Abstract
The soil bacterium Pseudomonas protegens Pf-5 (previously called P. fluorescens Pf-5) produces two siderophores, enantio-pyochelin and a compound in the large and diverse pyoverdine family. Using high-resolution mass spectroscopy, we determined the structure of the pyoverdine produced by Pf-5. In addition to producing its own siderophores, Pf-5 also utilizes ferric complexes of some pyoverdines produced by other strains of Pseudomonas spp. as sources of iron. Previously, phylogenetic analysis of the 45 TonB-dependent outer membrane proteins in Pf-5 indicated that six are in a well-supported clade with ferric-pyoverdine receptors (Fpvs) from other Pseudomonas spp. We used a combination of phylogenetics, bioinformatics, mutagenesis, pyoverdine structural determinations, and cross-feeding bioassays to assign specific ferric-pyoverdine substrates to each of the six Fpvs of Pf-5. We identified at least one ferric-pyoverdine that was taken up by each of the six Fpvs of Pf-5. Functional redundancy of the Pf-5 Fpvs was also apparent, with some ferric-pyoverdines taken up by all mutants with a single Fpv deletion but not by a mutant having deletions in two of the Fpv-encoding genes. Finally, we demonstrated that phylogenetically related Fpvs take up ferric complexes of structurally related pyoverdines, thereby establishing structure-function relationships that can be employed in the future to predict the pyoverdine substrates of Fpvs in other Pseudomonas spp.
INTRODUCTION
Pseudomonas is a genus of gammaproteobacteria known for its ubiquity in natural habitats and striking ecological, metabolic, and biochemical diversity. Within the genus, members of the Pseudomonas fluorescens group are common inhabitants of soil and plant surfaces, and certain strains function in the biological control of plant disease, protecting plants from infection by soilborne and aerial plant pathogens. The soil bacterium Pseudomonas protegens Pf-5 (previously called Pseudomonas fluorescens Pf-5) (1) is a well-characterized biological control strain, distinguished by its prolific production of secondary metabolites, including a spectrum of antibiotics that suppress plant-pathogenic fungi (2, 3). Pf-5 also produces two siderophores that function in iron acquisition by the bacterium, enantio-pyochelin (4) and a pyoverdine.
Pyoverdines are a group of siderophores produced by the fluorescent pseudomonads, with over 70 structures identified (5, 6). Many strains of Pseudomonas spp. produce another siderophore, in addition to a pyoverdine; these secondary siderophores are diverse in structure but typically bind iron with a lower affinity than the primary pyoverdine siderophores (7). Pyoverdines are composed of a dihydroxyquinoline chromophore, which is responsible for diffusible green fluorescence; an acyl side chain (either dicarboxylic acid or amide) bound to the amino group of the chromophore; and a peptide chain of variable length and composition (6 to 14 amino acids). The structural differences distinguishing pyoverdines are primarily found in the peptide chain, but the chromophore and acyl side chains also can vary (5). Iron is bound through interactions with the catechol unit of the chromophore and hydroxamate- or hydroxy acid-containing amino acids of the peptide chain (5).
Like other siderophores, the ferric-pyoverdines are bound and transported into the bacterial cell by TonB-dependent outer membrane proteins (TBDPs) called ferric-pyoverdine outer membrane proteins (Fpvs). The structural characteristics of Fpvs include a 22-stranded β barrel that forms a channel for transport of the ferric-pyoverdine complex through the outer membrane, extracellular loops, and a plug domain to block the channel formed by the β barrel (8, 9). Ferric-pyoverdines are moved via Fpvs across the outer membrane into the periplasm, where the iron is released from the pyoverdine (10). Transport of ferric-siderophore complexes by TBDPs requires energy, which is provided by proton motive force via TonB-ExbB-ExbD complexes in the inner membrane (11). In FpvAI, a well-characterized Fpv in P. aeruginosa, a 6-amino-acid motif called the TonB box is required for the interaction with TonB (9, 12). Most Fpvs also have an N-terminal signaling domain that interacts with a regulatory protein (anti-sigma factor), which controls the expression of an extracytoplasmic function (ECF) sigma factor (13). Together, the TBDP, ECF sigma factor, and anti-sigma factor constitute a cell surface signaling system that functions in environmental sensing and signal relay into the cytoplasm.
The ferric-pyoverdine/Fpv interaction is best understood in P. aeruginosa PAO1, where the structural components and key binding residues of FpvAI have been characterized (8, 13, 14). Collectively, strains of P. aeruginosa produce pyoverdines having three distinct structures (type I, II, or III), with each strain producing one pyoverdine and the corresponding FpvA variant (FpvAI, FpvAII, and FpvAIII). The ferric complex of the type I pyoverdine produced by PAO1 is bound by specific amino acids located in the plug, extracellular loops, and the β barrel of FpvAI. These amino acid residues primarily interact with the pyoverdine chromophore and the hydroxamate-containing amino acids of the peptide chain (15). The specificity of Fpvs in binding and transport of cognate pyoverdines is well established (9, 15), but Fpvs can also function in the uptake of ferric complexes of heterologous pyoverdines having similar peptide chain sequences (15–17). For example, FpvAI can bind and take up ferric-pyoverdines produced by several Pseudomonas spp., albeit with varied affinities. The first three residues of the pyoverdine peptide chain are the most critical determinants of affinity, but other factors, such as isomerization of the amino acid residues, also play a role in binding and transport by FpvAI (15).
The capacity to utilize siderophores produced by other microorganisms is thought to impart a selective advantage to bacteria, providing a mechanism to acquire iron that would otherwise be unavailable (18). Gram-negative bacteria commonly have multiple TBDPs in their outer membranes, some of which function in uptake of ferric complexes of siderophores produced by other organisms (19). For example, the soil bacterium P. protegens Pf-5 has 45 TBDPs, many of which have predicted functions in the uptake of ferric complexes of heterologous siderophores such as enterobactin, aerobactin, citrate, ferrioxamine, or ferrichrome (20). Phylogenetic analysis of the 45 TBDPs in the Pf-5 proteome indicated that 6 TBDPs fall into a well-supported clade with known Fpvs from other Pseudomonas spp. (20). Like many other Pseudomonas spp. (15, 21–25), Pf-5 can utilize the ferric complexes of many structurally distinct pyoverdines as iron sources (20).
The goal of the current study was to characterize the six putative Fpvs of Pf-5. We constructed six mutants, each having a deletion in one unique Fpv-encoding gene (fpv), and assessed their utilization of specific pyoverdines as iron sources. Each of the six mutants lacked the capacity to utilize ferric complexes of one or more pyoverdines, enabling the assignment of specific pyoverdines to each of the six Fpvs in the Pf-5 genome. One Fpv (FpvZ) recognized the ferric complex of the pyoverdine produced by Pf-5, which was structurally characterized by high-resolution mass spectroscopy (MS) and found to be identical to the previously described pyoverdine produced by P. protegens CHA0 (26). These results demonstrate that the capacity of P. protegens Pf-5 to utilize a diverse spectrum of ferric-pyoverdines as iron sources is achieved through six Fpvs that recognize structurally distinct pyoverdines.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Pseudomonas strains were grown routinely on King's medium B (KMB) (27) or in KMB broth at 27°C. Escherichia coli was grown in LB medium (10 g Bacto tryptone [Becton, Dickinson and Company, Sparks, MD], 5 g Bacto yeast extract [Becton, Dickinson and Company], and 10 g NaCl, pH 6.8, per liter) (28) or on solidified LB at 37°C. The following antibiotics were used at the indicated concentrations: gentamicin at 40 μg/ml (P. protegens) and 12.5 μg/ml (E. coli), kanamycin at 50 μg/ml, streptomycin at 100 μg/ml, and tetracycline at 200 μg/ml (P. protegens) and 20 μg/ml (E. coli).
The pyoverdine-producing strains of Pseudomonas spp. used in this study are listed in Table 1. Pyoverdine-deficient mutants of some strains were obtained as follows: mutant A506-1 from P. fluorescens A506 (51) was from Steven Lindow, mutant WCS374-02 from P. fluorescens WCS374 (41) was from Peter Bakker, and a pvdE mutant of P. aeruginosa 7NSK2 was from Isabelle Schalk. Derivatives of Pf-5 with mutations in pvdI (which encodes a nonribosomal synthetase required for pyoverdine production), in the enantio-pyochelin biosynthesis gene pchA or pchC, or in both pyoverdine and enantio-pyochelin biosynthesis genes (i.e., ΔpvdI-pchC and ΔpvdI-pchA mutants) were described previously (20). Deletions in the six fpv genes of Pf-5 were introduced into Pf-5 or the ΔpvdI-pchC or ΔpvdI-pchA mutant background as described by Hassan et al. (52) using overlap-extension PCR methods (53) with primers specific to each gene (see Table S1 in the supplemental material). The resulting deletion is associated with an 85-bp FLP recombinase target scar that does not exhibit any known polar effects on downstream genes (53).
Table 1.
