Abstract
The adaptive immune system comprising CRISPR (clustered regularly interspaced short palindromic repeats) arrays and cas (CRISPR-associated) genes has been discovered in a wide range of bacteria and archaea and has recently attracted comprehensive investigations. However, the subtype I-B CRISPR-Cas system in haloarchaea has been less characterized. Here, we investigated Cas6-mediated RNA processing in Haloferax mediterranei. The Cas6 cleavage site, as well as the CRISPR transcription start site, was experimentally determined, and processing of CRISPR transcripts was detected with a progressively increasing pattern from early log to stationary phase. With genetic approaches, we discovered that the lack of Cas1, Cas3, or Cas4 unexpectedly resulted in a decrease of CRISPR transcripts, while Cas5, Cas6, and Cas7 were found to be essential in stabilizing mature CRISPR RNA (crRNA). Intriguingly, we observed a CRISPR- and Cas3-independent inhibition of a defective provirus, in which the putative Cascade (CRISPR-associated complex for antiviral defense) proteins (Cas5, Cas6, Cas7, and Cas8b) were indispensably required. A sequence carried by a proviral transcript was found to be homologous to the CRISPR repeat RNA and vulnerable to Cas6-mediated cleavage, implying a distinct interference mechanism that may account for this unusual inhibition. These results provide fundamental information for the subtype I-B CRISPR-Cas system in halophilic archaea and suggest diversified mechanisms and multiple physiological functions for the CRISPR-Cas system.
INTRODUCTION
The CRISPR-Cas system has been recently recognized as a prokaryotic adaptive and inheritable immune system that is capable of defending against invasive genetic elements (1–4). CRISPR structures are long arrays of direct repeats that are separated by unique but regularly sized inserts known as spacers. A set of genes that are physically associated with the CRISPR arrays are termed cas (CRISPR-associated) genes (5). Although extensive divergence has been observed for this widespread and fast-evolving system, a unifying and flexible classification scheme has been proposed (6). According to this scheme, three main types (types I to III) are tentatively, but elaborately, defined. Type I is defined by the presence of the cas3 gene and further divided into six subtypes (I-A, -B, -C, -D, -E, and -F), while types II and III each consist of two subtypes (II-A and -B and III-A and -B) (6).
The immunity function of the CRISPR-Cas system is achieved through three stages: the adaption, CRISPR RNA (crRNA) biogenesis, and interference stages (3). During adaption, new spacers derived from invading genetic elements are integrated into CRISPR loci (7–10), yet detailed mechanisms of this stage have been less characterized than those of the other two stages. A usually AT-rich upstream region, namely, the leader sequence, contains promoter elements that initiate CRISPR transcription, and the transcripts are processed by Cas proteins, producing individual invader targeting sequences (11–14). These sequences are subsequently incorporated into the Cascade (CRISPR-associated complex for antiviral defense) complex and guide the target interference (15, 16).
Though extreme diversity has been observed for the Cas machineries, in most cases, Cas6 family endonucleases are responsible for pre-crRNA processing (11, 12, 17). Recent structural studies of Cas6 homologues, including Pyrococcus furiosus Cas6 (PfCas6) (12, 18, 19), Thermus thermophilus Cas6e/Cse3 (20–22), and Pseudomonas aeruginosa Cas6f/Csy4 (17), have revealed evident structural similarities; however, distinct structural elements and detailed mechanisms prove to be involved in processing their differently structured substrates. For instance, P. furiosus Cas6 recognizes unstructured repeat RNA through a wraparound mechanism (19), while Cas6e and Cas6f interact with the major groove of their stem-loop-structured repeats, respectively, by a β-hairpin and an α-helix (17, 21, 22). For Cas6f, interactions with the RNA major groove have been shown to be highly sensitive to the helical geometry of the stem-loop structure (23). Therefore, these endonucleases have probably coevolved with their cognate repeat RNAs to achieve a high degree of substrate specificity and to avoid recognizing noncognate cellular RNAs (17, 21–23).
Cas6 cleavages consistently generate a 5′ handle of 8 nucleotides (nt) (the last 8 nucleotides from the previous repeat) and a variable 3′ terminal tag (the 5′ fragment of the next repeat) flanking the spacer sequence (11, 12, 17, 21, 22, 24). After cleavage, the endonuclease remains tightly bound to the 3′ terminus (17, 18, 22), possibly acting as a nucleation point for the assembly of the Cascade complex. Structural study of Escherichia coli Cascade has revealed that it has a seahorse-shaped architecture with a stoichiometry of 6:1:1:2:1 (Cas7-Cas5-Cse1-Cse2-Cas6e) and that crRNA serves as a template for the complex assembly and, in return, gets stabilized while preserving the ability of base pairing (16). In P. aeruginosa PA14, subtype-specific Cas proteins, Cas6f and Csy1-3, assemble into a 350-kDa complex carrying mature crRNA molecules, and the revealed crescent-shaped structure shows a striking resemblance to the architecture of E. coli Cascade (25).
These extensive studies on the well-characterized subtypes have provided plenty of information for our understanding how Cas proteins generate and stabilize mature crRNAs. However, less information is available for other poorly studied subtypes. Therefore, we investigated the subtype I-B system from haloarchaea that thrive in habitats with a high salinity with a focus on crRNA biogenesis and Cas members involved in this process. In Haloferax volcanii, a plasmid-based invader assay revealed that multiple protospacer adjacent motif (PAM) sequences could be utilized, indicating unusual features for this subtype (26). Using Haloferax mediterranei as a model strain, we experimentally identified the CRISPR transcription start site and the precursor cleavage site. Interestingly, in the absence of Cas6, precursors were still processed but with different patterns, suggesting some nonspecific degradation. Deficiency of other Cas proteins, like Cas1, Cas3, and Cas4, evidently impaired crRNA production, while Cas5h, Cas7, and the endonuclease Cas6 seemed indispensably required in stabilizing the mature products in vivo. We also reported an interesting CRISPR- and Cas3-independent regulatory effect on behaviors of a defective provirus, which was probably achieved through Cas6-mediated cleavage that directly occurs on proviral transcripts.
MATERIALS AND METHODS
Strains and growth conditions.
