Abstract
DUX4 , a homeobox-containing gene present in a tandem array, is implicated in facioscapulohumeral muscular dystrophy (FSHD), a dominant autosomal disease. New findings about DUX4 have raised as many fundamental questions about the molecular pathology of this unique disease as they have answered. This review discusses recent studies addressing the question of whether there is extensive FSHD-related transcription dysregulation in adult-derived myoblasts and myotubes, the precursors for muscle repair. Two models for the role of DUX4 in FSHD are presented. One involves transient pathogenic expression of DUX4 in many cells in the muscle lineage before the myoblast stage resulting in a persistent, disease-related transcription profile (‘Majority Rules’), which might be enhanced by subsequent oscillatory expression of DUX4. The other model emphasizes the toxic effects of inappropriate expression of DUX4 in only an extremely small percentage of FSHD myoblasts or myotube nuclei (‘Minority Rules’). The currently favored Minority Rules model is not supported by recent studies of transcription dysregulation in FSHD myoblasts and myotubes. It also presents other difficulties, for example, explaining the expression of full-length DUX4 transcripts in FSHD fibroblasts. The Majority Rules model is the simpler explanation of findings about FSHD-associated gene expression and the DUX4-encoded homeodomain-type protein.
Keywords: expression profiling, FSHD, muscular dystrophy, myogenesis
BACKGROUND ABOUT FSHD
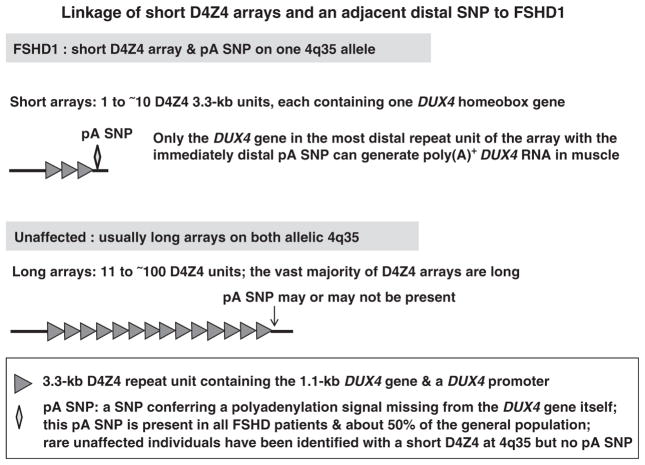
Facioscapulohumeral muscular dystrophy (FSHD), an autosomal dominant disease, is unique in its linkage to contraction of a tandem repeat array (D4Z4) consisting of large repeat units.1 Patients with FSHD1, the predominant form of the disease found in diverse ethnic populations,2,3 almost always have a short D4Z4 array in the subtelomeric region of 4q (4q35). Short arrays contain 1–10 copies of the 3.3-kb repeat unit, instead of ~11–100. Despite recent insights into the genetics of FSHD,4–6 there are still great challenges for understanding the role of dysregulation of gene expression in this disease and the frequent lack of genotype:phenotype correlations.7,8
Inside each D4Z4 repeat unit is DUX4, which encodes a transcription factor containing two homeodomains. However, only the most distal copy of DUX4 at 4q35 (Figure 1) is likely to be involved in this painful, debilitating and slowly progressive disease.4–6 Diagnosis of FSHD involves molecular analysis of D4Z4 arrays.9–11 Usually, the clinical presentation of FSHD is in the second or third decade, with symptoms mostly limited to a small set of skeletal muscles, often exhibiting an asymmetrical distribution of affected muscles.7,12 Patients with very small D4Z4 arrays (only one D4Z4 repeat unit) may present in very early childhood.13,14 Unlike Duchenne’s muscular dystrophy, cardiomyopathy is noted only rarely upon clinical presentation.15 Respiratory insufficiency and mild, high-frequency hearing loss are sometimes observed, especially in patients with moderate to severe FSHD.16–19 There are frequent associations with asymptomatic retinal telangiectasias.7,12,20 No effective treatment is available.
Figure 1.
A cartoon illustrating the linkage of short array D4Z4 arrays and an immediately distal SNP to FSHD1.
At subtelomeric 10q (10q26), there is a D4Z4 array that is almost identical to that at 4q35, with DUX4 in each repeat unit, and that array is similarly polymorphic in size. Much controversy has surrounded the issue of which 4q35 gene or non-genic DNA sequence is responsible for the linkage of FSHD to contraction of a D4Z4 array at 4q35 and not at 10q26.21–30 FRG1, the gene that is the nearest to 4q35 D4Z4 and absent from 10q26, has been reported to be very strongly upregulated in FSHD muscle;24 however, this was not observed in studies from four other labs.25,30–32 Genetic evidence indicates that a common single-nucleotide polymorphism (SNP) in the D4Z4-adjacent sequence at the distal end of the array at 4q35, but not at 10q26, is responsible for the 4q35 linkage (Figure 1). This SNP, which is present in about half of the general population,33–35 confers a polyadenylation signal (pA) to the last copy of DUX4 at 4q35 and is almost never found distal to 10q D4Z4.4,5 No pA is within the DUX4 gene itself, although this gene has a promoter.26 The evolutionary conservation of a coding function for DUX4-related sequences36 and the absence of any observed FSHD patients with a deletion of the entire D4Z4 array37 support FSHD disease models based upon D4Z4 shortening causing inappropriate, gain-of-function expression of DUX4. This gain of function requires the disease-permissive SNP immediately downstream of the last repeat unit in the contracted D4Z4 array. DUX4 is further implicated in FSHD pathogenesis through the discovery of a rare family of FSHD patients with the pA SNP in cis to a short D4Z4 array at 10q26, rather than at 4q35.5
Because this misexpressed DUX4 in the FSHD muscle lineage encodes a trans-acting protein, a transcription factor,27 any dysregulation of 4q35 gene expression other than at the D4Z4 array is likely to be a secondary effect comparable to DUX4-initiated FSHD-associated dysregulation of gene expression elsewhere in the genome. This conclusion is re-enforced by the above-mentioned finding of the rare FSHD family with normal-sized 4q35 D4Z4 arrays but with an exceptional pA SNP at 10q26 in cis to a contracted 10q26 D4Z4 array.5 However, there may well be normal myogenesis-associated changes in chromatin structure at 4q35.238 related to the muscle-lineage specific nature of FSHD that could impact the D4Z4 array.
