Abstract
Bacterial persistence is characterized by the ability of a subpopulation within bacterial cultures to survive exposure to antibiotics and other lethal treatments. The surviving persisters are not the result of genetic changes but represent epigenetic variants that are in a physiological state where growth is inhibited. Since characterization of persisters has been performed mainly in Escherichia coli K-12, we sought to identify mechanisms of persistence in the pathogen Salmonella enterica serovar Typhimurium. Isolation of new highly persistent mutants revealed that the shpAB locus (Salmonella high persistence) imparted a 3- to 4-order-of-magnitude increase in survival after ampicillin exposure throughout its growth phase and protected the population against exposure to multiple antibiotics. Genetic characterization revealed that shpAB is a newly discovered toxin-antitoxin (TA) module. The high-persistence phenotype was attributed to a nonsense mutation in the 3′ end of the shpB gene encoding an antitoxin protein. Characteristic of other TA modules, shpAB is autoregulated, and high persistence depends on the Lon protease.
INTRODUCTION
The phenomenon of bacterial persistence has been observed for decades, following the observation by Joseph Bigger that a small subpopulation of growing Staphylococcus aureus cells consistently survived treatment with penicillin. These “persisters,” as coined by Bigger, represented cells that did not inherit antibiotic resistance, as drug sensitivity was regained by the progeny of the surviving bacteria (1). Subsequent studies have shown that most, if not all, bacteria can enter into a physiological state that renders them tolerant of antimicrobial drugs, as well as other lethal stresses (2–7). Even though persisters are not drug resistant, their occurrence has been implicated in the failure of antibiotic therapy, as well as contributes to the inherent drug tolerance of biofilms (2, 6, 8, 9).
Persisters represent a small subpopulation within a bacterial culture that is phenotypically distinct (7, 10). In Escherichia coli, for example, persistent cells typically comprise only between 10−5 and 10−6 of the total population (11). The relatively low frequency of persistent cells has hampered the identification of the mechanisms responsible for antibiotic tolerance.
An initial attempt to understand the genetic basis for persistence was initiated by Moyed and Bertrand, who isolated highly persistent mutants of E. coli where a significantly greater proportion of the population (∼1,000-fold) had entered into a drug-tolerant state compared to cultures of wild-type bacteria (11). To isolate hip (high-persistence) mutants, Moyed and Bertrand treated cultures with a chemical mutagen and exposed the cells to lethal doses of penicillin to enrich for survivors. The resulting mutants showed a significant increase in persistence in response to β-lactam antibiotics, in addition to showing increased tolerance to other stresses, such as elevated temperature and deprivation of thymine and diaminopimelic acid (11). The locus responsible for this phenotype led to the identification of a two-gene operon named hipBA (11, 12). hipA7 is a gain-of-function mutation, as deletion of hipBA did not change the levels of persistence (11, 13).
More recent characterization of the hipA7 mutant used microfluidics to monitor the growth of individual cells. From this study, hipA7 mutants were classified as type I persisters, signifying that while persistent cells arise during stationary phase, a state of dormancy was not achieved until 1.5 h after transfer to fresh medium (10, 14). Apparently, signals triggering persistence are generated in stationary phase, but differentiation into persisters requires additional time for signal processing. Similar characterization of another highly persistent mutant, hipQ (15), revealed it to be a type II persister, which was continuously generated during the exponential growth phase (10). Despite its distinction from hipA, the function of hipQ remains unknown.
Genetic analysis of hipBA revealed it to encode a toxin-antitoxin (TA) module (13). Although TA modules were originally discovered as plasmid-borne genes responsible for plasmid partitioning by postsegregational killing (16, 17), they are now also known to reside throughout the chromosome of most bacteria (18–22). Consistent with the structure and function of most type II TA modules, hipBA form an operon of two tightly linked genes (12, 23). The product of hipA is a toxin, while that of hipB is an antitoxin that interacts directly with HipA (24, 25). Also in common with most other TA module family members are the findings that HipA cannot be expressed in cells without HipB (12, 13) and that hipBA autoregulate their own transcription (24). Biochemical studies indicated that HipA is a kinase that phosphorylates the translation factor EF-Tu, suggesting a mechanism whereby hipBA induce a state of dormancy in E. coli by interfering with translation (25).
In addition to hipBA, other evidence has emerged that TA modules are important contributors to persistence (26, 27). While initial studies to delete multiple TA modules from E. coli revealed no obvious phenotypes (28), a more extensive analysis recently revealed that persistence decreased when a sufficient number of genes was deleted (29). The functional redundancy of the multiple TA modules encoded throughout the bacterial chromosome apparently masks the contribution of individual genes to persistence. Another indirect link between TA modules and persistence comes from microarray analysis, which showed that genes encoding TA components, including mazEF and relBE (7), were among the genes whose expression was upregulated following an antibiotic challenge.
Other genetic approaches have included screening for insertion mutations that decrease persistence, as well as identifying genes whose products increase persistence when overexpressed. The results of these studies consistently showed that, in addition to TA modules, multiple cellular processes can contribute to persistence and that functional redundancy is prevalent. For example, the Keio collection of E. coli deletion mutants was screened in an attempt to find genes required for persistence (30). While no insertion mutations completely abolished persistence, 150 mutations affecting a variety of cellular functions, including chaperones and global transcriptional regulators, were found to decrease persistence (30).
Transposon mutant libraries have also been screened to identify mutations that cause either decreased or increased persistence in E. coli (31–33) and Pseudomonas aeruginosa (34). In general, these attempts revealed genes that likely only indirectly affect persistence. More recently, a high-density library of transposon insertion mutants was screened to identify new high persisters (31). Consistent with other studies, several different genes with diverse functions answered the selection for antibiotic tolerance (31).
In addition to toxins, including HipA (26, 27, 35–39), elevated expression of multiple gene products that inhibit E. coli growth can also elevate persistence (36). For example, an expression library screen yielded high persisters resulting from overexpression of GlpD with sn-glycerol-3-phosphate dehydrogenase activity. Deletion of glpD also decreased persistence in stationary phase (33).
In general, these studies reveal that multiple metabolic pathways can contribute to persistence in bacteria (3). Despite the challenge to identify specific mechanisms of persistence, the phenomenon requires additional study in efforts to improve the effectiveness of antibiotics (40). Since genetic and physiological studies of persistence have been performed primarily in E. coli K-12 strains, we sought to understand persistence in a bacterial pathogen. Salmonella enterica serovar Typhimurium is an important pathogen of humans and animals. It is the causative agent of a common form of food-borne illness and causes significant health and economic impacts each year in the United States (41). Salmonella strains with multidrug resistance are also emerging, with outbreaks of these strains occurring at a higher frequency (42).