The H. mediterranei strains used in this study are listed in Table 1. H. mediterranei DF50 (27) and the mutant strains constructed based on DF50 were cultured at 37°C in AS-168L medium (per liter, 150 g of NaCl, 20 g of MgSO4 · 7H2O, 2 g of KCl, 3 g of trisodium citrate, 1 g of sodium glutamate, 50 mg of FeSO4 · 7H2O, 0.36 mg of MnCl2 · 4H2O, 5 g of Bacto Casamino Acids, 5 g of yeast extract, pH 7.2) plus uracil (50 mg/liter). When needed, 5-fluoroorotic acid (5-FOA) was added to a final concentration of 250 mg/liter. AS-168SYL (AS-168L medium with yeast extract subtracted) was used to cultivate the transformants carrying expression vectors. E. coli JM109 was used for cloning and was cultured in Luria-Bertani (LB) medium. When needed, ampicillin was added to a final concentration of 100 mg/liter.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| H. mediterranei strains | ||
| DF50 | ΔpyrF strain of H. mediterranei ATCC33500 | 27 |
| Δcas1 strain | cas1 deletion mutant of DF50 | This study |
| Δcas2 strain | cas2 deletion mutant of DF50 | This study |
| Δcas3 strain | cas3 deletion mutant of DF50 | This study |
| Δcas4 strain | cas4 deletion mutant of DF50 | This study |
| Δcas5h strain | cas5h deletion mutant of DF50 | This study |
| Δcas6 strain | cas6 deletion mutant of DF50 | This study |
| Δcas7 strain | cas7 deletion mutant of DF50 | This study |
| Δcas8b strain | cas8b deletion mutant of DF50 | This study |
| CRF | CRISPR-free mutant of DF50 | This study |
| CRF Δcas6 | cas6 deletion mutant of CRF | This study |
| Plasmids | ||
| pHFX | 4.0 kb; suicide vector containing pyrF and its native promoter | 27 |
| pWL502 | 7.9 kb; expression vector containing pyrF and its native promoter | 28 |
| pDCAS1 | 5.1 kb; modified pHFX for knockout of cas1 | This study |
| pDCAS2 | 5.1 kb; modified pHFX for knockout of cas2 | This study |
| pDCAS3 | 5.2 kb; modified pHFX for knockout of cas3 | This study |
| pDCAS4 | 5.1 kb; modified pHFX for knockout of cas4 | This study |
| pDCAS5 | 5.0 kb; modified pHFX for knockout of cas5h | This study |
| pDCAS6 | 5.2 kb; modified pHFX for knockout of cas6 | This study |
| pDCAS7 | 5.1 kb; modified pHFX for knockout of cas7 | This study |
| pDCAS8 | 5.1 kb; modified pHFX for knockout of cas8b | This study |
| pDC2 | 5.2 kb; modified pHFX for knockout of C2 array | This study |
| pDC18 | 5.1 kb; modified pHFX for knockout of C18 array | This study |
| pDC22 | 5.2 kb; modified pHFX for knockout of C22 array | This study |
| pDC26 | 5.1 kb; modified pHFX for knockout of C26 array | This study |
| pDP12 | 5.2 kb; modified pHFX for knockout of P12 array | This study |
| pDP23 | 5.2 kb; modified pHFX for knockout of P23 array | This study |
| pWL502-cas6 | 9.0 kb; modified pWL502 containing cas6 and its native promoter | This study |
| pC2 | 8.3 kb; modified pWL502 carrying a complete C2 array | This study |
| pC2-37 | 8.2 kb; modified pWL502 carrying a C2 array with a 59-bp leader | This study |
| pC2-31 | 8.2 kb; modified pWL502 carrying a C2 array with a 53-bp leader | This study |
| pVC2 | 8.3 kb; modified pWL502 carrying a C2 array without the leader motif | This study |
| pNCG | 8.3 kb; modified pWL502 carrying a modified C2 array with its first direct repeat replaced by the proviral repeat-like sequence | This study |
Gene knockout and complementation.
To generate the knockouts of each cas gene (or CRISPR locus), upstream and downstream fragments (∼600 bp) were separately amplified with UF/UR or DF/DR primers. These two fragments for each gene were linked together by bridge PCR with UF/DR primers (see Table S1 in the supplemental material), inserted into a pGEM-T vector (pGEM-T Easy; Promega), and validated by DNA sequencing. These linked fragments (∼1.2 kb) were then cut out of the pGEM-T vectors and inserted into the suicide plasmid pHFX (27). The modified suicide plasmids were then transformed into H. mediterranei DF50 using a polyethylene glycol-mediated transformation method (29). The mutants were screened as previously described (27). Each mutant was verified by sequence analysis with UF/DR primers.
For complementation of the cas6 gene, the coding region and a 297-bp preceding region containing its native promoter were amplified with Cas6-F and Cas6-R primers (see Table S1). The product was digested and inserted into the expression plasmid pWL502 (28). The reconstructed plasmid was transformed into the Δcas6 strain.
For complementation of the CRISPR C2 arrays (where C indicates location on the chromosome and 2 is the repeat number) (see Fig. 1) with differently truncated leader sequences, the C2 array was amplified with a common reverse primer (C2-R) and different forward primers (C2-139, -37, or -31) (see Table S1 in the supplemental material). To test the function of the leader motif AT(C/T)GG(G/C)CATG, a 3′ truncated leader sequence and the repeat-spacer structure were separately amplified with C2-139/VC2-UR and VC2-DF/C2-R primer pairs and concatemerized by bridge PCR with C2-139/C2-R primers, utilizing the complementary sequences designed on the 5′ ends of VC2-UR and VC2-DF primers (see Table S1). Replacement of the first repeat with a spurious repeat was similarly performed with the C2-139/NCG-UR and NCG-DF/C2-R primer pairs.
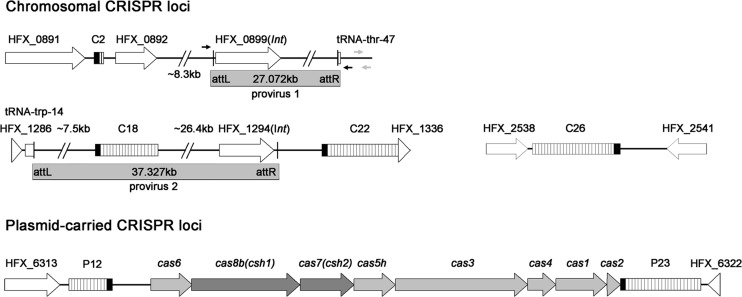
Fig 1.