Growth of Fpv− mutants of Pf-5 on an iron-limited medium in the presence of cross-feeding strains of Pseudomonas spp.
| Cross-feeding strain | Growth of Pf-5 deletion mutanta: |
Peptide chain compositionb | Reference(s) or source | |||||
|---|---|---|---|---|---|---|---|---|
| fpvZ | fpvU | fpvY | fpvW | fpvX | fpvV | |||
| P. protegens Pf-5e | − | + | + | + | + | + | Asp-FOHOrn-Lys-(Thr-Ala-Ala-FOHOrn-Lys) | This study |
| P. protegens CHA0e | − | + | + | + | + | + | Asp-FOHOrn-Lys-(Thr-Ala-Ala-FOHOrn-Lys) | 26 |
| P. chlororaphis DTR133 | − | + | + | + | + | + | Asp-FOHOrn-Lys-(Thr-Ala-Ala-FOHOrn-Ala) | 29 |
| P. aeruginosa PAO1e | + | − | + | + | + | + | Ser-Arg-Ser-FOHOrn-(Lys-FOHOrn-Thr-Thr) (type I pyoverdine) | 30 |
| P. fluorescens SBW25e | + | − | + | + | + | + | Ser-Lys-Gly-FOHOrn-(Lys-FOHOrn-Ser) | 31 |
| P. fluorescens ATCC 13525 | + | − | + | + | + | + | Ser-Lys-Gly-FOHOrn-(Lys-FOHOrn-Ser) | 32 |
| P. chlororaphis ATCC 9446 | + | − | + | + | + | + | Ser-Lys-Gly-FOHOrn-(Lys-FOHOrn-Ser) | 33 |
| P. rhodesiae CFML 92-104 | + | + | − | + | + | + | Ser-Lys-FOHOrn-Ser-Ser-Gly-(Lys-FOHOrn-Ser–Ser)c | 6, 34 |
| P. rhodesiae DSM14020 | + | + | − | + | + | + | Ser-Lys-FOHOrn-Ser-Ser-Gly-(Lys-FOHOrn-Ser-Ser)c | 6, 34 |
| P. marginalis pv. alfalfa NCPPB 2644 | + | + | − | + | + | + | Unknown | 35 |
| P. marginalis pv. marginalis NCPPB 667 | + | + | − | + | + | + | Unknown | 35 |
| P. marginalis pv. pastinacae NCPPB 806 | + | + | − | + | + | + | Unknown | 35 |
| Pseudomonas sp. B10 | + | + | + | − | + | + | εLys-OHAsp-Ala-aThr-Ala-cOHOrn | 36 |
| P. lini DLE411J | + | + | + | − | + | + | Lys-OHAsp-Ala-Thr-Ala-OHOrnc | 34, 37 |
| P. fluorescens ATCC 17513 | + | + | + | − | + | + | Unknown | 38 |
| P. putida CS111 | + | + | + | + | − | + | Ala-Lys-Thr-Ser-OHOrn-OHOrnc | 39 |
| Pseudomonas sp. SB8.3 | + | + | + | + | − | + | Ala-Lys-Thr-Ser-AcOHOrn-cOHOrn | 5 |
| P. putida Bn7 | + | + | + | + | + | − | Unknown | 40 |
| P. fluorescens A506e | + | + | + | + | + | + | Ser-Lys-Gly-FOHOrn-(Lys-FOHOrn-Ser)d | This study |
| P. fluorescens WCS374e | + | + | + | + | + | + | Ser-Lys-Gly-FOHOrn-(Lys-FOHOrn-Ser)c | 41 |
| P. fluorescens CLR711f | + | + | + | + | + | + | Ser-AcOHOrn-Ala-Gly-aThr-Ala-cOHOrn | 42 |
| P. costantinii CFBP 5705 | + | + | + | + | + | + | Ser-AcOHOrn-Gly- Thr-aThr-Gln-Gly-Ser-cOHOrn | 43 |
| P. fluorescens CTRp112f | + | + | + | + | + | + | Lys-AcOHOrn-Ala-Gly-aThr-Ser-cOHOrn | 42 |
| Pseudomonas sp. A6 | + | + | + | + | + | + | Lys-AcOHOrn-Gly-aThr-Thr-Gln-Gly-Ser-cOHOrn | 44 |
| P. putida CFML90-40 | + | + | + | + | + | + | Asp-Ala-Asp-AcOHOrn-Ser-cOHOrn | 39, 45 |
| P. aeruginosa ATCC 27853 | + | + | + | + | + | + | Ser-FOHOrn-Orn-Gly-aThr-Ser-cOHOrn (type II pyoverdine) | 46 |
| P. aeruginosa 7NSK2 | + | + | + | + | + | + | Ser-FOHOrn-Orn-Gly-aThr-Ser-cOHOrn (type II pyoverdine) | 47 |
Growth of Fpv− mutants in a ΔpvdI-pchC background of Pf-5 on an iron-limited medium (KMB amended with 400 or 600 μM 2,2′-dipyridyl) in the presence of the cross-feeding strain: +, growth; −, no growth. The ΔpvdI-pchC mutant of Pf-5 did not grow on the iron-limited medium in isolation but did grow in the presence of all cross-feeding strains listed. Values represent results from two replicate plates of at least two independent experiments.
Composition of the peptide chain of the pyoverdine produced by the cross-feeding strain. An underline denotes a d-amino acid. Parentheses define cyclic residues. cOHOrn, cyclo-hydroxy-ornithine; εLys, Lys linked by its ε-NH2; OHAsp, threo-β-hydroxy-aspartic acid; aThr, allo-Thr; AcOHOrn, δN-acetyl-δN-hydroxy-ornithine.
Incomplete structures lacking stereochemistry. Italicized peptide chains are inferred from siderotyping analysis (6). These pyoverdines are in the same siderotype as a pyoverdine with a chemically determined structure. Predicted pyoverdine structures do not show amino acid modifications or stereochemistry, as these are unknown.
Structures predicted from bioinformatic analysis of the NRPS sequences in the pyoverdine gene clusters in the genomes of these strains.
Strains known to produce secondary siderophores, in addition to pyoverdines: P. protegens Pf-5 and CHA0, enantio-pyochelin (4); P. aeruginosa PAO1 and 7NSK2, pyochelin (7, 48); P. fluorescens SBW25, ornicorrugatin (7); P. fluorescens WCS374 and A506, pseudomonine (41, 49). Pf-5 can utilize enantio-pyochelin as an iron source, but it cannot utilize pyochelin (50) and lacks the TBDPs for ornicorrugatin and pseudomonine (E. W. Davis II, S. L. Hartney, and J. E. Loper, unpublished data).
Strain CTRp112 is the same as strain PL8 and strain CLR711 is the same as strain PL7 (J. M. Meyer, personal communication), and the structures of pyoverdines produced by strains PL8 and PL7 are known (42).
Iron-limited growth.
Six mutants of Pf-5, each having a deletion in one of the six Fpv-encoding genes, were tested for iron-limited growth as described by Hartney et al. (20). Briefly, bacterial cells from overnight cultures grown in KMB broth were suspended in water to an optical density at 600 nm (OD600) of 0.1. Five microliters of 100-fold-diluted cell suspension was placed on KMB agar containing the iron chelator 2,2′-dipyridyl (Sigma-Aldrich, St. Louis, MO) at 0, 100, 200, 400, 600, and 800 μM. Bacterial growth was observed following 24 h of incubation at 27°C. Each strain was tested in at least two experiments, each evaluating two replicate plates.
Fpv sequence alignment and structure prediction.
The multiple-sequence-alignment tool T-Coffee (54) was used to align the amino acid sequences and FpvAI of P. aeruginosa. The PSIPRED GenThreader method (55) and a β-barrel prediction model (56) were used to predict the secondary structure of the six Fpvs in the Pf-5 proteome. Homology modeling of the six Fpvs was done using the SWISS-MODEL server and Deepview program (57, 58). The homology models were constructed using a structure-based sequence alignment with the crystal structure of FpvAI from P. aeruginosa PAO1 (8) as the template.
Bioinformatic prediction of pyoverdine peptide chain composition.
The amino acid composition of the peptide chain of pyoverdines with unknown structures was predicted from the sequences of genes encoding the corresponding nonribosomal peptide synthetases (NRPSs) using the NRPS/polyketide synthase predictor (59) and the NRPS predictor (http://www-ab.informatik.uni-tuebingen.de/software/NRPSpredictor) (60, 61).
Pyoverdine purification.
The pyoverdine from P. protegens Pf-5 was obtained from cultures of a ΔpchA mutant of Pf-5 grown in Difco minimal broth Davis without dextrose (Becton, Dickinson and Company) supplemented with 20 mM glycerol at 24°C with shaking at 140 rpm. After 72 h of incubation, bacterial cells were removed by centrifugation. Amberlite XAD-4 adsorbent resin was added to the supernatant (50 g/liter), and the mixture was agitated at 120 rpm for 24 h. The XAD-4 resin was then passed through a fritted funnel, and the bound metabolites were eluted with acetonitrile (MeCN)-H2O (75:25) under agitation for 24 h. The eluate, which contained the pyoverdine, was dried using a rotary evaporator and redissolved in methanol (MeOH)-H2O (50:50) for high-performance liquid chromatography (HPLC) analysis. For reverse-phase HPLC separation of this crude XAD-4 extract (Phenomenex aqua column; 4.6 by 250; particle size, 5 μm; flow rate, 1.0 ml/min; UV monitoring at 380 nm), we used isocratic elution at 2% MeCN in H2O for 10 min, followed by gradient elution from 2 to 10% MeCN in H2O over 10 min, followed by isocratic elution at 10% MeCN in H2O for an additional 10 min. This yielded the purified pyoverdine of Pf-5 in both the iron-chelated and the deferrated forms at an approximately 1:1 ratio, as determined by MS and UV analysis.
Pyoverdines from the other strains of Pseudomonas spp. were obtained from cultures grown in succinate medium (62) at 25°C with shaking at 200 rpm. After 72 h of incubation, bacterial cells were harvested by centrifugation. The supernatant was adjusted to pH 6.0, centrifuged again (5,200 × g, 30 min), and passed through a column of Amberlite XAD-4 (63). The pyoverdines were eluted with 100% methanol, and the eluate was dried in a rotary evaporator, resuspended in 5 ml of deionized water, and incubated at 4°C overnight. A second chromatography step was performed on a column of LiChroprep RP-18 (particle size, 40 to 63 μm; Merck, Whitehouse Station, NJ). After loading, the column was rinsed with 0.1 M EDTA, followed by pH 4.0 acidified water (1% formic acid), and pyoverdines were eluted with 50% methanol. After use, the column was washed with methanol, regenerated with 1% HCl, rinsed with deionized water, and stored at 4°C. The resultant partially purified pyoverdine preparations, which were composed almost entirely of the iron-free ligand, were concentrated and lyophilized prior to storage at 4°C in the dark.
Pyoverdines were isolated from the partially purified preparations by reverse-phase HPLC using a linear gradient of 2:98 to 100:0 MeOH-H2O over a period of 30 min, followed by isocratic elution with 100% MeOH for 10 min (Macherey-Nagel Nucleodur PolarTec column; 250 by 4.6 mm; particle size, 5 μm; flow rate, 1 ml/min; UV monitoring at 210, 254, and 370 nm). This procedure yielded the pyoverdines of P. rhodesiae CFML92-104 and Pseudomonas sp. strain SB8.3 in a highly purified form and, in the case of P. aeruginosa PAO1, P. putida Bn7, and Pseudomonas sp. strain B10, two to three pyoverdine isoforms. The mass of each pyoverdine was determined by high-resolution electron spray ionization (ESI)-MS.
Cross-feeding assays and utilization of heterologous pyoverdines by Pf-5.
