeled phage. The gathered phage titer was 107 PFU/ml at a minimum. Bacterial cultures grown to an OD600 of 0.1 were mixed 1:1 with phage, incubated for approximately 10 min, and then centrifuged at 10,000 × g for 3 min. The precipitate was resuspended with 50 μl of PBS. Phage VP4 adsorbed on the surface of bacteria was examined with a fluorescence microscope (BX51; Olympus, Japan) at a magnification of ×1,000 with blue-light (450 to 490 nm) excitation. For observation by confocal laser scanning microscopy (CLSM) (FV500; Olympus, Japan), the sample was mixed with an equal volume of 0.5% agarose and dropped on a slide in 1-μl aliquots.
Antiserum agglutination assay.
Clones grown to mid-logarithmic phase on LB plates were picked up with a toothpick and mixed thoroughly into a drop of O1 antiserum (S&A Reagents Lab, Bangkok, Thailand). Bacteria expressing O1 antigen produced a granular agglutination within seconds, while O1-antigen-negative bacteria stayed in homogenous suspension.
LPS pattern analysis.
Strains N16961-Sm, N16961-Sm ΔwbeU, and N16961-Sm ΔmanB, as well as complemented strains N16961-Sm ΔwbeU/CwbeU and N16961-Sm ΔmanB/CmanB, were cultured in LB broth for 12 h. A 100-ml sample of each culture was used for LPS extraction as described previously (49). The purified LPS were separated on a sodium dodecyl sulfate-polyacrylamide gel (18.5 cm by 20.0 cm by 1.0 mm) containing 4% acrylamide in the stacking gel and 12% acrylamide in the separating gel. Electrophoresis was performed at 10 mA in the stacking gel and 30 mA in the separating gel until the tracking dye ran to the end of the gel. The gel was visualized by silver staining using the method described by Tsai and Frasch (50).
Sequencing of the wb* cluster in the naturally VP4-resistant and -sensitive strains.
Thirteen primer pairs (wbe-1F/wbe-1R to wbe-13F/wbe-13R) (Table 2) were designed to PCR amplify the wb* cluster. Each amplicon was approximately 1,800 bp in size and shared a 200-bp overlapping region with the adjacent amplicons. The wb* gene clusters of naturally VP4-resistant strains and some genes of VP4-sensitive strains were sequenced, including four resistant strains (strain 367, strain V07-16, strain V06-92, and strain 95001) and four sensitive strains (strain 465, strain 470, strain 417, and strain 136) isolated from two different areas at different times. Among these strains, only 95001 carries the cholera toxin genes. The sequence data were assembled using DNAStar SeqMan (version 5.01; DNASTAR Inc.) and aligned using BLAST with Vector NTI Advance 10 (version 10; Invitrogen).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the wb* gene clusters of strains 95001, V07-16, V06-92, and 367 are KC152957, KC152956, KC152955, and KC152954, respectively.
RESULTS
Transposon insertion in the wb* gene cluster and recJ cause resistance to VP4 infection.
In order to search for the possible gene(s) related to resistance to VP4 infection, we first screened a transposon insertion library which was generated by plasmid pSC123, containing approximately 12,000 mutants. Nine mutants (427-2A1, 427-2H5, 514-3F2, 514-2G8, 514-4C2, 516-8B8, 506-2B11, 506-1C4, and 513-2E4) (Table 1) showed growth rates when coincubated with phage VP4 that were comparable to that of the control strain (N16961-Sm) without VP4, suggesting that they were resistant to VP4 infection. The resistances were further confirmed by a double-layer plaque assay. Seven transposon insertion mutants were confirmed, and the insertion sites were identified using arbitrary PCR. The manC gene was interrupted in two mutants (427-2A1 and 514-2G8), and insertions in wbeN were found in mutant strains 514-3F2 and 513-2E4. Mutants 516-8B8, 506-1C4, 514-4C2, 427-2H5, and 506-2B11 had single insertions in the genes manB, wbeG, gmd, wbeU, and recJ, respectively (Fig. 1A).
Fig 1.
