Abstract
Silicibacter sp. strain TM1040, a member of the Roseobacter clade, forms a symbiosis with unicellular phytoplankton, which is inextricably linked to the biphasic “swim or stick” lifestyle of the bacteria. Mutations in flaC bias the population toward the motile phase. Renewed examination of the FlaC− strain (HG1016) uncovered that it is composed of two different cells: a pigmented type, PS01, and a nonpigmented cell, PS02, each of which has an identical mutation in flaC. While monocultures of PS01 and PS02 had few motile cells (0.6 and 6%, respectively), coculturing the two strains resulted in a 10-fold increase in the number of motile cells. Cell-free supernatants from coculture or wild-type cells were fully capable of restoring motility to PS01 and PS02, which was due to increased fliC3 (flagellin) transcription, FliC3 protein levels per cell, and flagella synthesis. The motility-inducing compound has an estimated mass of 226 Da, as determined by mass spectrometry, and is referred to as Roseobacter Motility Inducer (RMI). Mutations affecting genes involved in phenyl acetic acid synthesis significantly reduced RMI, while defects in tropodithietic acid (TDA) synthesis had marginal or no effect on RMI. RMI biosynthesis is induced by p-coumaric acid, a product of algal lignin degradation. When added to algal cultures, RMI caused loss of motility, cell enlargement, and vacuolization in the algal cells. RMI is a new member of the roseobacticide family of troponoid compounds whose activities affect roseobacters, by shifting their population toward motility, as well as their phytoplankton hosts, through an algicidal effect.
INTRODUCTION
Dense populations of unicellular algae, often called harmful algal blooms (HABs), affect humans by producing toxins that accumulate in shellfish and fish, and the environment through an invasive biomass that affects other organisms and food webs. Marine algae (phytoplankton) interact with a diverse community of heterotrophic bacteria numerically dominated by species of the Roseobacter clade of Alphaproteobacteria (1). Roseobacters are rapid colonizers of surfaces and often inhibit other bacteria, establishing dominance over other bacteria in this niche (2–6). This dominance is in part due to the ability of roseobacters to utilize dimethylsulfoniopropionate (DMSP; see Fig. S1 in the supplemental material), the major organic sulfur compound produced by marine unicellular algae (7). Metabolism of DMSP leads to the production of dimethylsulfide (DMS), which eventually affects cloud formation and global climate (8, 9).
Our laboratory uses Silicibacter sp. strain TM1040 (henceforth TM1040) as a model to study the molecular mechanisms underlying roseobacter interactions with marine phytoplankton. TM1040 was isolated from a laboratory microcosm consisting of the heterotrophic DMSP-producing dinoflagellate Pfiesteria piscicida, Rhodomonas salina algae, and over 30 bacterial species (10, 11). TM1040 is chemotactic toward dinoflagellate products, particularly DMSP (12), and uses chemotaxis and flagellar rotation to swim into close proximity to its host (13). When in the vicinity of the algae, the bacteria stop swimming and form a biofilm on phytoplankton, thus initiating the symbiosis (10, 14). The symbiosis is facilitated by a bacterial biphasic “swim or stick” lifestyle that involves motile as well as sessile phases. Mutants of TM1040 that lose motility have attenuated ability to initiate the symbiosis, implying a major role of roseobacterial motility in the interaction, which is likely to play a role in phytoplankton bloom dynamics in the ocean (13).
In addition to its chromosome harboring >40 open reading frames (ORFs) required for motility and chemotaxis (15, 16), the genome of TM1040 consists of three large plasmids: pSTM1 (823 kb), pSTM2 (131 kb), and pSTM3 (135 kb) (16, 17). pSTM3 escaped initial sequencing attempts and is not part of the published genome, but subsequent analyses indicate that it harbors at least six genes required for biosynthesis of the antibiotic and quorum signaling molecule tropodithietic acid (TDA; see Fig. S1 in the supplemental material), which is pivotal to the TM1040-algal symbiosis (16–18). TDA is a troponoid molecule derived from phenyl acetic acid (PAA; see Fig. S1 in the supplemental material), containing twin sulfur atoms, and is synthesized primarily in the sessile phase (14, 19). Pigment production in roseobacters is correlated with TDA production and, along with biofilm formation, is a major hallmark of the sessile phase (3). TDA not only autoregulates its own production but also plays a putative role as an activator of sessile-phase gene expression (17, 18). In contrast, when grown under vigorous shaking, TM1040 cells are flagellated, are highly motile, and produce very low levels of TDA and pigment (17, 20).
We have previously reported on genes involved in the biosynthesis and regulation of flagella and control of the motile phase (13, 15). The TM1040 genome contains homologs of Caulobacter cckA and ctrA that encode a two-component master regulator of flagella and motility genes (13, 16). Mutations in either of the genes result in defects in TM1040 swimming (13). In addition, random transposon mutagenesis uncovered three novel transcriptional regulators of motility: FlaB, FlaC, and FlaD (15). The deduced sequence of FlaB suggests it is a histidine phosphotransferase, with homology to Caulobacter ChpT, and is involved in the phosphorelay between CckA and CtrA in C. crescentus (15, 21). FlaC is a response regulatory protein, and FlaD has a MarR-like DNA-binding domain, suggesting it functions as a transcriptional regulator (15).
FlaC is a 25.8-kDa protein possessing an N-terminal CheY-like receiver domain (22) and a C-terminal DNA-binding domain (23), suggesting that it functions as a two-component signal transduction transcriptional response regulator (24). Previously, we reported that a strain (HG1016), with a transposon insertion in flaC, is phenotypically biased toward the motile phase, i.e., in broth culture the population of HG1016 cells is highly enriched for motile cells that produce negligible TDA and pigment. We infer from this that FlaC is a positive regulator of the sessile phase.
HG1016 motility is odd: while producing robust motility in broth, when placed in semisolid motility agar, these cells barely move outward, and in our initial phenotypic screenings were considered Mot− (defect in swimming motility). In the present study, we focused on the odd swimming behavior. We found that HG1016 is composed of two different Mot− mutants, PS01 and PS02, each of which has an identical flaC mutation, that when in coculture (mixed incubation) in 2216 broth are highly motile. We discovered that the coculture supernatant contained a small molecule inducer of motility that we call roseobacter motility inducer (RMI), which acts at the transcriptional level to induce flagellar gene expression and flagellum biosynthesis. RMI is a troponoid molecule of 226 Da that uses phenylacetic acid as a precursor and may share some enzymes in common with the TDA biosynthetic pathway. RMI acts as an algicide, and marine unicellular algal species exposed to the compound quickly lose motility, become highly enlarged, and ultimately lyse. These characteristics indicate that RMI is a member of the newly discovered roseobacticide family of compounds (25, 26). More importantly, our findings suggest that RMI is both an inducer of roseobacter motile-phase and a killer of the host alga, features that could influence the roseobacter-alga symbiosis and phytoplankton population dynamics.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains and plasmids used in the present study are listed in Table 1. TM1040 was routinely grown in 2216 marine broth (Becton Dickinson, Franklin Lakes, NJ) at 30°C or on 2216 agar (2216 marine broth with 1.5% agar). Kanamycin and tetracycline were added to final concentrations of 120 and 15 μg/ml, respectively, as required. Escherichia coli strains were grown in Luria-Bertani (LB) broth (10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl per liter) or on LB agar (LB broth with 1.5% agar).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype and/or phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | F− endA1 hsdR17(rK− mK−)supE44 thi-1 recA1 gyrA96 relA1 ϕ80dlacZΔM15 | 27 |
| S17-1λpir | thi pro recA hsd(r− m+) RP4-2 Tc::Mu-Km::Tn7 Smr λpir | 28 |
| Roseobacter | ||
| Silicibacter sp. strain TM1040 | Wild type | 29 |
| HG1016 | flaC::EZ-Tn5; Kanr | 15 |
| HG1006 | flaB::EZ-Tn5; Kanr | 15 |
| HG1038 | ctrA::EZ-Tn5; Kanr | 17 |
| HG1310 | tdaA::EZ-Tn5; Kanr | 17 |
| HG1015 | tdaB::EZ-Tn5; Kanr | 17 |
| HG1265 | tdaE::EZ-Tn5, Kanr | 17 |
| HG1050 | tdaF::EZ-Tn5, Kanr | 17 |
| HG1244 | tdaH::EZ-Tn5; Kanr | 17, 18 |
| PS01 | flaC pigmented | This study |
| PS02 | flaC nonpigmented | This study |
| Plasmids | ||
| pEA203 | cdhA′::lacZ Ori pC2A Ampr Purr Ori R6K | 30 |
| pRK415 | Conjugative expression shuttle vector for Silicibacter sp. strain TM1040 and E. coli; Plac; Tetr | 31 |
| pPS1 | 300 bp upstream of fliC3 fused to promoterless lacZ gene in pRK415 | This study |
| pRSI506 | pRK415 vector with 1,022 bp of genomic DNA containing 235 bp upstream and 100 bp downstream of flaC | 15 |
Kanr, kanamycin resistance; Smr, streptomycin resistance; Tetr, tetracycline resistance; Ampr, ampicillin resistance; Purr, puromycin resistance.