Depiction of the genomic contexts of CRISPR loci interspersed within the H. mediterranei genome. Each CRISPR locus is indicated by an array of white bars (each bar represents a repeat-spacer unit); the black bars represent the leader sequence. The gray rectangles represent two proviruses, and the small black and gray arrows around provirus 1, respectively, indicate the test or control primers used for detecting its excision from the chromosome. The proviral genes (int) encoding integrases are shown.
Northern analysis and primer extension analysis.
Total RNA was extracted from H. mediterranei cultures during the stationary phase (unless otherwise specified) using TRIzol (Invitrogen) according to the standard protocol. The RNA concentration was determined using a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific). For Northern blotting of small RNAs, 20 μg of RNA was mixed with an equal volume of formamide loading buffer (80% [wt/vol] deionized formamide, 1 mg/ml xylene cyanol, 1 mg/ml bromophenol blue, 10 mM EDTA, pH 8.0) and run on an 8% polyacrylamide gel (7.6 M urea, 89 mM Tris, 89 mM boric acid, 2 mM EDTA, pH 8.3). A 50- to 1,000-nt RNA marker (Low Range ssRNA Ladder; NEB) and the RNA samples were coelectrophoresed and cotransferred onto Hybond N+ nylon membranes (GE Health Care) using a Trans-Blot SD Cell (Bio-Rad). RNA on the nylon membranes was stained and visualized with methylene blue dye solution (0.3 M sodium acetate [NaAc], 1 mM methylene blue). The band of 5S rRNA (visualized by staining) or 7S RNA (detected by 7S probe) was used as internal control. To detect larger intermediates, a 500- to 9,000-nt RNA marker (ssRNA Ladder; NEB) was coseparated on a 1.2% agarose gel and transferred by capillary blotting, and the 7S RNA band (detected by 7S probe) was used as a control. The oligonucleotide probes (see Table S1 in the supplemental material) were labeled with [γ-32P]ATP and T4 polynucleotide kinase (NEB), while the 7S probe was labeled with [α-32P]dCTP by PCR incorporation. Prehybridization, hybridization, and washing procedures were performed as previously described (14). The exposure time varied from 1 h to 4 days, depending on the radioactivity.
To determine the transcription start site of the P12 array, the primer P12-seq complementary to its first spacer was labeled at the 5′ end with [γ-32P]ATP and used for primer extension as previously described (30).
Detection of attA site restoration.
The chromosomal region encoding phage integrases and PhiH-like regulators, as well as multiple hypothetical proteins, and carrying tandem repeats (attL and attR) on the two ends was considered a putative provirus. To detect the attA site restoration of provirus 1, test primers PV1-testF and PV1-testR were designed upstream and downstream of the provirus, while both control primers PV1-ctrlF and PV1-ctrlR were designed downstream of the provirus (see Table S1). For each PCR, 8 pmol of each test primer and 2 pmol of each control primer were used, and ∼200 ng of genomic DNA extracted from exponential-phase cultures was used as a template. The PCR procedure consisted of 30 cycles, with 30 s at 95°C for denaturation, 30 s at 60°C for annealing, and 30 s at 72°C for elongation in each cycle.
CR-RT-PCR analysis.
To analyze the 5′ end and potential processing site of phiH transcript, RNA samples of H. mediterranei DF50 and Δcas6 strains were separately subjected to the circularized RNA reverse transcription-PCR (CR-RT-PCR) analysis (31). Briefly, RNA was 5′-to-3′ end ligated by T4 RNA ligase (NEB), followed by reverse transcription using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega) with the gene-specific primer phiH-PCR1. The cDNA was then amplified with the divergent primers phiH-PCR1/PCR2, followed by a second PCR with the nested primers phiH-NES1/NES2 (see Table S1 in the supplemental material). Similarly, to verify the absence of a 3′ trimming process, the RNA sample was self-ligated, followed by general RT-PCR procedures with a pair of divergent primers, P12S3-RTF and P12S3-RTR (see Table S1). The PCR products, which contained sequences corresponding to the 5′ and 3′ regions of the RNA samples, were directly sequenced with nested primers or cloned into pGEM-T vectors (Promega) and then sequenced.
CRISPR structural analysis.
A search for H. mediterranei CRISPRs was performed with the CRISPRfinder tool (32) against the complete genome. For each array, the orientation was determined according to the conserved leader sequence, and the repeat number was calibrated considering repeat degeneration at the distal end. The secondary structure of repeat RNA was predicted using the RNAfold server (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi).
RESULTS
Genome contexts of CRISPR arrays.
In this study, H. mediterranei was taken as a model to investigate the subtype I-B system from halophiles because this strain exhibits a highly abundant CRISPR content. Six arrays, namely, C2, C18, C22, C26, P12, and P23 (P or C indicates the location on the chromosome or a plasmid and is followed by the repeat number), are interspersed within the H. mediterranei genome (Fig. 1). C18, C22, C26, and P23 carry identical repeats, whereas P12 and C2 carry repeats with one or two base changes (Fig. 2A). P12 and P23 are divergently located on the pHM500 megaplasmid and physically linked by a shared cas operon, which consists of four core cas genes (cas1-cas4), the endonuclease-encoding gene (cas6), and three subtype-specific genes (cas5 [cas5h], cas7 [csh2], and cas8b [csh1]) (Fig. 1). The chromosomal arrays are not associated with any cas genes and thus should be operated by the plasmid-encoded Cas proteins.
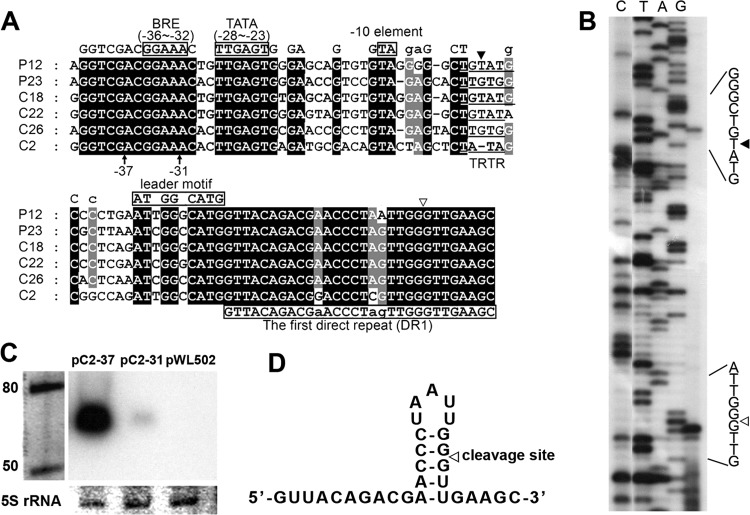
Fig 2.
