Abstract
The Lyme disease spirochete controls production of its OspC and Erp outer surface proteins, repressing protein synthesis during colonization of vector ticks but increasing expression when those ticks feed on vertebrate hosts. Early studies found that the synthesis of OspC and Erps can be stimulated in culture by shifting the temperature from 23°C to 34°C, leading to a hypothesis that Borrelia burgdorferi senses environmental temperature to determine its location in the tick-mammal infectious cycle. However, borreliae cultured at 34°C divide several times faster than do those cultured at 23°C. We developed methods that disassociate bacterial growth rate and temperature, allowing a separate evaluation of each factor's impacts on B. burgdorferi gene and protein expression. Altogether, the data support a new paradigm that B. burgdorferi actually responds to changes in its own replication rate, not temperature per se, as the impetus to increase the expression of the OspC and Erp infection-associated proteins.
INTRODUCTION
Vector-borne pathogens, such as the Lyme disease spirochete, Borrelia burgdorferi, have overcome the challenges of persisting within two very different animal environments and possess mechanisms to efficiently transmit back and forth between vertebrate hosts and arthropod vectors. To facilitate this complex life-style, the bacterium produces specific proteins appropriate for each step in the infectious cycle. Considerable effort has been expended to identify the mechanisms by which B. burgdorferi senses its environment and accordingly regulates gene expression (1, 2). Such information both provides insight into pathogenic mechanisms and identifies new targets for preventative and curative therapies.
One of the first studies to delve into the mechanisms of B. burgdorferi gene regulation focused on OspC (outer surface protein C). The exact function of that protein remains to be elucidated, but it is necessary for the establishment of mammalian infection (3–10). In a landmark study in 1995, Schwan et al. demonstrated that synthesis of OspC is repressed in bacteria within unfed ticks, yet OspC production is induced as those ticks begin to feed (11). Furthermore, they showed that regulation of OspC synthesis can be recapitulated in culture: the protein is poorly expressed by bacteria cultured at 23°C but is abundantly expressed by bacteria that are first grown at 23°C, diluted into fresh medium, and then cultured at 34°C to 37°C (11).
Shortly thereafter, it was demonstrated that increasing the culture temperature from 23°C to 34°C enhances the production of several other antigenic proteins (12). Among these are a paralogous family of outer surface lipoproteins designated Erp (OspE/OspF-related proteins) (12–15). Erp proteins are expressed throughout vertebrate infection, adhere to various host factors, and appear to play roles in dissemination and colonization (16–25). As with ospC, erp transcription is repressed during tick colonization and induced during tick feeding and transmission (1, 2, 20, 26).
Since those initial reports, numerous studies have determined that a substantial number of B. burgdorferi genes can be regulated during cultivation by altering the incubation temperature (e.g., see references 2, 27, and 28). The premise behind examining culture temperature effects was that such changes were hypothesized to mimic conditions within a colonized tick: 23°C was thought to represent the ambient temperature of an unfed tick, and the change from 23°C to 34°C is comparable to that experienced by bacteria when a tick feeds on a warm-blooded animal (1, 11, 29, 30). As yet, there are no validated mechanisms by which B. burgdorferi can directly sense environmental temperature.
Increasing the culture temperature from 23°C to 34°C has profound effects on B. burgdorferi metabolism, a point that has largely been overlooked. Most notably, borreliae cultured at 34°C grow much faster than do those cultured at 23°C (12, 31). Several recent studies have linked B. burgdorferi metabolic activity with regulation of gene expression. As examples, acetate and the mevalonate pathway are involved in regulating the production of OspC and several other mammal-specific proteins (32, 33), while cellular concentrations of the erp antirepressor, EbfC, increase with the rate of bacterial replication (34, 35).
Prompted by those observations, we developed culture conditions that cause changes in bacterial growth rate independently of temperature. A new approach to studying the effects of a culture temperature shift was also employed. Results indicate that the rate of B. burgdorferi growth, rather than temperature, influences the expression of the OspC and Erp proteins.
MATERIALS AND METHODS
Bacteria and growth conditions.
All studies utilized B. burgdorferi strain B31-MI-16, an infectious clone of the sequenced type strain B31 (20). Unless otherwise stated, B. burgdorferi bacteria were grown in complete Barbour-Stoenner-Kelly II (BSK-II) medium, which contains 6% (vol/vol) rabbit serum (36, 37). All media were preconditioned by incubation at 23°C or 34°C for 24 h prior to inoculation. In our experience, Lyme disease spirochetes grow and divide at optimal rates in artificial media when incubated near 34°C.
Densities of bacterial cultures were determined by microscopic enumeration using a Petroff-Hausser counting chamber (38). At each time point, cultures were counted in quadruplicate, and an average and standard deviation from this mean were calculated.
Temperature shift experiments from 23°C to 34°C were performed as described previously (11, 12). Briefly, B. burgdorferi was first cultured to late exponential phase (approximately 108 bacteria/ml) in BSK-II medium at either 23°C or 34°C. An aliquot of such a culture was diluted 1:100 into fresh BSK-II medium and then incubated at 23°C. Upon this culture attaining late exponential phase, an aliquot was diluted 1:100 into fresh medium and then incubated at 34°C. Late-exponential-phase cultures of the constant 23°C and the 23°C-to-34°C-shifted bacteria were harvested for analyses.
The effects of culture medium composition were assessed by using essentially the same technique as described above. Bacteria were grown to late exponential phase at 34°C in an incomplete medium. An aliquot of that culture was then diluted 1:100 into fresh, complete BSK-II medium and then incubated at 34°C. Both cultures were harvested at late exponential phase.
Two deficient media were used, each of which contains suboptimal concentrations of one or more essential metabolites. Complete BSK-II medium contains 6% rabbit serum, which provides the bacteria with lipids. Experiments were conducted to determine a concentration of rabbit serum that impaired borrelial growth yet permitted replication at 34°C at a rate comparable to that seen in complete medium at 23°C. As described here, B. burgdorferi bacteria were found to divide at a greatly reduced rate when cultured at 34°C in medium that contains only 1.2% (vol/vol) rabbit serum. BSK-II medium is a very complex, rich medium. Trials were undertaken with various concentrations of BSK-II medium diluted with phosphate-buffered saline (PBS) to determine a composition that facilitated slow borrelial growth. Severely reduced division rates were observed at 34°C in medium consisting of 25% (vol/vol) BSK-II medium diluted in PBS and containing 6% (vol/vol) rabbit serum.
To further test the hypothesis that an increased borrelial growth rate, rather than actual temperature, is responsible for the enhanced production of OspC and Erp proteins, B. burgdorferi bacteria were moved from −80°C to 23°C in complete BSK-II medium. Such temperature shifts were performed by diluting bacteria that had been frozen for >30 days at −80°C into fresh BSK-II medium and then culturing to late exponential phase at 23°C. To produce the frozen bacteria, glycerol was added to mid-exponential-phase cultures at a final concentration of 25%, and aliquots were then placed in a −80°C freezer.