Characterization of HG1016 (flaC) cell types.
HG1016 colonies were isolated on 2216 agar using the streak plate method, and the resulting colonies screened for pigment production and motility. Strains PS01 and PS02, derived from HG1016, were identified for further analysis. Pigment production after static culturing in 2216 marine broth for 48 h at 30°C was measured as the absorbance at 398 nm using a cell-free supernatant obtained by pelleting the bacteria by centrifugation at 10,000 × g for 10 min (3). To measure swimming motility, 5 μl of an overnight culture was stab inoculated into the center of marine motility agar (2216 marine broth supplemented with 0.3% agar per liter) and incubated at 30°C for 48 h, after which the outermost edge of the swimming colony was recorded.
DNA preparation.
Total DNA was purified from bacteria using the DNeasy blood and tissue kit (Qiagen, Valencia, CA). Plasmid DNA was extracted and purified using a QIAprep miniprep kit (Qiagen).
Complementing flaC mutation.
The flaC mutation in PS01 and PS02 was determined by PCR and DNA sequencing using primers FlaC-F (5′-CAGTCCCATCCTCAGATCCACTC-3′) and FlaC-R (5′-GACAGGGAGGATGCATATCGTGAC-3′) (15). PS01 and PS02 were transformed with pPRSI506 harboring an intact copy of flaC, as previously described (15). Complementation of the flaC defect in PS01 and PS02 was assessed by examining swimming motility.
Motion analysis.
Wet mounts of overnight cultures of bacteria grown in 2216 broth were prepared to determine the percentage of motile cells in the population. Ten fields of 5 s each were obtained using phase-contrast microscopy (Olympus BX60), a Qicam Fast 1394 camera, and Volocity software (Perkin-Elmer; v6.1.1). Prism 4.0 (GraphPad) statistical software was used to determine the mean and standard deviation of each parameter.
Flagellin detection.
Flagellin protein was purified from as described previously by Belas et al. (15). The protein content in each sample was determined by using a bicinchoninic acid protein assay (Pierce, Rockford, IL). Western blot analysis was carried out using rabbit polyvalent antisera against FliC3 (major flagellin) protein (United States Biological, Swampscott, MA). In short, 3 μg of total protein was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis using a 4 to 20% polyacrylamide gel (Ready-Gel; Bio-Rad, Hercules, CA). After electrophoresis, the proteins were transferred to a polyvinylidene difluoride membrane (Hybond-PGE; Healthcare Biosciences, Pittsburg, PA), followed by rinsing with 1× phosphate-buffered saline (PBS; 10 mM NaPO4, 150 mM NaCl, 3 mM KCl [pH 7.4]). The membrane was blocked overnight in 1× PBS (pH 7.3) containing 0.1% Tween 20 and 5% skim milk powder and then rinsed twice in 1× PBS plus 0.1% Tween 20. The FliC3 antibody was added at a 1:2,500 dilution and allowed to bind for 1 h at room temperature, after which it was rinsed from the gel, and horseradish peroxidase-conjugated goat anti-rabbit IgG (Cell Signaling Technology, Danvers, MA) was added at a final dilution of 1:25,000. After 1 h, the gel was rinsed in 1× PBS plus 0.1% Tween 20 and labeled bands detected using an ECL detection kit (Amersham, Pittsburg, PA).
Construction of a fliC3p::lacZ transcriptional reporter plasmid.
A transcriptional fusion between the fliC3 promoter (fliC3p: 300-bp segment 5′ to the start site of fliC3) and a promoterless copy of lacZ (30) was constructed as follows. Primers FliC3-F and FliC3-R (Table 2) were used to amplify a 300-bp fragment of TM1040 genomic DNA, introducing a PstI and SphI site, respectively. Similarly, PCR of pEA203 (30) template DNA using primers LacZ-F and LacZ-R introduced SphI and HindIII sites to the ends of DNA bearing the lacZ coding region. The amplified products were digested with PstI/SphI or SphI/HindIII, respectively, and the resulting products were ligated into pRK415 which had been previously digested with PstI and HindIII. The resulting plasmid, pPS1 (fliC3p::lacZ), was then transferred to E. coli DH5α by electroporation, with selection for tetracycline resistance. The correct construction was verified by PCR using primers FliC3-F and LacZ-R, followed by transfer of pPS1 to TM1040 strains via electroporation.
Table 2.
Primers used in the study
| Primer | Sequence (5′–3′) |
|---|---|
| FlaC-F | CAGTCCCATCCTCAGATCCACTC |
| FlaC-R | GACAGGGAGGATGCATATCGTGAC |
| LacZ-F | GCATGCGGAAACAGCTATGACCATGATTAC |
| LacZ-R | TGATAAGCTTTTATTTTTGACACCAGACCAACTGG |
| FliC3-F | AGTACTGCAGAAAGGGTCTGGATGTCTACAAT |
| FliC3-R | GCATGCCATAGCATGAGTTCCTTTGATTTG |
Measurements of fliC3 expression and induction.
Expression of fliC3p::lacZ reporter plasmid in PS02 (PS02/pPS1) was used to test the ability of various chemicals to induce transcription of fliC3 as follows. Cell-free supernatants were obtained from a coculture of PS01 and PS02 in 2216 marine broth at 30°C for 18 h under shaking conditions and with the respective chemicals added. PS02/pPS1 cells were resuspended in each supernatant and, after 90 min, fliC3 expression was measured using the Miller β-galactosidase assay (32). Compounds tested were p-coumaric acid (final concentration, 100 μM), phenylalanine (final concentration, 50 μM), and histidine (final concentration, 50 μM). A cell-free supernatant from coculture of PS01 and PS02 in 2216 broth, without the addition of any chemical, was used as a control. Means and standard deviations were determined and compared by using the Student t test (Prism 4.0). Each measurement was repeated at least three times.
Dose dependency and time course experiments.
Dose dependency experiments were performed by collecting cell-free supernatant from a culture of TM1040 inoculated in 2216 broth incubated at 30°C for 18 h. The supernatant was serially diluted in 2216 marine broth and used to resuspend the reporter strain, PS02/pPS1. Expression of fliC3 was measured using the Miller assay.
For the time course assay, PS02/pPS1 cells were incubated at 30°C in cell-free supernatants from a culture of TM1040 inoculated in 2216 broth incubated at 30°C for 18 h. At fixed time points (up to 7 h), 100 μl of the sample was collected and stored on ice. All samples were processed simultaneously using the Miller assay.
Biofilm measurement.
Overnight cultures of PS01, PS02, and TM1040 were diluted 1:100 with a cell-free supernatant from a culture of TM1040 grown in 2216 broth. Then, 200 μl of the dilution was then dispensed into 96-well plates and allowed to incubate for 48 h (static) at 30°C, after which wells were rinsed with 1× PBS. The biofilm formed on the surface of the wells was measured using a crystal violet assay (33), with. PS01, PS02, and TM1040 cells diluted in 2216 marine broth serving as controls.
Fluorescence microscopy.