Determination of the transcription start site (TSS; solid triangles) and pre-crRNA cleavage site (PCS; empty triangles). (A) Multiple alignments of partial leader sequences (consensus sequence on the top) and the first repeats (consensus sequence on the bottom) of the H. mediterranei arrays. Black or grey shading indicates the same nucleotide at this position in all or most sequences, respectively. The putative promoter motifs (BRE motif, TATA box, and −10 element) and the AT(C/T)GG(G/C)CATG leader motif (immediately preceding the first repeat), are boxed in black. The TRTR element containing the TSS is underlined. The black arrows at −37 and −31 indicate the 5′ ends of two truncated C2 arrays used in the experiment shown in panel C. (B) The primer extension result with a primer specific to the first spacer of P12. (C) Northern blot analysis with a repeat probe validating the prediction of promoter motifs. pC2-37 and pC2-31 are pWL502 vectors carrying C2 arrays with differently truncated 5′ leaders (shown in panel A). These plasmids were transformed into a CRISPR-free (CRF) strain, and total RNA was sampled at stationary phase. Values to the left indicate RNA size markers, in nucleotides. (D) Predicted secondary structure of the repeat RNA from P12 using the RNAfold server.
Intriguingly, the minimal array C2 is located near (∼8 kb) a provirus designated provirus 1, while C18 and C22 are situated, respectively, within and right beside (∼800 bp) another provirus designated provirus 2 (Fig. 1). The provirus-carried array C18 has an intact leader, suggesting that it may be utilized by the carrier to prevent superinfection by other mobile elements and meanwhile be spread through horizontal gene transfer (HGT) events.
Determination of the CRISPR transcription start site and precursor cleavage site.
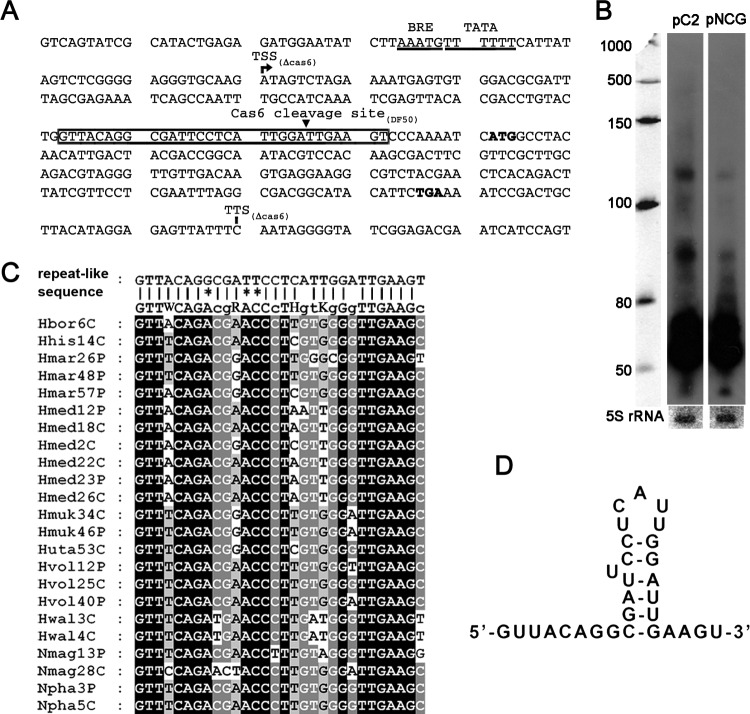
To facilitate further investigation, the CRISPR transcription start site (TSS) was experimentally determined. Primer extension analysis was performed with a primer complementary to the first spacer of P12 (Fig. 2B). Transcription initiation was revealed at the second thymine (T) within the TGTATG sequence, 21 bp upstream from the first repeat (Fig. 2A and B). Although this sequence is not common to other H. mediterranei arrays, a convincingly conserved TRTR (R stands for a purine) motif was observed with an also conserved distance (∼18 bp) to the first repeat (Fig. 2A). This conservation can be extended to nearly all haloarchaeal CRISPR arrays (data not shown); thus, we propose that this common TRTR motif generally contains at least one transcription start site for each CRISPR array. Preceding the transcription start site, evident haloarchaeal promoter motifs (33) (i.e., BRE motif, TATA box, and −10 element) were predicted (Fig. 2A). These motifs were confirmed by the nearly total absence of transcripts from a modified C2 array with a truncated BRE motif (Fig. 2C).
The primer extension result also revealed another functional site, the CRISPR RNA precursor cleavage site (PCS) within the first repeat (Fig. 2A and B). The PCS is 8 nt upstream of the following spacer and located in the middle of the 5-bp stem of the repeat RNA structure (Fig. 2D). Hence, cleavage at this site will result in a conserved 8-nt 5′ handle on mature crRNA, which is consistent with studies on other subtypes (11, 12, 17, 21, 22, 24). The conserved 5′ handle has been shown to be structurally essential to the assembly of the Cascade complex (16) and functionally important during the interference stage (34).
Growth phase-dependent processing of CRISPR transcripts.