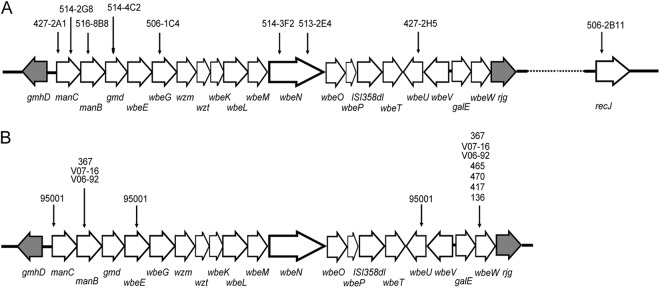
Mutations in the transposon insertion and VP4-resistant natural isolates. (A) Strains and transposon insertion sites in the wb* gene cluster and recJ gene. The large white arrows represent the wb* cluster, and the recJ gene and the transposon insertion positions are marked with small black arrows. The gmhD and the rjg junction genes are indicated by gray arrows. (B) Locations of mutations in the wb* gene clusters of VP4-resistant and -sensitive natural isolates. The large white arrows represent the wb* gene cluster, and the mutation sites are marked with small black arrows. The gmhD and the rjg junction genes are indicated by gray arrows.
Six of the genes from the screen (all except recJ) are located within the wb* cluster, which consists of ORFs between VC0240 (gmhD) and VC0264 (rjg) (41) in El Tor strain N16961 and is responsible for the biosynthesis of the O antigen. In this study, we focused on the wb* cluster. To further confirm the correlation between wb* gene mutations and VP4 infection resistance, the manB and wbeU genes were selected for in-frame deletion by homologous recombination (mediated by the suicide plasmid). These new manB and wbeU mutants lost sensitivity to VP4 infection as well. When complemented with the intact genes (manB and wbeU), these deletion mutants regained the VP4-sensitive phenotype.
Deletions of the wb* gene and transposon insertions cause LPS change and O-antigen loss.
Based on the results from the transposon mutation screen, we further constructed in-frame deletions of some wb* genes. The manB gene was assumed to be responsible for the biosynthesis of perosamine, which composes the backbone of the O antigen (51). Insertion mutations in the wbeU gene may cause O-antigen loss (52). The mutants N16961-Sm ΔwbeU and N16961-Sm ΔmanB showed a rough colony phenotype and were not agglutinated by anti-O1 antiserum (data not shown). When these strains were analyzed by LPS electrophoresis, as shown in Fig. 2, LPS from N16961-Sm ΔwbeU and N16961-Sm ΔmanB strains were missing the O-antigen ladder with the highest molecular weight (53). In contrast, when complemented with wbeU and manB, the corresponding deletion mutants had the smooth colony phenotype and agglutination with anti-O1 antiserum restored, and their LPS regained the same ladders as with N16961-Sm (Fig. 2). Similar O side chain loss was observed in the transposon mutant strains with insertions in manC, gmd, wbeG, and wbeN (data not shown).
Fig 2.
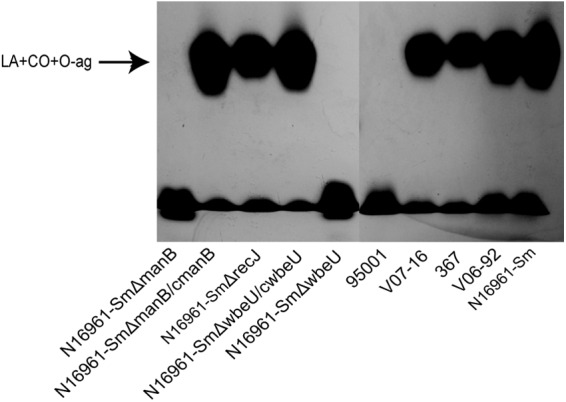
SDS-PAGE and silver staining of LPS from strain N16961-Sm and its mutants and the naturally VP4-resistant isolates. The top ladder indicates the integrated LPS. LA, lipid A; CO, core oligosaccharide; O-ag, O polysaccharide.
Loss of O antigen causes nonadsorption of VP4 to the mutants.
To observe the adsorption of VP4 to the wild-type strain and the mutants lacking O antigen, we performed quantitative phage adsorption assays and qualitatively observed labeled phage absorbing on the bacterial surfaces by microcopy and CLSM. The results are shown in Fig. 3. After the phages were incubated with N16961-Sm ΔwbeU and N16961-Sm ΔmanB, VP4 infection titers in the supernatants were similar to those after incubation with LB and much higher than those seen with strain N16961-Sm. When VP4 was incubated with the complemented strains of N16961-Sm ΔwbeU and N16961-Sm ΔmanB, its titers declined sharply, although they remained slightly higher than that with N16961-Sm. These data indicate that the deletion mutants are deficient for VP4 adsorption, while the complemented strains have the phage's ability to adsorb restored. In addition, when SYBR gold-stained phage VP4 was incubated with N16961-Sm and both of the complemented strains, the cells were coated with green spots visible under the fluorescence microscope (data not shown). A similar result was more clearly observed under CLSM, which showed VP4 adsorption on the surface of the wild-type strain and complemented strains of N16961-Sm ΔwbeU and N16961-Sm ΔmanB (Fig. 4). In contrast, no fluorescence was observed around the cells of the N16961-Sm ΔwbeU and N16961-Sm ΔmanB mutants (Fig. 4). All of these results suggest that the phage VP4 adheres to the sensitive strains by binding to the O antigen and using it as a cell surface receptor.