Flagella immunostaining was performed using the procedure of Smith et al. (34), with minor modification. The cells were cultured in accordance with the experimental requirements and fixed using 100 μl of formaldehyde/glutaraldehyde fixative (500 μl of 16% paraformaldehyde,1 μl of 25% glutaraldehyde; Electron Microscope Science, Hatfield, PA) plus 20 μl of sodium phosphate buffer (pH 7.4) to a final concentration of 2.6% paraformaldehyde and 0.04% glutaraldehyde. After gentle mixing, the fixed cells were pipetted onto poly-l-lysine-treated coverslips (Electron Microscope Science) and allowed to incubate for 20 min at room temperature. The coverslips were washed with 1× PBS (pH 7.4) three times, followed by incubation with 2% bovine serum albumin (BSA fraction V; Sigma) in 1× PBS for 1 h. Next, 20 μl of anti-FliC3 primary antibody was added at a dilution of 1:100, and the coverslips were placed in a moist chamber overnight at 4°C. Each coverslip was washed with 1× PBS eight times and with 2% BSA-PBS once. Goat anti-rabbit IgG conjugated to Alexa Fluor 586 (1:1,000 titer; Invitrogen, Carlsbad, CA) was added as the secondary antiserum to the coverslips, followed by incubation for 3 h in the dark. The coverslips were rinsed in 1× PBS and slow-fade equilibration buffer (Invitrogen) once. At this step, 1 μl of 1 mg of FM1-43 (Invitrogen)/ml and 2 μl of 2 μg of DAPI (4′,6′-diamidino-2-phenylindole; Invitrogen)/ml were added to stain the bacterial membrane and DNA, respectively. After an incubation period of 1 h, the coverslips was rinsed with SlowFade (Invitrogen) equilibration buffer and allowed to dry. Next, 5 μl of antifade glycerol (Invitrogen, Carlsbad, CA) was then added, and the coverslips were mounted on glass microscope slides. The slides were stored at 4°C and observed using a Zeiss Axio Observer Z1, a Hamamatsu ORCA R2 camera, and Volocity software for image acquisition, deconvolution, and analysis.
Purification and chemical characterization RMI.
TM1040 incubated in 2216 broth at 30°C for 18 h was used for the purification of RMI. After incubation, the cells were pelleted by centrifugation (10,000 × g for 10 min) to obtain a cell-free supernatant. Each batch of supernatant (500 ml) was extracted three times with 150 ml of acidified ethyl acetate (0.2% ethyl acetate). The extract was dried under nitrogen and resuspended in 9 ml of 9:1 acidified acetonitrile, which was applied to a C18 column (Oasis Max; Waters). Four fractions were eluted, and their biological activity was determined by their ability to induce fliC3 expression. Further purification was achieved with high-pressure liquid chromatography (HPLC; Agilent 1100) using a Phenomenex (Torres, CA) Curosil PFP column (150-cm length, 2-mm diameter, and 3-μm pore size) and an acidified acetonitrile gradient. Mass spectroscopy of the active peak was carried out by direct sample analysis (DSA) source on a Perkin-Elmer AxION TOF instrument. The sample was run in a positive mode, with the drying gas set at zero, APCI heater at 300, nebulizer pressure at 80 lb/in2, and corona needle set to 2,000 V.
Algal experiments.
Cultures of Tetraselmis striata and T. chuii were obtained from the IMET algal facility and propagated in f/2 medium at 25°C with a light/dark photoperiod of 14 h-10 h. Algae (106 cells/ml) were exposed to 10 μM HPLC-purified RMI for 48 h at 25°C. Differential interference contrast microscopy was used to record changes in morphology and swimming behavior of RMI-treated algae, using the same microscope equipment as used for fluorescent specimens. Algal strains incubated with DMSO were maintained as solvent controls. More than five fields consisting at least 50 cells each were analyzed, and representative data were recorded. Morphometric analysis of cell volume and size was done using Volocity software.
RESULTS
HG1016 (flaC) is composed of two cell types.
Strain HG1016 (flaC::EZ-Tn) has an unusual phenotype: in broth culture the population is highly skewed toward motile cells, and yet in semisolid motility agar the cells are Mot− (15). In reexamining the unusual motility phenotype, we discovered that HG1016 is composed of two colony types (called PS01 and PS02) differing in pigmentation. PS01 produced near wild-type pigment, with an optical density at 398 nm (OD398) of 0.47, whereas PS02 was almost white (OD398 = 0.05), with more than four times less pigment. The difference in pigmentation is not due to the transposon, as both strains have an identical transposon insertion in flaC (see Fig. S2 in the supplemental material), as originally reported by Belas et al. (15).
PS01 and PS02 are Mot− (Table 3). When grown in broth culture under shaking conditions, PS01 cultures have 6% motile cells, whereas PS02 populations are composed of <1% motile cells. Surprisingly, when PS01 was cocultured with PS02, the resulting population of cells contained ca. 65% motile cells, more than twice the motile cells found in wild-type population (15). While HG1016 is complemented by wild-type flaC in trans (15), attempts to complement PS01 and PS02 with a plasmid bearing flaC (pRSI506) failed to restore the wild-type motility (data not shown), making it unlikely that the flaC mutation (by itself) caused the loss of motility. Indeed, coculturing PS01 and PS02, each complemented by flaC+ in trans, resulted in a motile population. We infer from this that PS01 and PS02 carry separate second site mutations, with the secondary mutation in PS02 involved in pigment biosynthesis.
Table 3.
Motility of PS01, PS02, and a coculture of PS01 plus PS02a
| Strain | Mean no. of cells ± SD | Mean no. of motile cells ± SD | Avg % of motile cells |
|---|---|---|---|
| PS01 | 210 ± 45 | 11 ± 8 | 6.0 |
| PS02 | 221 ± 44 | 2 ± 1 | 0.6 |
| Coculture | 216 ± 57 | 137 ± 50 | 65 |
Bacteria were incubated in 2216 broth at 30°C for 18 h with shaking. The means and standard deviations of 10 fields of view are presented.
Cell-free supernatants from a PS01-PS02 coculture induce motile phase, flagellin biosynthesis, and fliC3 transcription.
We reasoned that the simplest way coculturing could induce a motile-phase population from two Mot− strains was through the action of an extracellular compound. Cell-free supernatants from monocultures of either PS01 or PS02 had no effect on either Mot− strain. However, exposure of either PS01 or PS02 cells to a cell-free coculture supernatant resulted in populations composed of ca. 65% motile cells, based on visual inspection with phase-contrast microscopy. The active material in the cell-free supernatant passed through a 3.5-kDa filter, indicating that it is likely due to a small molecule.
A shift to motile phase is predicted to be accompanied by an increase in flagellin per bacterium that can be quantified by anti-flagellin immunoblots and immunofluorescence microscopy. As shown in Fig. 1A, flagellin (FliC3) was not detected in PS01 or PS02, but when either strain was exposed to a cell-free coculture supernatant, a strong flagellin band became apparent. Similarly, flagella were rarely seen (<1 filament in 50 fields on average) in monocultures of PS01 and PS02 (Fig. 1B; PS01 used as an example), but, upon exposure to a cell-free coculture supernatant (Fig. 1C), flagellar filaments were present on most cells and in all fields examined. As can be seen in Fig. 1C, when in the presence of a cell-free coculture supernatant, many detached filaments were also present. This may suggest that the supernatant induced flagellin synthesis above wild-type thresholds, forcing the cells to shed excess filaments.
Fig 1.