Inspection of the S. Typhimurium strain LT2 genome reveals no obvious orthologues to hipBA (43). However, the pathogen shares many other genes, including a host of TA family members, implicated in E. coli persistence. Therefore, to better understand the mechanisms of persistence in a bacterial pathogen, we isolated mutants that show elevated levels of persistence. This report describes the identification and characterization of a mutant that results from a single base pair change in a newly discovered locus that we term shpAB, for Salmonella high persistence.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains are listed in Table 1. E. coli DH5α (NEB5α; New England BioLabs, Ipswich, MA) was used as a host for cloning experiments, and E. coli EPI400 (Epicentre, Madison, WI) was used to clone shpA, whose expression is toxic to bacteria.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| NEB5α | fhuA2Δ(argF-lacZ)U169 phoA glnV44 ϕ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | New England Biolabs |
| LMG194 | F− Δ(lacIPOZY)X74 galE galK thi rpsL ΔphoA ara714 | Invitrogen |
| EPI400 | F− mcrAΔ(mrr-hsdRMS-mcrBC)ϕ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139Δ(ara leu)7697 galU galKλ− rpsL(Strr) nupG trfA tonA pcnB4 dhfr | Epicentre |
| BW25113 | F− mcrAΔ(mrr-hsdRMS-mcrBC)ϕ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139Δ(ara leu)7697 galU galKλ− rpsL(Strr) nupG trfA tonA pcnB4 dhfr | Coli Genetics Stock Center |
| JW0429-1 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB3), Δlon-725::kan λ− rph-1 Δ(rhaD-rhaB)568 hsdR514 | Coli Genetics Stock Center |
| S. enterica | ||
| LT2 | Wild type | Salmonella Genetics Stock Center |
| AS17 | LT2 shpB1 | This study |
| AS26 | LT2 hsdM::Tn10d shpB1 | This study |
| AV1 | LT2 hsdM::Tn10d shpB1 | This study |
| AV7 | LT2 ΔrelA::kan | This study |
| AV9 | AS17 ΔrelA::kan | This study |
| AV47 | LT2 Δ(STM4528-STM4534)::kan | This study |
| AV53 | LT2 Δ(STM4528-STM4529)::kan | This study |
| Plasmids | ||
| pKD46 | repA(Ts) Ampr (expression of λ Red genes) | 44 |
| pNK972 | ColE1 ori Tetr Ampr (Tn10d mutagenesis) | 45 |
| pBAD-TOPO | ColE1 ori Ampr (ParaBAD-regulated gene expression) | Invitrogen |
| pTrcHisA | ColE1 ori Ampr (Ptrcgene-regulated expression, ColE1 ori Ampr) | Invitrogen |
| pJPC12 | pSC101 ori Camr (pSC101-derivative cloning vector) | 46 |
| pKRP13 | ColE1 ori Spcr Ampr (source of Spcr cassette) | 47 |
| pSMART-LCKan | ColE1 (pBR322) Kanr (cloning vector) | Lucigen |
| pSMART-shpAB | pSMART-LCKan shpAB+ | This study |
| pSMART-shpAB1 | pSMART-LCKan shpAB1 | This study |
| pBADshpAB | pBAD-TOPO shpAB+ | This study |
| pBADshpAB1 | pBAD-TOPO shpAB1 | This study |
| pBADshpB | pBAD-TOPO shpB+ | This study |
| pBADshpB1 | pBAD-TOPO shpB1 | This study |
| pBADshpA | pBAD-TOPO shpA | This study |
| pJPC-Trc-shpB | pJPC12 (Ptrc-shpB) | This study |
Bacterial growth media and chemicals.
All bacteria were grown on LB agar plates or in LB broth unless otherwise noted (48). LBEDO broth (49) was used for bacteriophage P22 transduction experiments. Mueller-Hinton II plates (Becton, Dickinson and Co., Franklin Lakes, NJ) were used for MIC measurements using AB Biodisk (Solna, Sweden) strips. Antibiotics and penicillinase (Pen) were purchased from Sigma Chemical Co., St. Louis, MO, and were used at the following concentrations: kanamycin (Kan) at 50 μg/ml, tetracycline (Tet) at 20 μg/ml, ampicillin (Amp) at 100 μg/ml, ofloxacin (Ofx) at 5 μg/ml, piperacillin (Pip) at 200 μg/ml, and chloramphenicol (Cam) at 20 μg/ml. The Pen stock was made at 2,500 U/ml in 50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol (DTT) in 20% glycerol. One hundred microliters of Pen stock was diluted in 4.9 ml LB to prepare a working solution. Restriction enzymes, T4 DNA ligase, and thermostable polymerases were obtained from New England BioLabs, Ipswich, MA.
PCR primers.
The sequences of the PCR primers used in this study are shown in Table 2. All primers were obtained from Integrated DNA Technologies (Coralville, IA).
Table 2.
DNA primers used in this study
| Primer name | Sequence (5′-3′)a |
|---|---|
| Tn10U | ACCAACCATTTGTTAAATCAGTTTTTGTTGTGA |
| IS10R | CAAGATGTGTATCCACCTTAACTTAATGATTTT |
| ARB1 | GGCCAGGCCTGCAGATGATG |
| ARB2 | GGCCACGCGTCGACTAGTACNNNNNNNNNNACGC |
| ARB3 | GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC |
| ARB4 | GGCCACCCGTCGACTAGTAC |
| STrelA-KD4.S | ATG GTC GCG GTA AGA AGT GCA CAT ATT AAT AAA GCT GGT GAA TTT GAT GTG TAG GCT GGA GCT GCT TC |
| STrelA-KD4.AS | ACC GAG CAC CCG GCC CAG CAC CTG CAG GTT GTA GAT CTC GAT GGT CAT ATG AAT ATC CTC CTT AG |
| STM4518-KD4.S | AGG CGA TAG AAA CAG CGG CTG CGG ATG TAC TGA ATT TCA TGT AAT TGT GTA GGC TGG AGC TGC TTC G |
| STM4518-KD4.AS | AGA ACG CGT CTC TTC CTG TCT GTG TTT TCA CCG GCA GCG TTT TTG CAT ATG AAT ATC CTC CTT AG |
| STmrr-KD4.S | ATT ACT TTG AGT AGT GGT AAC GCT GGA ACC TGA TTG CGG GTA ATT TGT GTA GGC TGG AGC TGC TTC G |
| STmrr-KD4.AS | TTC TGG GCG AGA GGA AAC ACA GGT GCC CAC AAC GTG GTC CAT CTC CAT ATG AAT ATC CTC CTT AG |
| STrelA-KD4.S | ATGGTCGCGGTAAGAAGTGCACATATTAATAAAGCTGGTGAATTTGATGT GTA GGC TGG AGC TGC TTC G |
| STrelA-KD4.AS | ACCGAGCACCCGGCCCAGCACCTGCAGGTTGTAGATCTCGATGGTCAT ATG AAT ATC CTC CTT AG |
| STM4535-KD4.S | TTC CCT CTG TTA ACC CGC ATC TTT TTT CGG CGC GCC ACT TTT TCT TGT GTA GGC TGG AGC TGC TTC G |
| STM4535-KD4.AS | TCT AAA ACA CAT TCC ACT GTG ATA AAA GAG AGC AAG CGG CGT GCC CAT ATG AAT ATC CTC CTT AG |
| STM4529-KD4.AS | CAT TAA GCG CTT ATC TGG CCT GGC GGG AAT TTG TAG GCC GGA TAA CAT ATG AAT ATC CTC CTT AG |
| STM4528.S | GGC GTA TCC TCA CGT TTA CTC AAG |
| STM4529.AS | GGG AAT TTG TAG GCC GGA TAA G |
| STM4528-ara.S | ACC ATG GAG TTT GAA TGG GAT GCG AAC |
| STM4528-ara.AS | GAT GCG TTG CCG CGT TTA TGT T |
| STM4529-ara.S | ACC ATG GGA AGC ATG GTT AAA CAT AAA CGC GGC |
| PshpAB-NheI.S | GCT AGC AAA GTT GAA GGT CTG GTG CTG GTG |
| PshpAB-NheI.AS | GCT AGC CTT GTT CGC ATC CCA TTC AAA CTC |
The underlined sequences represent the portion of the primers that anneal to pKD3 and pKD4 for recombineering.
Isolation of high-persistence mutants.