Protein expression analyses.
Cultures were harvested by centrifugation, washed at least twice with PBS, and then resuspended in PBS containing 1 mM phenylmethanesulfonyl fluoride (PMSF). Bacteria were lysed by immersion in boiling water for approximately 2 min. Total cellular proteins were separated by SDS-polyacrylamide gel electrophoresis. Specific proteins were detected by immunoblot analyses using previously described antibodies (34, 39–43). Murine monoclonal antibodies against B. burgdorferi RpoS were obtained from F. Yang (Indiana University, Indianapolis, IN) (43), and rat polyclonal antiserum against RpoS was obtained from M. Caimano and J. Radolf (University of Connecticut, Farmingdale, CT). The constitutively expressed FlaB protein served as a reference (12, 44). OspC, ErpA, ErpM, ErpY, BpaB, EbfC, OspA, and OspB immunoblot band intensities under each condition were quantified relative to the FlaB signal, using ImageJ (http://rsbweb.nih.gov/ij/).
Quantitative reverse-transcription PCR.
Total RNA was extracted from cultured B. burgdorferi cells, and transcript levels were determined by quantitative reverse transcription-PCR (Q-RT-PCR), as described previously by Miller (45). Oligonucleotide primers are described in Table 1 (14, 46, 47). Each analysis was performed in triplicate, and means and standard deviations were determined. Levels of mRNA for ospC, erpG, and rpoS were standardized to levels of the constitutively expressed flaB message. A two-tailed Student t test was used to calculate statistically significant differences between culture conditions. GraphPad Prism v.5 (GraphPad, La Jolla, CA) was used to generate all graphs.
Table 1.
Oligonucleotide primers used for Q-RT-PCR
| Transcript | Primer | Sequence (5′–3′) | Reference |
|---|---|---|---|
| ospC | OSPCF-7 | CAGGGAAAGATGCGAATACATCTGC | 46 |
| ospC | OSPCR-8 | TAAGCTAAAGCTAACAATGATCC | 46 |
| erpG | E-33 | TGCAAGATTGATGCG | 14 |
| erpG | E-34 | ATTTTGAGGCTCTGC | 14 |
| rpoS | 88-111 | CTTGCAGGACAAATACAAAGAGGC | 47 |
| rpoS | 245-220 | GCAGCTCTTATTAATCCCAAGTTGCC | 47 |
| flaB | FLA3 | GGGTCTCAAGCGTCTTGG | 46 |
| flaB | FLA4 | GAACCGGTGCAGCCTGAG | 46 |
RESULTS
Effects of culture medium composition on bacterial division rates.
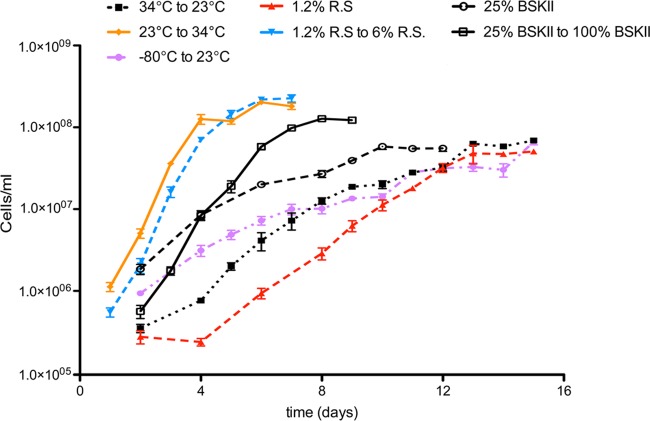
B. burgdorferi cells cultivated at 34°C in complete BSK-II medium with 6% rabbit serum divide approximately once every 12 h (Fig. 1). In contrast, bacteria grown in this medium at 23°C double approximately once every 32 h. We hypothesized that these differences in growth rate are responsible for the differences in B. burgdorferi expression patterns, independently of temperature. To test this prediction, culture media that lowered the rate of growth at 34°C were devised. The reduced genome of the obligately parasitic bacterium B. burgdorferi encodes very few anabolic enzymes, and the Lyme disease spirochete is an auxotroph for nearly all macromolecules (48). BSK-II medium is a very rich medium containing a mixture of defined and undefined ingredients (36, 37, 49). Complete BSK-II medium contains 6% rabbit serum, which provides sufficient fatty acids for optimal growth (36, 37, 49, 50). Reducing the amount of rabbit serum in the medium to 1.2% extended the doubling time to approximately 32 h (Fig. 1). Dilution of the components of BSK-II medium to one-quarter strength, with rabbit serum kept at 6%, impaired growth to approximately one doubling every 40 h (Fig. 1). These two media, designated “1.2% serum” and “25% BSK-II,” respectively, were then used for studies on the effects of the B. burgdorferi growth rate on gene and protein expression. To do so, bacteria were first cultured at 34°C in either 25% BSK-II or 1.2% serum medium and then diluted 1:100 into complete BSK-II medium and cultured at 34°C. All bacteria passaged in complete medium grew with a doubling time of approximately 12 h (Fig. 1). These culture medium formulations then permitted the following studies on the effects of different growth rates at a constant temperature.
Fig 1.
Effects of culture temperature and medium composition on B. burgdorferi growth. Culture densities were determined by enumerating cultured bacteria using a Petroff-Hausser counting chamber. Cultures were counted in quadruplicate at each time point. Culture conditions were as follows (unless otherwise noted, BSK-II medium contains 6% rabbit serum): 34°C to 23°C, grown to late exponential phase in BSK-II medium at 34°C, diluted into BSK-II medium, and then grown at 23°C; 23°C to 34°C, grown to late exponential phase in BSK-II medium at 23°C, diluted into BSK-II medium, and then grown at 34°C; −80°C to 23°C, stored at −80°C for >1 month, diluted into BSK-II medium, and then grown at 23°C; 1.2% R.S., grown to late exponential phase in BSK-II medium at 34°C, diluted into BSK-II medium plus 1.2% rabbit serum, and then grown at 34°C; 1.2% R.S. to 6% R.S., grown to late exponential phase in BSK-II medium plus 1.2% rabbit serum at 34°C, diluted into BSK-II medium, and then grown at 34°C; 25% BSK-II, grown to late exponential phase in BSK-II medium plus rabbit serum at 34°C, diluted into 25%-strength BSK-II medium, and then grown at 34°C; 25% BSK-II to 100% BSK-II, grown to late exponential phase in 25%-strength BSK-II medium at 34°C, diluted into full-strength BSK-II medium, and then grown at 34°C.
Effects of bacterial division rates on OspC and Erp protein and mRNA levels.