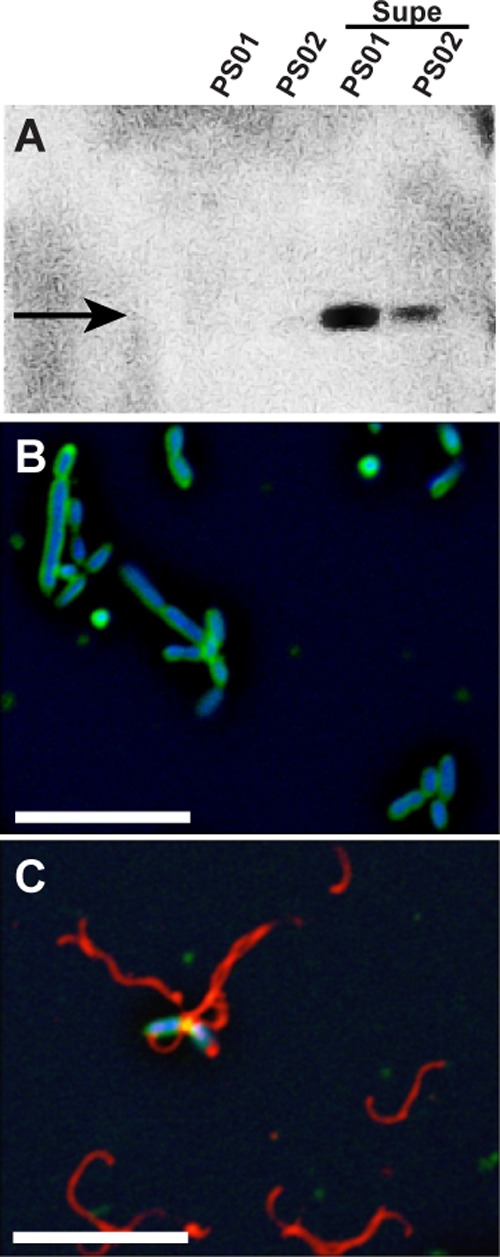
Effect of RMI on flagella biosynthesis. (A) Immunoblot with anti-flagellin antiserum. Lanes, from left to right: PS01, untreated; PS02, untreated; PS01, treated with RMI supernatant; PS01, treated with RMI supernatant. The arrow indicates the major flagellin, FliC3. (B and C) Immunofluorescence microscopy of untreated PS01 cells (B) and RMI supernatant-treated PS01 cells (C). A cell-free RMI supernatant (Supe) was obtained from a coculture of PS01 and PS02 cells, and PS01 cells were treated by resuspending the cells in RMI supernatant and incubating for 90 min at 30°C. In panels B and C, flagella are false-colored red, membranes are green, and DNA is blue. Scale bar, 5 μm.
Changes in expression of the major flagellin gene, fliC3, were also measured using a fliC3p::lacZ reporter plasmid (pPS1), and in keeping with flagellin protein measurements, when exposed to a cell-free coculture supernatant, fliC3 expression increased >2-fold (20.5 versus 55.1 U) when PS02 was exposed to a cell-free coculture supernatant (see Fig. 3). As expected, cell-free supernatant from monocultures of PS01 or PS02, or a mixture of the two monoculture supernatants (rightmost bar in Fig. 2) did not affect fliC3 expression. Thus, the action of the cell-free coculture supernatant is at the level of transcription, i.e., it is an inducer of flagellin expression. We call this unknown motility-enhancing substance RMI (Roseobacter Motility Inducer) and will use “RMI supernatant” when referring to a cell-free coculture supernatant, as used above.
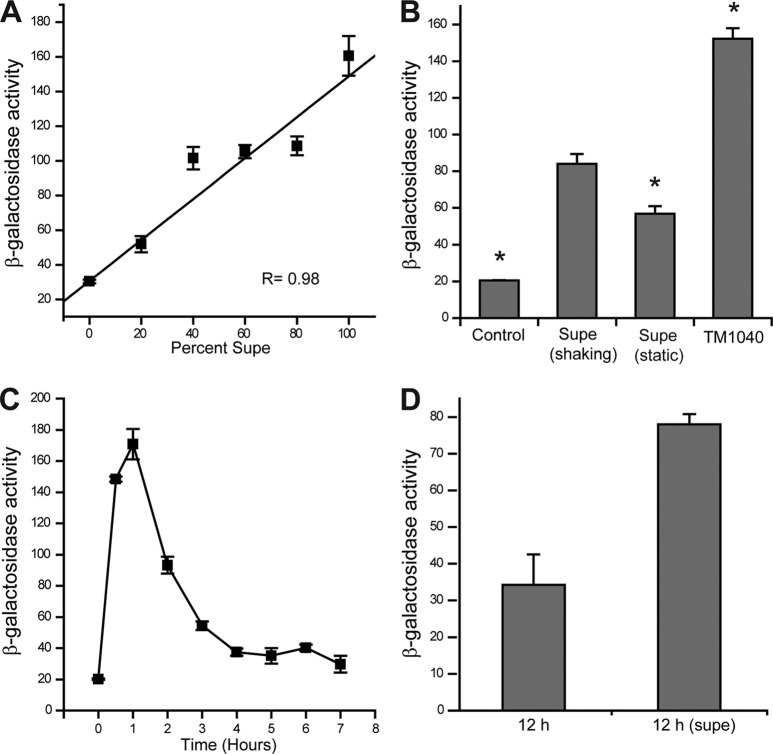
Fig 3.
RMI characteristics. A fliC3p::lacZ reporter strain, PS02/pPS1, was used to measure RMI activity, which is expressed in Miller units. (A) Dose dependence. Increasing amount of RMI supernatant, as a percentage of the culture medium, results in a linear increase in fliC3 expression. The linear regression coefficient for the plotted line is 0.98. (B) Culture of the cells in shaking nutrient broth (i.e., 2216) enhances RMI activity in a PS01-PS02 coculture, as well as in wild-type cells (TM1040). An asterisk above a bar indicates that the mean is statistically different from the supernatant collected from shaking cocultures of PS01 and PS02, labeled “Supe (shaking)”. (C) Time course of RMI induction of fliC3 expression. PS02/pPS1 cells were incubated in the presence of RMI cell-free supernatant at 30°C over the time period shown, with the maximum expression of fliC3 observed between 1 and 1.5 h. The β-galactosidase activity (fliC3 expression) fell thereafter to a basal level (4 h and beyond). (D) The addition of RMI after fliC3 expression falls reinduces transcription. Cell-free RMI supernatant was added to PS02/pPS1 cells 12 h after—“12 h (supe)”—an initial exposure to an RMI-containing supernatant. The same cells exposed to broth served as a control (“12 h”). The 12-h delay was chosen as preliminary data showed the greatest change in fliC3p::lacZ response occurred at this time. The means and standard deviations are shown.
Fig 2.
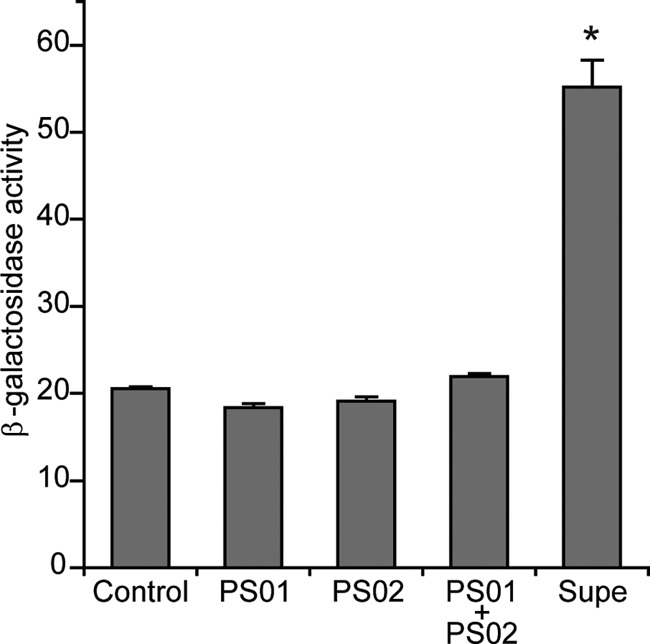
RMI is extracellular and requires coculturing of PS01 and PS02. Cell-free supernatants were obtained from PS01, PS02, and a coculture of both strains (labeled “Supe”) grown in 2216 broth. RMI activity was measured using a fliC3p::lacZ transcriptional fusion reporter strain (PS02/pPS1) and is reported as the β-galactosidase activity in Miller units. Controls: medium-only blank (Control) and monoculture supernatants from PS01 and PS02 separately and mixed 1:1 (PS01+PS02). The means plus the standard deviations are shown from three independent experiments. The asterisk indicates the cocultured supernatant is statistically significant from the controls, as determined by using the Student t test (P < 0.001).
RMI activity is dose dependent.