Inappropriate expression of DUX4 to give very low levels of full-length transcript (DUX4-fl RNA) in FSHD myotubes and myoblasts generally requires both array contraction and the pA SNP.4–6 One exception to the requirement for array contraction is the rare variant (~5% of cases) of FSHD called FSHD2 (or FSHD1B),39,40 which needs to be carefully differentiated from other dystrophies that sometimes mimic its clinical symptoms.41 Although generation of DUX4-fl RNA in FSHD2 myoblast or myotube nuclei is not linked to contraction of the D4Z4 array, it is still linked to the pA SNP at 4q35.6 In addition to DUX4-fl RNA, many shorter transcripts from both strands of D4Z4 are generated in low abundance,42 which is not surprising in view of recent findings that a large portion of the human genome is transcribed to generate noncoding RNAs,43 especially from DNA repeats.44,45 The two DUX4-fl RNA isoforms that have been found associated with FSHD vary only in the 3′ untranslated region, have no identified functional distinction,6,27 and will both be referred to as DUX4-fl RNA.
Another exception to the need for array contraction to generate full-length polyadenylated transcripts from DUX4 was seen in multiple normal testis samples, which generate DUX4-fl RNA from both 10q and 4q DUX4 at D4Z4.6 In testis, the last copy of DUX4 apparently uses either the common pA SNP (at 4q35) or a far distal, constitutively present pA (at 4q35 and 10q26) to provide for polyadenylation of the transcript. In addition, an induced pluripotent stem cell culture derived from control fibroblasts was positive for DUX4-fl RNA before, but not after, induction of differentiation.6 However, this result awaits confirmation from more samples.
FSHD1, FSHD2 and the germ lineage appear to confer a loose, transcription-conducive conformation to D4Z4 chromatin. This is evidenced by changes in histone modification;46 partial but variable hypomethylation of D4Z4 in FSHD1; and more extensive hypomethylation in FSHD2.39 There is yet more hypomethylation in normal sperm.47 D4Z4 hypomethylation does not suffice for the disease as seen in the absence of muscular dystrophy symptoms in patients with ICF (immunodeficiency, centromeric region instability and facial anomlies), the rare, unrelated DNA hypomethylation-associated disease in which D4Z4 is strongly hypomethylated.47,48
A MAJOR COMPLICATION IN UNDERSTANDING THE RELATIONSHIP OF DUX4 TO PATHOGENESIS
DUX4 protein is a transcription factor that can regulate expression of other genes.27 Moreover, it contains two homeodomains,49,50 domains that are found characteristically in proteins regulating early stages of differentiation,51 and localizes to the nucleus.27 Therefore, it is easy to envision its inappropriate expression in the muscle lineage leading to pathogenesis. However, a major complication in understanding the relationship of DUX4-fl transcripts to FSHD pathogenesis is their extraordinarily low average abundance in FSHD1 and FSHD2 myoblasts, myotubes, and muscle.52 Detection of DUX4-fl RNA in FSHD biomaterials requires nested PCR6 or unusually high amounts of cDNA template (400 ng).5,53 The need for these non-quantitative conditions for RT-PCR raises fundamental questions about the nature of the causal association of DUX4-fl and the disease pathology. Such PCR conditions also require unusual vigilance to prevent false positives.
It has been estimated that about 1 in 1000 FSHD myoblasts or nuclei in myotubes are positive for DUX4-fl transcripts (by RT–PCR of highly diluted FSHD myoblast cultures) or the corresponding protein in contrast to control cultures, which are negative.6 Therefore, the very low levels of FSHD-associated DUX4-fl RNA or protein in cell populations could be accounted for by only a very small percentage of DUX4-fl+ nuclei. Moreover, DUX4-fl transcripts were not detectable in some preparations of FSHD myotubes and, especially, FSHD myoblasts.6,53
TWO MODELS TO EXPLAIN DUX4-FL PATHOGENICITY
At clinical presentation, affected muscles in FSHD patients have heterogeneous and rather non-specific histological findings.13,54 Although Reed et al.55 found a significant increase in the distance between the sarcolemma and the underlying contractile apparatus in unfixed FSHD skeletal muscle, the basic contractile apparatus was normal by immunofluorescence and confocal microscopically. We will consider models of pathogenesis focused on abnormal regenerative repair of muscle56 in FSHD. This focus is supported by the typically slow progress of the disease and the usual presentation of symptoms after the first decade of life.
Satellite cells, which account for only~2–6% of the nuclei in adult skeletal muscle, are the main source of stem cells for repair of postnatal skeletal muscle through regenerative myogenesis, induced by muscle wear-and-tear, injury, or disease-related atrophy.56–58 During regenerative myogenesis, satellite cells are induced to proliferate and form myoblasts. These mononuclear cells then differentiate and fuse with the damaged myofiber or with other myoblasts to form multinucleated myotubes. A key question for understanding the biological implications of the very infrequent DUX4-fl+ nuclei (at the RNA or protein level) in FSHD myoblast and myotube cultures derived from affected muscle is whether a large fraction of these cells has a disease-associated expression phenotype.