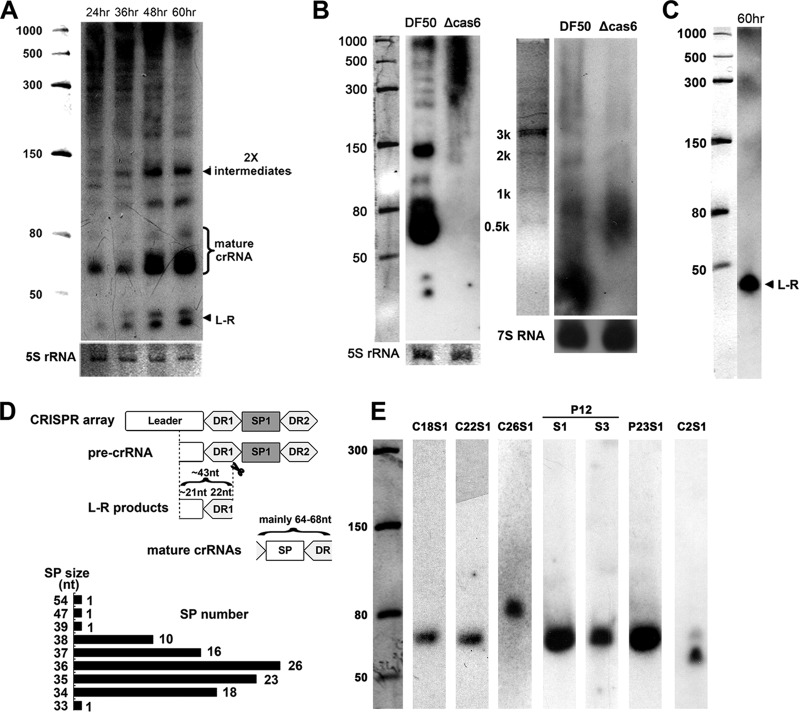
According to the growth curve measured by counting CFU, culture samples were harvested at a series of time points, i.e., 24, 36, 48, and 60 h, respectively, representing the early log, middle log, late log, and stationary phases. Total RNA extracted from these samples was analyzed by Northern blotting with a repeat-specific probe, and the result revealed an evident phase dependence for crRNA biogenesis (Fig. 3A). In early exponential phase, a limited number of mature products were generated; however, CRISPR structures appeared quite efficiently transcribed because lots of larger intermediates were also observed. As the culture entered stationary phase, these larger intermediates were progressively processed into smaller ones and finally mature crRNAs. Given that Cas6 has proved to be a single-turnover catalyst (22), the progressively increased processing indicates that the endonuclease was gradually accumulated and highly expressed in stationary phase. Moreover, the larger bands seemed obviously obscured by some irregular products, which may result from nonspecific degradations. To test this hypothesis, we investigated crRNA maturation in a cas6 mutant and discovered that large transcripts were degraded into products of sizes ranging from 300 to 1,000 nt (Fig. 3B). Therefore, our data support the competition between specific processing and nonspecific degradation revealed in E. coli cells (35). Studies in another archaeon, Sulfolobus acidocaldarius, have revealed a similar growth dependence (36), but the stationary-phase product pattern is very different, and major transcripts range from 3,000 to 3,500 nt (14), indicating a complex transcription pattern (like premature termination) or a more active degradation system in S. acidocaldarius.
Fig 3.
Analysis of crRNA biogenesis pattern. (A) Northern analysis against H. mediterranei DF50 total RNA sampled at different time points with a repeat probe. Smaller processed products are labeled. (B) Nonspecific degradation was detected when the endonuclease Cas6 was missing. The RNA samples were separated by an 8% PAGE gel (left) or a 1.2% agarose gel (right). (C) Northern analysis of a stationary-phase RNA sample with a mixture of probes against the transcribed leader regions. (D) Schematic diagram showing the generation of smaller products and their sizes; the distribution of various spacer (SP) sizes is also shown. (E) Transcriptional analysis of each CRISPR locus with spacer-specific probes. For P12, probes against the first (S1) and third (S3) spacers were used for two separate experiments. The product pattern of the one-spacer array C2 was different, maybe due to the degeneration of the second repeat. Values to the left indicate RNA size markers, in nucleotides.
The most abundant products were basically consistent with the single repeat-spacer unit (mainly of 64 to 68 nt), given the constant repeat size (of 30 nt) and the spacer size varying from 34 to 38 nt (with four exceptions of 33, 39, 47, and 54 nt) (Fig. 3D). Interestingly, products smaller than 50 nt were also detected by the repeat probe (Fig. 3A) and were assumed to be the processed transcripts extending from the TSS to the first PCS (∼43 nt). This assumption was confirmed by another Northern analysis with a mixture of six probes against these transcribed leader regions (Fig. 3C), and these products were thus designated leader-repeat (L-R) products. In addition, Northern analyses with spacer-specific probes were also performed against all CRISPR loci. A sole band corresponding to the single repeat-spacer unit was observed for most arrays (Fig. 3E), suggesting the absence of an exonucleolytic trimming process. We further performed RT-PCR analysis against 5′- to 3′-end-ligated RNAs with divergent primers (P12S3-F/R) specific to the third spacer of P12 array (see Materials and Methods), and the result revealed that nearly all products, including the intermediates and mature crRNAs, could be cyclized with their two ends perfectly forming an intact repeat (see Fig. S1 in the supplemental material), thus reaffirming the absence of the trimming process.
Multiple Cas proteins are involved in crRNA biogenesis.
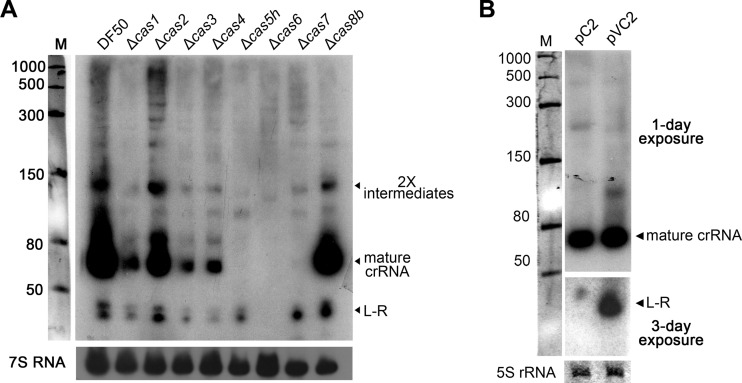
To investigate the roles of core or subtype-specific Cas proteins in crRNA biogenesis, we constructed single mutants for each cas gene. Total RNA was sampled from their stationary cultures, and Northern blot analysis was performed with the repeat probe (Fig. 4A). As expected, deletion of cas6 resulted in extinction of nearly all regularly processed products. Unexpectedly, the intermediates and mature products obviously decreased in cas1, cas3, and cas4 mutants (Fig. 4A). Because Cas1, Cas3, and Cas4 are believed to be involved in either adaption or interference stages, it was implied that crRNA biogenesis might be coordinated with the other two stages through unrevealed mechanisms. A similar decrease was observed for cas5 and cas7 mutants, but their product patterns are rather special in that mature crRNA completely disappeared while the L-R product unexpectedly persisted (Fig. 4A). The persistence of the L-R product indicated that neither CRISPR transcription nor endonucleolytic processing was blocked by the lack of Cas5 or Cas7. Thus, the result underlined the essential roles of Cas5 and Cas7 in stabilizing mature crRNA, which is consistent with their in vitro revealed interactions with the conserved 5′ handle (16, 37).
Fig 4.