A dose-dependent response is often a sign of specificity of the reaction. We measured RMI activity using the fliC3 reporter plasmid over a dilution series that gave a range of RMI supernatant concentrations (Fig. 3A). The result was a straight line, dose-dependent response.
Shaking culture conditions enhance RMI activity.
Culture conditions affect the “swim-or-stick” switch: static broth culturing favors the sessile phase, while shaking cultures are biased toward motile cells (14). Similarly, we observed changes in RMI activity depended upon whether bacteria were incubated in shaking or static conditions (Fig. 3B). RMI activity (as measured by the fliC3 reporter plasmid) in RMI supernatants from shaking cultures was >145% higher than the activity from static culturing (84 ± 5 compared to 57 ± 4 Miller units). In contrast, the TDA activity is nearly absent when TM1040 is cultured under shaking conditions and maximal in static broths (17).
Given the results from PS01 and PS02, we predicted that wild-type cells should also produce RMI. Cell-free supernatants were obtained from TM1040 grown in shaking broth culture, and these were monitored for RMI activity using the fliC3p::lacZ reporter strain (PS02/pPS1). The results are shown in Fig. 3B (rightmost bar). TM1040 produced 152 U of activity, nearly twice the activity (1.8-fold) produced from RMI supernatant. We conclude that RMI is produced by wild-type cells, with maximal production during shaking broth culture conditions. Hence, cell-free supernatant obtained from culturing TM1040 in 2216 broth under shaking conditions were used for future experiments.
RMI decreases biofilm formation.
Since RMI-containing supernatant from TM1040 cultures induced the motile-phase phenotype, we sought to determine whether it also reduced the sessile phase phenotype and specifically biofilm formation. As shown in Table 4, RMI containing supernatant inhibited the formation of biofilms by PS01, PS02, and wild-type cells, reducing the amount of biofilm formed by each of them by >70% (Table 4). We conclude that RMI inhibits biofilm formation and propose that its effect extends to other sessile-phase phenotypes.
Table 4.
RMI effects on biofilm formation
| Strain | Mean avg biofilm (A600) ± SDa |
% Decrease in biofilm formation | |
|---|---|---|---|
| Pretreatment | Posttreatment | ||
| PS01 | 1.18 ± 0.12 | 0.18 ± 0.05* | 84.74 |
| PS02 | 1.21 ± 0.10 | 0.24 ± 0.06* | 80.16 |
| TM1040 | 1.27 ± 0.11 | 0.37 ± 0.04* | 70.86 |
Pretreatment refers to biofilm formation in 2216 broth. Posttreatment refers to biofilm formation in the presence of cell-free RMI supernatant collected from a culture of TM1040 inoculated in 2216 broth and incubated at 30°C for 18 h under shaking conditions. An asterisk indicates a statistically significant difference from the pretreated cultures using the Student t test (P < 0.001).
Our results implicate RMI as a signaling molecule, and this predicts that the effect of RMI should be transient and diminish with time due to adaptation and/or metabolic breakdown of RMI. To test this, a time course of RMI induction was conducted (Fig. 3C). RMI activity, as measured by fliC3 expression, increased to a peak (170 Miller units) 1.5 h after initial exposure, followed by a quick decline and leveling off to a baseline of <40 U after 4 h. Fresh RMI supernatant added 12 h after the start of the time course resulted in a 2-fold increase in fliC3 expression from <0 to 78 Miller units (Fig. 3D). Although the reexposure to RMI supernatant did not result in as much fliC3 expression as the initial exposure, the decreased induction is likely due to the growth phase of the cells, i.e., cells in late exponential phase have less fliC3 expression than those from mid-exponential phase of growth. These results support the hypothesis that RMI functions as a signaling molecule but are inconclusive as to whether metabolic breakdown of RMI or bacterial sensory adaptation to the effects of RMI are involved in signaling.
Identification of genes involved in RMI activity.
The results from PS01 and PS02 (Fig. 1 and see Fig. S2 in the supplemental material) suggest that FlaC, a putative transcriptional regulatory protein involved in maintaining sessility, is not involved directly in RMI activity, since cells with a defect in flaC respond to RMI induction of motility. These strains most likely have a separate second site mutation that prevents them from synthesizing RMI independently. What genes are involved in RMI activity? To answer this, we used our collection of TM1040 mutants, starting with genes known to be involved in regulating the “swim-or-stick” switch and its component pathways.
RMI activity was measured in strains bearing defects in two putative regulators of motility, CtrA and FlaB (13). HG1038 (ctrA::EZ-Tn5)/pPS1 and HG1006 (flab::EZ-Tn5)/pPS1 cells were separately exposed to RMI containing supernatant, and the fliC3 expression was determined. TM1040/pPS1 served as a positive control in these experiments. RMI-containing supernatant did not induce fliC3 expression in either ctrA or flaB mutants (see Table S1 in the supplemental material). One interpretation of these results is that RMI activity requires the flagellar “master regulatory circuit”—CckA, CtrA, and FlaB (ChpT)—in its induction of fliC3 expression. This means that RMI exerts its effects above the CckA-CtrA-FlaB circuit. Alternatively, RMI could function downstream of the circuit but upstream of a more direct regulator of fliC3.
In parallel experiments, RMI activity was measured in mutants with defects in the TDA pathway (Table 5). Supernatant from cells with mutation in the PAA pathway (paaI, paaJ, and paaK) had much less RMI activity than a control (averaging less than half the activity), suggesting that RMI synthesis requires a PAA precursor. Mutations in genes whose functions are dedicated to the TDA pathway, i.e., tdaA, tdaB, tdaE, tdaF, and tdaH, had little effect on RMI activity, although it is worth noting that both tdaA and tdaB mutants had consistently higher levels of RMI activity than the control (Table 5). Since TDA biosynthesis also requires a PAA precursor, the subtle increase in RMI activity may be due to more PAA becoming available for RMI synthesis when tdaA or tdaB is mutated. The common molecular origin of both TDA and RMI hints that RMI may share structural similarities to TDA.
Table 5.
RMI activity in paa and tda mutant backgrounds
| Strain | Function | Mean β-Galactosidase activitya | % β-Galactosidaseb |
|---|---|---|---|
| paaI mutant | Phenyl oxygenase | 26 ± 2.10 | 39 |
| paaJ mutant | Phenyl oxygenase | 42 ± 1.05 | 63 |
| paaK mutant | Phenyl oxidoreductase | 39 ± 0.33 | 59 |
| tdaA mutant | LysR substrate binding domain protein | 72 ± 0.70 | 109 |
| tdaB mutant | β-Etherase, GST | 80 ± 2.30 | 121 |
| tdaE mutant | Acyl-coenzyme A dehydrogenase (ACAD) | 58 ± 1.60 | 88 |
| tdaF mutant | Phosphopantothenoylcysteine decarboxylase | 60 ± 1.40 | 91 |
| tdaH mutant | Sulfite oxidase domain protein | 67 ± 1.20 | 100 |
| Control | 66 ± 2.20 | 100 |
RMI activity was measured using a fliC3p::lacZ reporter strain, PS02/pPS1, and is expressed as the β-galactosidase activity in Miller units. The means obtained from three independent experiments are presented.
That is, the percent increases from control (cell-free RMI supernatant obtained from TM1040 culture in 2216 broth).
The chemical nature of RMI.
We developed a purification scheme for RMI based on our method of TDA purification that relies on C18 solid-phase extraction, followed by further HPLC purification (17). We obtained four fractions upon applying a step gradient of increasing acetonitrile to water, and these were dried and tested for RMI activity using PS02/pPS1. The fourth (9:1 [acetonitrile-water]) fraction contained the most RMI activity, based on our fliC3 reporter plasmid. Purification of the active fraction by HPLC resulted in three peaks (see Fig. S3A in the supplemental material), with peak 2 yielding RMI activity. Based on mass spectrometry analysis (see Fig. S3B in the supplemental material) of HPLC peak 2, the mass of RMI is ca. 226 Da (227.16 M+H [see Materials and Methods]), which is very close to the mass of TDA (molecular weight of 212) and strengthens the hypothesis that RMI and TDA are structurally related.