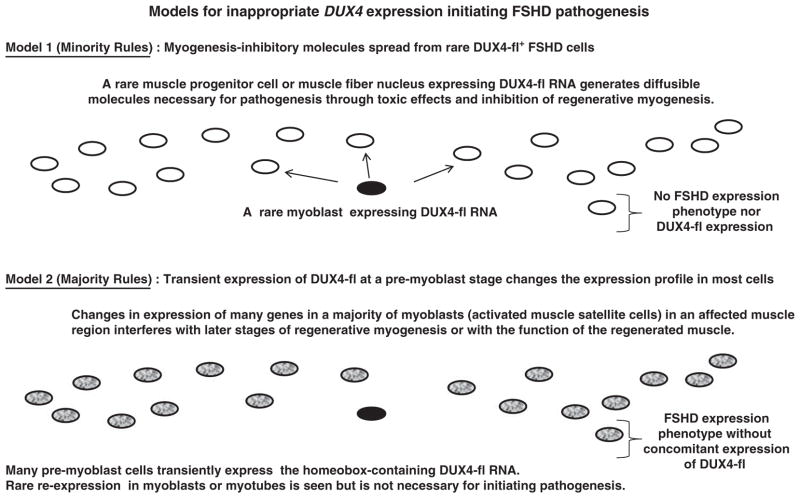
We consider two models inferred from recent articles about FSHD.4–6,53,59–61 Model 1 (‘Minority Rules’) involves undefined molecules spreading toxicity initiated by DUX4-fl protein in the tiny percentage of FSHD myoblasts or nuclei in FSHD myotubes that are positive for DUX4-fl. In this model, there is no need for a disease-linked expression profile in the vast majority of the cells in the population (Figure 2). Currently, there is an emphasis in the literature on the toxic and pro-apoptotic effects of DUX4 as central to pathogenesis, as deduced from experiments involving the introduction of plasmid or viral DNA constructs encoding moderate to high amounts of DUX4-fl RNA in various cell types.4,6,59,62 Even at low, non-toxic concentrations of experimentally induced DUX4-fl RNA in mouse myoblasts, there is downregulation of Myod1 RNA levels and concomitant inhibition of myotube formation and expression.59 When myoblasts are undergoing differentiation to myotubes, DUX4-fl constructs are less toxic but, nonetheless, upregulate atrophy-associated genes (FOXO32/MURF1 and TRIM63/ATROGIN-1) and lead to decreased yields of myotubes or myotubes of very abnormal morphology upon differentiation of myoblasts.59,63
Figure 2.
Two models for inappropriate DUX4 expression initiating FSHD pathogenesis. Both the Minority Rules model (Model 1) and the Majority Rules model (Model 2) involve only certain skeletal muscles being affected and generally slow progression with age. A variant of Model 2 would have transient oscillating expression of DUX4-fl at the myotube stage reinforcing an FSHD expression phenotype established at a pre-myoblast stage.
In contrast to Model 1, Model 2 (‘Majority Rules’) is predicated on a large fraction of FSHD myoblasts (and myotube nuclei) exhibiting a disease-associated expression phenotype in the absence of detectable DUX4-fl RNA in >99% of the nuclei (Figure 2). The disease-associated expression phenotype would include genes whose abnormal up- or downregulation leads to defects in muscle regeneration and function. Model 2 explains the discrepancy between DUX4-fl protein expression in only a tiny fraction of FSHD myoblasts and a large percentage of these cells having disease-linked alterations in expression as a result of transient, non-toxic expression of DUX4-fl RNA in a large fraction of the cells previous to the myoblast stage. This short-term inappropriate expression of DUX4 would initiate an irreversible cascade of gene dysregulation, similar to the way that other homeobox genes direct gene expression profiles by early transient expression during differentiation.51 The expression of DUX4-fl RNA at the myoblast (and myotube or later) stages in a tiny fraction of nuclei in FSHD cells would be a stochastic event that is peripheral to the establishment of pathogenesis. Moreover, DUX4-fl expression in the very small fraction of myoblast or myotube nuclei expressing the gene might be toxic but it would not be toxic at the pre-myoblast stage, according to this model.
A variant of Model 2 (not shown) would involve oscillating, non-toxic generation of DUX4-fl transcripts throughout the FSHD myotube population such that, at any one time, <1% of the nuclei are generating the transcript. Such oscillating expression, which is seen for genes encoding certain transcription regulatory factors in stem cells,64–66 might reinforce an FSHD expression phenotype set up at a pre-myoblast stage and might avoid inhibition of myotube formation if the expression were very transient to give much lower intracellular levels of DUX4-fl protein than in transfection and transduction experiments.4,59,63 DUX4-fl constructs have been reported to be less toxic at the myotube stage.59,63 However, strong induction of expression of DUX4-fl in differentiating or differentiated myoblasts transduced or transfected with DNA constructs still resulted in some cell death, gave very abnormal-looking myotubes, and increased levels of expression of genes associated with muscle atrophy.59,63
In the above-described models, stochastic effects determining the generation of DUX4-fl RNA and protein could contribute to the lack of consistent phenotype–genotype correlations, the asymmetry in affected skeletal muscle upon clinical presentation, and the finding that FSHD symptoms are usually not seen earlier than the second decade.7,8 Moreover, there may be more of a range of expression levels of DUX4-fl RNA and protein per FSHD myogenic cell or nucleus than is currently appreciated. In addition, the levels or timing of inappropriate expression of DUX4-fl in vivo in myogenic precursors (or the extent of dysregulation of downstream genes) may be different than in vitro, just as is the case for the reprogramming factor Zcan4 in embryos vs embryonal stem cells67,68 (see below). Nonetheless, FSHD muscle appears to have much lower levels of DUX4-fl expression than cultured FSHD myotubes and is sometimes undetectable in affected FSHD muscle biopsies,42,52,61 a finding that is consistent with the models based upon the central role of myogenic precursor cells in disease pathogenesis.