Cross-feeding assays were performed as described by Hartney et al. (20). Briefly, cells from test strains and indicator strains were suspended in water to an OD600 of 0.1. Ten microliters of the test strain suspension was placed at the center of a petri plate containing an iron-limited medium (KMB amended with 2,2′-dipyridyl at 400 μM or 600 μM). For test strains that did not grow on the iron-limited medium, an agar plug (6 mm) obtained from a 48-h culture on KMB was substituted for the cell suspension. Cell suspensions of the indicator strains (OD600 of 0.1) were diluted 100-fold in sterile water, and 5 μl of the diluted suspension was placed 10 mm away from the test strain or pyoverdine on the agar surface. Indicator strains were derivatives of Pf-5 deficient in the production of both pyoverdine and enantio-pyochelin (i.e., ΔpvdI-pchC and ΔpvdI-pchA) with deletions in genes encoding one or more of the six Fpvs of Pf-5. The indicator strains could not grow on KMB amended with 2,2′-dipyridyl at 400 μM unless they could use the ferric complex of a siderophore produced by the test strain (20). Plates were incubated at 27°C, and growth of the indicator strain was observed at 2 days.
Tests evaluating the utilization of heterologous pyoverdines by Pf-5 were done following a procedure similar to that used for the cross-feeding assays, with 5 μl of a pyoverdine solution substituting for the test strain. Concentrations of the pyoverdine solutions were determined from the mass of the dried pyoverdine samples. Due to variability in the purity of the partially purified preparations, we estimate that the pyoverdine concentration in those samples ranged from 5 to 8 μM.
Phylogenetic analysis.
Amino acid sequences of the six Fpvs from Pf-5 were submitted to the NCBI database of nonredundant protein sequences to identify the five best hits for each using the PSI-BLAST algorithm (64). These sequences and those of Fpvs having known ferric-pyoverdine substrates were aligned using the ClustalW program (65) with a gap open penalty of 15 and a gap extension penalty of 0.3. A phylogenetic analysis of the aligned sequences was done using the neighbor-joining method available through the MEGA (version 5.0.5) package (66).
RESULTS AND DISCUSSION
Structural analysis of ferric-pyoverdine outer membrane proteins.
Protein structure analysis using PSIPRED GenThreader matched all six of the putative Pf-5 Fpvs to FpvAI (PA2398) of P. aeruginosa PAO1. Due to their similarities to FpvAI, we adopted the naming convention established for P. aeruginosa; here, the six pyoverdine uptake proteins in Pf-5 are called FpvU (PFL_2391), FpvV (PFL_2527), FpvW (PFL_2293), FpvX (PFL_3315), FpvY (PFL_3485), and FpvZ (PFL_4092).
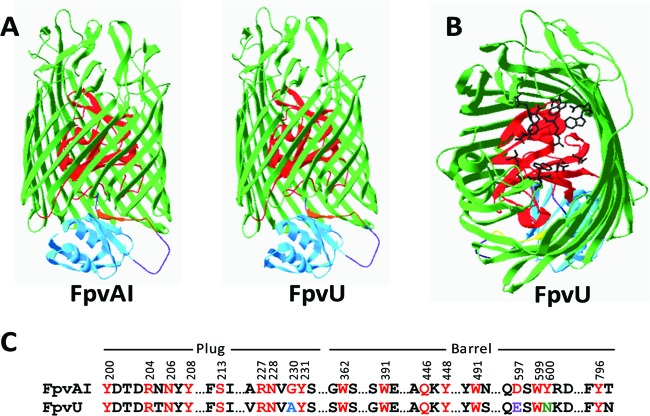
Homology modeling of the six Pf-5 Fpvs was done using the known crystal structure of FpvAI (Protein Data Bank accession number 2w16A) as a template (Fig. 1A; see Fig. S1 in the supplemental material). A model of FpvW could not be generated due to its divergence from FpvAI, but root mean square (RMS) values were calculated for backbone residues of the other five Fpvs in Pf-5: FpvU, 0.173Å; FpvY, 0.40Å; FpvX, 0.55Å; FpvZ, 0.626Å; and FpvV, 1.51Å. Comparison of the six Fpvs of Pf-5 to FpvAI identified secondary structural similarities of the proteins, which were especially evident in the β strands of the β barrel, the plug domain involved in binding and movement of substrate (9), and the N-terminal signaling domain involved in the signaling cascade to regulate genes involved in uptake of ferric-pyoverdine complexes (13) (Fig. 1A; see Fig. S1 in the supplemental material).
Fig 1.
(A) Homology model of FpvU from P. protegens Pf-5 compared to FpvAI from P. aeruginosa PAO1 showing the structural components, with the β barrel in green, plug in red, N-terminal signaling domain in blue, connecting loop in purple, and TonB box in brown. (B) Positions of amino acid residues in the plug and β-barrel domains of FpvU corresponding to the residues of FpvAI that are involved in pyoverdine binding. Amino acid side chain structures are shown in black. (C) Alignment of FpvAI and FpvU. Amino acid residues of FpvAI that interact with the PAO1 pyoverdine (8, 13) are numbered and shown in red font. Identical residues in FpvU are in red, conservative substitutions are in blue, semiconservative substitutions are in purple, and nonconservative substitutions are in green.
The amino acid sequences of the TonB boxes of the Pf-5 Fpvs are divergent but could putatively be identified on the basis of their locations in the β strand and the presence of bordering leucine residues (Fig. 1A; see Fig. S1 and S2 in the supplemental material). FpvU shares four of six residues with the well-characterized TonB box of FpvAI (8, 12), whereas the TonB boxes of the other five Fpvs of Pf-5 share only one to three of the six residues (see Fig. S2 in the supplemental material). The lack of sequence conservation in TonB boxes was previously noted for the 45 TBDPs of Pf-5 (20) and for an extensive set of TBDPs characterized from other bacteria (11). The latter study concluded that divergence in the amino acid sequence of the TonB box has little effect on substrate binding, as long as the surrounding residues forming a β strand are conserved. Rather than the primary amino acid sequence, the secondary protein structure, specifically, the interaction of a β strand encompassing the TonB box motif of the membrane protein with the β-sheet structure of TonB, is thought to determine binding to TonB (11). Homology models of five of the Pf-5 Fpvs clearly indicate the presence of a β-strand structure overlapping the TonB box (Fig. 1A; see Fig. S1 in the supplemental material), which is consistent with the contention that the secondary structure of the TonB box region is required for interaction with TonB.
The RMS values and sequence alignments of the Pf-5 Fpvs suggest that FpvU is the most closely related to FpvAI (68% identical at the amino acid level). Of the 18 amino acids located in the binding pocket of FpvAI that interact with the pyoverdine of PAO1 (8, 13), 15 are also present in the binding pocket of FpvU (Fig. 1B and C). Homology modeling between FpvAI and FpvU indicated that the binding residues are in similar locations in both proteins: the channels of the β barrels and the plug domains (Fig. 1B). Two of the altered binding residues are conservative substitutions (G230 to A237 in the plug domain and D597 to E606 in the β barrel), and one is a nonconservative substitution (Y600 to N609 in the β barrel) (Fig. 1C). Putative pyoverdine binding residues of FpvZ, FpvV, FpvW, FpvX, and FpvY could not be identified from homology models or by alignment due to their divergence from FpvAI (see Fig. S2 in the supplemental material).
Iron-limited growth of Pf-5 mutants with an fpv deletion (i.e., Fpv− mutants).
To determine the functions of the six Fpvs in ferric-pyoverdine uptake, a deletion in each fpv was introduced into wild-type Pf-5, which produces a pyoverdine and enantio-pyochelin (4, 20). Deletions in five of the fpv genes (fpvU, fpvV, fpvW, fpvX, and fpvY) had no detectable effect on iron-limited growth when introduced into the wild-type background (data not shown). These five mutants exhibited wild-type levels of fluorescence and, like the parental strain Pf-5, grew on an iron-limited medium (KMB amended with up to 800 μM the chelator 2,2′-dipyridyl). In contrast, the fpvZ mutant, like the pvdI mutant of Pf-5, which lacks a nonribosomal synthetase required for pyoverdine biosynthesis, was not fluorescent and grew only on KMB containing ≤400 μM 2,2′-dipyridyl. The lack of growth of the fpvZ mutant on the iron-limited medium was not surprising because fpvZ is located in one of the pyoverdine biosynthesis gene clusters of Pf-5 (20) (Fig. 2A) and therefore was expected to function in the uptake of ferric complexes of the pyoverdine synthesized by Pf-5, as was demonstrated in experiments described below. The involvement of the cognate Fpv in the regulation of pyoverdine biosynthesis genes is well established in the literature (67, 68), so the lack of fluorescence of the fpvZ mutant was likely due to reduced expression of pyoverdine biosynthesis genes.
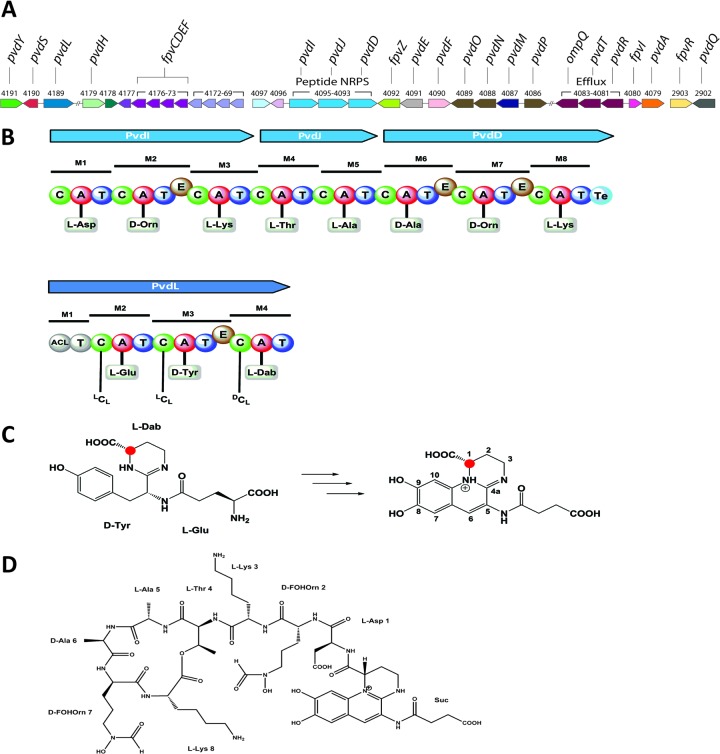
Fig 2.