Salmonella enterica serovar Typhimurium strain LT2 was mutagenized with N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) as described previously (48). This included growing cells to a concentration of 108 CFU/ml, followed by exposure to 50 μg/ml of MNNG for 10 min. Cells were then washed twice with 8.5% NaCl and resuspended to a concentration of 109 CFU/ml and incubated overnight.
High persisters were isolated as described by Moyed and Bertrand (11), with modifications. A saturated overnight culture of mutagenized cells was pelleted and resuspended in sterile saline. Forty microliters of cells was used to inoculate 40 ml of LB in a 250-ml baffled flask. The flask was incubated with shaking for 1 h at 37°C before addition of Amp, followed by continued growth for an additional 3 h. The cells were then centrifuged and resuspended in 40 ml of fresh LB, and the enrichment was repeated a second time. After the second round of Amp treatment, cells were washed with saline, plated on LB plates containing Amp, and incubated at 37°C. After 24 h, the plates were sprayed with the Pen working solution delivered by a martini mister and incubated again at 37°C for an additional 24 h.
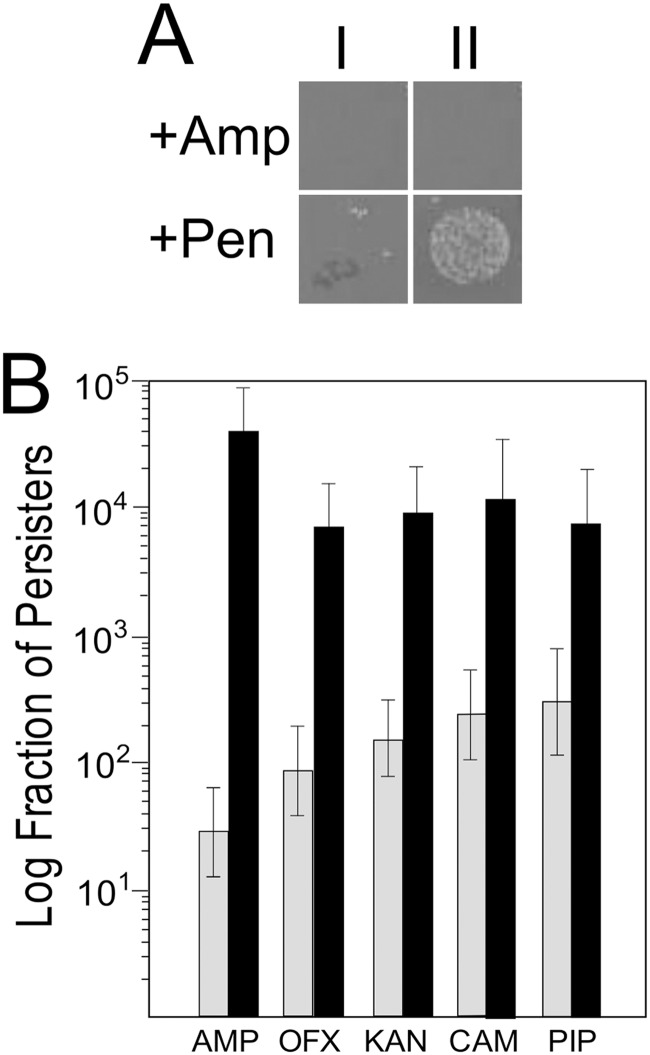
Mutants were scored for increased tolerance to Amp by transferring individual colonies, along with unmutagenized control colonies, into 100 μl of LB broth with Amp in 96-well microtiter plates. The plates were covered with Parafilm and incubated overnight with shaking at 37°C. The contents of each well were diluted 1:1,000 with fresh LB before spotting 5 to 10 μl onto 100-mm-square LB-plus-Amp plates. After overnight growth at 37°C, the plates were sprayed with Pen and again cultured overnight at 37°C. The next morning, the plates were inspected, and highly persistent mutants were identified as having confluent growth, while controls yielded only a few colonies (Fig. 1A).
Fig 1.
Phenotypes of higher persisters. (A) Wild-type stain LT2 (I) and highly persistent mutant AS17 (II) were spotted onto LB-Amp plates and incubated for 24 h (+Amp). Plates were sprayed with penicillinase and incubated for another 24 h (+Pen). (B) The levels of persistence of strains LT2 (wild type, gray bars) and AS17 (shpB1, black bars) were measured following exposure to the antibiotics Amp, Ofx, Kan, Cam, and Pip.
Assay for persistence.
To quantify persistence using Amp, overnight cultures of high persisters and controls were diluted 10−3, 10−5, and 10−6 in LB. One hundred microliters of the 10−3 and 10−5 dilutions was plated on LB with Amp, while the 10−6 dilution was plated on LB. Cultures were plated in triplicate and grown overnight at 37°C before being sprayed with Pen, followed by overnight incubation. The number of colonies on each plate was counted, and the level of persistence was calculated as the percentage of surviving cells.
To measure tolerance to antibiotics in broth culture, 20 μl from 5-ml overnight cultures was transferred to 125-ml baffled flasks containing 20 ml of LB. After 1 h incubation at 37°C with shaking, 0.5 ml was transferred to culture tubes containing 0.5 ml of LB medium containing Ofx, Cam, Kan, or Pip. Serial dilutions were also plated on LB. The tubes were incubated for 4.5 h at 37°C, after which cells were collected by centrifugation and resuspended in fresh LB to minimize transfer of antibiotics. Serial dilutions were plated onto LB plates to measure viability. The plates were sealed with Parafilm and incubated at 37°C for up to 5 days to allow cells with delayed growth to form colonies. Each assay was performed in triplicate, and the results were averaged.
Time course experiments were also performed to determine the effect of growth phase on persistence. For this, cultures were prepared as just described using Amp and Ofx. At regular intervals, samples were removed from the cultures and serial dilutions were plated on LB plates to assay the numbers of CFU/ml. When assaying for Amp persistence, dilutions were plated onto LB-Amp plates, as described above.
To measure the levels of persistence of E. coli K-12 strains, the plasmid pSMART-shpAB1 (Table 1) expressing shpAB1 was transformed into BW25113 and the lon derivative JW0429-1. Tolerance to Amp was measured as described above.
MIC determination.
Overnight cultures were spread evenly onto Mueller-Hinton II plates (150 by 15 mm; Becton, Dickinson and Co., Franklin Lakes, NJ) with sterile cotton swabs. Antibiotic test strips representing 5 different antibiotics (see Table 3) were transferred to each plate per the manufacturer's instruction, and the plates were incubated overnight at 37°C. MIC values were determined by recording the antibiotic concentration on the strip that intersected with the point of visible inhibition of bacterial growth.
Table 3.
Measurement of MICs
| Strain | MIC (μg/ml) |
||||
|---|---|---|---|---|---|
| Ampicillin | Piperacillin | Amoxicillin | Chloramphenicol | Ofloxacin | |
| LT2 | 1 | 1.5–2 | 1.5–2 | 3–4 | 0.125–0.20 |
| AS17 | 1 | 1.5–2 | 3 | 3–4 | 0.125–0.20 |
Growth rates measurements.
Twenty microliters of overnight cultures was transferred to 20 ml of LB contained in a 125-ml baffled flask and incubated at 37°C with shaking. The optical density at 600 nm (OD600) reading was recorded every 30 min over the course of several hours.
To measure the delay in the onset of growth, overnight cultures were diluted and plated on LB plates and sealed with Parafilm. Colonies were counted at regular intervals following growth at 37°C. These values were compared to the number of colonies observable after 80 h.
Isolation and mapping of Tn10d markers linked to high persistence.