The borrelial ospC and erp genes are carried on distinct genetic elements and are regulated through different mechanisms (31, 51). The ospC locus is carried on a small circular replicon, cp26, while erp operons are located on episomal prophages named cp32s (13, 52–55). Three different Erp proteins were assayed, which are encoded by separate operons, each on a different cp32 prophage: ErpA, encoded by two identical erpAB operons, one each on cp32-1 and cp32-8; ErpM, encoded by erpLM on cp32-7; and ErpY, encoded by erpHY on cp32-4 (54). Previous analyses demonstrated that Erp protein levels are regulated at the level of transcription and that cellular erp mRNA levels are directly proportional to Erp protein levels (14). Rather than be repetitive, transcript levels of a fourth erp locus, erpG, encoded on cp32-3, were examined by Q-RT-PCR. To enable comparisons of the current studies with previous reports, steady-state protein and mRNA levels were determined by immunoblotting and Q-RT-PCR.
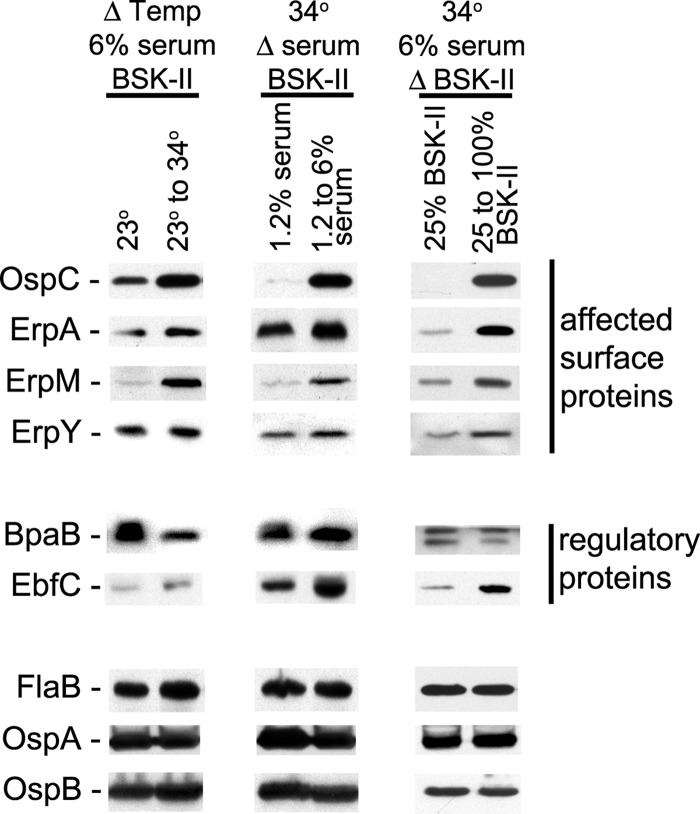
Temperature shift experiments recapitulated previously reported results (11, 12, 14), with B. burgdorferi cultivated at 23°C producing less OspC and Erp proteins than did bacteria shifted from 23°C to 34°C (Fig. 2). In the analysis illustrated in Fig. 2, OspC protein levels were 1.5-fold higher in the bacteria shifted from 23°C to 34°C than in the bacteria maintained at 23°C. As previously observed, relative expression levels of different Erp proteins varied between alleles, apparently due to differences in promoter and operator strengths and the proportional utilization of the housekeeping RpoD and alternative RpoS sigma factors (14, 26, 56–60). For the experiment illustrated in Fig. 2, bacteria shifted in temperature produced 1.4-fold more ErpA, 3.4-fold more ErpM, and a slight, but statistically insignificant, increase in the ErpY level. Q-RT-PCR analyses mirrored protein results, with significantly higher levels of ospC and erpG mRNAs being produced by bacteria shifted from 23°C to 34°C than by those maintained at 23°C (Fig. 3).
Fig 2.
Increases in B. burgdorferi growth rate correlate with increased production of OspC and Erp proteins. Effects of changing culture conditions on B. burgdorferi protein expression profiles were examined by immunoblotting. For each column, two conditions were kept constant, while a third was varied. For all studies, bacteria were first cultured to late exponential phase under a condition that impaired growth (complete BSK-II medium at 23°C, BSK-II medium containing only 1.2% rabbit serum at 34°C, or 25%-strength BSK-II medium with 6% serum at 34°C) and then diluted 1:100 into fresh, complete BSK-II medium and cultured at 34°C. All cultures were harvested at late exponential phase. The constitutively expressed flagellar component FlaB served as a control, and fold changes of other proteins were calculated relative to the FlaB band intensities (44). Illustrated data for each condition are from analyses of the same paired bacterial lysates.
Fig 3.
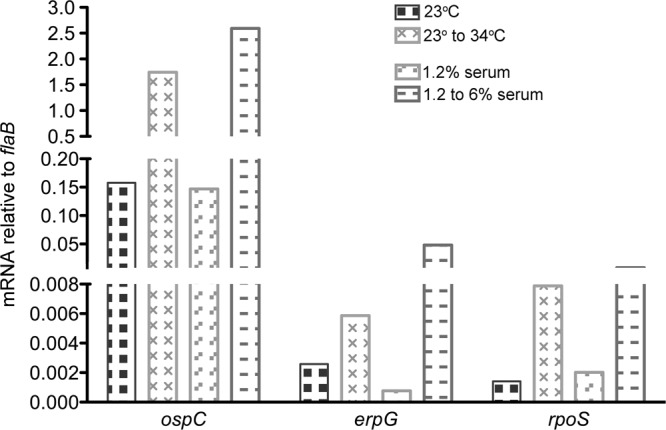
Further indications that changes in borrelial growth rates lead to alterations in gene expression. Shown are data from quantitative reverse transcription-PCR (Q-RT-PCR) analyses of bacterial ospC, erpG, and rpoS mRNAs. Results are presented relative to mRNA levels of the constitutively expressed flaB gene (45). All analyses were performed in triplicate. Error bars (±1 standard deviation) are below the resolution of the figure. Differences within paired conditions were all statistically significant (P < 0.001).
Disassociation of bacterial growth rate and temperature was achieved by using the 1.2% serum and 25% BSK-II formulations of culture medium. B. burgdorferi cells were cultured at 34°C in either of the incomplete media and then diluted into complete medium and grown further at 34°C. Immunoblot analyses demonstrated differential protein expression patterns comparable to those observed with temperature shifts. In the experiment illustrated in Fig. 2, shifting bacteria from 1.2% serum to complete medium increased OspC levels 14-fold. B. burgdorferi bacteria cultured in 25% BSK-II medium did not produce detectable levels of OspC, whereas those shifted to complete BSK-II medium expressed high levels of that protein. The expression of Erp proteins was similarly induced by shifts from either 1.2% serum or 25% BSK-II medium to complete medium (Fig. 2). For the illustrated experiments, shifting B. burgdorferi from 1.2 to 6% serum increased ErpA levels 1.3-fold, ErpM 3.8-fold, and ErpY 1.3-fold. Changing from 25% to full-strength BSK-II medium increased ErpA levels 4.5-fold, ErpM 1.8-fold, and ErpY 1.9-fold. Q-RT-PCR of bacteria shifted from 1.2% to complete medium also showed that greatly enhanced ospC and erpG transcript levels accompanied an enhanced bacterial growth rate (Fig. 3). These differences in protein and mRNA levels are in line with analyses of temperature shift effects (see above) (11, 12, 14, 26).