Dissection of the roles of Cas proteins in crRNA biogenesis. (A) Northern blot of total RNA from the cas mutants and their parental strain (DF50) with the repeat probe. After exposure and stripping, a second hybridization was performed with the 7S RNA-specific probe serving as the internal control. (B) Northern blot with a repeat probe analyzing the products from the modified pWL502 vectors carrying a wild-type C2 array (pC2) or a variant array (pVC2) lacking the AT(C/T)GG(G/C)CATG leader motif (Fig. 2A). These plasmids were transformed into a CRISPR-free strain. Values to the left indicate RNA size markers, in nucleotides.
The blotting result also indicates that none of the Cas proteins, except Cas6, could be responsible for stabilizing the L-R product (Fig. 4A), and we propose that it may have been stabilized by Cas6, given the in vitro observation that Cas6 remains tightly bound to the 3′ terminus of products after cleavage (17, 18, 22). However, we also noticed a conserved AU(U/C)GG(G/C)CAUG motif carried by the L-R product (Fig. 2A), suggesting another possibility that other cellular proteins may specifically protect the L-R product by binding to this motif. We constructed a variant C2 array with the motif omitted, and, unexpectedly, the shortened L-R product became more abundant (Fig. 4B); this result definitely ruled out the latter possibility and indicated the Cas6-mediated stabilization for this product, which seemed to be negatively related to the size of the 5′ free end of the L-R product. This negative correlation is consistent with the different Cas6-mediated stabilization effects on mature crRNA (64 to 68 nt) and the L-R product (∼43 nt) in cas5h and cas7 mutants (Fig. 4A).
The putative Cascade proteins mediate inhibition of a defective provirus.
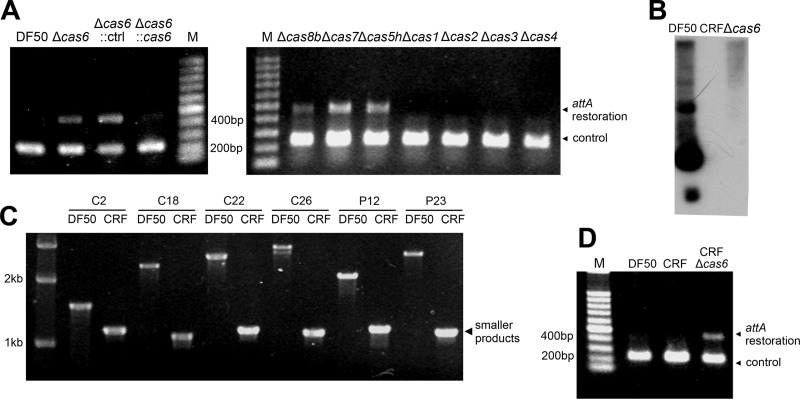
As mentioned above, in the H. mediterranei genome, most chromosomal CRISPR loci are physically related to proviruses (Fig. 1). To test whether the disruption of the CRISPR-Cas system has any effects on proviral behaviors, test primers were designed based on the chromosomal sequences surrounding provirus 1 (Fig. 1). When the provirus excises itself from the chromosome, a small PCR product consisting of the upstream and downstream sequences will be observed with an intact attA (attachment site of archaea) site restored.
Genomic DNA samples from various cas mutant strains were used separately as templates for PCRs. Interestingly, a 471-bp PCR product was observed in the absence of cas6, cas5h, cas8b, or cas7 (Fig. 5A), and this product was consistent with the predicted uninterrupted chromosomal sequence. This result indicated that the provirus had been derepressed and excised from the chromosome in these cas gene environments. However, no virus particles were observed, suggesting that the provirus may be corrupted and defective for propagation even though it was active in an integration-excision process. Complementation of a plasmid-carried cas6 gene in the cas6 mutant rescued the wild-type phenotype (Fig. 5A), thus confirming that the observed inhibition was a result of the coexistence of these proteins, which seemed to be components of the Cascade complex. Interestingly, for the single mutants of any core cas gene, including cas3, attA site restoration was rarely observed. This result was extremely surprising because the Cas3 protein has been shown to be indispensable for the type I interference pathway (11, 38). Therefore, a Cas3-independent mechanism is suggested for the Cascade ribonucleoprotein complex.
Fig 5.
Detection of attA site restoration of provirus 1 in strains with various CRISPR-Cas backgrounds. The PCR product indicating attA site restoration and a control product were labeled in panels A and D, and the corresponding primers are diagramed in Fig. 1. (A) Detection of attA site restoration in strains with different cas gene environments. The Δcas6::cas6 and Δcas6::ctrl strains are Δcas6 strains complemented with a pWL502 vector carrying the gene cas6 or not, respectively. (B) Northern blot analysis with a repeat probe confirming the extinction of crRNA in a CRF strain. (C) Validation of the genotype of the CRISPR-free (CRF) strain. Forward and backward primers were ∼600 bp upstream or downstream from each deleted CRISPR array; thus, smaller products (∼1.2 kb) for the CRF strain indicate the absence of these arrays. (D) The effect of CRISPR deficiency on attA site restoration. Lanes M, molecular size markers.
To determine which CRISPR locus is responsible for this inhibition, we successively knocked out all six CRISPR loci (Fig. 5B and C). Unexpectedly, for the final CRISPR-free (CRF) strain, attA site restoration was still not observed (Fig. 5D). When cas6 was further deleted, attA site restoration was reestablished, implying that the inhibition was relieved again. These results indicate either the presence of an undetected CRISPR array or the independence on CRISPR arrays. However, Northern blotting of crRNA production in the CRF strain with the repeat probe did not detect any processed or unprocessed transcripts (Fig. 5B). Therefore, we propose that the observed inhibitory effect on this provirus is mediated by the Cascade protein members independently of CRISPR.
Cas6-mediated cleavage occurs against a proviral transcript.
Based on the observations above, we infer that Cas6-mediated processing and/or the recognition by other Cascade proteins must have occurred on some non-CRISPR RNA substrates. A logical hypothesis may be that Cas6 directly targeted some proviral transcripts. Therefore, using the HMMER package (39), a hidden Markov model constructed from the direct repeat alignments was employed to search for putative cleavage sites within the proviral genome. The best candidate sequence was found at 9 bp upstream of the coding region of HFX_0906, which encodes a PhiH-like regulator (Fig. 6A).
Fig 6.