Fig 3.
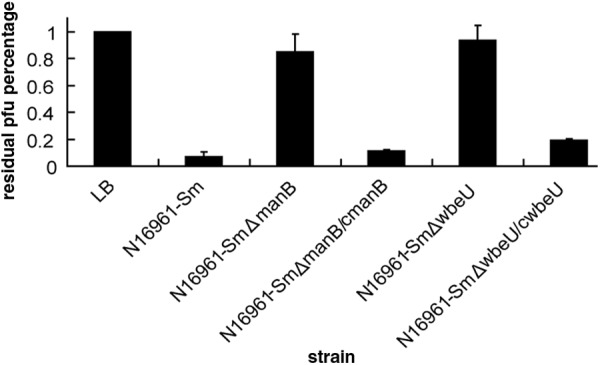
VP4 adsorption to manB and wbeU gene deletion mutants and complemented strains, shown as residual PFU percentages. LB was used as the no-bacterium control, and the phage titer in the control supernatant was set to 100%. Error bars indicate ranges.
Fig 4.
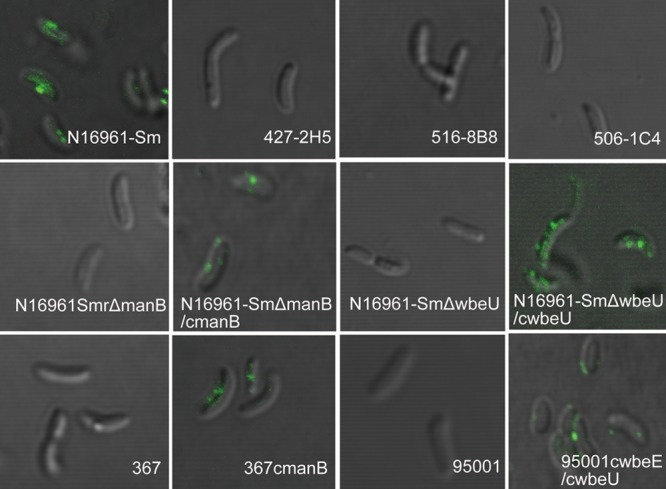
Binding of SYBR gold-labeled VP4 on the surfaces of different strains observed by CLSM.
Mutations in the wb* gene clusters of the naturally resistant isolates.
To investigate the mechanism by which natural V. cholerae strains are resistant to VP4 infection, four resistant strains (strain 367, strainV07-16, strain V06-92, and strain 95001) and four sensitive strains (strain 465, strain 470, strain 417, and strain 136) were selected for analysis. Their wb* gene clusters between gmhD and rjg were amplified to overlapping PCR fragments that were sequenced. Variances of tens of nucleotide were found in the wb* cluster for each strain, but most were silent mutations. With the exception of strain 95001, only the genes manB and wbeW had amino acid residue changes in these strains. However, all four sensitive strains had three amino acid residues that varied compared to those in the control strain N16961 (Table 3 and Fig. 1B), indicating that these residue changes have no effect on VP4 adsorption and infection. These residue changes in wbeW were also found in the four VP4-resistant strains that we analyzed. We did not pursue this gene any further.
Table 3.
Mutations and gene complementation of natural isolates resistant or sensitive to phage VP4
| Strain | Sensitivitya | Amino acid substitution in wb* cluster genes | Gene(s) used to complement | Sensitivity after complementation |
|---|---|---|---|---|
| 367 | R | manB, I72V, I77L, Y91H; wbeW, A13V, A118V, S122A | manB | Restored |
| V07-16 | R | manB, I72V, I77L, Y91H; wbeW, A13V, A118V, S122A | manB; manB+wbeW | Not restored |
| V06-92 | R | manB, I72V, I77L, Y91H; wbeW, A13V, A118V, S122A | manB; manB+wbeW | Not restored |
| 95001 | R | wbeU, 12-base fragment deletion of ntb 1670–1681; wbeE, single-nucleotide deletion of nt 393, codon TAA occurs in nt 413; manC, S54P | wbeE+wbeU | Restored |
| 465 | S | wbeW, A13V, A118V, S122A | ||
| 470 | S | wbeW, A13V, A118V, S122A | ||
| 417 | S | wbeW, A13V, A118V, S122A | ||
| 136 | S | wbeW, A13V, A118V, S122A |
R, resistant to VP4; S, sensitive to VP4.
nt, nucleotide.