Control studies indicated constitutive expression of the flagellin subunit FlaB under all culture conditions (Fig. 2). The OspA and OspB surface proteins were not affected to any significant extents by either changes in the culture temperature or BSK-II concentrations. However, a decrease in the OspA, but not the OspB, level was occasionally observed following an increase of the serum concentration from 1.2 to 6% (Fig. 2). Previous studies have shown mixed results for OspA regulation in culture, with some reports describing no effects on OspA following a temperature shift (11, 12) and others noting changes in culture (61, 62). The mechanisms controlling the ospAB operon are evidently complex and will require substantial further experimentation to unravel.
Effects of bacterial division rates on regulatory factors.
OspC and Erp protein levels are regulated at the level of transcription (14, 63). Transcription of erp genes is controlled through two DNA-binding proteins and, to various extents, the RpoS sigma factor (34, 59, 60). BpaB represses erp transcription, while EbfC competes against BpaB for binding to the erp operator and functions as the antirepressor (34, 64). In contrast, transcription of ospC is dependent primarily upon RpoS, with possible contributions from DNA supercoiling and an additional, as-yet-unidentified DNA-interacting factor(s) (62, 65–67).
Immunoblot analyses indicated that cellular levels of the erp repressor, BpaB, were decreased following changes from slow- to fast-growth conditions. In the studies illustrated in Fig. 2, BpaB levels decreased 2.2-fold, 1.1-fold, or 1.3-fold as a result of a shift to complete medium at 34°C from 23°C, 1.2% serum, or 25% BSK-II medium, respectively. These same changes in culture conditions also led to 1.3-fold, 1.7-fold, or 3.5-fold increases, respectively, in cellular levels of the erp antirepressor, EbfC (Fig. 2). These changes correspond with the known functions of BpaB and EbfC in regulating erp transcription (34).
RpoS is apparently expressed at relatively low levels under all examined conditions and could not be detected by immunoblotting using any of the anti-RpoS monoclonal or polyclonal antibodies. Although the cellular concentration of RpoS in actively growing B. burgdorferi bacteria is not known, Escherichia coli may contain between 0 and 100 molecules of each alternative sigma factor per cell (68). However, rpoS mRNA was detectable by Q-RT-PCR, which indicated that bacteria shifted from 1.2% to 6% serum increased rpoS mRNA levels by 4-fold (Fig. 3).
Actual culture temperature does not affect B. burgdorferi OspC or Erp protein levels.
Previous 23°C-versus-34°C temperature shift studies used actively growing cultures to inoculate the 23°C culture (e.g., see references 11, 12, 14, and 69). Regardless of whether inoculating bacteria are first grown at 23°C or at 34°C, B. burgdorferi will produce low levels of OspC and Erp proteins when it is cultured at 23°C (11, 12, 14, 69). Considering the effects of the bacterial growth rate changes described above, and the observation that a temperature shift from 23°C to 34°C enhances the growth rate, we evaluated what the effect would be if 23°C cultures were started with bacteria that initially had even less metabolic activity. To do so, cultures that had been frozen at −80°C for over 1 month were used as inocula for cultures that were then incubated at 23°C. The hypothesis that environmental temperature directly influences OspC and Erp expression predicts that levels of those proteins will be the same in all 23°C cultures, independent of conditions previously experienced by the inoculum. Growth curve analyses of B. burgdorferi bacteria cultured at 23°C in complete medium indicated the same doubling times regardless of whether inocula were previously grown at 34°C or passaged directly from −80°C (Fig. 1).
Contrary to predictions of the temperature hypothesis, bacteria passaged from −80°C to 23°C produced appreciably larger amounts of OspC and Erp proteins than did bacteria passaged from 34°C to 23°C (Fig. 4). Comparing the cultures grown at −80°C to 23°C with those grown at 34°C to 23°C, the illustrated studies found 1.7-fold more OspC, 1.3-fold more ErpA, 4.6-fold more ErpM, and 1.8-fold more ErpY.
Fig 4.
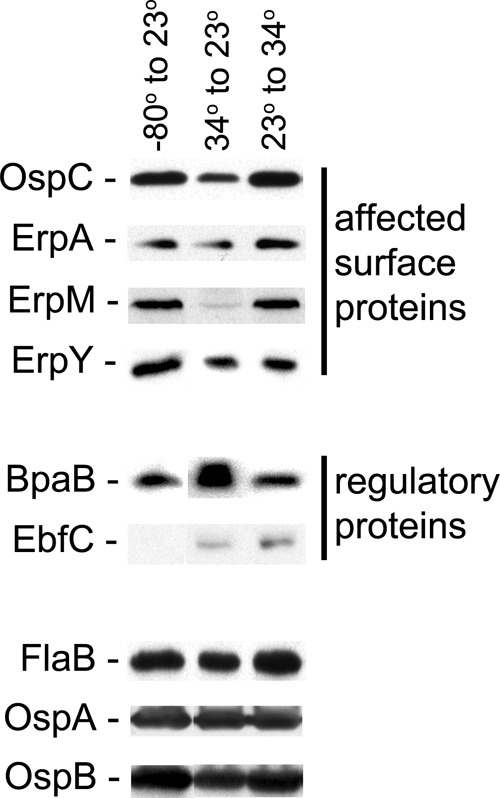
Absolute temperature does not control production of OspC or Erp proteins. B. burgdorferi bacteria were cultured in complete BSK-II medium under various temperature regimens, and protein contents were examined by immunoblotting. Conditions examined were cultures inoculated with bacteria that had been frozen for >1 month at −80°C and then cultured at 23°C (left lanes), cultures inoculated with bacteria that had been grown to late exponential phase at 34°C and then cultured at 23°C (center lanes), and cultures inoculated with bacteria that had been grown to late exponential phase at 23°C and then cultured at 34°C (right lanes). All cultures were harvested at late exponential phase. The constitutively expressed flagellar component FlaB served as a control, and fold changes of other proteins were calculated relative to the FlaB band intensities (44). Illustrated data for each condition are from analyses of the same paired bacterial lysates.
Cellular levels of BpaB, the erp repressor, were 1.6-fold higher in bacteria shifted from 34°C to 23°C than in bacteria shifted from −80°C to 23°C (Fig. 4). EbfC protein and rpoS mRNA were produced at low levels under both growth conditions (Fig. 3 and 4 and data not shown).
Control studies indicated that neither FlaB, OspA, nor OspB levels were affected by shifts from −80°C to 23°C or 34°C to 23°C (Fig. 4).