Based on the known characteristics of RMI, we searched for other similar compounds produced by roseobacters and turned up two possibilities. The first one is a pair of newly discovered N-acylhomoserine lactones (AHLs) that induce motility in Ruegeria sp. strain KLH11 (35), while the second possibility is one of the recently discovered roseobacticides (25, 26) that share two features with RMI: a requirement for PAA and their size (ca. 250 to 500 Da). One interesting feature of roseobacticides is that their synthesis is induced by algal lignin breakdown products, such as p-coumaric acid (pCA) (36). We therefore measured the effect of pCA on RMI activity.
RMI supernatant was obtained from TM1040 grown under shaking broth culture conditions with 100 μM pCA (see Materials and Methods). The results are shown in Table 6. In the presence of pCA, the RMI activity increased ca. 1.6× (from 105 to 165 Miller units), whereas pCA on its own had no effect on fliC3 transcription. Other compounds tested (listed in Materials and Methods) had no effect on RMI production. Induction of RMI activity by pCA is in keeping with a hypothesis that RMI is a newly discovered member of the roseobacticide family of compounds.
Table 6.
pCA effect on RMI activity
| Condition | Mean β-galactosidase activity ± SDa |
|---|---|
| pCA | 167 ± 5 |
| Control | 105 ± 5 |
100 μM p-coumaric acid (pCA) was added to a mid-log-phase culture of PS02/pPS1 (fliC3p::lacZ). The control is RMI cell-free supernatant from TM1040 culture in 2216 broth incubated under shaking conditions. The RMI activity is expressed as the β-galactosidase activity in Miller units. Values indicate the means of three independent experiments.
Roseobacticides get their name because they harm unicellular algal cells (25, 26). We exposed two species of the marine algae, Tetraselmis striata and T. chuii, to 10 μM RMI (HPLC purified) and measured changes in the algal cells (Fig. 4). After 48 h of exposure, both species showed obvious physiological and morphological changes. These included loss of motility, a 2.5-fold-increased overall size of the cell (239 versus 95 μm2 for T. striata; Fig. 4), and increased numbers and sizes of intracellular vacuoles. Longer exposures to RMI resulted in cell lysis. These results are similar to those reported by Seyadsayamdost et al. on the effect of Roseobacticide A (see Fig. S1 in the supplemental material) on Emiliania huxleyi (26). The algicidal effect of RMI supports the hypothesis that RMI is chemically related to the roseobacticides.
Fig 4.
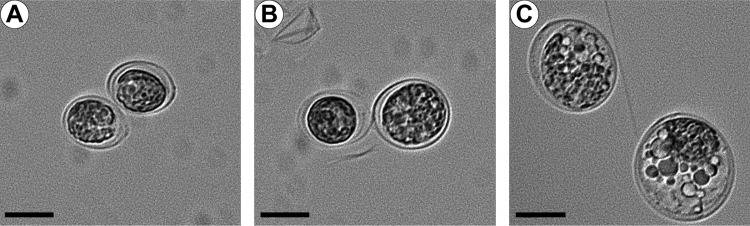
Effect of HPLC-purified RMI on unicellular algae. HPLC-purified RMI was added to cultures of Tetraselmis striata and T. chuii (not shown) at a final concentration of 10 μM. After 48 h, T. striata with no addition (A), or exposed to DMSO (B; solvent control) showed no changes in morphology. Algae exposed to RMI developed difficulty in swimming in < 6 h postexposure. (C) By 48 h, RMI-exposed T. striata ceased dividing, and their cells swelled, resulting in an overall enlargement and rounding of the algae; this was accompanied by an increase in the number of intracellular vacuoles. Similar results occurred with T. chuii exposed to RMI. Bar, 10 μm.
DISCUSSION
We encountered a fortunate stroke of serendipity in the discovery of RMI, when experiments designed to understand the unusual motility phenotype of HG1016 (flaC::EZ-Tn5) uncovered that it was composed of two separate strains, PS01 and PS02, with the same flaC mutation, but each having an independent secondary mutation. In hindsight, the flaC background was instrumental in identifying RMI, as the mutation destroyed the “stick” part of the switch and favored observation of the motile phase (when RMI is present). Without the flaC mutation, the effects of RMI are more subtle, being modulated in part by the action of FlaC and undoubtedly other, currently unknown factors. Moreover, the phenotype of PS01 and PS02 tell us that FlaC activity is not required for RMI activity, since both mutants respond to RMI induction. Interestingly, a FlaC mutant constructed in our laboratory, via Campbell-type plasmid integration has no defect in RMI production and is motile, with a greater percentage of motile cells in its population, clearly indicating that FlaC is not required for RMI production. The most parsimonious explanation for this is FlaC is not part of the RMI-induced upregulation of the motile phase, rather FlaC is part of the sessile-phase pathway.
RMI has characteristics of a quorum-signaling molecule: it is a small (mass of 226 Da [Da]) extracellular compound that acts at the transcriptional level to shift the population toward the motile phase, while inhibiting biofilm formation in TM1040, a member of the ubiquitous Roseobacter clade of marine Alphaproteobacteria. TDA, a similar small (212 Da) quorum-signaling molecule found in TM1040, is distinct from RMI. Unlike RMI whose activity increases in shaking culture conditions, the activity of TDA is maximal when the bacteria are grown in static broth cultures. Additionally, TDA was found to elute in the third fraction on a C18 column (see Materials and Methods), whereas maximum RMI activity was observed in the fourth fraction on the same column, indicating that the two molecules have different chemical properties. RMI is also not likely to be an AHL quorum-sensing signaling molecule, such as those recently described to affect motility and biofilm formation in the sponge-associated roseobacter, Ruegeria sp. strain KLH11 (35), because TM1040 does not produce (known) AHLs (14), nor are luxI homologs present in the annotated genome (16). Since the published genome lacks genes harbored on pSTM3, a large 135-kb plasmid that escaped annotation (17), one may speculate that a novel AHL may be synthesized from one or more of the plasmid genes. However, loss of pSTM3 has no effect on RMI activity (data not shown), which is supported by our results showing that mutations in tda genes (harbored on pSTM3) have little or no effect on RMI activity. This makes it highly improbable that an undiscovered luxI homolog residing on pSTM3 might be responsible for RMI.
Instead, RMI has characteristics in common with the roseobacticides, as recently reported by Seyedsayamdost et al. for the roseobacter, Phaeobacter gallaeciensis BS107 (25, 26). RMI is a potent algicide, resulting in a loss of motility, vacuolization, cell swelling, and ultimately lysis of unicellular marine algal cells. Also, RMI is synthesized in response to pCA from PAA. These features of RMI are very similar to those described for the roseobacticides and, specifically, for Roseobacticide A, a troponoid compound (26). None of the known roseobacticides has a mass of 226 Da; their masses are larger, hinting that RMI is a new member of this family of compounds. Significantly, our results show that RMI acts to induce the motile phase, thereby affecting both the bacterium and its host. This feature has not been reported for other roseobacticides, but if it is widely shared among this group of molecules, it suggests that their activities impact both members of the roseobacter/algal symbiosis. The molecular mechanism underlying pCA induction of RMI is not known, but degradation products of pCA are presumably scavenged and used in the PAA pathway providing additional precursor molecules for RMI synthesis. RMI may act above the CckA regulatory circuit—CckA, CtrA, and FlaB (ChpT)—that we have previously shown to be involved in regulating flagellar gene expression in TM1040 (13).
The ultimate outcome of RMI induction is an increase in transcription of fliC3, with a commensurate increase in the major flagellin protein (15). Indeed, we were able to exploit this to construct a reporter plasmid, pPS1 (fliCp::lacZ), to use as a proxy to measure RMI activity. Although very useful for this purpose, RMI induction of fliC3 expression offers little information about the role RMI plays in regulating flagellar gene expression, flagellum biosynthesis, and motility. It remains unresolved whether RMI induces the expression of other flagellar biosynthetic or regulatory genes, chemotaxis genes, or the multitude of genes encoding both membrane-bound and cytoplasmic chemoreceptor proteins.