Bioinformatic and structural analysis of the pyoverdine produced by P. protegens Pf-5. (A) Pyoverdine biosynthesis gene clusters of Pf-5 (80). (B) NRPSs in the pyoverdine gene cluster. For each module (M) in the NRPSs, domains (C, condensation; A, adenylation; T, thiolation; E, epimerization; Te, thioesterase) and the specific amino acid predicted from the sequence of each adenylation domain are shown. The NRPS encoded by pvdI, pvdJ, and pvdD synthesizes the peptide chain, and the NRPS encoded by pvdL synthesizes the chromophore. (C) Scheme for the pyoverdine chromophore biosynthesis. The linear tripeptide-intermediate l-Glu–d-Tyr–l-Dab undergoes hydroxylation, dehydration, and cyclization reactions to give the completed quinoline chromophore. The red dot indicates that the absolute configuration at C-1 of the chromophore results from the absolute configuration of the integrated amino acid Dab, which is in turn determined by PvdL. (D) Structure of the Pf-5 pyoverdine determined from high-resolution MS analysis. Suc, succinic acid.
Characterization of the pyoverdine of P. protegens Pf-5.
We began our analysis of the substrate specificity of FpvZ by using a combination of bioinformatics and analytical chemistry to determine the structure of the Pf-5 pyoverdine. The pyoverdine gene clusters of Pf-5 contain four NRPS-encoding genes (20) (Fig. 2A). One of these, PFL_4189 (pvdL), presumably governs the synthesis of the pyoverdine chromophore, as it shares 75% predicted amino acid identity with the chromophore NRPS-encoding gene pvdL of P. aeruginosa PAO1. Likewise, the contiguous genes PFL_4093 to PFL_4095 are expected to code for the peptidic side chain of the Pf-5 pyoverdine. From an in silico analysis of the corresponding adenylation and epimerization domains thereof, the following peptide sequence was deduced: l-Asp1, d-Orn2, l-Lys3, l-Thr4, l-Ala5, d-Ala6, d-Orn7, l-Lys8 (Fig. 2B). Due to the presence of orthologs of pvdA (PFL_4079, 76% amino acid identity to PAO1 PvdA) and pvdF (PFL_4090, 73% amino acid identity to PAO1 PvdF) (Fig. 2A), which encode an ornithine hydroxylase and an ornithine transformylase, respectively, we assume that Orn2 and Orn7 are present as δN-formyl-δN-hydroxy-ornithine (FOHOrn) residues. The resulting peptide sequence thus conforms exactly to the peptide chain of the pyoverdine of P. fluorescens CHA0 (26), which is additionally cyclized between Thr4 and Lys8. The bioinformatic prediction is corroborated by a previously reported isoelectric focusing (IEF) analysis (22), which confirmed the peptide sequence and the cyclization scheme. The absolute configuration of the chromophore was not determined experimentally; however, since all chromophores obtained from related pyoverdines possess an S configuration at this center, we assume that the chromophore residue in the pyoverdine of Pf-5 also has an S configuration. Support for this hypothesis is also provided by the in silico analysis of pvdL (PFL_4189): module 4 of PvdL determines the absolute configuration of the amino acid residue 2,4-diaminobutyric acid (Dab) and, therefore, the configuration at C-1 of the chromophore (69). The C domain of module 4 of PvdL clearly groups with C domains of the dCl type, hence linking an l-configured Dab residue to the growing peptide chain ending with d-tyrosine. The lack of an epimerization domain in module 4 is consistent with a 1S configuration for the chromophore of the Pf-5 pyoverdine (Fig. 2C). This left the nature of the dicarboxylic side chain to be established. The molecular mass of the Pf-5 pyoverdine was detected by ESI-MS mainly as a double-charged quasimolecular ion at m/z 644.7941 [M + 2H]2+ (see Fig. S3 in the supplemental material), indicating the molecular formula C55H81N15O21 (1,287 Da, calculated for m/z 644.7938 [M + 2H]2+). If the C, N, and O atoms accounted for by the eight identified amino acid residues and the chromophore moiety are subtracted from the molecular formula of the Pf-5 pyoverdine, the remaining fragment of the molecule must contain four carbon, five hydrogen, and three oxygen atoms, which corresponds to succinic acid. The structure of the Pf-5 pyoverdine was thus determined to be succinic acid–chromophore–l-Asp–d-FOHOrn–l-Lys-(l-Thr–l-Ala–d-Ala–d-FOHOrn–l-Lys) (Fig. 2D) and is therefore identical to that of the pyoverdine of P. protegens CHA0 (26), with the absolute configuration of the alanine residues now clarified.
Cross-feeding of Pf-5 and Fpv− derivatives by siderophore-producing strains of Pseudomonas spp.
Previously, we reported that mutants of Pf-5 lacking both the NRPS-encoding pvdI and a gene for enantio-pyochelin biosynthesis (i.e., ΔpvdI-pchC or ΔpvdI-pchA mutants) are deficient in siderophore production and cannot grow on an iron-limited medium unless an exogenous ferric-siderophore complex that can be used by Pf-5 is provided (20). We also identified strains of Pseudomonas spp. that cross-fed the ΔpvdI-pchC mutant, thereby enabling growth of the mutant on the iron-limited medium. Here, we followed up on that study by testing representative strains of Pseudomonas spp. that cross-fed the ΔpvdI-pchC mutant of Pf-5 for cross-feeding of the six Fpv− mutants (the fpvU, fpvV, fpvW, fpvX, fpvY, and fpvZ mutants) in the ΔpvdI-pchC background (Table 1). Because there is residual enantio-pyochelin production by the ΔpvdI-pchC mutant of Pf-5 (20), a subset of the strains was also tested for cross-feeding of Fpv− mutants in a ΔpvdI-pchA background. The ΔpvdI-pchA mutant does not produce detectable levels of pyoverdine or enantio-pyochelin (20). Cross-feeding results did not differ between a given Fpv− mutant in the two mutant backgrounds (Table 1; see Table S3 in the supplemental material).
As predicted from its location within a pyoverdine biosynthesis gene cluster (20), FpvZ was necessary for cross-feeding by wild-type Pf-5. FpvZ was also necessary for cross-feeding by P. protegens CHA0, which produces the identical pyoverdine as Pf-5, and P. chlororaphis DTR133 (Table 1). Like Pf-5 and CHA0, strain DTR133 produces a pyoverdine having 8 amino acids in the peptide chain, with the first 7 amino acids (Asp-FOHOrn-Lys-Thr-Ala-Ala-FOHOrn) being in common (Table 1).
Based upon the high level of conservation in the binding residues between FpvU of Pf-5 and FpvAI of P. aeruginosa PAO1, we expected that the two proteins would recognize similar pyoverdines. FpvAI recognizes the type I pyoverdine produced by P. aeruginosa strain PAO1 (70), and a deletion in fpvU eliminated cross-feeding of Pf-5 by strain PAO1, as expected. FpvU was also required for cross-feeding by P. fluorescens SBW25 and P. fluorescens ATCC 17518 (Table 1). The three pyoverdines recognized by FpvU have 7 or 8 amino acids in the peptide chain, with d-Ser in the first position, Arg or Lys in position 2, and a small residue (Ser or Gly) in position 3, followed by FOHOrn-Lys-FOHOrn. Thus, the first 4 amino acids of these peptide chains are identical to those found in pyoverdines that bind with high affinity to FpvAI (15).
FpvY was required for cross-feeding by two strains of P. rhodesiae and three strains of P. marginalis. The structure of the pyoverdine(s) produced by the strains of P. marginalis is unknown, but both strains of P. rhodesiae produce a pyoverdine having a peptide chain composed of 10 amino acids (Table 1). FpvW was required for cross-feeding by Pseudomonas sp. B10, P. lini DLE411J, and P. fluorescens ATCC 17513. Of these strains, only strain B10 produces a pyoverdine of known structure, although the same pyoverdine is produced by P. lini DLE411J, as determined by siderotyping (34) (Table 1). FpvX was required for cross-feeding by Pseudomonas sp. SB8.3 and P. putida CS111, which produce the same pyoverdine (Table 1). FpvV was required for cross-feeding by P. putida Bn7, which produces a pyoverdine of unknown structure (Table 1).
In summary, each of the six Fpvs was required by Pf-5 for cross-feeding by one or more strains of Pseudomonas spp. Collectively, the six Fpvs were required for cross-feeding by 18 strains that produce at least eight different pyoverdines, with seven having known structures (Table 1). Therefore, the cross-feeding assays identified candidate pyoverdines that may be recognized by each of the six Fpvs of Pf-5.
Specific Fpv proteins are required for the utilization of purified pyoverdines as iron sources.
To confirm that the candidate pyoverdines were recognized by the specific Fpvs identified above, we purified representative pyoverdines from culture supernatants of cross-feeding strains of Pseudomonas spp. and tested the purified compounds. A single pyoverdine isoform was detected in culture supernatants of P. protegens Pf-5, P. rhodesiae CFML92-104, and Pseudomonas sp. strain SB8.3 (Table 2). In contrast, two or more pyoverdine isoforms were detected in culture supernatants of P. aeruginosa PAO1, Pseudomonas sp. B10, and P. putida Bn7. In each of the last three strains, differences in mass of the pyoverdine isoforms could be explained by variations in acyl side chains (see Text S1 in the supplemental material). Pyoverdine isoforms, having an identical chromophore and peptide chain but differing in the nature of the dicarboxylic side chain, are known to be produced by many pseudomonads (6). The purified pyoverdines were tested to determine if they could be utilized as iron sources by Pf-5 and the six mutants lacking an Fpv (i.e., the fpvU, fpvV, fpvW, fpvX, fvpY, and fvpZ mutants) (Fig. 3). In all cases, the purified pyoverdine isoforms and the partially purified pyoverdines from a given strain were identical to one another in stimulating the growth of Pf-5 on the iron-limited medium (Table 2). These results conform to those of Meyer et al. (72), who reported that the succinic acid, succinamide, and α-ketoglutaric acid isoforms of the P. aeruginosa ATCC 15692 pyoverdine display identical efficiencies in iron uptake, suggesting that the modification of the acyl chain does not affect the biological function of these compounds. In the present study, each purified pyoverdine failed to stimulate the growth of one of the six Fpv− mutants on the iron-limited medium, and the Fpv recognizing each purified pyoverdine (Table 2) was identical to that identified in assays evaluating cross-feeding by the pyoverdine-producing strains (Table 1). Therefore, a specific pyoverdine corresponding to each of the Fpvs of Pf-5 was assigned, and pyoverdines of known structure were matched to five of the six Fpvs.
Table 2.