Analysis of a noncognate repeat-like sequence on the proviral phiH transcript. (A) The repeat-like sequence (boxed) on the phiH transcript. The transcription start (TSS) and termination (TTS) sites determined in a cas6 mutant strain are labeled. The putative BRE and TATA elements are underlined, and the start and stop codons are in bold. The Cas6 cleavage site within the repeat-like sequence determined in the DF50 strain is indicated with a small black triangle. (B) Northern analysis of crRNA production from a modified C2 array (pNCG) whose first repeat has been replaced by the proviral noncognate sequence, with a probe against the spacer sequence. Values to the left indicate RNA size markers, in nucleotides. (C) Alignment of the noncognate repeat of the phiH transcript to the consensus sequence of the haloarchaeal CRISPR repeats (Hbor6C to Npha5C: Hbor, Halogeometricum borinquense; Hhis, Haloarcula hispanica; Hmar, Haloarcula marismortui; Hmed, H. mediterranei; Hmuk, Halomicrobium mukohataei; Hwal, Haloquadratum walsbyi; Hvol, Haloferax volcanii; Huta, Halorhabdus utahensis; Nmag, Natrialba magadii; Npha, Natronomonas pharaonis). Mismatches are indicated by asterisks. P or C represents the location on a plasmid or a chromosome, respectively, and the number denotes the repeat number. Black or grey shading indicates the same nucleotide at this position in all or most sequences, respectively. (D) Secondary structure of the repeat-like RNA sequence predicted by the RNAfold server.
The gene HFX_0906 shares a high sequence similarity with two other coexisting phiH homologues (HFX_0905 and HFX_1329). To circumvent its low transcription level and noises from other homologues, we constructed a modified C2 array with its first repeat unit replaced by this noncognate sequence and detected its transcripts in the CRF strain with a spacer-specific probe (Fig. 6B). Interestingly, this sequence was shown to be efficiently processed by Cas6, despite three mismatches at conserved positions with respect to the repeat consensus sequence and an altered stem-loop structure (Fig. 6C and D). Additionally, we employed a recently developed high-sensitivity method, namely, CR-RT-PCR (22), to determine the 5′ extremity of phiH transcript (see Materials and Methods). The phiH transcript in the Δcas6 strain initiated 121 bp upstream from the start codon, while in the DF50 strain the 5′ end was determined within the repeat-like sequence (Fig. 6A), explicitly demonstrating the occurrence of Cas6-mediated cleavage. This cleavage site was generally consistent with the PCS identified in the pre-crRNA (Fig. 2), with a 1-nt discrepancy which possibly resulted from the RNA sequence and structural variations (Fig. 6C and D). Cleavage at this spurious repeat sequence produced a spurious 5′ handle, possibly recognized by other Cascade proteins, which may impede the translation process and decrease the dose of PhiH-like regulator that may regulate proviral behaviors. Therefore, we propose that the interactions between Cascade proteins and the repeat-like sequence may account for the observed CRISPR-independent inhibition (Fig. 5).
DISCUSSION
The haloarchaeal Cas system seems to have undergone a long-term independent evolution based on the following observations: (i) in the phylogenetic tree constructed from the alignment of haloarchaeal Cas1 (the most common Cas) proteins and their most closely related homologues, the haloarchaeal members gathered into an isolated group (see Fig. S2 in the supplemental material); (ii) subtype I-B is defined by the signature protein Cas8b, while a BLAST search with H. mediterranei Cas8b (HFX_6315) as the query sequence revealed full-length homology only to proteins from haloarchaeal genomes and not to any other Cas8b homologues or TM1802 family proteins (data not shown). Given this evolutionary independence, distinct mechanisms could have evolved for this special ecological group, for example, the multiple PAM sequences utilized for interference (26).
Indeed, as to the crRNA biogenesis pathway, we have observed significant features that distinguish the haloarchaeal systems from other subtype I-B CRISPR-Cas systems. In the bacteria Clostridium thermocellum and the archaeon Methanococcus maripaludis C5, subtype I-B systems have been recently characterized with respect to crRNA biogenesis, and a 3′ trimming process was detected for them (24). However, in our study, the subtype I-B CRISPR RNA was only subjected to an endonucleolytic process (Fig. 3E; see also Fig. S1 in the supplemental material). Furthermore, the haloarchaeal CRISPR-Cas system seems rather different from that of M. maripaludis, not only in repeat sequence but also in cas operon architecture, and the homology of their Cas6 proteins could hardly be detected even using the most sensitive PSI-BLAST algorithm. Though one of the two C. thermocellum Cas6 proteins (Cthe_2303) shows considerable homology to H. mediterranei Cas6 (HmCas6), their cognate repeat sequences are rather different. Therefore, although collectively defined as I-B subtypes, the haloarchaeal systems are very different from those of methanogens or Clostridia, not only in components but also in molecular mechanisms.
Exquisite structural studies have focused on interactions between three Cas6 homologues and their cognate substrates (17, 19, 21, 22). Cas6e and Cas6f utilize different structural elements to interact with the major grove of the stem-loop structure on repeat RNAs (17, 22), while a distinct wrap-around mechanism has been proposed for P. furiosus Cas6 (PfCas6) in processing an unstructured RNA substrate (19). HmCas6 exhibits a higher sequence homology (with an identity of 16%) to PfCas6 than the other two homologues. The predicted secondary structure of HmCas6 also shows a highly similar arrangement to that of PfCas6 (see Fig. S3 in the supplemental material) although the revealed nucleolytic triad for PfCas6 (18) is not conserved in HmCas6. In addition, in contrast to the sequence- and structure-specific processing mechanism revealed for Cas6f (17, 23), the HmCas6-mediated cleavage seems able to tolerate more variations in substrate sequence and in the helical geometry of the stem-loop structure, as in the case of the proviral repeat-like sequence (Fig. 6). Therefore, we propose that HmCas6 may adopt a wraparound mechanism similar to that of PfCas6 though H. mediterranei repeat sequences exhibit a high probability of forming a stem-loop structure (Fig. 2D).
Given the considerable tolerance of sequence variations, physiological roles other than processing pre-crRNA may have evolved for HmCas6, and the observed inhibition of a defective provirus may be one of them. In this inhibition, CRISPR RNA and Cas3, both recognized as key elements involved in the type I interference stage (11, 38), were not required (Fig. 5). Instead, the putative Cascade proteins, Cas5h, Cas6, Cas7, and Cas8b, proved to be indispensably involved, suggesting a distinct mechanism in which these RNA-interacting proteins should have interacted with some non-CRISPR RNA substrates. As expected, a phiH-like transcript was found containing a repeat-like sequence and was efficiently processed by Cas6 (Fig. 6), suggesting that the transcript could be directly targeted by this endonuclease. Although cleavage at this site does not disrupt the coding region, the cleavage itself and/or subsequent binding of other Cascade proteins to the conserved 5′ handle may impede the translation process. The altered dose of PhiH regulator will certainly influence proviral behaviors, like the excision process. Therefore, we propose that the spurious recognition and cleavage of this proviral transcript should be responsible for the repression of this provirus though detailed mechanisms need to be further investigated. According to our knowledge, this is the first report of a naturally occurring interaction between Cas6 endonuclease and a non-CRISPR RNA molecule. It is also implied that Cas6 may process other cellular RNA candidates and that the Cascade proteins may be involved in other physiological pathways. In fact, we observed other physiological alterations for these cas mutants, like the altered population growth parameters (data not shown).