RMI does more than upregulate motility, it represses biofilm formation (Table 4). It would be surprising if this repression did not occur at the level of transcription, similar to fliC3. Given that biofilm formation is a component of the larger sessile phase phenotype, which in addition includes rosette formation, as well as TDA and pigment biosynthesis, we predict that RMI negatively regulates genes involved in these pathways as well. This hypothesis is currently being tested in our laboratory. If true, the effect of RMI would be opposite that exerted by FlaC, a putative regulator and activator of sessile phase.
Why does coculturing of PS01 and PS02 lead to RMI activity? Both strains lack the ability to independently produce RMI, so coculturing must involve the secretion of the missing compound and cross-feeding. The phenotypes of PS01 and PS02 provide hints as to the nature of the second site mutation in each of these strains. The production of TDA and pigment are positively and absolutely correlated in TM1040 (17). This suggests that nonpigmented PS02 has a mutation in one of the TDA biosynthesis genes. We hypothesize that the second site defect in PS02 must lie in one of the early genes in the pathway, maybe in one of the paa genes. This is currently being tested with experiments designed to cross-feed and complement the second site mutation. On the other hand, PS01 cells are pigmented; therefore, it is unlikely that the second mutation in this strain is involved in TDA. Rather, it is more probable that the second site mutation in PS01 is in a currently unknown gene required for RMI biosynthesis or function.
As indicated earlier, RMI biosynthesis requires PAA as a precursor, which is similar to the PAA requirement of the roseobacticides (26). As shown in Table 5, we observed a significant reduction in RMI activity in supernatants obtained from cells with defects in the gene paaI, paaJ, or paaK. These mutants are also defective in TDA biosynthesis (17), establishing PAA as a central precursor for both RMI and TDA. We saw small but reproducible drops in RMI activity with two other genes involved in the TDA pathway, hinting that RMI biosynthesis may also utilize some of the enzymes of the TDA pathway, particularly TdaE and TdaF, encoding a putative acyl-coenzyme A dehydrogenase (ACAD) and an aldehyde dehydrogenase, respectively (17). Mutations in either of these genes reduced, but did not eliminate, RMI activity, so they appear to play a minor role. Also notable, is the >20% increase in RMI activity in cells with defects in TdaA, the major transcriptional regulator of tda expression (37). This may be due to shunting PAA from TDA to RMI biosynthesis, thereby providing more precursors for RMI synthesis.
RMI activity also extends to the host algae, with exposure to RMI resulting in loss of motility, cellular swelling, vacuolization, and ultimately death. RMI-induced algal cell death has characteristics in common with programmed cell death (PCD) in phytoplankton defined as “an autocatalytic cell-suicide mechanism in which an endogenous biochemical pathway leads to cellular demise with apoptotic morphological changes” (38). PCD has been identified in the marine alga, E. huxleyi (39), and is accompanied by morphological changes in the cell, resulting in rapid internal degradation of cellular components, reduction in the photosynthetic efficiency, upregulation of metacaspase protein expression, and an induction of caspase-like activity (39). Our laboratory is currently developing a transcriptomic approach, using the annotated genome of E. huxleyi, to determine how RMI affects marine algae at the molecular level.
The discovery of small molecule modulators of bacterial-algal interactions, e.g., TDA and RMI, offers new insights into the inner workings of the cell-to-cell communications that occur during the symbiosis and how these compounds may influence algal bloom dynamics. It is fascinating to consider that these molecules affect the physiology of both the bacteria and the host and do so in very profound ways: TDA upregulates sessile-phase tda gene expression and serves as an antibiotic that may protect the alga from harmful pathogens, whereas RMI upregulates motile-phase fliC3 expression and acts as an algicide. Since both molecules have characteristics suggestive of quorum-signaling compounds and since laboratory evidence supports the role of TDA as a quorum-signaling autoinducer (18), their effects may be felt beyond the immediate niche on a given algal cell and could disperse through a larger community of algae. Thus, TDA and RMI may be considered the lingua franca that helps coordinate bacterial activities with those of the algal host.
A model may be developed interconnecting the roseobacterial swim-or-stick lifestyle, algal bloom dynamics, and the production and activity of TDA and RMI (Fig. 5). In our model, motile roseobacters swim toward extracellular compounds secreted by the algae (particularly DMSP), using chemotaxis behavior and flagellar rotation to drive them near the surface of their host, where they utilize DMSP as a nutrient and source of sulfur for TDA (and quite possibly RMI). We posit that the environment immediately adjacent to the alga has properties in common with laboratory static culture conditions, known to induce TDA biosynthesis. Thus, as the roseobacter comes into proximity with the alga, TDA is produced and begins its action as an autoinducer of its own synthesis. TDA is a quorum signal that induces the sessile-phase phenotype, throwing the “switch” toward biofilm development on the surface of the host. As TDA accumulates, its antibacterial activity prevents other nonroseobacters from establishing a foothold on the algal, permitting the roseobacters to monopolize the niche. Production of PAA, a known plant hormone (40), and other molecules (such as indole acetic acid [IAA]) may also benefit the algae.
Fig 5.
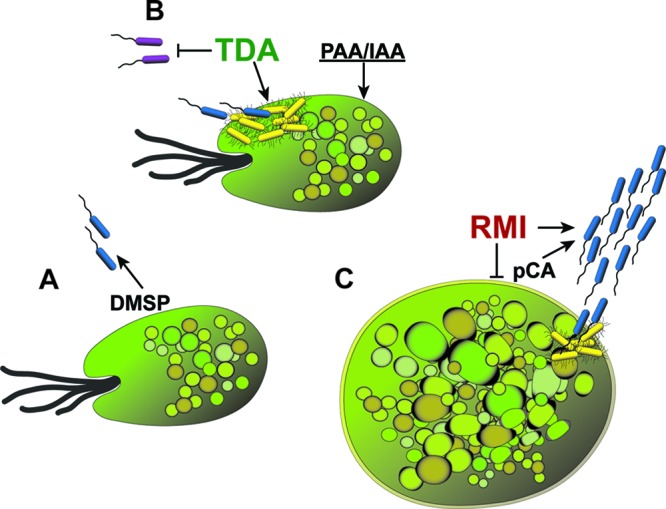
RMI and TDA: the lingua franca of the roseobacter-algal symbiosis? Shown is a diagram of our model describing the roles of TDA and RMI in the roseobacter-algal symbiosis. (A) Roseobacters in their motile phase (blue bacteria) swim toward algae, directed by chemotaxis to attractants such as DMSP. DMSP is metabolized by the bacteria, which use its sulfur for synthesis of bioactive compounds, including TDA (and most likely roseobacticides). Proximity to the host and TDA itself induces the sessile phase (yellow bacteria). (B) TDA acts as a quorum signal to induce its own synthesis and initiate sessile phase in other roseobacters. TDA also acts as an antibiotic, inhibiting nonroseobacters (purple bacteria). As the roseobacter biofilm develops and matures on the algal surface, PAA, IAA, and other algal growth-promoting compounds are synthesized by the bacteria. (C) As the symbiosis matures, algal cell death increases, resulting in an increase in lignin and its breakdown products, including pCA, which induces RMI synthesis. RMI flips the biphasic switch toward the motile phase, breaking the symbiosis, which is further harmed by the algicidal action of RMI. The swim-or-stick switch is reset, permitting the roseobacters to renew its symbiosis with a fresh algal host.
As the algal population increases in density, the number of dead and dying algae also increases, and with this comes a release of lignin breakdown products, including pCA (36). The increased concentration of pCA combined with changes in the algal surface environment reminiscent of those in laboratory shaking cultures are hypothesized to cause an increase in RMI synthesis, which results in a shift to the motile phase. At the same time, elevated RMI acting as an algicide, kills the host, and increases pCA release, each of which ultimately leads to a collapse of the algal population and an end to the bloom.
Obviously, this model is naive and does not consider the multitude of biotic and abiotic factors that affect algal populations in the marine ecosystem. Nonetheless, it provides a working framework for how TDA and RMI may affect this symbiosis. Understanding the molecular mechanisms underlying RMI and TDA actions are therefore important, especially when one considers that symbioses between marine bacteria and their algal hosts are critical to marine nutrient cycles and may play a role in the maintenance of global climate (8).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by National Science Foundation award IOS-0842331.