DISCUSSION
In order for the Lyme disease spirochete to infect a human or another vertebrate host, it is critical that bacteria within a tick detect when the arthropod is feeding on blood and then synthesize proteins appropriate for transmission and infection. In nature, Ixodes sp. ticks fast for several months between blood meals. Thus, B. burgdorferi bacteria within the midgut of an unfed tick experience a nutrient-poor environment, and the bacteria probably do not grow or divide to any great extent (2). However, as the tick begins to feed on a host, nutrients in the ingested blood enable bacterial division at rates of approximately 1 to 2 h per cycle (1, 2, 70–74). This sudden change from low-level metabolism to rapid multiplication occurs only during transmission from tick to vertebrate host.
The initial hypothesis to explain the regulated expression of the ospC and erp operons proposed that bacteria sense environmental temperature, which changes from ambient to blood temperature during tick feeding (1, 11, 12). Cultivation at 23°C was hypothesized to represent the environment of an unfed tick, while a change in temperature from 23°C to 34°C represented the incoming warm blood meal. Results of the present studies contradict that notion and suggest that it be replaced with a new paradigm. The effects of changing culture conditions from −80°C to 23°C were similar to the effects of shifting temperatures from 23°C to 34°C, demonstrating that temperature per se cannot not be the primary cue controlling ospC and erp expression. However, both tested changes in culture temperatures increased the rate of bacterial growth and cell division. OspC and Erp production was also increased following shifts in culture medium composition that stimulated the growth rate, without any changes in temperature. Relating these observations to the natural infectious cycle, we hypothesize that B. burgdorferi uses changes in its own metabolic activity to determine when its vector tick is feeding on a vertebrate host. Since the change from slow to fast metabolism occurs only during the tick-to-vertebrate transmission stage, it would be an appropriate signal that B. burgdorferi must start producing factors involved in transmission through the tick and establishment of mammalian infection.
While the molecular mechanisms by which B. burgdorferi controls ospC and erp expression have not been completely elucidated, all current data support the hypothesized connection between levels of bacterial metabolism and gene expression. Transcription of ospC is dependent primarily upon the alternative sigma factor RpoS, which, in turn, is controlled by the Rrp2 response regulator, the Fur-like BosR DNA-binding protein, the RNA-binding protein CsrA, and an antisense RNA (47, 65, 75–84). Activation of Rrp2 is sensitive to levels of the metabolite acetate (32, 33). BosR responds to oxidizing conditions (79–81, 83, 85–87). The mechanisms controlling the antisense RNA or CsrA are not yet known, although in other bacterial species, CsrA is responsive to carbon source availability (82, 84). In addition, rpoS mutant B. burgdorferi bacteria are unable to transmit from feeding ticks to mammals, apparently due to defects in energy production (88). Borrelial erp transcription utilizes both RpoS and the housekeeping sigma factor RpoD, with some operons being dependent primarily upon RpoS and others not being detectably affected by this sigma factor (59, 60). erp transcription is also controlled by two DNA-binding proteins that compete for the erp operator: the repressor BpaB and the antirepressor EbfC (34, 58, 64, 89–91). ebfC is cotranscribed with dnaX, which encodes subunits of DNA polymerase, and both are positively regulated by the rate of bacterial multiplication (35). The present studies indicated that cytoplasmic concentrations of BpaB are inversely correlated with rates of bacterial replication (Fig. 2 and 4). Borrelial growth leads to production of 4,5-dihydroxy-2,3-pentanedione, also known as autoinducer-2, which positively affects production of Erp and other infection-associated B. burgdorferi proteins (42, 92–97). Taken together, these data support the hypothesis that the bacterial metabolic level is a key factor in the regulation of B. burgdorferi ospC and erp gene expression. Additional tests of this hypothesis are under way, including metabolic analyses of borreliae under various in vitro and in vivo conditions.
B. burgdorferi differentially expresses many proteins during its vertebrate-tick infectious cycle. We caution that bacterial metabolism levels may not be involved in the regulation of all borrelial proteins. Each gene and protein will need to be tested experimentally.
Evidence is accumulating that other pathogens control production of infection-associated factors in response to changes in their metabolism. As examples, Salmonella enterica senses its levels of ATP to control a virulence locus (98), while Listeria monocytogenes and Legionella pneumophila regulate virulence factor production in response to metabolites such as fatty acids and amino acids (99–101). The demonstrated effects of various metabolic activities in B. burgdorferi and other pathogens indicate that previously reported effects of temperature on other bacteria should be reexamined to determine the actual regulatory cue(s) (30).
ACKNOWLEDGMENTS
This work was funded by U.S. National Institutes of Health grant R01-AI044254 and a University of Kentucky College of Medicine bridge award to Brian Stevenson.
We thank Melissa Caimano, Ming He, Justin Radolf, and Frank Yang for providing antibodies and Alex Bobrov, Grant Jones, and Wolfram Zückert for constructive comments on the work and manuscript.
Footnotes
Published ahead of print 7 December 2012
REFERENCES
- 1. Stevenson B, von Lackum K, Riley SP, Cooley AE, Woodman ME, Bykowski T. 2006. Evolving models of Lyme disease spirochete gene regulation. Wien. Klin. Wochenschr. 118: 643–652 [DOI] [PubMed] [Google Scholar]
- 2. Radolf JD, Caimano MJ, Stevenson B, Hu LT. 2012. Of ticks, mice, and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat. Rev. Microbiol. 10: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grimm D, Tilly K, Byram R, Stewart PE, Krum JG, Bueschel DM, Schwan TG, Policastro PF, Elias AF, Rosa PA. 2004. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc. Natl. Acad. Sci. U. S. A. 101: 3142–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pal U, Yang X, Chen M, Bockenstedt LK, Anderson JF, Flavell RA, Norgard MV, Fikrig E. 2004. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J. Clin. Invest. 113: 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ramamoorthi N, Narasimhan S, Pal U, Bao F, Yang XF, Fish D, Anguita J, Norgard MV, Kantor FS, Anderson JF, Koski RA, Fikrig E. 2005. The Lyme disease agent exploits a tick protein to infect the mammalian host. Nature 436: 573–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tilly K, Krum JG, Bestor A, Jewett MW, Grimm D, Bueschel D, Byram R, Dorward D, Vanraden MJ, Stewart P, Rosa P. 2006. Borrelia burgdorferi OspC protein is required exclusively in a crucial early stage of mammalian infection. Infect. Immun. 