EVIDENCE FOR A DISEASE-ASSOCIATED TRANSCRIPTION PHENOTYPE IN FSHD MYOBLASTS AND MYOTUBES: SUPPORT FOR THE ‘MAJORITY RULES’ MODEL
Expression microarray profiling of FSHD vs control muscle biopsy samples has been done, but with little consistency in the results.31,32,69–71 This is probably due partly to the use of very different types of microarrays, none of which were the recently improved, exon-based microarrays. Moreover, studies of muscle are complicated by various extents of contamination with non-muscle cells. In addition, myogenesis-specific changes in muscle tissue would be obscured by the very low percentage of satellite cells in muscle tissue. Recently, four multi-gene expression studies were reported for FSHD and control myoblasts and myotubes that used either exon-based microarrays53,72 or panels of genes for qRT-PCR.60,61 Although different experimental methods or types of samples were used, which probably contribute to considerable differences between the gene expression profiles, these studies present evidence for disease-linked dysregulation of many genes in FSHD myoblasts and myotubes.
First, we summarize results from our expression profiling of immunocytochemically characterized myoblast and myotube preparations53 using myoblast cultures that were ~70% confluent or that had been in differentiation medium for 4–6 days. Of the ~17 000 analyzed genes on the exon-based microarray, 295 and 797 were significantly dysregulated in FSHD vs control myoblasts and myotubes, respectively (fold change >2; adjusted P<0.01). Many genes that displayed disease-related dysregulation (for example, genes for muscle structure, mitochondrial function and signal transduction) exhibited a dampening, but importantly, not the absence of normal myogenesis-specific expression changes. Some critical myogenesis-associated genes (for example, MYOD1 and MYOG) displayed normal levels of expression in FSHD myoblasts and myotubes, consistent with the normal growth and differentiation of the FSHD myoblasts and normal appearance of FSHD myotubes. However, other regulatory genes (for example, MEF2A and all four of the Argonaute genes) that could have widespread effects on gene expression were dysregulated in FSHD myogenic cells.53 That 60 genes showed about 4–16-fold downregulated RNA levels in FSHD vs control myotube preparations indicates a high percentage of the nuclei displaying FSHD-associated dysregulation of transcription.
Our findings suggest that FSHD-related changes in gene expression contribute to abnormalities in muscle function and structure in FSHD. Among the most overrepresented functional terms associated with upregulated genes in FSHD vs control myotubes were inflammation, fatty acid elongation in mitochondria and extracellular matrix.53 These could be relevant to the inflammation, fatty acid infiltration or fibrosis that has been observed in a varying percentages of FSHD muscle biopsies.18,73–76 For example, expression of the pro-inflammatory genes IL6, IL8, IL18R1, BDKRB1, CCL2, CCL20, TNFAIP6 and TNFRSF12A was upregulated in FSHD myotubes (fold change >2; adjusted P<0.01). The fibrosis-associated CTGF, which was found to be upregulated at the RNA level in FSHD vs control muscle32 and at the RNA and protein level in muscle fibers from patients with other muscular dystrophies,77 was upregulated threefold in FSHD vs control myotubes.53
Secondly, in an expression profiling study similar to ours, Cheli et al.72 concluded that there was specific dysregulation in myoblasts and myotubes from FSHD patients vs. controls. However, their data are unconvincing because of the lack of assessment of the quality of their myoblast and myotube preparations by immunocytochemistry. Indeed, paradoxically, they reported no muscle-related terms among 177 functional terms associated with genes differentially expressed in control myoblast vs control myotube preparations. In contrast, as expected, in our microarray study all six of the top functional terms for genes displaying differential expression in control myoblasts vs control myotubes had the term ‘muscle’ in them.53 In addition, Cheli et al. reported <4% overlap between several hundred genes with dysregulation in FSHD vs control cells at the myoblast stage and those dysregulated at the myotube stage, unlike in our study in which there was 48% overlap between myoblasts and myotubes for the genes with FSHD dysregulation (fold-change >2; P<0.01).
Thirdly, Homma et al.60 analyzed by qRT-PCR the relative expression of 64 test genes and 11 control genes in well-characterized FSHD and control myoblasts at five time points before or after induction of differentiation to myotubes.60 Their subjects were cohorts of affected and unaffected family members, with myoblasts generated from the deltoid and bicep biopsies of each subject. A hierarchical clustering analysis demonstrated that there were strong cohort-related groupings among the expression profiles and that most of the correlations in expression profiles between samples from closely related patients were stronger than the correlations between pairs of FSHD samples or pairs of control samples. The authors then analyzed the average differences in expression between the FSHD and control samples at each time point using standard t-tests and concluded that there were ‘no consistent, overall differences in mRNA expression patterns or levels’ between FSHD and control myoblasts. However, this analysis was statistically flawed in two critical respects: (1) the measurements at each time point were treated as independent rather than as repeated measures from each subject and (2) there was no adjustment for the variation associated with the sample cohorts. To remedy these deficiencies and thereby greatly increase the power to detect statistically significant FSHD-related differences, we fit mixed-effects models to predict the PCR-derived Ct value as a function of time and/or sample type for their data from the last three differentiation time points (2, 4 and 7 days in differentiation medium). We used nested random intercepts to account for varying baseline levels among and within each cohort, and modeled the effect of FSHD status as an additive term and, where statistically significant, as an adjustment to the time-related slope coefficient. By this re-analysis, we identified 13 genes with significant FSHD-related dysregulation of expression from their data (CXCL11, KLF4 and FRG2B, upregulated; ACTN3, DES, MYH5, MYH6, MYH14, PGK1, SULF2, FBXO32/ATROGIN1, TRIM63/MURF1 and SLC25A4/ANT1, downregulated). The SLC25A4 downregulation is contrary to previous findings,24,30,78 and the specificity of FRG2B probes remains to be demonstrated for this gene, which has very similar sequences throughout the genome. In our expression profiling, six of the 64 genes common to the study of Homma et al.60 and ours53 displayed significant FSHD-related downregulation of at least twofold (DES, MYF6, MYH6, MYH7, TRIM63/MURF1). Three of these were also significantly downregulated in our re-evaluation of the data of Homma et al. (DES, MYH6 and TRIM63). Of the 64 genes, 50, including FSHD candidate gene PITX1,24,27 displayed no significant differential expression in either study. The P-value for the relationship between the direction of differential expression between the two studies was 0.09 (Fisher’s exact test), providing some evidence of an association, although not at a statistically significant level.