We thank members of the Belas laboratory for helpful suggestions and encouragement, Haifeng Geng (Johns Hopkins University) for help with TDA purification and HPLC analyses, and Joshua Wilhide (UMBC Molecular Characterization & Analysis Facility) for assistance with mass spectrometry.
Footnotes
Published ahead of print 16 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01777-12.
REFERENCES
- 1. Gonzalez JM, Moran MA. 1997. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brinkhoff T, Bach G, Heidorn T, Liang L, Schlingloff A, Simon M. 2004. Antibiotic production by a Roseobacter clade-affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Appl. Environ. Microbiol. 70:2560–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bruhn JB, Nielsen KF, Hjelm M, Hansen M, Bresciani J, Schulz S, Gram L. 2005. Ecology, inhibitory activity, and morphogenesis of a marine antagonistic bacterium belonging to the roseobacter clade. Appl. Environ. Microbiol. 71:7263–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dang H, Lovell C. 2000. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 66:467–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cole JJ. 1982. Interactions between bacteria and algae in aquatic ecosystems. Annu. Rev. Ecol. Syst. 13:291–314 [Google Scholar]
- 6. Doucette G. 1995. Interactions between bacteria and harmful algae: a review. Natural Toxins 3:65–74 [DOI] [PubMed] [Google Scholar]
- 7. Gage DA, Rhodes D, Nolte KD, Hicks WA, Leustek T, Cooper AJ, Hanson AD. 1997. A new route for synthesis of dimethylsulfoniopropionate in marine algae. Nature 387:891–894 [DOI] [PubMed] [Google Scholar]
- 8. Howard EC, Henriksen JR, Buchan A, Reisch CR, Burgmann H, Welsh R, Ye W, Gonzalez JM, Mace K, Joye SB, Kiene RP, Whitman WB, Moran MA. 2006. Bacterial taxa that limit sulfur flux from the ocean. Science 314:649–652 [DOI] [PubMed] [Google Scholar]
- 9. Grover JP. 2000. Resources competition and community structure in aquatic micro-organisms: experimental studies of algae and bacteria along a gradient of organic carbon to inorganic phosphorus supply. J. Plank. Res. 22:1591–1610 [Google Scholar]
- 10. Alavi M, Miller T, Erlandson K, Schneider R, Belas R. 2001. Bacterial community associated with Pfiesteria-like dinoflagellate cultures. Environ. Microbiol. 3:380–396 [DOI] [PubMed] [Google Scholar]
- 11. Miller TR, Belas R. 2003. Pfiesteria piscicida, P. shumwayae, and other Pfiesteria-like dinoflagellates. Res. Microbiol. 154:85–90 [DOI] [PubMed] [Google Scholar]
- 12. Miller TR, Hnilicka K, Dziedzic A, Desplats P, Belas R. 2004. Chemotaxis of Silicibacter sp. TM1040 toward dinoflagellate products. Appl. Environ. Microbiol. 70:4692–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miller TR, Belas R. 2006. Motility is involved in Silicibacter sp. TM1040 interaction with dinoflagellates. Environ. Microbiol. 8:1648–1659 [DOI] [PubMed] [Google Scholar]
- 14. Bruhn JB, Gram L, Belas R. 2007. Production of antibacterial compounds and biofilm formation by roseobacter species are influenced by culture conditions. Appl. Environ. Microbiol. 73:442–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Belas R, Horikawa E, Aizawa SI, Suvanasuthi R. 2009. Genetic determinants of Silicibacter sp. TM1040 motility. J. Bacteriol. 191:4502–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moran MA, Belas R, Schell MA, Gonzalez JM, Sun F, Sun S, Binder BJ, Edmonds J, Ye W, Orcutt B, Howard EC, Meile C, Palefsky W, Goesmann A, Ren Q, Paulsen I, Ulrich LE, Thompson LS, Saunders E, Buchan A. 2007. Ecological genomics of marine roseobacters. Appl. Environ. Microbiol. 73:4559–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geng H, Bruhn JB, Nielsen KM, Gram L, Belas R. 2008. Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters. Appl. Environ. Microbiol. 74:1535–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geng H, Belas R. 2010. Expression of tropodithietic acid biosynthesis is controlled by a novel autoinducer. J. Bacteriol. 192:4377–4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cane DE, Wu Z, Van Epp JE. 1992. Thiotropocin biosynthesis: shikimate origin of a sulfur-containing tropolone derivative. J. Am. Chem. Soc. 114:8479–8483 [Google Scholar]
- 20. Bruhn JB, Juul Haagensen JA, Bagge-Ravn D, Gram L. 2006. Culture conditions of Roseobacter 27-4 affect its attachment and biofilm formation, as measured by a novel real-time PCR method. Appl. Environ. Microbiol. 72:3011–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. 2006. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444:899–904 [DOI] [PubMed] [Google Scholar]
- 22. Pao GM, Saier MH., Jr 1995. Response regulators of bacterial signal transduction systems: selective domain shuffling during evolution. J. Mol. Evol. 40:136–154 [DOI] [PubMed] [Google Scholar]
- 23. Martinez-Hackert E, Stock AM. 1997. The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure 5:109–124 [DOI] [PubMed] [Google Scholar]
- 24. Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 3:e334 doi:10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seyedsayamdost MR, Carr G, Kolter R, Clardy J. 2011. Roseobacticides: small molecule modulators of an algal-bacterial symbiosis. J. Am. Chem. Soc. 133:18343–18349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seyedsayamdost MR, Case RJ, Kolter R, Clardy J. 2011. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3:331–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28. De Lorenzo M, Herrero M, Jakubzik U, Timmis KN. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miller TR, Belas R. 2004. Dimethylsulfoniopropionate (DMSP) metabolism by Pfiesteria-associated Roseobacter spp. Appl. Environ. Microbiol. 70:3383–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Apolinario EE, Jackson KM, Sowers KR. 2005. Development of a plasmid-mediated reporter system for in vivo monitoring of gene expression in the archaeon Methanosarcina acetivorans. Appl. Environ. Microbiol. 71:4914–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kiene RP, Linn LJ, Gonzalez J, Moran MA, Bruton JA. 1999. Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl. Environ. Microbiol. 65:4549–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 33. Sule P, Wadhawan T, Carr NJ, Horne SM, Wolfe AJ, Pruss BM. 2009. A combination of assays reveals biomass differences in biofilms formed by Escherichia coli mutants. Lett. Appl. Microbiol. 49:299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wozniak CE, Chevance FF, Hughes KT. 2010. Multiple promoters contribute to swarming and the coordination of transcription with flagellar assembly in Salmonella. J. Bacteriol. 192:4752–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zan J, Cicirelli EM, Mohamed NM, Sibhatu H, Kroll S, Choi O, Uhlson CL, Wysoczinski CL, Murphy RC, Churchill ME, Hill RT, Fuqua C. 2012. A complex LuxR-LuxI type quorum sensing network in a roseobacterial marine sponge symbiont activates flagellar motility and inhibits biofilm formation. Mol. Microbiol. 85:916–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schaefer AL, Greenberg EP, Oliver CM, Oda Y, Huang JJ, Bittan-Banin G, Peres CM, Schmidt S, Juhaszova K, Sufrin JR, Harwood CS. 2008. A new class of homoserine lactone quorum-sensing signals. Nature 454:595–599 [DOI] [PubMed] [Google Scholar]
- 37. Geng H, Belas R. 2011. TdaA regulates tropodithietic acid synthesis by binding to the tdaC promoter region. J. Bacteriol. 193:4002–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bidle KD, Falkowski PG. 2004. Cell death in planktonic, photosynthetic microorganisms. Nat. Rev. Microbiol. 2:643–655 [DOI] [PubMed] [Google Scholar]
- 39. Bidle KD, Haramaty L, Barcelos ERJ, Falkowski P. 2007. Viral activation and recruitment of metacaspases in the unicellular coccolithophore, Emiliania huxleyi. Proc. Natl. Acad. Sci. U. S. A. 104:6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wightman F, Lighty DL. 1982. Identification of phenylacetic acid as a natural auxin in the shoots of higher plants. Physiologia Plantarum 55:17–24 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.