The manB genes in strain 367, strain V07-16, and strain V06-92 had three amino acid residue changes in common compared to N16961 and the four assayed sensitive strains (Table 3 and Fig. 1B). CLSM showed that no fluorescence appeared around the cells of these strains when SYBR gold-stained VP4 phage were incubated with them (Fig. 4) (only strain 367 is presented as an example). When the plasmid pACYC184-c0242, which carries a copy of manB from N16961, was complemented into these strains, strain 367cmanB showed sensitivity to VP4 infection, and VP4 adsorption to this strain was also observed under CLSM (Fig. 4). Nevertheless, strains V07-16 and V06-92 that were complemented with pACYC184-c0242 still showed resistance to VP4 infection, suggesting that additional mutations that cause resistance to VP4 infection may be present in both strains.
In strain 95001, amino acid changes were found within genes manC, wbeE, and wbeU. One residue change was found in manC (Table 3 and Fig. 1B). A single-nucleotide deletion within wbeE resulted in a frameshift and truncated translation of this gene. Two independent single-nucleotide deletions and a 10-bp fragment deletion in wbeU caused frameshifts leading to truncated translation of this gene as well (Table 3 and Fig. 1B). No VP4 adsorption to 95001 was observed with CLSM (Fig. 4). We complemented genes wbeE and wbeU first with the individual gene plasmids and then with both. Individual complementation of either wbeE or wbeU did not convert 95001 to VP4 sensitivity, but when exogenous copies of both genes were provided, the resulting strain (95001 CwbeE/CwbeU) was sensitive to VP4, and VP4 adsorption to the surface of this strain was observed in the CLSM assay (Fig. 4). Wild-type strain 95001 is the rough type, but it became smooth when complemented with the combination of wbeE and wbeU. It could also be agglutinated with serogroup O1 antiserum, showing that its LPS structure could bind effectively to VP4 and the O1 serogroup antibody.
DISCUSSION
Phages have important roles in microbial genetics and evolution as well as applications in the diagnosis of disease, the treatment of infection, the food industry, and biotechnology. Phages are also used in bacterial subtyping because they are able to specifically recognize and infect their host bacteria. In this study, we found that V. cholerae El Tor typing phage VP4 uses the O antigen as its receptor, and we further found that some naturally VP4-resistant El Tor strains have variation in the O-antigen genes and cause failure of adsorption of VP4 to the host cell. These findings suggest that the resistance of these natural isolates to VP4 infection is related to some aspect of the receptor, possibly structure changes.
Some phages that infect V. cholerae and other bacterial species use LPS as their receptors. These glycosyl structures are the sites with which RBPs of phage interact. Genes within the wb* cluster are responsible for the biosynthesis of the O antigens. Through transposon mutations and in-frame deletions, we identified the wb* genes whose mutations had resulted in O-antigen changes and thereby caused failure of adsorption of phage VP4.
LPS chemically consists of O antigen, core oligosaccharide, and lipid A. The wb* gene cluster (and a newly recognized gene important for perosamine biosynthesis) and a complex pathway are required for O1 antigen synthesis (54). It could be speculated that some mutations in these genes may result in the changes or loss of O antigen, including the chemical group modification on the residues caused by some of the wb* gene mutations, and then the phages may fail to bind and infect the strains or may reduce the infection efficiencies. An example is the finding that O1 antigen variation mediated by three homonucleotide tracts in two genes in V. cholerae results in a lower molecular weight of O1 antigen and infection failure of O1-specific phage ICP1 afterwards, or lower ICP1 infection efficiency without macroscopic change in LPS pattern, possibly because of less O1 antigen-substituted LPS in such a case (54). In our study, the naturally VP4-resistant strain 367 is still of the O1 serogroup and has an O-antigen pattern similar to that of the VP4-sensitive O1 strain, and complementation of its mutant manB converted its sensitivity to VP4, suggesting that some group modifications of O antigen are responsible for the phage infection (binding) but do not affect recognition of O1 antiserum. In V. cholerae O1, serotypes Ogawa and Inaba differ by the presence of a 2-O-methyl group in the nonreducing terminal sugar of the Ogawa O antigen but its absence in that of Inaba (55). V. cholerae O1 strains sensitive to phage VP4 include both Ogawa and Inaba serotypes; therefore, the 2-O-methyl group may not be the binding site of VP4.