Given the lack of normalization of the qRT-PCR data from test genes to standard genes by Homma et al. and their unconventional use of a pre-amplification for 14 cycles before the real-time PCR, the finding of significant downregulation for three genes in both of these studies is noteworthy. Moreover, all three of these genes are strongly upregulated in control myotubes vs 19 different non-muscle cell type,53 and were seen as upregulated in myoblasts vs non-myogenic cells in RNA-seq (http://genome.ucsc.edu/ENCODE/, Tom Gingeras, Cold Spring Harbor). This is consistent with our finding that one of the most prominent classes of genes to be dysregulated in FSHD myotubes was genes normally upregulated during myogenesis. In summary, the conclusion of Homma et al. that FSHD and control myogenic precursors have indistinguishable patterns of gene expression is not supported by our re-analysis of their data.
In the last of these recent expression studies, Geng et al.61 transduced human myoblast cultures with DUX4-fl or control constructs and profiled differential gene expression with an expression microarray 24 h after transduction. The short time of incubation was probably intended to minimize the contribution of toxic effects of induced DUX4-fl expression to myoblasts, although it is likely that such effects still altered expression of many genes. Among the more than 1000 genes that were significantly dysregulated (fold change >2; false discovery rate <0.01) by the DUX4-fl construct was a small group of genes strongly upregulated due to transduction with DUX4-fl and expressed specifically in germ cells or during early development. By qRT-PCR, Geng et al. showed that six of these genes (ZSCAN4, KHDC1, PRAMEF1, RFPL2, MBD3L2 and TRIM43) were expressed at moderate levels in normal testis and confluent FSHD myoblasts. The steady–state levels of their RNAs were usually much lower in FSHD skeletal muscle samples, and they displayed little or no expression in control muscle or confluent control myoblasts. These results are consistent with roles for these genes in gametogenesis and abnormal regenerative myogenesis in FSHD.
The only one of these six genes with a known function is ZSCAN4/Zscan4, which is expressed specifically at the late two-cell stage and promotes the normal progression of mouse embryos to the four-cell stage.67 The transient expression of this gene regulates pluripotency, genome stability, and telomere stability and can upregulate expression of several hundred genes during the late stages of induced pluripotent stem cell formation with major changes in phenotypic outcome.79 We noticed that among the genes regulated by Zscan4 are the murine homologs of KHDC1, TRIM43 and PRAMEF7,79 all of which were found to be upregulated by DUX4-fl transduction.61 The first two of these genes were also analyzed in human samples and shown to be testis- and FSHD-associated.61 The 1.9-kb enhancer and promoter region of ZSCAN4 contains four binding sites for DUX4-fl protein and was responsive to strong upregulation by transduced DUX-fl in a reporter gene assay using a human rhabdosarcoma cell line.61 Given its ability to upregulate many genes during early differentiation, this gene might be one of the earliest to be dysregulated by inappropriate expression of DUX4-fl in the FSHD muscle lineage and could have a major role in establishing the FSHD transcription phenotype.
Four of the above six testis/FSHD muscle-lineage genes, ZSCAN4, PRAMEF1, KHDC1 and RFPL2, were included in our expression array study.53 We found that ZSCAN4, PRAMEF1 and KHDC1 were upregulated ~4-, 5- and 2-fold (adjusted P=3×10−6, 10−4 and 10−4), respectively, in FSHD vs control myotubes with only about 1.5-fold upregulation in FSHD cells at the myoblast stage. Some of the hundreds of changes in gene expression from myoblasts to myotubes53 may be responsible for our observing a stronger FSHD-associated upregulation of levels of these transcripts at the myotube stage. We used myoblasts from 70% confluent cultures, which are not committed to myotube formation, unlike Geng et al.61 who used confluent myoblast cultures, and this may account for their higher FSHD-specific upregulation of these genes at the myoblast stage. The testis association of these genes probably reflects the finding that the only normal postnatal tissue shown to express DUX4-fl RNA and protein is testis.6 Another testis-associated gene, CCNA1, which encodes a meiosis-associated cyclin, was strongly upregulated in control myoblasts transduced with a DUX4-fl expression construct in the study of Geng et al.61 Analogously, we found strong upregulation of this gene, 3.6- and 24-fold, in FSHD vs control myoblasts and myotubes, respectively (adjusted P<0.01 and 10−6).53 In summary, several studies indicate that multiple genes are dysregulated in normal-appearing FSHD myoblasts and myotubes.