Growth of Fpv− mutants of Pf-5 on an iron-limited medium in the presence of specific pyoverdines
| Peptide chain composition of pyoverdine testeda | Source strain of pyoverdine | Pyoverdine preparationb | Growth of Pf-5 deletion mutantc: |
|||||
|---|---|---|---|---|---|---|---|---|
| fpvZ | fpvU | fpvY | fpvW | fpvX | fpvV | |||
| Asp-FOHOrn-Lys-(Thr-Ala-Ala-FOHOrn-Lys) | P. protegens Pf-5 | Partially purified | − | + | + | + | + | + |
| Purified | − | + | + | + | + | + | ||
| Ser-Arg-Ser-FOHOrn-(Lys-FOHOrn-Thr-Thr) (type I pyoverdine) | P. aeruginosa PAO1 | Partially purified | + | − | + | + | + | + |
| Pyoverdine D | + | − | + | + | + | + | ||
| Pyoverdine C | + | − | + | + | + | + | ||
| Ser-Lys-Gly-FOHOrn-(Lys-FOHOrn-Ser) | P. fluorescens ATCC 13525 | Partially purifiedd | + | − | + | + | + | + |
| Ser-Lys-FOHOrn-Ser-Ser-Gly-(Lys-FOHOrn-Ser-Ser) | P. rhodesiae CFML92-104 | Partially purified | + | + | − | + | + | + |
| Purified | + | + | − | + | + | + | ||
| εLys-OHAsp-Ala-aThr-Ala-cOHOrn | Pseudomonas sp. B10 | Partially purified | + | + | + | − | + | + |
| B10-1034 | + | + | + | − | + | + | ||
| B10-1018 | + | + | + | − | + | + | ||
| Pseudobactin | + | + | + | − | + | + | ||
| Ala-Lys-Thr-Ser-AOHOrn-cOHOrn | Pseudomonas sp. SB8.3 | Partially purified | + | + | + | + | − | + |
| Purified | + | + | + | + | − | + | ||
| Unknown | P. putida Bn7 | Partially purified | + | + | + | + | + | − |
| Bn7-1162 | + | + | + | + | + | − | ||
| Bn7-1046 | + | + | + | + | + | − | ||
| Bn7-1133 | + | + | + | + | + | − | ||
| Ser-FOHOrn-Orn-Gly-aThr-Ser-cOHOrn (type II pyoverdine) | P. aeruginosa ATCC 27853 | Partially purified | + | + | + | + | + | + |
| Ser-FOHOrn-Orn-Gly-aThr-Ser-cOHOrn (type II pyoverdine) | P. aeruginosa 7NSK2 | Partially purified | + | + | + | + | + | + |
Underline denotes a d-amino acid. Parentheses define cyclic residues. cOHOrn, cyclo-hydroxy-ornithine; εLys, Lys linked by its ε-NH2; OHAsp, threo-β-hydroxy-aspartic acid; aThr, allo-Thr. Italicized structures were predicted by siderotyping and lack stereochemistry (Table 1).
Partially purified pyoverdines were obtained following liquid chromatography of culture supernatants from the designated strains of Pseudomonas spp. Purified samples were obtained following HPLC fractionation of the partially purified samples. Structures of purified pyoverdines were determined by high-resolution ESI-MS (see Text S1 in the supplemental material), and m/z values were consistent with the peptide chain, as shown in all cases. Acyl side chains for the pyoverdine isoforms were identified as follows: P. aeruginosa PAO1, pyoverdine C (α-ketoglutaric acid) and pyoverdine D (succinic acid) (71); Pseudomonas sp. B10, pseudobactin (succinic acid) (36), B10-1018 (possibly glutamic acid), and B10-1034 (possibly a hydroxylated congener of B10-1018 [e.g., OH-glutamate, 5-OH-chromophore, exchange of Ala with Ser]). The structure of the pyoverdine produced by P. putida Bn7 is unknown, and high-resolution ESI-MS analysis revealed three isoforms with [M + H]+ ions at m/z 1,163.4956, 1,047.441 and 1,134.4751. The difference of 29 mass units between Bn7-1133 and Bn7-1162 may correspond to acyl side chains composed of succinic acid and glutamic acid, respectively.
Growth of Fpv− mutants in a ΔpvdI-pchA background of Pf-5 on an iron-limited medium (KMB amended with 600 μM 2,2′-dipyridyl) in the presence of the pyoverdine sample: +, growth; −, no growth. The ΔpvdI-pchA mutant of Pf-5 did not grow on the iron-limited medium in isolation but did grow in the presence of all pyoverdine samples listed. Values represent results from two replicate plates in each of two experiments evaluating each partially purified sample. Purified isoforms were evaluated in only one experiment due to the limited amounts available.
The pyoverdine from ATCC 13525 was a gift from Isabelle Schalk and Valérie Geoffroy.
Fig 3.
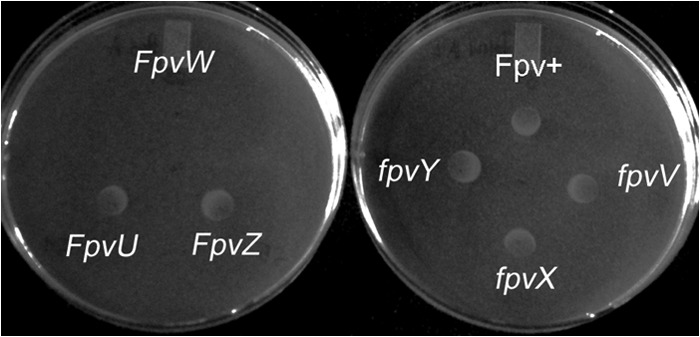
Utilization of a ferric-pyoverdine complex by a siderophore-deficient mutant of Pf-5. Five microliters of an 8 μM solution of pyoverdine B10-2 (from Pseudomonas sp. strain B10) was spotted in the center of each plate containing an iron-limited medium (KMB amended with 600 mM 2,2′-dipyridyl). Ten microliters of a cell suspension (∼106 CFU/ml) of a pvdI-pchA mutant of Pf-5 (Fpv+) or the pvdI-pchA mutant with a deletion in one of six fpv genes (fpvU, fpvV, fpvX, fpvY, or fpvZ) was spotted 1 cm from the center of the plate. The plate was incubated at 27°C for 2 days. Growth of all strains with the exception of the pvdI-pchA-fpvW mutant was observed, indicating that FpvW is required for uptake of pyoverdine B10-2.
Phylogenetic analysis of Fpvs.
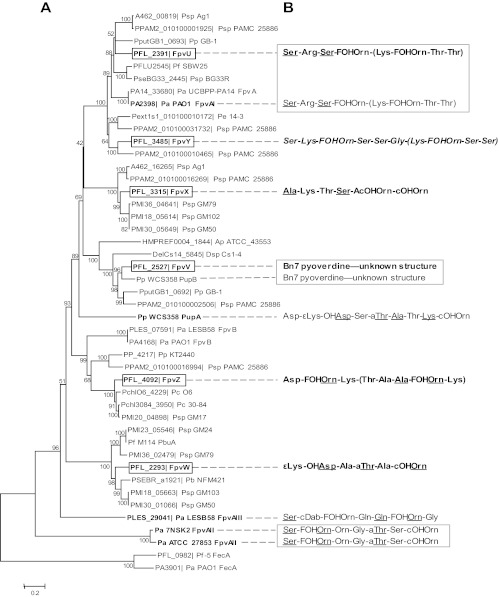
The six Fpvs of Pf-5, their closest orthologs, and characterized Fpvs with known substrates from other Pseudomonas spp. were aligned and subjected to phylogenetic analysis (Fig. 4A). FpvV is in the same clade as PupB from P. putida WCS358 (40), and both FpvV and PupB recognize the pyoverdine of unknown structure produced by P. putida Bn7 (Fig. 4B). Neither FpvX, FpvY, nor FpvZ, the TBDP for the uptake of Pf-5's own pyoverdine, has an ortholog with known substrates. To our knowledge, Fpvs recognizing the pyoverdine structures assigned here to FpvX, FpvY, or FpvZ have not been reported in the literature. FpvU is in a well-supported clade with FpvAI (70), and both proteins take up the type I pyoverdine produced by P. aeruginosa PAO1. FpvW and PbuA from Pseudomonas sp. strain M114 (73) are in the same clade; both proteins can take up pseudobactin, the pyoverdine produced by Pseudomonas sp. B10. Pf-5 does not have an ortholog to PupA (Fig. 4A), which recognizes the pyoverdine of P. putida WCS358 (74). This observation is consistent with our earlier finding that strain WCS358 does not cross-feed Pf-5 (20). By comparing our phylogenetic analysis with observations from cross-feeding experiments (Fig. 4), we conclude that phylogenetically related Fpvs recognized structurally related pyoverdines.
Fig 4.
(A) Neighbor-joining analysis of Fpv proteins. Pf-5 Fpvs are shown in bold boxed font, and Fpvs having known substrates are shown in bold font; TBDPs that take up ferric citrate (FecA) are included as an outgroup. Abbreviations for species represented in the tree are as follows: PFL_, P. protegens Pf-5; Ap ATCC_43553, Achromobacter piechaudii ATCC 43553; Dsp, Delftia sp. strain Cs1-4; Pa PAO1, Pa ATCC 27853, Pa LESB58, Pa UCBPP-PA14, and Pa 7NSK2, P. aeruginosa PAO1, ATCC 27853, LESB58, UCBPP-PA14, and 7NSK2, respectively; Pb NFM421, P. brassicacearum subsp. brassicacearum NFM421; Pc O6 and Pc 30-84, Pseudomonas chlororaphis O6 and 30-84, respectively; Pe 14-3, Pseudomonas extremaustralis 14-3; Pf SBW25, P. fluorescens SBW25; Pp GB-1, Pp KT2440, and Pp WCS358, P. putida GB-1, KT2440, and WCS358, respectively; Psp Ag1, Psp BG33R, Psp GM17, Psp GM24, Psp GM50, Psp GM79, Psp GM102, Psp GM103, and Psp PAMC 25886, Pseudomonas sp. strains Ag1, BG33R, GM17, GM24, GM50, GM79, GM102, GM103, and PAMC 25886, respectively. The consensus tree was inferred from 1,000 replicates, and branches corresponding to partitions reproduced in less than 30% bootstrap replicates were collapsed. The percentage of replicate trees in which the proteins clustered together in the bootstrap test is shown next to each branch. (B) Pyoverdine peptide chain sequences recognized by the adjacent Fpvs. See Table 2 for an explanation of abbreviations and formatting. Pyoverdines associated with Fpvs of Pf-5 in this study (Table 2) are in bold font. Boxes surround structures that are recognized by more than one Fpv in the corresponding clade shown in panel A. The following Fpvs were associated previously with specific ferric-pyoverdine complexes: PupB (40), PupA (74), FpvA (15), FpvB (78), PbuA (73), and FpvAII (47).