In these VP4-resistant and -sensitive strains, most of the observed nucleotide variation in the wb* cluster led to silent mutations, showing the conservation of O-antigen biosynthesis genes. In this study, manB gene mutations occurred in three wild-type VP4-resistant strains, preventing VP4 adsorption. manB which is predicted to produce phosphomannomutase, as well as manA, manC, manB, and gmd, is involved in GDP-perosamine biosynthesis (56, 57). The manB of the natural strain 367 has only three amino acid mutations but not the gene deletion; no obvious change was found in its LPS staining (Fig. 2), and this strain still agglutinates with O1 antiserum. These observations suggest that small modifications or other changes have occurred in its O antigen, but these changes still effectively block the binding of VP4. Additionally, amino acid mutations occurred in three O-antigen biosynthesis genes in resistant strain 95001, including obvious mutations in genes wbeE and wbeU. Restoration of both genes demonstrated that its VP4 resistance mechanism results from an O-antigen deficiency.
Phages specifically recognize and infect a narrow range of hosts. The resistance mechanisms of bacteria have evolved in the multiple steps of phage infection. These strategies include blocking receptors or altering receptor structure to prevent phage adsorption, preventing phage DNA entry, digesting phage nucleic acid, inhibiting replication of the phage genome, causing abortive phage infection, etc. (15). In this study, we identified receptor genes and another gene (recJ) in a V. cholerae O1 strain that are related to VP4 infection, also showing that multiple mechanisms have evolved to provide VP4 resistance. Two other strains, V07-16 and V06-92, have the same amino acid mutations within their wb* clusters as strain 367, and no obvious change was found in the LPS staining (Fig. 2), but complementation of manB did not restore the sensitivity to VP4 infection, suggesting that some other mechanism may be involved in VP4 resistance, possibly in a different step of infection. In fact, in addition to the O-antigen receptor genes, we found that recJ may also be related to VP4 infection, because complementing the transposon mutant of N16961 with intact recJ restored sensitivity to VP4 infection (data not shown). The recJ gene is recognized as being involved in base excision repair, mismatch repair, homologous recombination (58), and the rescue of arrested replication forks (59), and it is also the only 5′-3′ exonuclease specific for single-stranded DNA (ssDNA) in most prokaryotic species (60, 61). In Acinetobacter baylyi, the recJ DNase strongly suppresses short foreign DNA fragments from integrating into genomic DNA (62). The resistance may be related to its role in restriction of VP4 DNA. Further studies are necessary to clarify the VP4 resistance mechanism relevant to recJ.
Phage infection is host specific; different sensitivities to the phage of strains that belong to the same species and serogroups can be determined by their genetic differences, such as whether a receptor exists or not, different receptor structures of these strains, or other differences which play roles in multiple steps of phage infection. Understanding infection mechanisms of the typing phages can lead to the determination of genomic differences between phage-sensitive and -resistant strains. Additionally, the evolution of resistance mechanisms against phages may enhance the ability of bacteria to survive in various environments. The phage resistance may protect against phage predation in environment, emergence of new clones of pathogens, and subsequent epidemics. From the aspect of biology, studies have shown that phages in the environment may lead to the collapse of cholera epidemics because phage predation leads to the decline of the sensitive bacteria. Moreover, this process can also lead to the emergence of a new clone that is resistant to a phage (12, 13), although this change does not happen frequently in the wild (63). Therefore, it may be speculative to suggest that the strains that have VP4 resistance (similar to strain 367) have a stronger ability to survive in an environment containing VP4 phages.
In summary, we identified the O antigen as the receptor of the V. cholerae El Tor typing phage VP4, and we confirmed that mutations in the O-antigen biosynthesis genes were responsible for VP4 adsorption failure in some wild-type resistant strains. This may be due to interaction between and coevolution of phage and host bacteria. Moreover, the study of phage resistance mechanisms can also be a strategy employed to find genomic and functional genome differences between sensitive and resistant strains, not only differences in their receptors, but also differences in other functions for which the host bacteria have evolved, such as preventing phage DNA entry, blocking DNA replication, or causing abortive infection.
ACKNOWLEDGMENTS
This work was supported by the National Basic Research Priorities Program (2009CB522604) from the Ministry of Scientific Technology, China, and by the Priority Project on Infectious Disease Control and Prevention (2012ZX10004215 and 2008ZX10004-008) from the Ministry of Health, China.