OTHER EVIDENCE FAVORS THE ‘MAJORITY RULES’ MODEL
Because DUX4 is implicated in FSHD and is a homeobox gene, it is important to consider the nature of homeobox genes in evaluating models for DUX4 pathogenicity. The expression of homeobox genes is tightly regulated temporally as well as being highly specific for cell type, in accord with their ability to select developmental fates.51,80–82 Although some homeobox genes are expressed in specific adult cell populations in which they have maintenance or cell-survival functions, generally, expression of this class of genes in higher eukaryotes is most prominent in the early stages of cellular differentiation.82–86 Expression of many homeobox genes depends on their long-range chromatin epigenetic environment and is facilitated by their frequent clustering.82–84,87 Their expression is also a function of the presence of other early differentiation-associated proteins82,83 as well as post-transcriptional and post-translational control.88,89
Model 2 (Majority Rules, Figure 2) involves DUX4-fl protein, at certain developmental stages, not inducing acute toxicity, but rather causing aberrant modulation of the transcription program. A parallel to this hypothesis is the finding that even typical homeobox genes like HOXA5 and PITX1 can cause p53-mediated apoptosis upon aberrant upregulation in certain cell types.90,91 Specific epigenetic and transcription factor determinants of expression of homeobox genes and their association with early stages of development could explain why DUX4-fl RNA might be generated transiently from a high percentage of FSHD muscle precursor cells only at a pre-myoblast, probably satellite-cell stage. Similarly, the biological activity of the homeodomain proteins encoded by homeobox genes is highly dependent on cell type. This includes the need for stage-specific DNA-binding partners and proteins that affect the activity of homeodomain proteins once they are bound to their DNA target.92,93
Evidence that DUX4-fl is not toxic at certain cell stages is seen in FSHD fibroblast-derived induced pluripotent stem cell cultures.6 These differentiate to give embryoid bodies of normal appearance and cell composition, despite the evidence for a higher abundance of DUX4-fl RNA in these embryoid bodies than in FSHD myoblasts.6 Moreover, the finding of long DUX4-fl RNA and corresponding protein encoded at both 4q35 and 10q26 DUX4 in normal testis6 suggests a genetically programmed function for D4Z4-derived DUX4 protein at some stage(s) in normal human development that is yet to be determined.
In addition to the tandem arrays at 4q35 and 10q26, representatives of paralogous groups of mammalian DUX-type double-homeobox sequences are present in many locations in the human genome.94 The only DUX-related gene with evidence as to its specific function is mouse Duxbl, whose expression pattern implicates it in gametogenesis, thymocyte maturation and prenatal myogenesis in mice.95,96 Knockout of this gene confirmed its role in the production of murine CD4+/CD8+ thymocytes.96 However, Leidenroth and Hewitt36 concluded that Duxbl is not an otholog of DUX4. Therefore, it is unclear whether functions of Duxbl are relevant to DUX4. Moreover, they found that evolutionary descent of DUX4 from a DUXC-type precursor is more likely than from a DUXB precursor.36 They described a tandem repeat of the DUXC-related rodent Dux gene as the most likely functional equivalent of DUX4, and it is of unknown function.
With respect to models that propose that the toxicity of DUX4 expression in myoblasts or myotubes is central to pathogenesis (Minority Rules, Model 1), C2C12 cells transduced with PAX3 or PAX7 constructs plus a DUX4 construct can be spared the acute toxicity from DUX4-fl expression.59 PAX3 and PAX7 have important roles in embryonic and early postnatal myogenesis, and PAX7 RNA is persistently found in adult satellite cells.86 Because PAX7 RNA is also present in undifferentiated myoblasts, which are nonetheless susceptible to DUX4-fl toxicity, the relevance of PAX7 to DUX4 and FSHD pathogenicity is unclear. The sequence similarity of the homeodomains of PAX7 and DUX4 have been invoked in hypotheses about PAX7 and DUX4 competing for DNA binding.59 However, other homeodomain proteins in addition to PAX7 have similar homeodomains to those of DUX4 and so also may compete with DUX4-fl for binding to DNA.36 In any event, PAX3 and PAX7 offer paradigms for how the spatio-temporally limited presence of transcription factors can counteract DUX4-fl toxicity.
The hallmarks of DUX4-fl toxicity in myoblasts are the loss of cell viability, MYOD1/Myod1 downregulation, inhibition of myotube formation or formation of very abnormal-looking myotubes, and upregulation of atrophy-associated FBXO32/ATROGIN1 and TRIM63/MURF1.59,63 None of these changes were observed by us or by Homma et al.53,60 upon examination of many FSHD myoblast cell strains; see above for evidence of FSHD downregulation, rather than upregulation, of TRIM63. Both of these groups and that of Barro et al.53,60,97 found that FSHD and control myoblasts cannot be distinguished by viability, growth rates or rates of differentiation to myotubes. Although some differences in the shape of FSHD and control myotubes were reported,63,97 we and Homma et al. found the shape of myotubes to be variable among both control and FSHD cell strains with no disease association.53,60 The good growth and differentiation of FSHD myoblasts should reflect the population of satellite cells in vivo because FSHD myoblast cell strains from moderately affected muscle are no more difficult to generate than control myoblast cell strains and can undergo similar numbers of cell population doublings.53 However, it is possible that FSHD myoblasts from severely affected muscle, which are difficult to propagate, have more frequent expression of DUX4-fl RNA and protein.
Another finding favoring Model 2 over Model 1 is that FSHD fibroblasts contain DUX4-fl transcripts at very low levels, as do FSHD (but not control) myoblasts, myotubes and muscle tissue.6 FSHD is predominantly a skeletal muscle-specific disease. Therefore, it is likely that the inappropriate DUX4 expression that establishes pathogenesis is mostly specific to the muscle lineage and would not be shared with fibroblasts. Unlike Model 1, Model 2 with its postulated frequent expression of DUX4-fl at a muscle lineage-specific pre-myoblast stage obviates the difficulty posed by FSHD fibroblasts expressing DUX4-fl RNA.