Functional redundancy of Fpvs in P. protegens Pf-5.
Nine of the pyoverdine-producing strains of Pseudomonas spp. cross-fed all of the single Fpv− mutants (Table 1), indicating that cross-feeding by these strains was not mediated by a single Fpv in Pf-5. P. fluorescens strains WCS374 and A506 were among the nine strains, which was unexpected because the pyoverdine produced by WCS374 (41) has the same amino acids in the peptide chain as the pyoverdines produced by three strains (P. fluorescens SBW25, P. fluorescens ATCC 13525, and P. chlororaphis ATCC 9446) that cross-fed Pf-5 via FpvU. Bioinformatic analysis of the NRPSs (PvdI and PvdD) responsible for biosynthesis of the pyoverdine peptide chain in the genome of strain A506 predicted that the peptide chain of the A506 pyoverdine has the same amino acid composition as the pyoverdines from the other four strains (SBW25, ATCC 13525, ATCC 9446, and WCS374) (Fig. 5). WCS374 produces the secondary siderophore pseudomonine (41), and a pseudomonine biosynthesis gene cluster is also present in the A506 genome (49). Pyoverdine-deficient mutants of WCS374 and A506 did not cross-feed the Pf-5 ΔpvdI-pchC mutant, however, indicating that the pyoverdines, rather than pseudomonine or another secondary siderophore that may be produced by these strains, were responsible for the cross-feeding. Because there are similarities in both the N-terminal and C-terminal amino acids of the peptide chains of pyoverdines recognized by FpvU and FpvY (Table 2; Fig. 4), we reasoned that there may be overlap in pyoverdine uptake by these two Fpvs. To explore this possibility, we generated and tested a double fpvU-fpvY mutant in the Pf-5 ΔpvdI-pchC background and found that it was not cross-fed by strain WCS374 or A506. We also made derivatives of Pf-5 lacking two Fpvs in all combinations, and with the exception of the fpvU-fpvY mutant, all other double mutants were cross-fed by WCS374 and A506 (Table 3). Therefore, strains WCS374 and A506 cross-fed Pf-5 via both FpvU and FpvY, whereas three other strains (SBW25, ATCC 13525, and ATCC 9446) that produce pyoverdines having peptide sequences with an identical amino acid composition cross-fed Pf-5 via FpvU alone (Table 1). We attribute this apparent discrepancy to differences in the absolute configuration of the pyoverdines, as our bioinformatic analysis revealed that the NRPS encoded by PvdI and PvdD in the A506 genome lacks the epimerization domain in module 6 that is present in the SBW25 NRPS (Fig. 5). This difference is expected to result in the l configuration for the δN-formyl-δN-hydroxy-ornithine at position 6 in the peptide chain of the A506 pyoverdine versus the d configuration for this amino acid in the SBW25 pyoverdine. To determine if the WCS374 pyoverdine also has an l-δN-formyl-δN-hydroxy-ornithine at position 6, we designed PCR primers complementary to pvdD in A506 and SBW25 (see Table S2 in the supplemental material) and amplified and sequenced the region of the WCS374 genome between the thiolation domain of module 6 and the condensation domain of module 7 (Fig. 5). As with A506, no epimerization domain was detected in module 6 of the WCS374 PvdI/PvdD NRPS (data not shown). We suggest that a difference in stereochemistry, specifically, the configuration of the l-δN-formyl-δN-hydroxy-ornithine at position 6, is responsible for the differential uptake of these five pyoverdines by FpvY. This finding has precedence in the literature: FpvAI recognizes with high affinity only one of the possible stereoisomers of certain ferric-pyoverdine complexes and has a low affinity for pyoverdines that exist primarily in the incorrect conformation (15). Overlapping functions of TBDPs also are not uncommon in Pseudomonas spp. For example, P. aeruginosa has two TBDPs for enterobactin (75, 76) and two for ferrichrome (77), and FpvB is thought to function as a second receptor (in addition to FpvAI) for uptake of ferric complexes of the type I pyoverdine (78).
Fig 5.
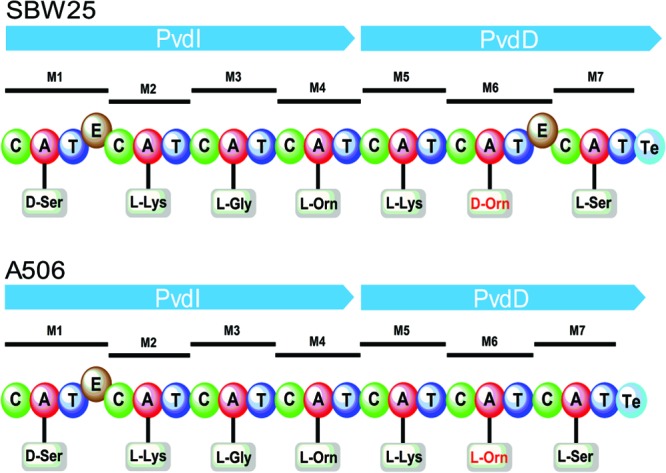
Bioinformatic analysis of the NRPSs for the biosynthesis of the pyoverdine peptide chain of P. fluorescens strains SBW25 and A506. For each module (M) in the NRPSs, domains (C, condensation; A, adenylation; T, thiolation; E, epimerization; Te, thioesterase) and the specific amino acid predicted from the sequence of each adenylation domain are shown. The predicted amino acid sequence and composition of the two peptide chains are identical, with the exception of the d versus l configuration of the ornithine residue of module 6 (shown in red font). PvdA and PvdF, which are encoded from the pyoverdine gene clusters of SBW25 (31) and A506 (49), confer the ornithine hydroxylase and ornithine transformylase activities needed to transform ornithine to δN-formyl-δN-hydroxy-ornithine residues.
Table 3.
Growth of double and single Fpv− mutants of Pf-5 on an iron-limited medium in the presence of P. fluorescens strain WCS374 or A506a
| Gene | Growth of mutant: |
|||||
|---|---|---|---|---|---|---|
| fpvZ | fpvU | fpvY | fpvW | fpvX | fpvV | |
| fpvZ | + | + | + | + | + | + |
| fpvU | + | − | + | + | + | |
| fpvY | + | + | + | + | ||
| fpvW | + | + | + | |||
| fpvX | + | + | ||||
| fpvV | + | |||||
Growth of Fpv− mutants in a ΔpvdI-pchC background of Pf-5 on an iron-limited medium (KMB amended with 600 μM 2,2′-dipyridyl) in the presence of the pyoverdine-producing (cross-feeding) strain WCS374 or A506: +, growth; −, no growth. The ΔpvdI-pchC mutant of Pf-5 did not grow on the iron-limited medium in isolation or in the presence of pyoverdine-deficient mutants of WCS374 or A506 but did grow in the presence of the wild-type pyoverdine-producing strain WCS374 or A506. Values represent results from two replicate plates in each of two experiments.
Due to the overlapping roles of FpvU and FpvY in pyoverdine uptake, we tested the possibility that other Pf-5 Fpvs also have overlapping functions. The remaining seven strains of Pseudomonas spp. that cross-fed all single Fpv− mutants (Table 2) were tested for cross-feeding of Pf-5 mutants lacking two of the six Fpvs in all combinations. Each of the seven strains (P. fluorescens CLR711 and CTRp112, P. costantinii CFBP 5705, Pseudomonas sp. strain A6, P. putida CFML90-40, and P. aeruginosa ATCC 27853 and 7NSK2) cross-fed all of the double Fpv− mutants of Pf-5 (data not shown). As described above, secondary siderophores produced by the strains could be responsible for cross-feeding of Pf-5. This possibility could be excluded only for strain 7NSK2, for which a mutant deficient in pyoverdine production was available. The pyoverdine-deficient mutant of 7NSK2 did not cross-feed the ΔpvdI-pchC mutant of Pf-5, indicating that the pyoverdine produced by 7NSK2 was responsible for cross-feeding. To verify these results, we partially purified the type II pyoverdines from culture supernatants of P. aeruginosa strains 7NSK2 and ATCC 27853 and tested them in cross-feeding experiments. Like the producing strains, the pyoverdines alone cross-fed all of the double Fpv− mutants of Pf-5 (data not shown). It is possible that 1 or more of the 39 TBDPs in the Pf-5 genome that were not evaluated here could contribute to the uptake of these ferric-pyoverdine complexes, even though those TBDPs do not show high levels of sequence identity to known Fpvs. Another possibility is that there is overlapping substrate recognition for the Fpvs, such that a given pyoverdine can be taken up by several of these receptors. Although Fpvs exhibit strict specificity in high-affinity pyoverdine uptake, previous studies have shown that pyoverdines can be transported into the cell with a lower affinity by Fpvs lacking strict specificity (15). Differences between high- and low-affinity uptake cannot be distinguished in cross-feeding experiments such as those done in this study, but future studies evaluating the uptake of labeled ferric-pyoverdine complexes by Pf-5 and derivative strains would be useful in exploring the possibility that these seven pyoverdines may be taken up by multiple Fpvs.
Conclusions.
For a soil bacterium like P. protegens Pf-5, which can establish in the rhizosphere where biologically available iron is limited, the ability to use heterologous pyoverdines can provide a competitive advantage (18, 24, 79). P. protegens Pf-5 produces and utilizes the same pyoverdine produced by P. protegens CHA0 as well as the lower-affinity siderophore enantio-pyochelin to provide itself with iron, yet it maintains an arsenal of TBDPs for the uptake of heterologous siderophores. In this study, we employed a combination of phylogenetics, bioinformatics, mutagenesis, pyoverdine structural determinations, and cross-feeding bioassays to assign specific ferric-pyoverdine substrates to each of the six Fpvs of Pf-5. We established that FpvZ serves as a component of the pyoverdine-mediated iron acquisition system of Pf-5, taking up the ferric complex of the strain's cognate pyoverdine, and all six Fpvs recognize pyoverdines produced by other strains of Pseudomonas spp. We also demonstrated that phylogenetically related Fpvs take up ferric complexes of structurally related pyoverdines, thereby establishing structure-function relationships that can be employed in the future to predict the pyoverdine substrates of Fpvs in other Pseudomonas spp.