Tn10d(Tc) libraries were prepared in strain LT2 as described previously (45, 49). Tet-resistant (Tetr) colonies were pooled and resuspended in 1.5 M NaCl for preparation of bacteriophage P22 lysates (49). Generalized transduction was used to transfer Tn10d insertions into the highly persistent mutant AS17 (Table 1) (49). Individual Tetr transductants were then scored for high persistence as described above.
Arbitrary PCR (50, 51) was used to map the locations of Tn10d elements in AS17 (Table 1). Two rounds of PCR were run using primers listed in Table 2. The first round of arbitrary PCR used the primers ARB3 or ARB1 and Tn10U (52), followed by a second round of PCR that used the primers ARB4 and IS10R. PCR products were gel purified and sequenced by the Iowa State University DNA Facility.
Recombineering.
S. Typhimurium was prepared for electroporation as described previously by washing mid-exponential-phase cells with 1 mM MOPS (morpholinepropanesulfonic acid) in 20% glycerol (53). Antibiotic resistance markers were generated by using pKD3 (Cam) or pKD4 (Kan) (44) as the PCR templates. Primers with ∼45 to 50 bases of homology to the targeted genome sequence were synthesized (Table 2). The PCR products were gel eluted using a Qiagen MinElute purification kit (Qiagen, Valencia CA). For recombineering, 1 μg of PCR product was used for electroporation into S. Typhimurium transformed with pKD46 (44). When appropriate, drug resistance cassettes were removed by site-specific recombination using pCP20 (44).
Recombinant plasmids.
shpAB and shpAB1 were cloned under the control of their native promoters and amplified by PCR using 5′-phosphorylated primers STM4528.S and STM4529.AS (Table 2). Amplification products were gel purified and ligated into pSMART-LCKan (Lucigen, Middleton, WI). The shp genes were also placed under the control of the araBAD promoter using primers STM4528-ara.S and STM4529.AS (shpAB or shpAB1), STM4528-ara.S and STM4528-ara.AS (shpB or shpB1), and STM4529-ara.S and STM4529.AS (shpA) (Table 2) and cloned into pBAD-TOPO (Invitrogen, Carlsbad, CA). Each sense primer included sequences to generate an NcoI restriction site. After cloning, plasmids were digested with NcoI to remove the leader sequence found on plasmid pBAD-TOPO before religating. The Ampr of the resulting plasmid was converted to spectinomycin (Spc) resistance by inserting a blunt-ended (SmaI) Spcr cassette from pKD13 (47) into the unique ScaI site within bla. All plasmid inserts were confirmed by DNA sequencing and were purified from E. coli NEB5α before introduction into Salmonella by electroporation. Gene expression was induced with l-arabinose at a final concentration of 0.01%.
The shpB gene was also expressed from the tac promoter by cloning an NcoI-PmeI fragment from pBADshpB into pTrcHisA (Invitrogen). A DNA fragment representing lacI-Ptrc-shpB was then inserted into the pSC101-derived cloning vector pJPC12 (46), generating pJPC-Trc-shpB (Table 1). Gene expression was induced with isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 0.1 mM.
The shpAB promoter region was cloned by PCR from LT2 genomic DNA using primers PshpAB-NheI.S and PshpAB-NheI.AS (Table 2). The resulting PCR product included the region upstream of shpAB and contained the putative promoter and ribosome binding site for the targeted genes. The PCR product was digested with NheI, gel purified, and ligated into the unique XbaI site of the lacZ fusion vector pLacZ2 (54), generating a translational gene fusion.
β-Galactosidase assays.
The pLacZ2-derivative plasmid expressing the Φ(shpA′-lacZ)Hyb gene fusion was introduced into strains LT2, AV53 (ΔshpAB), and AS17 (shpB1). The transformants were assayed for β-galactosidase activity (48) on cultures grown to mid-logarithmic phase or from saturated overnight cultures in LB at 37°C.
RESULTS
Isolation of highly persistent mutants.
Although S. Typhimurium does not contain an orthologue to hipBA, we nonetheless predicted that high-persistence mutants could be isolated in this serovar. Since the hipBA mutants found in E. coli K-12 were the result of specific base pair changes and not a complete-loss-of-function mutation (13), we mutagenized S. Typhimurium strain LT2 with the alkylating agent MNNG and screened for mutants that survived prolonged exposure to Amp (see Materials and Methods). We identified several dozen mutants that showed increased survival after Amp exposure while remaining antibiotic sensitive. Basic characterization of the mutants revealed that the majority had obvious growth defects, as revealed by formation of small colonies compared to the size of the colonies of wild-type LT2 (data not shown). These mutants were not characterized further. Here we describe the characterization of an shp (Salmonella highly persistent) mutant with elevated tolerance to Amp (Fig. 1A) and with a growth rate similar to that of wild type.
shp mutant characterization.
To determine if the shp mutant showed high tolerance to multiple antibiotics, we measured the mutant's ability to survive exposure to Amp, Ofx, Kan, Cam, and Pip. As shown in Fig. 1B, the mutant showed a greater than 2-order-of-magnitude increase in tolerance to Amp and a 20- to 30-fold increase in tolerance to Cam, Kan, and Pip. In contrast, the MICs of multiple antibiotics were not altered (Table 3).
To more accurately measure the growth of the shp mutant, we compared changes in the OD600 compared with that of wild-type strain LT2 over the course of several hours. As shown in Fig. 2A, the shp mutants showed growth rates comparable to the growth rate of wild-type LT2 during exponential growth phase (each with a mass doubling time of 34 min at 37°C) but exhibited a modest extension in lag phase. In plating the shp mutant, we also consistently observed that colony size was heterogeneous, especially after prolonged incubation. To better understand the basis for this, we measured the time that it took for cells to form visible colonies after plating in the absence of antibiotic selection. As shown in Fig. 2B, a significant number of the shp cells did not immediately initiate growth after plating; i.e., they remained dormant. An equivalent number of colonies eventually appeared after continued incubation.
Fig 2.
Characterization of the high persister. (A) Growth curves of strains LT2 (circles) and AS17 (squares). (B) Appearance of colonies on agar plates. Overnight cultures of LT2 (circles) and AS17 (squares) were diluted, plated onto LB plates, and incubated overnight at 37°C. (C) RelA is not required for formation of high persisters. A comparison between LT2 (white bar) and AV7 (LT2 relA::kan, light gray bar) on the left with the high persister AS17 (black bar) and its relA::kan derivative AV9 (dark gray bar) on the right is shown.
A previous report indicated that the high persistence of hipBA E. coli mutants requires a functional relA gene. Korch et al. showed that the persistence of the hipA7 mutant decreased by 3 orders of magnitude in the absence of relA (13). To determine if the highly persistent mutant had a similar requirement for relA, we constructed relA deletion mutants in wild-type and shp backgrounds (Table 1) and measured tolerance to Amp. In contrast to E. coli, no significant change in persistence was observed (Fig. 2C), indicating that shp-mediated high persistence was achieved in the absence of (p)ppGpp synthesis by RelA.
In E. coli, the fraction of the culture that is in a persistent state is dependent on the growth phase (55). For comparison, we determined the fraction of persisters in cultures of S. Typhimurium during different periods of cell growth. As shown in Fig. 3, the number of cells persistent in the presence of Amp tracked with the number of cells in the culture, reaching their maximum in stationary phase, indicating density dependence; i.e., quorum-sensing regulation is not involved in high persistence. Both wild type and the shp mutant followed the same trends, with the latter strain showing elevated persistence throughout each of the growth phases (Fig. 3). A similar profile was observed when tolerance to ofloxacin was measured (data not shown).