74: 3554–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stewart PE, Wang X, Bueschel DM, Clifton DR, Grimm D, Tilly K, Carroll JA, Weis JJ, Rosa PA. 2006. Delineating the requirement for the Borrelia burgdorferi virulence factor OspC in the mammalian host. Infect. Immun. 74: 3547–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tilly K, Bestor A, Jewett MW, Rosa P. 2007. Rapid clearance of Lyme disease spirochetes lacking OspC from skin. Infect. Immun. 75: 1517–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Antonara S, Ristow P, McCarthy J, Coburn J. 2010. Effect of Borrelia burgdorferi OspC at the site of inoculation in mouse skin. Infect. Immun. 78: 4723–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seemanapalli SV, Xu Q, McShan K, Liang FT. 2010. Outer surface protein C is a dissemination-facilitating factor of Borrelia burgdorferi during mammalian infection. PLoS One 5: e15830 doi:10.1371/journal.pone.0015830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwan TG, Piesman J, Golde WT, Dolan MC, Rosa PA. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. U. S. A. 92: 2909–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stevenson B, Schwan TG, Rosa PA. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63: 4535–4539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stevenson B, Tilly K, Rosa PA. 1996. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 178: 3508–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stevenson B, Bono JL, Schwan TG, Rosa P. 1998. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 66: 2648–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hefty PS, Jolliff SE, Caimano MJ, Wikel SK, Radolf JD, Akins DR. 2001. Regulation of OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 69: 3618–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hellwage J, Meri T, HeikkilÄ T, Alitalo A, Panelius J, Lahdenne P, SeppälÄ IJT, Meri S. 2001. The complement regulatory factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276: 8427–8435 [DOI] [PubMed] [Google Scholar]
- 17. Kraiczy P, Skerka C, Brade V, Zipfel PF. 2001. Further characterization of complement regulator-acquiring surface proteins of Borrelia burgdorferi. Infect. Immun. 69: 7800–7809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McDowell JV, Sung SY, Price G, Marconi RT. 2001. Demonstration of the genetic stability and temporal expression of select members of the Lyme disease spirochete OspF protein family during infection in mice. Infect. Immun. 69: 4831–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alitalo A, Meri T, Lankinen H, SeppälÄ I, Lahdenne P, Hefty PS, Akins D, Meri S. 2002. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 169: 3847–3853 [DOI] [PubMed] [Google Scholar]
- 20. Miller JC, von Lackum K, Babb K, McAlister JD, Stevenson B. 2003. Temporal analysis of Borrelia burgdorferi Erp protein expression throughout the mammal-tick infectious cycle. Infect. Immun. 71: 6943–6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stevenson B, Bykowski T, Cooley AE, Babb K, Miller JC, Woodman ME, von Lackum K, Riley SP. 2006. The Lyme disease spirochete Erp lipoprotein family: structure, function and regulation of expression, p 354–372 In Cabello FC, Godfrey HP, Hulinska D. (ed), Molecular biology of spirochetes. IOS Press, Amsterdam, Netherlands [Google Scholar]
- 22. Brissette CA, Haupt K, Barthel D, Cooley AE, Bowman A, Skerka C, Wallich R, Zipfel PF, Kraiczy P, Stevenson B. 2009. Borrelia burgdorferi infection-associated surface proteins ErpP, ErpA, and ErpC bind human plasminogen. Infect. Immun. 77: 300–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brissette CA, Verma A, Bowman A, Cooley AE, Stevenson B. 2009. The Borrelia burgdorferi outer-surface protein ErpX binds mammalian laminin. Microbiology 155: 863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hovis KM, Tran E, Sundy CM, Buckles E, McDowell JV, Marconi RT. 2006. Selective binding of Borrelia burgdorferi OspE paralogs to factor H and serum proteins from diverse animals: possible expansion of the role of OspE in Lyme disease pathogenesis. Infect. Immun. 74: 1967–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kenedy MR, Akins DR. 2011. The OspE-related proteins inhibit complement deposition and enhance serum resistance of Borrelia burgdorferi, the Lyme disease agent. Infect. Immun. 79: 1451–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hefty PS, Jolliff SE, Caimano MJ, Wikel SK, Akins DR. 2002. Changes in the temporal and spatial patterns of outer surface lipoprotein expression generate population heterogeneity and antigenic diversity in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 70: 3468–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Revel AT, Talaat AM, Norgard MV. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. U. S. A. 99: 1562–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, Barbour A, Jasinskas A, Benach J, Katona L, Radolf J, Caimano M, Skare J, Swingle K, Akins D, Schwartz I. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 71: 1689–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miller JF, Mekalanos JJ, Falkow S. 1989. Coordinate regulation and sensory transduction in the control of bacterial virulence. Science 243: 916–922 [DOI] [PubMed] [Google Scholar]
- 30. Konkel ME, Tilly K. 2000. Temperature-regulated expression of bacterial virulence genes. Microbes Infect. 2: 157–166 [DOI] [PubMed] [Google Scholar]
- 31. Babb K, El-Hage N, Miller JC, Carroll JA, Stevenson B. 2001. Distinct regulatory pathways control the synthesis of Borrelia burgdorferi infection-associated OspC and Erp surface proteins. Infect. Immun. 69: 4146–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu H, Caimano MJ, Lin T, He M, Radolf JD, Norris SJ, Gherardini F, Wolfe AJ, Yang XF. 2010. Role of acetyl-phosphate in activation of the Rrp2-RpoN-RpoS pathway in Borrelia burgdorferi. PLoS Pathog. 6: e1001104 doi:10.1371/journal.ppat.1001104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Van Laar TA, Lin YH, Miller CL, Karna SLR, Chambers JP, Seshu J. 2012. Effect of levels of acetate on the mevalonate pathway of Borrelia burgdorferi. PLoS One 7: e38171 doi:10.1371/journal.pone.0038171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jutras BL, Verma A, Adams CA, Brissette CA, Burns LH, Whetstine CR, Bowman A, Chenail AM, Zückert WR, Stevenson B. 2012. BpaB and EbfC DNA-binding proteins regulate production of the Lyme disease spirochete's infection-associated Erp surface proteins. J. Bacteriol. 194: 778–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jutras BL, Bowman A, Brissette CA, Adams CA, Verma A, Chenail AM, Stevenson B. 2012. EbfC (YbaB) is a new type of bacterial nucleoid-associated protein and a global regulator of gene expression in the Lyme disease spirochete. J. Bacteriol. 194: 3395–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barbour AG. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57: 521–525 [PMC free article] [PubMed] [Google Scholar]
- 37. Zückert WR. 2007. Laboratory maintenance of Borrelia burgdorferi. Curr. Protoc. Microbiol. Chapter 12: Unit 12C.1 doi:10.1002/9780471729259.mc12c01s4 [DOI] [PubMed] [Google Scholar]