Footnotes
Published ahead of print 7 December 2012
REFERENCES
- 1. Brüssow H, Hendrix RW. 2002. Phage genomics: small is beautiful. Cell 108: 13–16 [DOI] [PubMed] [Google Scholar]
- 2. Hood AM. 1953. Phage typing of Staphylococcus aureus. J. Hyg. (Lond.) 51: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pang S, Octavia S, Reeves PR, Wang Q, Gilbert GL, Sintchenko V, Lan R. 2012. Genetic relationships of phage types and single nucleotide polymorphism typing of Salmonella enterica serovar Typhimurium. J. Clin. Microbiol. 50: 727–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chakrabarti AK, Ghosh AN, Nair GB, Niyogi SK, Bhattacharya SK, Sarkar BL. 2000. Development and evaluation of a phage typing scheme for Vibrio cholerae O139. J. Clin. Microbiol. 38: 44–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chattopadhyay DJ, Sarkar BL, Ansari MQ, Chakrabarti BK, Roy MK, Ghosh AN, Pal SC. 1993. New phage typing scheme for Vibrio cholerae O1 biotype El Tor strains. J. Clin. Microbiol. 31: 1579–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jimenez Misas CA, Valdivia Alvarez JA, Mazon Zamora DM. 1989. Phage typing of Micobacterium tuberculosis in Cuba. Mem. Inst. Oswaldo Cruz 84: 271. [DOI] [PubMed] [Google Scholar]
- 7. Clavel-Sérès S, Clément F, Jimenez-Misas C. 1988. Distribution of bacteriophage types of Mycobacterium tuberculosis in France. Rev. Mal. Respir. 5: 577–581 [PubMed] [Google Scholar]
- 8. Jiménez Misas CA, Valdés Tejo E, Valdivia Alvarez JA, Ferrá Salazar C. 1992. Phage typing of Mycobacterium tuberculosis using the overlay technique. Preliminary study. Rev. Cubana. Med. Trop. 44: 189–192 [PubMed] [Google Scholar]
- 9. Kaper JB, Morris JG, Jr, Levine MM. 1995. Cholera. Clin. Microbiol. Rev. 8: 48–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feeley JC. 1965. Classification of Vibrio cholerae (Vibrio comma), including El Tor vibrios, by infrasubspecific characteristic. J. Bacteriol. 89: 665–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao S, Wu S, Liu B. 1984. Characteristics of typing phages of Vibrio cholerae biotype El Tor. Fu Huo Luan Zi Liao Hui Bian 4: 237–245 [Google Scholar]
- 12. Faruque SM, Islam MJ, Ahmad QS, Faruque AS, Sack DA, Nair GB, Mekalanos JJ. 2005. Self-limiting nature of seasonal cholera epidemics: role of host-mediated amplification of phage. Proc. Natl. Acad. Sci. U. S. A. 102: 6119–6124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jensen MA, Faruque SM, Mekalanos JJ, Levin BR. 2006. Modeling the role of bacteriophage in the control of cholera outbreaks. Proc. Natl. Acad. Sci. U. S. A. 103: 4652–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hyman P, Abedon ST. 2010. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 70: 218–248 [DOI] [PubMed] [Google Scholar]
- 15. Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8: 317–327 [DOI] [PubMed] [Google Scholar]
- 16. Tremblay DM, Tegoni M, Spinelli S, Campanacci V, Blangy S, Huyghe C, Desmyter A, Labrie S, Moineau S, Cambillau C. 2006. Receptor-binding protein of Lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site. J. Bacteriol. 188: 2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Merino S, Camprubi S, Tomas JM. 1990. Isolation and characterization of bacteriophage PM3 from Aeromonas hydrophila the bacterial receptor for which is the monopolar flagellum. FEMS Microbiol. Lett. 57: 277–282 [DOI] [PubMed] [Google Scholar]
- 18. Waldor MK, Mekalanos JJ. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272: 1910–1914 [DOI] [PubMed] [Google Scholar]
- 19. Chakrabarty AM, Niblack JF, Gunsalus IC. 1967. A phage-initiated polysaccharide depolymerase in Pseudomonas putida. Virology 32: 532–534 [DOI] [PubMed] [Google Scholar]
- 20. Lindberg AA. 1973. Bacteriophage receptors. Annu. Rev. Microbiol. 27: 205–241 [DOI] [PubMed] [Google Scholar]