Supplementary Material
ACKNOWLEDGMENTS
We thank Keith Poole for providing P. putida Bn7, Isabelle Schalk and Valérie Geoffroy for providing strains of Pseudomonas spp. and the P. fluorescens ATCC 13525 pyoverdine, Eric Bernaud for his technical assistance, Karl Hassan for assistance with Deepview, Virginia Stockwell, Teresa Kidarsa, and Jennifer Clifford for reviewing the manuscript, and Brenda Shaffer for assistance with the figures.
H. Gross and S. Mehnaz gratefully acknowledge the generous contribution of the Alexander von Humboldt Foundation, which provided financial support (Georg Forster Fellowship awarded to S. Mehnaz). This work was supported by grants 2006-35319-17427 and 2008-35600-18770 from the USDA National Institute of Food and Agriculture.
Footnotes
Published ahead of print 7 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01639-12.
REFERENCES
- 1. Ramette A, Frapolli M, Fischer-La Saux M, Gruffaz C, Meyer JM, Défago G, Sutra L, Moënne-Loccoz Y. 2011. Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst. Appl. Microbiol. 34: 180–188 [DOI] [PubMed] [Google Scholar]
- 2. Howell CR, Stipanovic RD. 1979. Control of Rhizoctonia solani in cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology 69: 480–482 [Google Scholar]
- 3. Loper JE, Paulsen IT, Kobayashi DY. 2007. The genomic sequence of Pseudomonas fluorescens Pf-5: insights into biological control. Phytopathology 97: 233–238 [DOI] [PubMed] [Google Scholar]
- 4. Youard ZA, Mislin GL, Majcherczyk PA, Schalk IJ, Reimmann C. 2007. Pseudomonas fluorescens CHA0 produces enantio-pyochelin, the optical antipode of the Pseudomonas aeruginosa siderophore pyochelin. J. Biol. Chem. 282: 35546–35553 [DOI] [PubMed] [Google Scholar]
- 5. Budzikiewicz H. 2004. Siderophores of the Pseudomonadaceae sensu stricto (fluorescent and non-fluorescent Pseudomonas spp.). Fortschr. Chem. Org. Naturst. 87: 81–237 [DOI] [PubMed] [Google Scholar]
- 6. Meyer JM, Gruffaz C, Raharinosy V, Bezverbnaya I, Schafer M, Budzikiewicz H. 2008. Siderotyping of fluorescent Pseudomonas: molecular mass determination by mass spectrometry as a powerful pyoverdine siderotyping method. Biometals 21: 259–271 [DOI] [PubMed] [Google Scholar]
- 7. Cornelis P. 2010. Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol. 86: 1637–1645 [DOI] [PubMed] [Google Scholar]
- 8. Cobessi D, Celia H, Folschweiller N, Schalk IJ, Abdallah MA, Pattus F. 2005. The crystal structure of the pyoverdine outer membrane receptor FpvA from Pseudomonas aeruginosa at 3.6 angstrom resolution. J. Mol. Biol. 347: 121–134 [DOI] [PubMed] [Google Scholar]
- 9. Nader M, Journet L, Meksem A, Guillon L, Schalk IJ. 2011. Mechanism of ferripyoverdine uptake by Pseudomonas aeruginosa outer membrane transporter FpvA: no diffusion channel formed at any time during ferrisiderophore uptake. Biochemistry 50: 2530–2540 [DOI] [PubMed] [Google Scholar]
- 10. Greenwald J, Hoegy F, Nader M, Journet L, Mislin GL, Graumann PL, Schalk IJ. 2007. Real time fluorescent resonance energy transfer visualization of ferric pyoverdine uptake in Pseudomonas aeruginosa. A role for ferrous iron. J. Biol. Chem. 282: 2987–2995 [DOI] [PubMed] [Google Scholar]
- 11. Krewulak KD, Vogel HJ. 2011. TonB or not TonB: is that the question? Biochem. Cell Biol. 89: 87–97 [DOI] [PubMed] [Google Scholar]
- 12. Shirley M, Lamont IL. 2009. Role of TonB1 in pyoverdine-mediated signaling in Pseudomonas aeruginosa. J. Bacteriol. 191: 5634–5640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schalk IJ, Lamont IL, Cobessi D. 2009. Structure-function relationships in the bifunctional ferrisiderophore FpvA receptor from Pseudomonas aeruginosa. Biometals 22: 267–268 [DOI] [PubMed] [Google Scholar]
- 14. Nader M, Dobbelaere W, Vincent M, Journet L, Adams H, Cobessi D, Gallay J, Schalk IJ. 2007. Identification of residues of FpvA involved in the different steps of Pvd-Fe uptake in Pseudomonas aeruginosa. Biochemistry 46: 11707–11717 [DOI] [PubMed] [Google Scholar]
- 15. Greenwald J, Nader M, Celia H, Gruffaz C, Geoffroy V, Meyer JM, Schalk IJ, Pattus F. 2009. FpvA bound to non-cognate pyoverdines: molecular basis of siderophore recognition by an iron transporter. Mol. Microbiol. 72: 1246–1259 [DOI] [PubMed] [Google Scholar]
- 16. Meyer JM, Geoffroy VA, Baysse C, Cornelis P, Barelmann I, Taraz K, Budzikiewicz H. 2002. Siderophore-mediated iron uptake in fluorescent Pseudomonas: characterization of the pyoverdine-receptor binding site of three cross-reacting pyoverdines. Arch. Biochem. Biophys. 397: 179–183 [DOI] [PubMed] [Google Scholar]
- 17. Meyer JM, Stintzi A, Poole K. 1999. The ferripyoverdine receptor FpvA of Pseudomonas aeruginosa PAO1 recognizes the ferripyoverdines of P. aeruginosa PAO1 and P. fluorescens ATCC 13525. FEMS Microbiol. Lett. 170: 145–150 [DOI] [PubMed] [Google Scholar]
- 18. Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8: 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hider RC, Kong X. 2010. Chemistry and biology of siderophores. Nat. Prod. Rep. 27: 637–657 [DOI] [PubMed] [Google Scholar]
- 20. Hartney SL, Mazurier S, Kidarsa TA, Quecine MC, Lemanceau P, Loper JE. 2011. TonB-dependent outer-membrane proteins and siderophore utilization in Pseudomonas fluorescens Pf-5. Biometals 24: 193–213 [DOI] [PubMed] [Google Scholar]
- 21. Koster M, Ovaa W, Bitter W, Weisbeek P. 1995. Multiple outer membrane receptors for uptake of ferric pseudobactins in Pseudomonas putida WCS358. Mol. Gen. Genet. 248: 735–743 [DOI] [PubMed] [Google Scholar]
- 22. Matthijs S, Laus G, Meyer JM, Abbaspour-Tehrani K, Schafer M, Budzikiewicz H, Cornelis P. 2009. Siderophore-mediated iron acquisition in the entomopathogenic bacterium Pseudomonas entomophila L48 and its close relative Pseudomonas putida KT2440. Biometals 22: 951–964 [DOI] [PubMed] [Google Scholar]
- 23. Mirleau P, Delorme S, Philippot L, Meyer J, Mazurier S, Lemanceau P. 2000. Fitness in soil and rhizosphere of Pseudomonas fluorescens C7R12 compared with a C7R12 mutant affected in pyoverdine synthesis and uptake. FEMS Microbiol. Ecol. 34: 35–44 [DOI] [PubMed] [Google Scholar]
- 24. Raaijmakers JM, van der Sluis L, Bakker PAHM, Schippers B, Koster M, Weisbeek PJ. 1995. Utilization of heterologous siderophores and rhizosphere competence of fluorescent Pseudomonas spp. Can. J. Microbiol. 41: 126–135 [Google Scholar]
- 25. Loper JE, Henkels MD. 1999. Utilization of heterologous siderophores enhances levels of iron available to Pseudomonas putida in the rhizosphere. Appl. Environ. Microbiol. 65: 5357–5363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wong-Lun-Sang S, Bernardini Hennard J-JC, Kyslik P, Dell A, Abdallah MA. 1996. Bacterial siderophores: structure elucidation, 2D 1H and 13C NMR assignments of pyoverdins produced by Pseudomonas fluorescens CHA0. Tetrahedron Lett. 37: 3329–3332 [Google Scholar]
- 27. King EO, Ward MK, Raney DE. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44: 301–307 [PubMed] [Google Scholar]
- 28. Atlas RM. 1993. Handbook of microbiological media. CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 29. Barelmann I, Fernandez DU, Budzikiewicz H, Meyer JM. 2003. The pyoverdine from Pseudomonas chlororaphis D-TR133 showing mutual acceptance with the pyoverdine of Pseudomonas fluorescens CHA0. Biometals 16: 263–270 [DOI] [PubMed] [Google Scholar]
- 30. Demange P, Wendenbaum S, Linget C, Mertx C, Cung M, Dell A, Abdallah MA. 1990. Bacterial siderophores: structure and NMR assignment of pyoverdins Pa, siderophores of Pseudomonas aeruginosa ATCC 15692. Biometals 3: 155–170 [Google Scholar]
- 31. Moon CD, Zhang XX, Matthijs S, Schafer M, Budzikiewicz H, Rainey PB. 2008. Genomic, genetic and structural analysis of pyoverdine-mediated iron acquisition in the plant growth-promoting bacterium Pseudomonas fluorescens SBW25. BMC Microbiol. 8: 7 doi:10.1186/1471-2180-8-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Linget C, Stylianou DG, Dell A, Wolff RE, Piémont Y, Abdallah MA. 1992. Bacterial siderophores: the structure of a desferriferribactin produced by Pseudomonas fluorescens ATCC 13525. Tetrahedron Lett. 33: 3851–3854 [Google Scholar]
- 33. Hohlneicher U, Hartmann R, Taraz K, Budzikiewicz H. 1995. Pyoverdin, ferribactin, azotobactin—a new triade of siderophores from Pseudomonas chlororaphis ATCC 9446 and its relation to Pseudomonas fluorescens ATCC 13525. Z. Naturforsch. C 50: 337–344 [Google Scholar]
- 34. Meyer JM. 2007. Siderotyping and bacterial taxonomy: a siderophore bank for a rapid identification at the species level of fluorescent and non-fluorescent Pseudomonas, p 43–65 In Varma A, Chincholkar SB. (ed), Soil biology: microbial siderophores, vol 12 Springer-Verlag, Berlin, Germany [Google Scholar]
- 35. Yamamoto S, Kasai H, Arnold DL, Jackson RW, Vivian A, Harayama S. 2000. Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146: 2385–2394 [DOI] [PubMed] [Google Scholar]
- 36. Teintze M, Hossain MB, Barnes CL, Leong J, van der Helm D. 1981. Structure of ferric pseudobactin, a siderophore from a plant growth promoting Pseudomonas. Biochemistry 20: 6446–6457 [DOI] [PubMed] [Google Scholar]