- 38. von Lackum K, Stevenson B. 2005. Carbohydrate utilization by the Lyme borreliosis spirochete, Borrelia burgdorferi. FEMS Microbiol. Lett. 243: 173–179 [DOI] [PubMed] [Google Scholar]
- 39. Sadziene A, Jonsson M, Bergström S, Bright RK, Kennedy RC, Barbour AG. 1994. A bactericidal antibody to Borrelia burgdorferi is directed against a variable region of the OspB protein. Infect. Immun. 62: 2037–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barbour AG, Hayes SF, Heiland RA, Schrumpf ME, Tessier SL. 1986. A Borrelia-specific monoclonal antibody binds to a flagellar epitope. Infect. Immun. 52: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. El-Hage N, Stevenson B. 2002. Simultaneous coexpression of Borrelia burgdorferi Erp proteins occurs through a specific, erp locus-directed regulatory mechanism. J. Bacteriol. 184: 4536–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bykowski T, Babb K, von Lackum K, Riley SP, Norris SJ, Stevenson B. 2006. Transcriptional regulation of the Borrelia burgdorferi antigenically variable VlsE surface protein. J. Bacteriol. 188: 4879–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boardman BK, He M, Ouyang Z, Xu H, Pang X, Yang XF. 2008. Essential role of the response regulator Rrp2 in the infectious cycle of Borrelia burgdorferi. Infect. Immun. 76: 3844–3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bono JL, Tilly K, Stevenson B, Hogan D, Rosa P. 1998. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology 144: 1033–1044 [DOI] [PubMed] [Google Scholar]
- 45. Miller JC. 2005. Example of real-time quantitative reverse transcription-PCR (Q-RT-PCR) analysis of bacterial gene expression during mammalian infection: Borrelia burgdorferi in mouse tissues. Curr. Protoc. Microbiol. Chapter 1D: Unit 1D.3 doi:10.1002/9780471729259.mc01d03s00 [DOI] [PubMed] [Google Scholar]
- 46. Bykowski T, Woodman ME, Cooley AE, Brissette CA, Brade V, Wallich R, Kraiczy P, Stevenson B. 2007. Coordinated expression of Borrelia burgdorferi complement regulator-acquiring surface proteins during the Lyme disease spirochete's mammal-tick infection cycle. Infect. Immun. 75: 4227–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, Schwartz I, Radolf JD. 2007. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol. Microbiol. 65: 1193–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb J-F, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidmann J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390: 580–586 [DOI] [PubMed] [Google Scholar]
- 49. Kelly R. 1971. Cultivation of Borrelia hermsi. Science 173: 443–444 [DOI] [PubMed] [Google Scholar]
- 50. Livermore BP, Bey RF, Johnson RC. 1978. Lipid metabolism of Borrelia hermsi. Infect. Immun. 20: 215–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang X, Popova TG, Goldberg MS, Norgard MV. 2001. Influence of cultivation media on genetic regulatory patterns in Borrelia burgdorferi. Infect. Immun. 69: 4159–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sadziene A, Wilske B, Ferdows MS, Barbour AG. 1993. The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect. Immun. 61: 2192–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marconi RT, Samuels DS, Garon CF. 1993. Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J. Bacteriol. 175: 926–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Casjens S, van Vugt R, Tilly K, Rosa PA, Stevenson B. 1997. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J. Bacteriol. 179: 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser C. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35: 490–516 [DOI] [PubMed] [Google Scholar]
- 56. Akins DR, Porcella SF, Popova TG, Shevchenko D, Baker SI, Li M, Norgard MV, Radolf JD. 1995. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Mol. Microbiol. 18: 507–520 [DOI] [PubMed] [Google Scholar]
- 57. Skare JT, Foley DM, Hernandez SR, Moore DC, Blanco DR, Miller JN, Lovett MA. 1999. Cloning and molecular characterization of plasmid-encoded antigens of Borrelia burgdorferi. Infect. Immun. 67: 4407–4417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Babb K, McAlister JD, Miller JC, Stevenson B. 2004. Molecular characterization of Borrelia burgdorferi erp promoter/operator elements. J. Bacteriol. 186: 2745–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Eggers CH, Caimano MJ, Radolf JD. 2004. Analysis of promoter elements involved in the transcription initiation of RpoS-dependent Borrelia burgdorferi genes. J. Bacteriol. 186: 7390–7402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Eggers CH, Caimano MJ, Radolf JD. 2006. Sigma factor selectivity in Borrelia burgdorferi: RpoS recognition of the ospE/ospF/elp promoters is dependent on the sequence of the −10 region. Mol. Microbiol. 59: 1859–1875 [DOI] [PubMed] [Google Scholar]
- 61. Yang X, Goldberg MS, Popova TG, Schoeler GB, Wikel SK, Hagman KE, Norgard MV. 2000. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol. Microbiol. 37: 1470–1479 [DOI] [PubMed] [Google Scholar]
- 62. Alverson J, Bundle SF, Sohasky CD, Lybecker MC, Samuels DS. 2003. Transcriptional regulation of the ospAB and ospC promoters of Borrelia burgdorferi. Mol. Microbiol. 48: 1665–1677 [DOI] [PubMed] [Google Scholar]
- 63. Tilly K, Casjens S, Stevenson B, Bono JL, Samuels DS, Hogan D, Rosa P. 1997. The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol. Microbiol. 25: 361–373 [DOI] [PubMed] [Google Scholar]
- 64. Burns LH, Adams CA, Riley SP, Jutras BL, Bowman A, Chenail AM, Cooley AE, Haselhorst LA, Moore AM, Babb K, Fried MG, Stevenson B. 2010. BpaB, a novel protein encoded by the Lyme disease spirochete's cp32 prophages, binds to erp operator 2 DNA. Nucleic Acids Res. 38: 5443–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hübner A, Yang X, Nolen DM, Popova TG, Cabello PC, Norgard MV. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. U. S. A. 98: 12724–12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang XF, Lybecker MC, Pal U, Alani SM, Blevins J, Revel AT, Samuels DS, Norgard MV. 2005. Analysis of the ospC regulatory element controlled by the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. J. Bacteriol. 187: 4822–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xu Q, McShan K, Liang FT. 2007. Identification of an ospC operator critical for immune evasion of Borrelia burgdorferi. Mol. Microbiol. 64: 220–231 [DOI] [PubMed] [Google Scholar]
- 68. Ishihama A. 2000. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 54: 499–518 [DOI] [PubMed] [Google Scholar]
- 69. Schwan TG, Piesman J. 2000. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J. Clin. Microbiol. 38: 382–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Piesman J, Oliver JR, Sinsky RJ. 1990. Growth kinetics of the Lyme disease spirochete (Borrelia burgdorferi) in vector ticks (Ixodes dammini). Am. J. Trop. Med. Hyg. 42: 352–357 [DOI] [PubMed] [Google Scholar]
- 71. Burkot TR, Piesman J, Wirtz RA. 1994. Quantitation of the Borrelia burgdorferi outer surface protein A in Ixodes scapularis: fluctuations during the tick life cycle, doubling times and loss while feeding. J. Infect. Dis. 170: 883–889 [DOI] [PubMed] [Google Scholar]