- 21. Cole ST, Chen-Schmeisser U, Hindennach I, Henning U. 1983. Apparent bacteriophage-binding region of an Escherichia coli K-12 outer membrane protein. J. Bacteriol. 153: 581–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu SL, Ko KL, Chen CS, Chang YC, Syu WJ. 2000. Characterization of the distal tail fiber locus and determination of the receptor for phage AR1, which specifically infects Escherichia coli O157:H7. J. Bacteriol. 182: 5962–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hazelbauer GL. 1975. Role of the receptor for bacteriophage lambda in the functioning of the maltose chemoreceptor of Escherichia coli. J. Bacteriol. 124: 119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rabsch W, Ma L, Wiley G, Najar FZ, Kaserer W, Schuerch DW, Klebba JE, Roe BA, Laverde Gomez JA, Schallmey M, Newton SM, Klebba PE. 2007. FepA- and TonB-dependent bacteriophage H8: receptor binding and genomic sequence. J. Bacteriol. 189: 5658–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Menichi B, Buu A. 1983. Integration of the overproduced bacteriophage T5 receptor protein in the outer membrane of Escherichia coli. J. Bacteriol. 154: 130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jesaitis MA, Goebel WF. 1952. The chemical structure and antiviral properties of the somatic antigen of phaseII Shigella sonei. J. Exp. Med. 96: 409–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Michiael JG. 1968. The surface antigen and phage receptors in Escherichia coli. B. Proc. Soc. Exp. Biol. 128: 434–438 [DOI] [PubMed] [Google Scholar]
- 28. Weidel W. 1958. Bacterial viruses(with particular reference to adsorbtion/penetration). Annu. Rev. Microbiol. 12: 24–48 [DOI] [PubMed] [Google Scholar]
- 29. Lindberg AA, Hellerqvist CG. 1971. Bacteriophage attachment sites, serological specificity, and chemical composition of the lipopolysaccharides of semirough and rough mutants of Salmonella typhimurium. J. Bacteriol. 105: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zinder ND. 1958. Lysogenization and superinfection immunity in Salmonella. Virology 5: 291–326 [DOI] [PubMed] [Google Scholar]
- 31. Camprubí S, Merino S, Benedí VJ, Tomás JM. 1991. Isolation and characterization of bacteriophage FC3-10 from Klebsiella spp. FEMS Microbiol. Lett. 67: 291–297 [DOI] [PubMed] [Google Scholar]
- 32. Al-Hendy A, Toivanen P, Skurnik M. 1992. Lipopolysaccharide O side chain of Yersinia enterocolitica O:3 is an essential virulence factor in an orally infected murine model. Infect. Immun. 60: 870–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kiljunen S, Datta N, Dentovskaya SV, Anisimov AP, Knirel YA, Bengoechea JA, Holst O, Skurnik M. 2011. Identification of the lipopolysaccharide core of Yersinia pestis and Yersinia pseudotuberculosis as the receptor for bacteriophage ΦA1122. J. Bacteriol. 193: 4963–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang J, Li W, Zhang Q, Wang H, Xu X, Diao B, Zhang L, Kan B. 2009. The core oligosaccharide and thioredoxin of Vibrio cholerae are necessary for binding and propagation of its typing phage VP3. J. Bacteriol. 191: 2622–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guidolin A, Manning PA. 1985. Bacteriophage CP-T1 of Vibrio cholerae. Identification of the cell surface receptor. Eur. J. Biochem. 153: 89–94 [DOI] [PubMed] [Google Scholar]
- 36. Holst Sørensen MC, van Alphen LB, Harboe A, Li J, Christensen BB, Szymanski CM, Brøndsted L. 2011. Bacteriophage F336 recognizes the capsular phosphoramidate modification of Campylobacter jejuni NCTC11168. J. Bacteriol. 193: 6742–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hossain MJ, Rahman KS, Terhune JS, Liles MR. 2012. An outer membrane porin protein modulates phage susceptibility in Edwardsiella ictaluri. Microbiology 158: 474–487 [DOI] [PubMed] [Google Scholar]
- 38. Riechmann L, Holliger P. 1997. The C-terminal domain of TolA is the coreceptor for filamentous phage infection of E. coli. Cell 90: 351–360 [DOI] [PubMed] [Google Scholar]
- 39. Silverman JA, Benson SA. 1987. Bacteriophage K20 requires both the OmpF porin and lipopolysaccharide for receptor function. J. Bacteriol. 169: 4830–4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vinga I, Baptista C, Auzat I, Petipas I, Lurz R, Tavares P, Santos MA, Sao-Jose C. 2012. Role of bacteriophage SPP1 tail spike protein gp21 on host cell receptor binding and trigger of phage DNA ejection. Mol. Microbiol. 83: 289–303 [DOI] [PubMed] [Google Scholar]
- 41. Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406: 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chiang SL, Mekalanos JJ. 2000. Construction of a Vibrio cholerae vaccine candidate using transposon delivery and FLP recombinase-mediated excision. Infect. Immun. 68: 6391–6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1: 784–791 [Google Scholar]