- 37. Delorme S, Lemanceau P, Christen R, Corberand T, Meyer Gardan J-ML. 2002. Pseudomonas lini sp. nov., a novel species from bulk and rhizospheric soils. Int. J. Syst. Evol. Microbiol. 52: 513–523 [DOI] [PubMed] [Google Scholar]
- 38. Stanier RY, Palleroni NJ, Doudoroff M. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43: 159–271 [DOI] [PubMed] [Google Scholar]
- 39. Meyer JM, Gruffaz C, Tulkki T, Izard D. 2007. Taxonomic heterogeneity, as shown by siderotyping, of strains primarily identified as Pseudomonas putida. Int. J. Syst. Evol. Microbiol. 57: 2543–2556 [DOI] [PubMed] [Google Scholar]
- 40. Koster M, van de Vossenberg J, Leong J, Weisbeek PJ. 1993. Identification and characterization of the pupB gene encoding an inducible ferric-pseudobactin receptor of Pseudomonas putida WCS358. Mol. Microbiol. 8: 591–601 [DOI] [PubMed] [Google Scholar]
- 41. Djavaheri M, Mercado-Blanco J, Versluis C, Meyer JM, Van Loon LC, Bakker PAHM. 2012. Iron-regulated metabolites produced by Pseudomonas fluorescens WCS374r are not required for eliciting induced systemic resistance against Pseudomonas syringae pv. tomato in Arabidopsis. MicrobiologyOpen 1: 311–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barelmann I, Taraz K, Budzikiewicz H, Geoffroy V, Meyer JM. 2002. The structures of the pyoverdins from two Pseudomonas fluorescens strains accepted mutually by their respective producers. Z. Naturforsch. C 57: 9–16 [DOI] [PubMed] [Google Scholar]
- 43. Fernandez DU, Fuchs R, Taraz K, Budzikiewicz H, Munsch P, Meyer JM. 2001. The structure of a pyoverdine produced by a Pseudomonas tolaasii-like isolate. Biometals 14: 81–84 [DOI] [PubMed] [Google Scholar]
- 44. Beiderbeck H, Taraz K, Meyer JM. 1999. Revised structures of the pyoverdins from Pseudomonas putida CFBP 2461 and from Pseudomonas fluorescens CFBP 2392. Biometals 12: 331–338 [DOI] [PubMed] [Google Scholar]
- 45. Jacques P, Ongena M, Gwose I, Seinsche D, Schrooten C, Delfosse P, Thonart P, Taraz K, Budzikiewicz H. 1995. Structure and characterization of isopyoverdin from Pseudomonas putida BTP1 and its relation to the biogenetic pathway leading to pyoverdins. Z. Naturforsch C 50: 622–629 [DOI] [PubMed] [Google Scholar]
- 46. Tappe R, Taraz K, Budzikiewicz H, Meyer JM, Lefevre JF. 1993. Structure elucidation of a pyoverdin produced by Pseudomonas aeruginosa ATCC27853. J. Prakt. Chem. 335: 83–87 [Google Scholar]
- 47. De Chial M, Ghysels B, Beatson SA, Geoffroy V, Meyer JM, Pattery T, Baysse C, Chablain P, Parsons YN, Winstanley C, Cordwell SJ, Cornelis P. 2003. Identification of type II and type III pyoverdine receptors from Pseudomonas aeruginosa. Microbiology 149: 821–831 [DOI] [PubMed] [Google Scholar]
- 48. Audenaert K, Pattery T, Cornelis P, Höfte M. 2002. Induction of systemic resistance to Botrytis cinerea in tomato by Pseudomonas aeruginosa 7NSK2: role of salicylic acid, pyochelin, and pyocyanin. Mol. Plant Microbe Interact. 15: 1147–1156 [DOI] [PubMed] [Google Scholar]
- 49. Loper JE, Hassan KA, Mavrodi DV, Davis EW, Lim IICK, Shaffer BT, Elbourne LDH, Stockwell VO, Hartney SL, Breakwell K, Henkels MD, Tetu SG, Rangel LI, Kidarsa TA, Wilson NL, van de Mortel JE, Song C, Blumhagen R, Radune D, Hostetler JB, Brinkac LM, Durkin AS, Kluepfel DA, Wechter WP, Anderson AJ, Kim YC, Pierson LS, Pierson EA, III, Lindow SE, Kobayashi DY, Raaijmakers JM, Weller DM, Thomashow LS, Allen AE, Paulsen IT. 2012. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 8: e1002784 doi:10.1371/journal.pgen.1002784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Youard ZA, Reimmann C. 2010. Stereospecific recognition of pyochelin and enantio-pyochelin by the PchR proteins in fluorescent pseudomonads. Microbiology 156: 1772–1782 [DOI] [PubMed] [Google Scholar]
- 51. Lindow SE. 1984. Integrated control and role of antibiosis in biological control of fire blight and frost injury, p 83–115 In Windels C, Lindow SE. (ed), Biological control on the phylloplane. APS Press, Minneapolis, MN [Google Scholar]
- 52. Hassan KA, Johnson A, Shaffer BT, Ren Q, Kidarsa TA, Elbourne LDH, Hartney S, Duboy R, Goebel NC, Zabriskie TM, Paulsen IT, Loper JE. 2010. Inactivation of the GacA response regulator in Pseudomonas fluorescens Pf-5 has far-reaching transcriptomic consequences. Environ. Microbiol. 12: 899–915 [DOI] [PubMed] [Google Scholar]
- 53. Choi KH, Schweizer HP. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5: 30 doi:10.1186/1471-2180-5-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302: 205–217 [DOI] [PubMed] [Google Scholar]
- 55. McGuffin LJ, Bryson K, Jones DT. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16: 404–405 [DOI] [PubMed] [Google Scholar]
- 56. Bigelow HR, Petrey DS, Liu J, Przybylski D, Rost B. 2004. Predicting transmembrane beta-barrels in proteomes. Nucleic Acids Res. 32: 2566–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- 58. Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. 2009. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 4: 1–13 [DOI] [PubMed] [Google Scholar]
- 59. Bachmann BO, Ravel J. 2009. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 458: 181–217 [DOI] [PubMed] [Google Scholar]
- 60. Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. 2005. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res. 33: 5799–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stachelhaus T, Mootz HD, Marahiel MA. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6: 493–505 [DOI] [PubMed] [Google Scholar]
- 62. Meyer JM, Abdallah MA. 1978. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. J. Gen. Microbiol. 107: 319–328 [Google Scholar]
- 63. Carson KC, Meyer JM, Dilworth MJ. 2000. Hydroxamate siderophores of root nodule bacteria. Soil Biol. Biochem. 32: 11–21 [Google Scholar]
- 64. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lamont IL, Beare PA, Ochsner U, Vasil AI, Vasil ML. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 99: 7072–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shen J, Meldrum A, Poole K. 2002. FpvA receptor involvement in pyoverdine biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 184: 3268–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gross H, Loper JE. 2009. Genomics of secondary metabolite production by Pseudomonas spp. Nat. Prod. Rep. 26: 1408–1446 [DOI] [PubMed] [Google Scholar]
- 70. Poole K, Neshat S, Krebes K, Heinrichs DE. 1993. Cloning and nucleotide sequence analysis of the ferripyoverdine receptor gene fpvA of Pseudomonas aeruginosa. J. Bacteriol. 175: 4597–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Briskot G, Taraz K, Budzikiewicz H. 1989. Pyoverdin-type siderophores from Pseudomonas aeruginosa. Liebigs Ann. Chem. 1989: 375–384 [Google Scholar]
- 72. Meyer J, Hallé F, Hohnadel D, Lemanceau P, Ratefiarivelo H. 1987. Siderophores of Pseudomonas—biological properties, p 188–205 In Winkelmann G, Van der Helm D, Neilands JB. (ed), Iron transport in microbes, plants and animals. VCH, Weinheim, Germany [Google Scholar]
- 73. Morris J, Donnelly DF, O'Neill E, McConnell F, O'Gara F. 1994. Nucleotide sequence analysis and potential environmental distribution of a ferric pseudobactin receptor gene of Pseudomonas sp. strain M114. Mol. Gen. Genet. 242: 9–16 [DOI] [PubMed] [Google Scholar]
- 74. Bitter W, Marugg JD, de Weger LA, Tommassen J, Weisbeek PJ. 1991. The ferric-pseudobactin receptor PupA of Pseudomonas putida WCS358: homology to TonB-dependent Escherichia coli receptors and specificity of the protein. Mol. Microbiol. 5: 647–655 [DOI] [PubMed] [Google Scholar]
- 75. Dean CR, Poole K. 1993. Cloning and characterization of the ferric enterobactin receptor gene (pfeA) of Pseudomonas aeruginosa. J. Bacteriol. 175: 317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ghysels B, Ochsner U, Mollman U, Heinisch L, Vasil M, Cornelis P, Matthijs S. 2005. The Pseudomonas aeruginosa pirA gene encodes a second receptor for ferrienterobactin and synthetic catecholate analogues. FEMS Microbiol. Lett. 246: 167–174 [DOI] [PubMed] [Google Scholar]
- 77. Hannauer M, Barda Y, Mislin GL, Shanzer A, Schalk IJ. 2010. The ferrichrome uptake pathway in Pseudomonas aeruginosa involves an iron release mechanism with acylation of the siderophore and recycling of the modified desferrichrome. J. Bacteriol. 192: 1212–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ghysels B, Dieu BT, Beatson SA, Pirnay JP, Ochsner UA, Vasil ML, Cornelis P. 2004. FpvB, an alternative type I ferripyoverdine receptor of Pseudomonas aeruginosa. Microbiology 150: 1671–1680 [DOI] [PubMed] [Google Scholar]
- 79. Loper JE, Henkels MD. 1997. Availability of iron to Pseudomonas fluorescens in rhizosphere and bulk soil evaluated with an ice nucleation reporter gene. Appl. Environ. Microbiol. 63: 99–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gross H, Stockwell VO, Henkels MD, Nowak-Thompson B, Loper JE, Gerwick WH. 2007. The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chem. Biol. 14: 53–63 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.