Fig 3.

Levels of persistence correlate with growth phase. Strains LT2 (wild type) (A) and AS17 (high persister) (B) were inoculated into fresh LB from overnight cultures, and the numbers of CFU (closed squares) and tolerance to Amp (open squares) were measured at 1-h intervals, as described in Materials and Methods.
Mapping the gene responsible for the Shp phenotype.
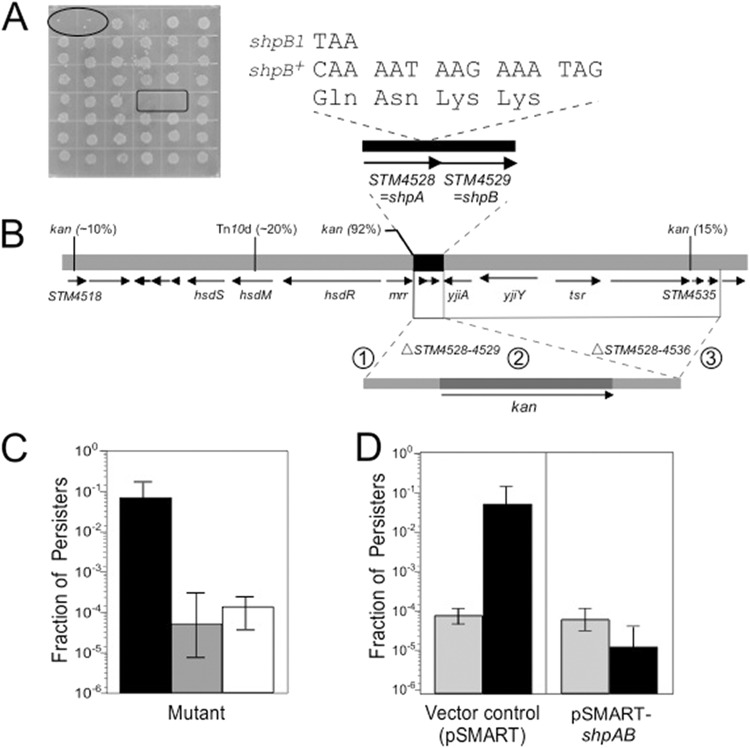
To identify the gene(s) responsible for high persistence in S. Typhimurium, we isolated Tn10d insertions linked to the shp locus. As described in Materials and Methods, we first constructed a random Tn10d library in LT2 and then transduced the Shp mutant AS17 to Tetr. Approximately 100 Tetr transductants were then tested for loss of tolerance to Amp. This screen, an example of which is shown in Fig. 4A, yielded two transductants with transposons linked to the shp mutation in AS17 (Table 1).
Fig 4.
Mapping and characterization of the locus responsible for high persistence in Salmonella. (A) Screen used to identify wild-type transductants, two of which are shown within the rectangle. Wild-type controls are shown within the oval. (B) Identification of wild-type and mutant shpAB on the Salmonella Typhimurium chromosome. The linear map in the middle shows the positions of shpAB and flanking genes, as well as the sites of insertion of Tn10d and kan markers. The transduction linkages to shpAB are given in parentheses. shpAB (STM4528 and STM4529) genes are shown in black. The locations of the kan insertion/deletion mutations are shown above the linear chromosome map. Deletion of STM4528 to STM4536 (AV47) corresponds to recombination events 1 and 3 (circled numbers), while deletion of STM4528 and STM4529 (AV53) corresponds to events 1 and 2. Nucleotide sequences and predicted amino acid sequences of the shpB+ and shpB1 alleles are shown above the linear map. (C) Deletion of shpAB results in loss of high persistence. Persistence of AS17 (shpB1, black bar) along with mutants with deletions of STM4528 to STM4536 (gray bar) and STM4528 and STM4529 (shpAB, white bar) is shown. (D) Complementation of the Shp phenotype. A ColE1-derivative plasmid (pSMART-LCKan) expressing shpAB+ (pSMART-shpAB), as well as the empty vector, was transformed into LT2 (gray bars) or AS17 (shpB1, black bars).
Bacteriophage P22 was then used to transduce wild-type LT2 to Tetr and high persistence, yielding AS26 and AV1 (Table 1). Characterization of the transductants revealed that they had the same growth characteristics and levels of persistence as AS17, indicating that high persistence could be attributed to the single mutation in shpB (data not shown). Arbitrary PCR was then used to map the location of these Tn10d insertions. DNA sequencing revealed that Tn10d inserted at different positions within hsdM (Fig. 4B), indicating that shp was in the vicinity of the hsdSMR locus.
To identify the shp locus, we used bacteriophage λ-mediated homologous recombination (recombineering) to introduce Kanr markers at different locations in the vicinity of hsdM (Fig. 4B). This effort resulted in isolation of three different Kanr insertions linked to shp, with an insertion in mrr being most tightly linked (92% cotransduction; Fig. 4B). Inspection of the genomic DNA sequence of LT2 (43) in the vicinity of mrr revealed multiple open reading frames of unknown function that were likely candidates for the shp locus. To find which of these open reading frames was responsible for generating high persisters, we constructed a derivative of AS17 by replacing the open reading frames distal to mrr (STM4528 to STM4536) with a kan cassette (Fig. 4B). After deletion of the kan cassette by site-specific recombination (44), persistence was measured and found to be significantly reduced (Fig. 4C).
Within the STM4528 to STM4536 deletion, we were intrigued that two of the predicted open reading frames, STM4528 and STM4529, were unusually small and appeared to compose a two-gene operon (Fig. 4B). Indeed, deletion of STM4528 and STM4529 also abolished high persistence in the AS17-derivative mutant (Fig. 4C). Since the open reading frames STM4528 and STM4529 were previously uncharacterized and are responsible for high persistence, they were named shpA and shpB, respectively.
To understand the basis for how the mutation in shpB imparts high persistence, we performed complementation tests by cloning shpAB+ onto pBR322-derivative, medium-copy-number plasmids (see Materials and Methods). This plasmid, along with an empty-vector control, was introduced into strain LT2 and the high persister AS17, and persistence was measured. As shown in Fig. 4D, expression of shpAB+ complemented the mutation in AS17 and restored persistence to the level observed in wild-type LT2.
Roles of shpA and shpB in conferring high persistence.
The shpAB locus was also cloned, and comparison of the DNA sequences with the sequence of the wild-type allele revealed a single transition mutation that changed a cytosine to a thiamine near the 3′ end of shpB, yielding the shpB1 allele. The predicted result of this change is a truncation of the last four amino acids of the ShpB gene product (Fig. 4B).