- 72. de Silva AM, Fikrig E. 1995. Growth and migration of Borrelia burgdorferi in Ixodes ticks during blood feeding. Am. J. Trop. Med. Hyg. 53: 397–404 [DOI] [PubMed] [Google Scholar]
- 73. Piesman J, Schneider BS, Zeidner NS. 2001. Use of quantitative PCR to measure density of Borrelia burgdorferi in the midgut and salivary glands of feeding tick vectors. J. Clin. Microbiol. 39: 4145–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Piesman J, Schneider BS. 2002. Dynamic changes in Lyme disease spirochetes during transmission by nymphal ticks. Exp. Appl. Acarol. 28: 141–145 [DOI] [PubMed] [Google Scholar]
- 75. Seshu J, Boylan JA, Gherardini FC, Skare JT. 2004. Dissolved oxygen levels alter gene expression and antigen profiles in Borrelia burgdorferi. Infect. Immun. 72: 1580–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Burtnick MN, Downey JS, Brett PJ, Boylan JA, Frye JG, Hoover TR, Gherardini FC. 2007. Insights into the complex regulation of rpoS in Borrelia burgdorferi. Mol. Microbiol. 65: 277–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lybecker MC, Samuels DS. 2007. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol. Microbiol. 64: 1075–1089 [DOI] [PubMed] [Google Scholar]
- 78. Smith AH, Blevins JS, Bachlani GN, Yang XF, Norgard MV. 2007. Evidence that RpoS (σS) in Borrelia burgdorferi is controlled directly by RpoN (σ54/σN). J. Bacteriol. 189: 2139–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hyde JA, Shaw DK, Smith R, Trzeciakowski JP, Skare JT. 2009. The BosR regulatory protein of Borrelia burgdorferi interfaces with the RpoS regulatory pathway and modulates both the oxidative stress response and pathogenic properties of the Lyme disease spirochete. Mol. Microbiol. 74: 1344–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ouyang Z, Kumar M, Kariu T, Haq S, Goldberg M, Pal U, Norgard MV. 2009. BosR (BB0647) governs virulence expression in Borrelia burgdorferi. Mol. Microbiol. 74: 1331–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hyde JA, Shaw DK, Smith R, Trzeciakowski JP, Skare JT. 2010. Characterization of a conditional bosR mutant in Borrelia burgdorferi. Infect. Immun. 78: 265–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Karna SLR, Sanjuan E, Esteve-Gassent MD, Miller CL, Maruskova M, Seshu J. 2011. CsrA modulates levels of lipoproteins and key regulators of gene expression critical for pathogenic mechanisms of Borrelia burgdorferi. Infect. Immun. 79: 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ouyang Z, Deka RK, Norgard MV. 2011. BosR (BB0647) controls the RpoN-RpoS regulatory pathway and virulence expression in Borrelia burgdorferi by a novel DNA-binding mechanism. PLoS Pathog. 7: e1001272 doi:10.1371/journal.ppat.1001272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sze CW, Li C. 2011. Inactivation of bb0184, which encodes carbon storage regulator A, represses the infectivity of Borrelia burgdorferi. Infect. Immun. 79: 1270–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Boylan JA, Posey JE, Gherardini FC. 2003. Borrelia oxidative stress response regulator, BosR: a distinct Zn-dependent transcriptional activator. Proc. Natl. Acad. Sci. U. S. A. 100: 11684–11689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Katona LI, Tokarz R, Kuhlow CJ, Benach J, Benach JL. 2004. The Fur homologue in Borrelia burgdorferi. J. Bacteriol. 186: 6443–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hyde JA, Seshu J, Skare JT. 2006. Transcriptional profiling of Borrelia burgdorferi containing a unique bosR allele identifies a putative stress regulon. Microbiology 152: 2599–2609 [DOI] [PubMed] [Google Scholar]
- 88. Dunham-Ems SM, Caimano MJ, Eggers CH, Radolf JD. 2012. Borrelia burgdorferi requires the alternative sigma factor RpoS for dissemination within the vector during tick-to-mammal transmission. PLoS Pathog. 8: e1002532 doi:10.1371/journal.ppat.1002532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Babb K, Bykowski T, Riley SP, Miller MC, DeMoll E, Stevenson B. 2006. Borrelia burgdorferi EbfC, a novel, chromosomally encoded protein, binds specific DNA sequences adjacent to erp loci on the spirochete's resident cp32 prophages. J. Bacteriol. 188: 4331–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Riley SP, Bykowski TT, Cooley AE, Burns LH, Babb K, Brissette CA, Bowman A, Rotondi M, Miller MC, DeMoll E, Lim K, Fried MG, Stevenson B. 2009. Borrelia burgdorferi EbfC defines a newly-identified, widespread family of bacterial DNA-binding proteins. Nucleic Acids Res. 37: 1973–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chenail AM, Jutras BL, Adams CA, Burns LH, Bowman A, Verma A, Stevenson B. 2012. Borrelia burgdorferi cp32 BpaB modulates expression of the prophage NucP nuclease and SsbP single-stranded DNA-binding protein. J. Bacteriol. 194: 4570–4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Stevenson B, Babb K. 2002. LuxS-mediated quorum sensing in Borrelia burgdorferi, the Lyme disease spirochete. Infect. Immun. 70: 4099–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Babb K, von Lackum K, Wattier RL, Riley SP, Stevenson B. 2005. Synthesis of autoinducer 2 by the Lyme disease spirochete, Borrelia burgdorferi. J. Bacteriol. 187: 3079–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Riley SP, Bykowski T, Babb K, von Lackum K, Stevenson B. 2007. Genetic and physiological characterization of the Borrelia burgdorferi ORF BB0374-pfs-metK-luxS operon. Microbiology 153: 2304–2311 [DOI] [PubMed] [Google Scholar]
- 95. von Lackum K, Babb K, Riley SP, Wattier RL, Bykowski T, Stevenson B. 2006. Functionality of Borrelia burgdorferi LuxS: the Lyme disease spirochete produces and responds to the pheromone autoinducer-2, and lacks a complete activated-methyl cycle. Int. J. Med. Microbiol. 296(Suppl 40): 92–102 [DOI] [PubMed] [Google Scholar]
- 96. von Lackum K, Ollison KM, Bykowski T, Nowalk AJ, Hughes JL, Carroll JA, Zückert WR, Stevenson B. 2007. Regulated synthesis of the Borrelia burgdorferi inner-membrane lipoprotein IpLA7 (P22, P22-A) during the Lyme disease spirochaete's mammal-tick infectious cycle. Microbiology 153: 1361–1371 [DOI] [PubMed] [Google Scholar]
- 97. Xavier KB, Bassler BL. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6: 191–197 [DOI] [PubMed] [Google Scholar]
- 98. Lee EJ, Groisman EA. 2012. Control of a Salmonella virulence locus by an ATP-sensing leader messenger RNA. Nature 486: 271–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Edwards RL, Dalebroux ZD, Swanson MS. 2009. Legionella pneumophila couples fatty acid flux to microbial differentiation and virulence. Mol. Microbiol. 71: 1190–1204 [DOI] [PubMed] [Google Scholar]
- 100. Lobel L, Sigal N, Borovok I, Ruppin E, Herskovits AA. 2012. Integrative genomic analysis identifies isoleucine and CodY as regulators of Listeria monocytogenes virulence. PLoS Genet. 8: e1002887 doi:10.1371/journal.pgen.1002887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Sun Y, Wilkinson BJ, Standiford TJ, Akinbi HT, O'Riordan MX. 2012. Fatty acids regulate stress resistance and virulence factor production for Listeria monocytogenes. J. Bacteriol. 194: 5274–5284 [DOI] [PMC free article] [PubMed] [Google Scholar]