- 44. Frost JA, Kramer JM, Gillanders SA. 1999. Phage typing of Campylobacter jejuni and Campylobacter coliand its use as an adjunct to serotyping. Epidemiol. Infect. 123: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Judson N, Mekalanos JJ. 2000. TnAraOut, a transposon-based approach to identify and characterize essential bacterial genes. Nat. Biotechnol. 18: 740–745 [DOI] [PubMed] [Google Scholar]
- 46. Tsou AM, Liu Z, Cai T, Zhu J. 2011. The VarS/VarA two-component system modulates the activity of the Vibrio cholerae quorum-sensing transcriptional regulator HapR. Microbiology 157: 1620–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Donnenberg MS, Kaper JB. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59: 4310–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chang AC, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134: 1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nesper J, Kapfhammer D, Klose KE, Merkert H, Reidl J. 2000. Characterization of Vibrio cholerae O1 antigen as the bacteriophage K139 receptor and identification of IS1004 insertions aborting O1 antigen biosynthesis. J. Bacteriol. 182: 5097–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tsai CM, Frasch CE. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119: 115–119 [DOI] [PubMed] [Google Scholar]
- 51. Chatterjee SN, Chaudhuri K. 2003. Lipopolysaccharides of Vibrio cholerae. I. physical and chemical characterization. Biochim. Biophys. Acta 1639: 65–79 [DOI] [PubMed] [Google Scholar]
- 52. Fallarino A, Mavrangelos C, Stroeher UH, Manning PA. 1997. Identification of additional genes required for O-antigen biosynthesis in Vibrio cholerae O1. J. Bacteriol. 179: 2147–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nesper J, Lauriano CM, Klose KE, Kapfhammer D, Kraiss A, Reidl J. 2001. Characterization of Vibrio cholerae O1 El Tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69: 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chatterjee SN, Chaudhuri K. 2004. Lipopolysaccharides of Vibrio cholerae II. genetics of biosynthesis. Biochim. Biophys. Acta 1690: 93–109 [DOI] [PubMed] [Google Scholar]
- 55. Wang J, Villeneuve S, Zhang J, Lei P, Miller CE, Lafaye P, Nato F, Szu SC, Karpas A, Bystricky S, Robbins JB, Kovác P, Fournier JM, Glaudemans CP. 1998. On the antigenic determinants of the lipopolysaccharides of Vibrio cholerae O:1, serotypes Ogawa and Inaba. J. Biol. Chem. 273: 2777–2783 [DOI] [PubMed] [Google Scholar]
- 56. Seed KD, Faruque SM, Mekalanos JJ, Calderwood SB, Qadri F, Camilli A. 2012. Phase variable O antigen biosynthetic genes control expression of the major protective antigen and bacteriophage receptor in Vibrio cholerae O1. PLoS Pathog. 8: e1002917 doi:10.1371/journal.ppat.1002917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stroeher UH, Karageorgos LE, Brown MH, Morona R, Manning PA. 1995. A putative pathway for perosamine biosynthesis is the first function encoded within the rfb region of Vibrio cholerae O1. Gene 166: 33–42 [DOI] [PubMed] [Google Scholar]
- 58. Han ES, Cooper DL, Persky NS, Sutera VJ, Whitaker RD, Montello ML, Lovett ST. 2006. RecJ exonuclease: substrates, products and interaction with SSB. Nucleic Acids Res. 34: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chow K, Courcelle J. 2007. RecBCD and RecJ/RecQ initiate DNA degradation on distinct substrates in UV-irradiated Escherichia coli. Radiat. Res. 168: 499–506 [DOI] [PubMed] [Google Scholar]
- 60. Lovett ST, Kolodner RD. 1989. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 86: 2627–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wakamatsu T, Kitamura Y, Kotera Y, Nakagawa N, Kuramitsu S, Masui R. 2010. Structure of RecJ exonuclease defines its specificity for single-stranded DNA. J. Biol. Chem. 285: 9762–9769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Harms K, Schön V, Kickstein E, Wackernagel W. 2007. The RecJ DNase strongly suppresses genomic integration of short but not long foreign DNA fragments by homology-facilitated illegitimate recombination during transformation of Acinetobacter baylyi. Mol. Microbiol. 64: 691–702 [DOI] [PubMed] [Google Scholar]
- 63. Zahid MS, Udden SM, Faruque AS, Calderwood SB, Mekalanos JJ, Faruque SM. 2008. Effect of phage on the infectivity of Vibrio cholerae and emergence of genetic variants. Infect. Immun. 76: 5266–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]