CONCLUSIONS
A large fraction of FSHD myoblasts and myotubes from moderately affected muscle displays an expression dysregulation phenotype. This and other findings support a model invoking transient, pathogenic expression of DUX4 in a large fraction of cells at the pre-myoblast stage (Figure 2, Model 2). Among the dysregulated targets of the hypothesized burst of DUX4 expression in this model could be the genes encoding the testis—and very early embryogenesis-specific reprogramming and telomere-stabilizing factor ZSCAN4 and the meiosis-associated cyclin CCNA1.79,98 These genes were upregulated by transduction of DUX4 constructs into control myoblasts.61 In FSHD patients, we propose that there is transient expression of DUX4-fl at a pre-myoblast stage in affected regions of skeletal muscle and possibly among certain subsets of muscle satellite cells.99,100 This could result in upregulation of expression of these and other genes and, during regenerative myogenesis, the dampening of expression of many muscle lineage-associated genes.53 This, in turn, could decrease the efficiency of the late stages of regenerative myogenesis or affect muscle function in a manner consistent with the usually slow progression of FSHD.
Acknowledgments
This article is dedicated to the memory of Carol A Perez, who helped to found the FSH Society, and worked with uncommon devotion and intelligence on behalf of FSHD patients. Supported in part by the FSHD Global Research Foundation and NIH Grant NS048859.
References
- 1.Hewitt JE, Lyle R, Clark LN, Valleley EM, Wright TJ, Wijmenga C, et al. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum Mol Genet. 1994;3:1287–1295. doi: 10.1093/hmg/3.8.1287. [DOI] [PubMed] [Google Scholar]
- 2.Matsumura T, Goto K, Yamanaka G, Lee J, Zhang C, Hayashi YK, et al. Chromosome 4q;10q translocations; comparison with different ethnic populations and FSHD patients. BMC Neurol. 2002;2:7. doi: 10.1186/1471-2377-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemmers RJ, van der Vliet PJ, van der Gaag KJ, Zuniga S, Frants RR, de Knijff P, et al. Worldwide population analysis of the 4q and 10q subtelomeres Identifies only four discrete interchromosomal sequence transfers in human evolution. Am J Hum Genet. 2010;86:364–377. doi: 10.1016/j.ajhg.2010.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kowaljow V, Marcowycz A, Ansseau E, Conde CB, Sauvage S, Matteotti C, et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord. 2007;17:611–623. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camano P, Dauwerse JG, et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snider L, Geng LN, Lemmers RJ, Kyba M, Ware CB, Nelson AM, et al. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tawil R. Facioscapulohumeral muscular dystrophy. Neurotherapeutics. 2008;5:601–606. doi: 10.1016/j.nurt.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scionti I, Fabbri G, Fiorillo C, Ricci G, Greco F, D’Amico R, et al. Facioscapulohumeral muscular dystrophy: new insights from compound heterozygotes and implication for prenatal genetic counselling. J Med Genet. 2011 doi: 10.1136/jmedgenet-2011-100454. [DOI] [PubMed] [Google Scholar]
- 9.Lemmers RJLF, van der Wielen M, Bakker E, van der Maarel S. Molecular diagnosis of FSHD. In: Upadhyaya M, Cooper DN, editors. FSHD Facioscapulohumeral Muscular Dystrophy: Clinical Medicine and Molecular Cell Biology. BIOS Scientific Publishers; New York, NY: 2004. pp. 211–234. [Google Scholar]
- 10.Goto K, Nishino I, Hayashi YK. Rapid and accurate diagnosis of facioscapulohumeral muscular dystrophy. Neuromuscul Disord. 2006;16:256–261. doi: 10.1016/j.nmd.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen K, Walrafen P, Bernard R, Attarian S, Chaix C, Vovan C, et al. Molecular combing reveals allelic combinations in facioscapulohumeral dystrophy. Ann Neurol. 2011;70:627–633. doi: 10.1002/ana.22513. [DOI] [PubMed] [Google Scholar]
- 12.Fitzsimons RB. Retinal vascular disease and the pathogenesis of facioscapulohumeral muscular dystrophy. A signalling message from Wnt? Neuromuscul Disord. 2011;21:263–271. doi: 10.1016/j.nmd.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Goto K, Lee JH, Matsuda C, Hirabayashi K, Kojo T, Nakamura A, et al. DNA rearrangements in Japanese facioscapulohumeral muscular dystrophy patients: clinical correlations. Neuromuscul Disord. 1995;5:201–208. doi: 10.1016/0960-8966(94)00055-e. [DOI] [PubMed] [Google Scholar]
- 14.Dorobek M, Kabzinska D. A severe case of facioscapulohumeral muscular dystrophy (FSHD) with some uncommon clinical features and a short 4q35 fragment. Eur J Paediatr Neurol. 2004;8:313–316. doi: 10.1016/j.ejpn.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Tsuji M, Kinoshita M, Imai Y, Kawamoto M, Kohara N. Facioscapulohumeral muscular dystrophy presenting with hypertrophic cardiomyopathy: a case study. Neuromuscul Disord. 2009;19:140–142. doi: 10.1016/j.nmd.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Wohlgemuth M, van der Kooi EL, van Kesteren RG, van der Maarel SM, Padberg GW. Ventilatory support in facioscapulohumeral muscular dystrophy. Neurology. 2004;63:176–178. doi: 10.1212/01.wnl.0000133126.86377.e8. [DOI] [PubMed] [Google Scholar]
- 17.Carter GT, Bird TD. Ventilatory support in facioscapulohumeral muscular dystrophy. Neurology. 2005;64:401. doi: 10.1212/wnl.64.2.401. [DOI] [PubMed] [Google Scholar]