In summary, our study provides fundamental information for the evolutionarily isolated haloarchaeal subtype I-B CRISPR-Cas systems concerning essential elements and mechanisms involved in the crRNA biogenesis pathway. It is also indicated that, after long-term independent evolution, unusual mechanisms and physiological functions have evolved for the haloarchaeal Cas machineries, for example, the putative interactions between Cascade proteins and noncognate RNAs, which will be further elucidated in the future.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (30830004, 30925001, and 31271334) and the Chinese Academy of Sciences (KSCX2-EW-G-2-4).
Footnotes
Published ahead of print 30 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01688-12.
REFERENCES
- 1. Karginov FV, Hannon GJ. 2010. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol. Cell 37:7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sorek R, Kunin V, Hugenholtz P. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6:181–186 [DOI] [PubMed] [Google Scholar]
- 3. van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. 2009. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 34:401–407 [DOI] [PubMed] [Google Scholar]
- 4. Wiedenheft B, Sternberg SH, Doudna JA. 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482:331–338 [DOI] [PubMed] [Google Scholar]
- 5. Jansen R, Embden JD, Gaastra W, Schouls LM. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43:1565–1575 [DOI] [PubMed] [Google Scholar]
- 6. Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9:467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 [DOI] [PubMed] [Google Scholar]
- 8. Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190:1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Horvath P, Romero DA, Coûté-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190:1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Erdmann S, Garrett RA. 2012. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol. Microbiol. 85:1044–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carte J, Wang R, Li H, Terns RM, Terns MP. 2008. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 22:3489–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lillestøl RK, Shah SA, Brügger K, Redder P, Phan H, Christiansen J, Garrett RA. 2009. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol. Microbiol. 72:259–272 [DOI] [PubMed] [Google Scholar]
- 15. Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, Beijer MR, Barendregt A, Zhou K, Snijders AP, Dickman MJ, Doudna JA, Boekema EJ, Heck AJ, van der Oost J, Brouns SJ. 2011. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 18:529–536 [DOI] [PubMed] [Google Scholar]
- 16. Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E. 2011. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477:486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329:1355–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. 2010. Binding and cleavage of CRISPR RNA by Cas6. RNA 16:2181–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang R, Preamplume G, Terns MP, Terns RM, Li H. 2011. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure 19:257–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ebihara A, Yao M, Masui R, Tanaka I, Yokoyama S, Kuramitsu S. 2006. Crystal structure of hypothetical protein TTHB192 from Thermus thermophilus HB8 reveals a new protein family with an RNA recognition motif-like domain. Protein Sci. 15:1494–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. 2011. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat. Struct. Mol. Biol. 18:688–692 [DOI] [PubMed] [Google Scholar]
- 22. Sashital DG, Jinek M, Doudna JA. 2011. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat. Struct. Mol. Biol. 18:680–687 [DOI] [PubMed] [Google Scholar]
- 23. Sternberg SH, Haurwitz RE, Doudna JA. 2012. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA 18:661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Richter H, Zoephel J, Schermuly J, Maticzka D, Backofen R, Randau L. 2012. Characterization of CRISPR RNA processing in Clostridium thermocellum and Methanococcus maripaludis. Nucleic Acids Res. 40:9887–9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, Doudna JA. 2011. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. U. S. A. 108:10092–10097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fischer S, Maier LK, Stoll B, Brendel J, Fischer E, Pfeiffer F, Dyall-Smith M, Marchfelder A. 2012. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J. Biol. Chem. 287:33351–33363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu H, Han J, Liu X, Zhou J, Xiang H. 2011. Development of pyrF-based gene knockout systems for genome-wide manipulation of the archaea Haloferax mediterranei and Haloarcula hispanica. J. Genet. Genomics 38:261–269 [DOI] [PubMed] [Google Scholar]
- 28. Cai S, Cai L, Liu H, Liu X, Han J, Zhou J, Xiang H. 2012. Identification of the haloarchaeal phasin (PhaP) that functions in polyhydroxyalkanoate accumulation and granule formation in Haloferax mediterranei. Appl. Environ. Microbiol. 78:1946–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cline SW, Lam WL, Charlebois RL, Schalkwyk LC, Doolittle WF. 1989. Transformation methods for halophilic archaebacteria. Can. J. Microbiol. 35:148–152 [DOI] [PubMed] [Google Scholar]
- 30. Sun C, Li Y, Mei S, Lu Q, Zhou L, Xiang H. 2005. A single gene directs both production and immunity of halocin C8 in a haloarchaeal strain AS7092. Mol. Microbiol. 57:537–549 [DOI] [PubMed] [Google Scholar]
- 31. Kuhn J, Binder S. 2002. RT-PCR analysis of 5′ to 3′-end-ligated mRNAs identifies the extremities of cox2 transcripts in pea mitochondria. Nucleic Acids Res. 30:439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35:W52–W57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brenneis M, Hering O, Lange C, Soppa J. 2007. Experimental characterization of Cis-acting elements important for translation and transcription in halophilic archaea. PLoS Genet. 3:e229 doi:10.1371/journal.pgen.0030229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marraffini LA, Sontheimer EJ. 2010. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463:568–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Djordjevic M, Severinov K. 2012. CRISPR transcript processing: a mechanism for generating a large number of small interfering RNAs. Biol. Direct 7:24 doi:10.1186/1745-6150-7-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lillestøl RK, Redder P, Garrett RA, Brügger K. 2006. A putative viral defence mechanism in archaeal cells. Archaea 2:59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, Sdano M, Peng N, She Q, Copié V, Young MJ, White MF, Lawrence CM. 2011. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J. Biol. Chem. 286:21643–21656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Westra ER, van Erp PB, Künne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, de Vos WM, Dame RT, de Vries R, Brouns SJ, van der Oost J. 2012. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell 46:595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14:755–763 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.