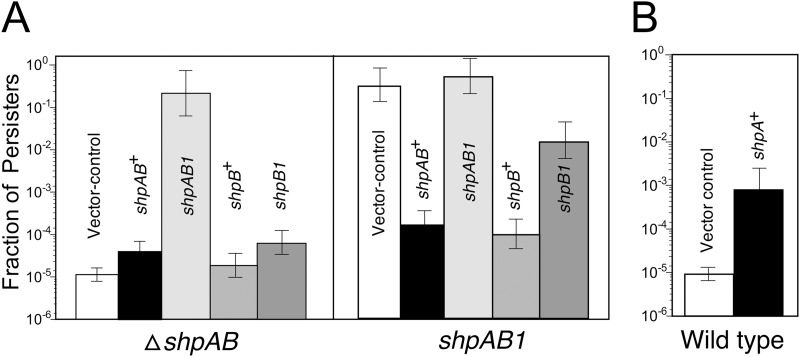
To understand how this mutation results in high persistence, we constructed additional plasmids by placing shpAB+, shpAB1, shpB+, and shpB1 under the control of the araBAD promoter. Each plasmid, along with an empty-vector control, was introduced into AS17 (shpB1) and the ΔshpAB mutant AV53, and persistence was measured. As shown in Fig. 5A, expression of shpAB1 in the ΔshpAB mutant imparted high persistence, even in the absence of l-arabinose. In the shpAB1 background, expression of shpAB+ or shpB+ alone complemented the mutation, while expression of shpB1 partially reduced persistence. In contrast, persistence remained high when shpAB1 was expressed in the shpAB1 background. These results reveal that while the shpB1 mutation can be fully complemented with shpB+, it is only partially complemented by shpB1, consistent with a defect in its gene product. Furthermore, we observed that shpA is required for high persistence, since ectopic expression of shpAB1 is capable of conferring high persistence.
Fig 5.
Effect of expression of shpAB on persistence. (A) Persistence of AV53 (ΔshpAB) or AS17 (shpB1) transformed with a plasmid expressing shpAB+, shpAB1, shpB+, or shpB1, as indicated; (B) persistence of LT2 (shpAB+) transformed with either an empty vector (vector control) or a plasmid expressing shpA+.
Despite repeated attempts, we were unable to clone shpA+ on a plasmid without shpB, except where shpA+ was under the control of the araBAD promoter. Transformants grown with l-arabinose were inviable, indicating that ShpA is a toxic gene product to both E. coli and Salmonella (data not shown). Expression of shpA+ alone elevated persistence in LT2 (Fig. 5B).
shpAB comprises a toxin-antitoxin module.
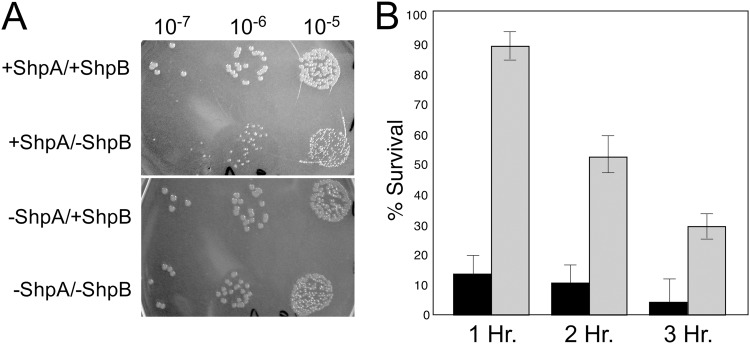
The result that ectopic expression of shpA is lethal unless shpB is also expressed and the genetic configuration in which the two small genes are tightly linked in an operon highly resemble the findings observed for most type II toxin-antitoxin (TA) systems. TA modules have also been linked to persistence (3, 29). To more directly test if shpAB comprises a previously undiscovered TA module in Salmonella, we expressed shpA and shpB separately from different compatible plasmids under the control of the araBAD and trc promoters, respectively, in E. coli. As shown in Fig. 6A, expression of shpB significantly improved the growth of transformants expressing shpA, while expression of shpB alone did not significantly affect growth. The relatively poor growth of the transformants expressing shpA from the araBAD promoter (Fig. 6A) was achieved even in the absence of l-arabinose, indicating that even low levels of the toxin are sufficient to inhibit growth. Likewise, expression of shpB from the trc promoter in the absence of IPTG was sufficient to negate the effects of ShpA expression.
Fig 6.
The detrimental effect of shpA expression on cell growth is overcome by expression of shpB. Plasmids expressing the shpA gene product from the araBAD promoter (pBAD) and shpB from the trc promoter (pTrc) were transformed into E. coli, as described in Materials and Methods. (A) Colonies expressing either shpA or shpB alone or in combination formed. Dilutions of transformants were grown in the absence of inducers (l-arabinose for pBAD and IPTG for pTrc). (B) Survival of cells following induction of shpA (black bars) by induction with l-arabinose, followed by addition of IPTG of shpB (gray bars), for the times indicated.
Another characteristic of many TA modules is that toxin expression is either bacteriostatic or lethal and growth inhibition can be relieved by expression of the cognate antitoxin (56). To determine if this is the case for ShpAB, we quantified the effect of shpA expression on cell viability both with and without shpB. While prolonged elevated expression of shpA dramatically reduced viability, growth inhibition could be overcome by elevated expression of shpB (Fig. 6B).
Type II TA modules are also known to be autoregulated, where the toxin-antitoxin complex functions as a transcriptional repressor (18). To determine if shpAB is autoregulated, we constructed a lacZ gene fusion so that the activity of the shpAB promoter could be measured by β-galactosidase activity (see Materials and Methods). The fusion was constructed on a ColE1-derivative plasmid and expressed in wild-type, ΔshpAB, and shpAB1 backgrounds. While β-galactosidase activity in LT2 was 330 (±50) Miller units, it was significantly elevated in the two shpB mutants: 1,550 (±150) Miller units in AV53 (ΔshpAB) and 3,100 (±450) Miller units in AS17 (shpB1).
There are numerous examples of cases where the antitoxin protein is a substrate of the Lon protease (29, 57–60). If ShpB is also degraded by Lon, we predicted that high persistence will be dependent upon a functional Lon protease. To test this, we expressed shpAB1 from a medium-copy-number plasmid with and without functional Lon. In E. coli, similar to LT2, ectopic expression of shpAB1 elevated the fraction of persisters in the culture (2 × 10−1). However, in the absence of lon, the fraction of persisters was significantly decreased (4 × 10−6).
With the recent characterization of several different TA modules, gene products have been classified into different families depending on the target of the toxin. One of the most common families of toxins includes mRNA nucleases, which exert their bacteriostatic effects by degrading mRNA transcripts, either by themselves or in association with the ribosome (20, 61). Recently, a comprehensive analysis of amino acid sequence alignments has revealed sequence signatures of RelE-like RNases and ribbon-helix-helix (RHH) DNA binding motifs common to many antitoxins (22). Comparison of the ShpA amino acid sequence with the sequences of other RelE-like members strongly suggests that, indeed, ShpA is another member of this RNase family (Fig. 7A). Also, analysis of the ShpB sequence revealed it to have an amino acid sequence signature of an RHH domain, consistent with the role of ShpB as an antitoxin (Fig. 7B) (22).
Fig 7.

ShpA and ShpB homology comparisons. (A) The amino acid sequence of ShpA (residues 1 to 95) is compared to the consensus amino acid properties of RelE-like RNases, as shown below the sequence. (B) The amino acid sequence (residues 59 to 95) representing the HRR region of ShpB antitoxin is shown above the consensus amino acid properties for HRR DNA binding proteins (22).
DISCUSSION
The results of this study are consistent with shpAB representing a newly discovered TA module. Despite the diversity of type II TA modules, in general, the systems include genes that encode a toxin protein along with its cognate antitoxin polypeptide. In complex, the antitoxin prevents the toxin from inhibiting one of several essential cellular processes, including translation, transcription, DNA replication, or cell division (18–20). TA modules were first recognized as determinants of plasmid stability (16, 62, 63) but have since been implicated in many other cellular processes (19, 64–66), as they are found to be encoded throughout chromosomes of free-living bacteria (67). Most antitoxins also have DNA binding activity that enables TA complexes to repress their own transcription (20). Disruption of the balance of the TA complex by cell division or by the activity of cellular proteases such as Lon destabilizes the antitoxin, leading to bacteriostasis or cell death (3, 20, 68).
Although multiple mechanisms can contribute to persistence (3), the role of TA modules is becoming well established (2, 69). The first locus responsible for high persistence, hipBA, also encodes a TA module, and ectopic expression of multiple toxin genes increases persistence (26, 27, 35–39). Deletion of the type I tisAB TA system decreased persistence under SOS-inducing conditions (26). Similarly, deletion of the mqsRA TA module reduces persistence, as well as affects biofilm formation and motility (27, 70). However, deletion of other TA modules, including hipBA, does not alter persistence levels (13, 29). This is explained by the high degree of redundancy of TA modules; genes representing different TA families are encoded throughout the genomes of most bacteria (64). Recently, this has been confirmed, since deletion of 10 TA modules predicted to encode toxins with mRNA nuclease activity decreased the level of persistence naturally occurring in E. coli (29).
Collectively, these studies suggest a model for bacterial persistence where increased toxin activity within a subpopulation of cells leads to inhibition of a cellular process necessary for growth, resulting in a state of dormancy that protects against the lethal effects of antibiotics (2, 3, 7, 66, 71). Since antitoxins can be degraded stochastically under normal growth conditions (72), as well as in stressed cells by increased proteolytic activity (73), persisters are continually present in bacterial populations.
We propose the following mechanism for shpAB-mediated persistence. The shpB1 mutation causes a truncation of four carboxy-terminal amino acids, two of which are lysine residues (Fig. 4). Consistent with other antitoxins, the carboxy terminus is typically unstructured and can serve as a substrate for Lon protease or is important for binding the toxin (57, 74). Many DNA binding proteins also have unstructured amino- or carboxy- terminal tails rich in lysine and arginine residues that assist the folded domain of the protein in contacting DNA (75–77). We propose that the product of shpB1, with its altered carboxy terminus, has reduced affinity for ShpA and, consequently, is not able to fully subdue toxin activity. In addition, elevated expression of shpA as a result of lost repressor activity likely contributes to increased toxin activity in the mutants. Together, the increased ShpA toxin expression and activity induce a state of dormancy in a larger subpopulation of cells, likely by inhibiting translation.
Although shpAB represents another example of a mutation in a TA module that causes high persistence, it is distinct from the E. coli hipA7 mutation. The hipA7 allele encodes an altered protein that is nontoxic to the cell (13). Since ectopic expression of the hipA7 product in a ΔhipBA background confers high-level persistence, the antitoxin HipB must play no part in persistence (37). Korch et al. also showed that relA and spoT are necessary for high persistence in the hipA7 mutant, leading them to propose that high persistence results from an increased rate of (p)ppGpp synthesis (13). In contrast, shpAB1-mediated persistence was independent of relA, indicating that shp is not acting through the (p)ppGpp signaling. Characterization of the shpAB1 mutant also revealed that the persister cells were generated throughout exponential growth (Fig. 3), while hipA7 high persisters occurred primarily at high cell densities (13).
Recently, another example of a mutation in a TA module yielding high persistence was reported (58). CcdAB, well established as a determinant of plasmid stability for the F factor (63), was also linked to persistence since a mutant CcdB toxin can cause release of wild-type toxin when expressed in diploid (58). The shpB1 mutant reported here represents the only example of a mutation in an antitoxin gene linked to high persistence.
One of the first reports identifying the STM4528 and STM4529 (shpAB) open reading frames was made by Sibley and Raleigh, who characterized the immigration control region (ICR) of E. coli and related bacteria (78). The ICR was originally defined as a 14-kb region near the origin of the E. coli K-12 genetic map encoding three distinct restriction-modification systems (79). Comparative genomics of this chromosomal region of several E. coli strains and S. Typhimurium LT2 revealed significant variability in genetic content. Genes within this region appear to be part of mobile cassettes that potentially integrate by site-specific recombination (78). Interestingly, shpAB is found in this highly variable region, but only in S. Typhimurium (78). TA modules similar to shpAB have also been found in bacteriophages and plasmids, suggesting that they may be widely distributed through horizontal gene transfer (22).
ACKNOWLEDGMENTS
This work was supported by the USDA (2003-35201-13889).
We thank Tom Hill and Jim Slauch for helpful suggestions and bacterial strains.
Footnotes
Published ahead of print 30 November 2012
REFERENCES
- 1. Bigger J. 1944. Treatment of staphylococcal infections with penicillin by intermittent treatment. Lancet 244:497–500 [Google Scholar]
- 2. Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48–56 [DOI] [PubMed] [Google Scholar]
- 3. Lewis K. 2010. Persister cells. Annu. Rev. Microbiol. 64:357–372 [DOI] [PubMed] [Google Scholar]
- 4. Dawson CC, Intapa C, Jabra-Rizk MA. 2011. “Persisters”: survival at the cellular level. PLoS Pathog. 7:e1002121 doi:10.1371/journal.ppat.1002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fauvart M, De Groote V, Michiels J. 2011. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J. Med. Microbiol. 60:699–709 [DOI] [PubMed] [Google Scholar]
- 6. Levin B, Rozen D. 2006. Non-inherited antibiotic resistance. Nat. Rev. Microbiol. 4:556–562 [DOI] [PubMed] [Google Scholar]
- 7. Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6:53 doi:10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lewis K. 2008. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322:107–131 [DOI] [PubMed] [Google Scholar]
- 9. Wang X, Wood T. 2011. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 77:5577–5583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Balaban N, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625 [DOI] [PubMed] [Google Scholar]
- 11. Moyed H, Bertrand K. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155:768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Black DS, Kelly AJ, Mardis MJ, Moyed HS. 1991. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 173:5732–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Korch S, Henderson T, Hill T. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50:1199–1213 [DOI] [PubMed] [Google Scholar]
- 14. Gefen O, Gabay C, Mumcuoglu M, Engel G, Balaban NQ. 2008. Single cell protein induction dynamics reveals a period of vulnerability to antibiotics in persister bacteria. Proc. Natl. Acad. Sci. U. S. A. 105:6145–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wolfson J, Hooper D, McHugh G, Bozza M, Swartz M. 1990. Mutants of Escherichia coli K-12 exhibiting reduced killing by both quinolone and beta-lactam antimicrobial agents. Antimicrob. Agents Chemother. 34:1938–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gerdes K, Rasmussen PB, Molin S. 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. U. S. A. 83:3116–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jensen RB, Gerdes K. 1995. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol. Microbiol. 17:205–210 [DOI] [PubMed] [Google Scholar]
- 18. Yamaguchi Y, Park JH, Inouye M. 2011. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45:61–79 [DOI] [PubMed] [Google Scholar]
- 19. Van Melderen L, De Bast MS. 2009. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 5:e1000437 doi:10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamaguchi Y, Inouye M. 2011. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 9:779–790 [DOI] [PubMed] [Google Scholar]
- 21. Hayes F, Van Melderen L. 2011. Toxins-antitoxins: diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 46:386–408 [DOI] [PubMed] [Google Scholar]
- 22. Makarova KS, Wolf YI, Koonin EV. 2009. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct 4:19 doi:10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moyed HS, Broderick SH. 1986. Molecular cloning and expression of hipA, a gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 166:399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Black D, Irwin B, Moyed H. 1994. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 176:4081–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. 2009. Tolerance and its neutralization by HipB molecular mechanisms of HipA-mediated multidrug. Science 323:396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dörr T, Vulić M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8:e1000317 doi:10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim Y, Wood TK. 2010. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 391:209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsilibaris V, Maenhaut-Michel G, Mine N, Van Melderen L. 2007. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J. Bacteriol. 189:6101–6108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maisonneuve E, Shakespeare LJ, Jørgensen MG, Gerdes K. 2011. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U. S. A. 108:13206–13211 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30. Hansen S, Lewis K, Vulić M. 2008. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob. Agents Chemother. 52:2718–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Girgis HS, Harris K, Tavazoie S.2012. Large mutational target size for rapid emergence of bacterial persistence. Proc. Natl. Acad. Sci. U. S. A. 109:12740–12745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu Y, Coates AR. 2005. Transposon mutagenesis identifies genes which control antimicrobial drug tolerance in stationary-phase Escherichia coli. FEMS Microbiol. Lett. 243:117–124 [DOI] [PubMed] [Google Scholar]
- 33. Spoering AL, Vulic M, Lewis K. 2006. GlpD and PlsB participate in persister cell formation in Escherichia coli. J. Bacteriol. 188:5136–5144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Groote VN, Verstraeten N, Fauvart M, Kint CI, Verbeeck AM, Beullens S, Cornelis P, Michiels J. 2009. Novel persistence genes in Pseudomonas aeruginosa identified by high-throughput screening. FEMS Microbiol. Lett. 297:73–79 [DOI] [PubMed] [Google Scholar]
- 35. Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vazquez-Laslop N, Lee H, Neyfakh AA. 2006. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J. Bacteriol. 188:3494–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Korch S, Hill T. 2006. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J. Bacteriol. 188:3826–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Falla TJ, Chopra I. 1998. Joint tolerance to beta-lactam and fluoroquinolone antibiotics in Escherichia coli results from overexpression of hipA. Antimicrob. Agents Chemother. 42:3282–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Correia FF, D'Onofrio A, Rejtar T, Li L, Karger BL, Makarova K, Koonin EV, Lewis K. 2006. Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli. J. Bacteriol. 188:8360–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Centers for Disease Control and Prevention 2011. Vital signs: incidence and trends of infection with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 1996-2010. MMWR Morb. Mortal. Wkly. Rep. 60:749–755 [PubMed] [Google Scholar]
- 42. Parry CM, Threlfall EJ. 2008. Antimicrobial resistance in typhoidal and nontyphoidal salmonellae. Curr. Opin. Infect. Dis. 21:531–538 [DOI] [PubMed] [Google Scholar]
- 43. McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 44. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Way JC, Davis MA, Morisato D, Roberts DE, Kleckner N. 1984. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32:369–379 [DOI] [PubMed] [Google Scholar]
- 46. Peterson J, Phillips GJ. 2008. New pSC101-derivative cloning vectors with elevated copy numbers. Plasmid 59:193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reece KS, Phillips GJ. 1995. New plasmids carrying antibiotic-resistance cassettes. Gene 165:141–142 [DOI] [PubMed] [Google Scholar]
- 48. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 49. Maloy S, Stewart V, Taylor R. 1996. Genetic analysis of pathogenic bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 50. Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103:2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rappleye CA, Roth JR. 1997. A Tn10 derivative (T-POP) for isolation of insertions with conditional (tetracycline-dependent) phenotypes. J. Bacteriol. 179:5827–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Altman E, Roth JR, Hessel A, Sanderson KE. 1996. Transposons currently in use in genetic analysis of Salmonella species. In Neidhardt FC, Curtiss R, III, Ingraham JL, Lin CC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 53. Murphy KC, Campellone KG. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 4:11 doi:10.1186/1471-2199-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jain C. 1993. New improved lacZ gene fusion vectors. Gene 133:99–102 [DOI] [PubMed] [Google Scholar]
- 55. Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13–18 [DOI] [PubMed] [Google Scholar]
- 56. Christensen-Dalsgaard M, Jørgensen MG, Gerdes K. 2010. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 75:333–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hansen S, Vulić M, Min J, Yen TJ, Schumacher MA, Brennan RG, Lewis K. 2012. Regulation of the Escherichia coli HipBA toxin-antitoxin system by proteolysis. PLoS One 7:e39185 doi:10.1371/journal.pone.0039185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tripathi A, Dewan PC, Barua B, Varadarajan R. 2012. Additional role for the ccd operon of F-plasmid as a transmissible persistence factor. Proc. Natl. Acad. Sci. U. S. A. 109:12497–12502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. U. S. A. 98:14328–14333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang X, Kim Y, Hong S, Ma Q, Brown B, Pu M, Tarone A, Benedik M, Peti W, Page R, Wood T. 2011. Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 7:359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yamaguchi Y, Inouye M. 2009. mRNA interferases, sequence-specific endoribonucleases from the toxin-antitoxin systems. Prog. Mol. Biol. Transl. Sci. 85:467–500 [DOI] [PubMed] [Google Scholar]
- 62. Jaffe A, Ogura T, Hiraga S. 1985. Effects of the ccd function of the F plasmid on bacterial growth. J. Bacteriol. 163:841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ogura T, Hiraga S. 1983. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. U. S. A. 80:4784–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Van Melderen L. 2010. Toxin-antitoxin systems: why so many, what for? Curr. Opin. Microbiol. 13:781–785 [DOI] [PubMed] [Google Scholar]
- 65. Gerdes K. 2000. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 182:561–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gerdes K, Christensen S, Lobner-Olesen A. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371–382 [DOI] [PubMed] [Google Scholar]
- 67. Pandey DP, Gerdes K. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33:966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pedersen K, Christensen SK, Gerdes K. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45:501–510 [DOI] [PubMed] [Google Scholar]
- 69. Gerdes K, Maisonneuve E. 2012. Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 13:103–123 [DOI] [PubMed] [Google Scholar]
- 70. Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. 2004. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64:515–524 [DOI] [PubMed] [Google Scholar]
- 71. Harrison JJ, Ceri H, Roper NJ, Badry EA, Sproule KM, Turner RJ. 2005. Persister cells mediate tolerance to metal oxyanions in Escherichia coli. Microbiology 151(Pt 10):3181–3195 [DOI] [PubMed] [Google Scholar]
- 72. Koh RS, Dunlop MJ. 2012. Modeling suggests that gene circuit architecture controls phenotypic variability in a bacterial persistence network. BMC Syst. Biol. 6:47 doi:10.1186/1752-0509-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kuroda A, Nomura K, Ohtomo R, Kato J, Ikeda T, Takiguchi N, Ohtake H, Kornberg A. 2001. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293:705–708 [DOI] [PubMed] [Google Scholar]
- 74. Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12:913–923 [DOI] [PubMed] [Google Scholar]
- 75. Crane-Robinson C, Dragan AI, Privalov PL. 2006. The extended arms of DNA-binding domains: a tale of tails. Trends Biochem. Sci. 31:547–552 [DOI] [PubMed] [Google Scholar]
- 76. Eliason JL, Weiss MA, Ptashne M. 1985. NH2-terminal arm of phage lambda repressor contributes energy and specificity to repressor binding and determines the effects of operator mutations. Proc. Natl. Acad. Sci. U. S. A. 82:2339–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jordan SR, Pabo CO. 1988. Structure of the lambda complex at 2.5Å resolution: details of the repressor-operator interactions. Science 242:893–899 [DOI] [PubMed] [Google Scholar]
- 78. Sibley MH, Raleigh EA. 2004. Cassette-like variation of restriction enzyme genes in Escherichia coli C and relatives. Nucleic Acids Res. 32:522–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Raleigh EA. 1992. Organization and function of the mcrBC genes of E. coli K-12. Mol. Microbiol. 6:1079–1086 [DOI] [PubMed] [Google Scholar]