- 18.Saito A, Higuchi I, Nakagawa M, Saito M, Uchida Y, Inose M, et al. An overexpression of fibroblast growth factor (FGF) and FGF receptor 4 in a severe clinical phenotype of facioscapulohumeral muscular dystrophy. Muscle Nerve. 2000;23:490–497. doi: 10.1002/(sici)1097-4598(200004)23:4<490::aid-mus6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 19.Balatsouras DG, Korres S, Manta P, Panousopoulou A, Vassilopoulos D. Cochlear function in facioscapulohumeral muscular dystrophy. Otol Neurotol. 2007;28:7–10. doi: 10.1097/01.mao.0000244362.39696.c8. [DOI] [PubMed] [Google Scholar]
- 20.Matsuzaka T, Sakuragawa N, Terasawa K, Kuwabara H. Facioscapulohumeral dystrophy associated with mental retardation, hearing loss, and tortuosity of retinal arterioles. J Child Neurol. 1986;1:218–223. doi: 10.1177/088307388600100308. [DOI] [PubMed] [Google Scholar]
- 21.Ehrlich M. Exploring hypotheses about the molecular etiology of FSHD: loss of heterochromatin spreading and other long-range interaction models. In: Cooper DN, Upadhyaya M, editors. FSHD Facioscapulohumeral Muscular Dystrophy: Molecular Cell Biology & Clinical Medicine. BIOS Scientific Pub; New York, NY: 2004. pp. 253–276. [Google Scholar]
- 22.van der Maarel SM, Frants RR, Padberg GW. Facioscapulohumeral muscular dystrophy. Biochim Biophys Acta. 2007;1772:186–194. doi: 10.1016/j.bbadis.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Alexiadis V, Ballestas ME, Sanchez C, Winokur S, Vedanarayanan V, Warren M, et al. RNAPol-ChIP analysis of transcription from FSHD-linked tandem repeats and satellite DNA. Biochem Biophys Acta. 2007;(1769):29–40. doi: 10.1016/j.bbaexp.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 25.Jiang G, Yang F, van Overveld PG, Vedanarayanan V, van der Maarel S, Ehrlich M. Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum Mol Genet. 2003;12:2909–2921. doi: 10.1093/hmg/ddg323. [DOI] [PubMed] [Google Scholar]
- 26.Gabriels J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 27.Dixit M, Ansseau E, Tassin A, Winokur S, Shi R, Qian H, et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci USA. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosnakovski D, Lamb S, Simsek T, Xu Z, Belayew A, Perlingeiro R, et al. DUX4c, an FSHD candidate gene, interferes with myogenic regulators and abolishes myoblast differentiation. Exp Neurol. 2008;214:87–96. doi: 10.1016/j.expneurol.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 29.Wuebbles RD, Hanel ML, Jones P. LFSHD region gene 1 (FRG1) is crucial for angiogenesis linking FRG1 to facioscapulohumeral muscular dystrophy-associated vasculopathy. Dis Model Mech. 2009;2:267–274. doi: 10.1242/dmm.002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klooster R, Straasheijm K, Shah B, Sowden J, Frants R, Thornton C, et al. Comprehensive expression analysis of FSHD candidate genes at the mRNA and protein level. Eur J Hum Genet. 2009;17:1615–1624. doi: 10.1038/ejhg.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winokur ST, Chen YW, Masny PS, Martin JH, Ehmsen JT, Tapscott SJ, et al. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum Mol Genet. 2003;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- 32.Osborne RJ, Welle S, Venance SL, Thornton CA, Tawil R. Expression profile of FSHD supports a link between retinal vasculopathy and muscular dystrophy. Neurology. 2007;68:569–577. doi: 10.1212/01.wnl.0000251269.31442.d9. [DOI] [PubMed] [Google Scholar]
- 33.Lemmers RJ, Wohlgemuth M, van der Gaag KJ, van der Vliet PJ, van Teijlingen CM, de Knijff P, et al. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2007;81:884–894. doi: 10.1086/521986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsumagari K, Chen D, Hackman JR, Bossler AD, Ehrlich M. FSH dystrophy and a subtelomeric 4q haplotype: a new assay and associations with disease. J Med Genet. 2010;47:745–751. doi: 10.1136/jmg.2009.076703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spurlock G, Jim HP, Upadhyaya M. Confirmation that the specific SSLP microsatellite allele 4qA161 segregates with fascioscapulohumeral muscular dystrophy (FSHD) in a cohort of multiplex and simplex FSHD families. Muscle Nerve. 2010;42:820–821. doi: 10.1002/mus.21766. [DOI] [PubMed] [Google Scholar]
- 36.Leidenroth A, Hewitt JE. A family history of DUX4: phylogenetic analysis of DUXA, B, C and Duxbl reveals the ancestral DUX gene. BMC Evol Biol. 2010;10:364. doi: 10.1186/1471-2148-10-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tupler R, Berardinelli A, Barbierato L, Frants R, Hewitt JE, Lanzi G, et al. Monosomy of distal 4q does not cause facioscapulohumeral muscular dystrophy. J Med Genet. 1996;33:366–370. doi: 10.1136/jmg.33.5.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu X, Tsumagari K, Sowden J, Tawil R, Boyle AP, Song L, et al. DNaseI hypersensitivity at gene-poor, FSH dystrophy-linked 4q35.2. Nucleic Acids Res. 2009;37:7381–7393. doi: 10.1093/nar/gkp833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Greef JC, Lemmers RJ, van Engelen BG, Sacconi S, Venance SL, Frants RR, et al. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum Mutat. 2009;30:1–11. doi: 10.1002/humu.21091. [DOI] [PubMed] [Google Scholar]
- 40.Yamanaka G, Goto K, Ishihara T, Oya Y, Miyajima T, Hoshika A, et al. FSHD-like patients without 4q35 deletion. J Neurol Sci. 2004;219:89–93. doi: 10.1016/j.jns.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 41.Leidenroth A, Sorte HS, Gilfillan G, Ehrlich M, Lyle R, Hewitt JE. Diagnosis by sequencing: correction of misdiagnosis from FSHD2 to LGMD2A by whole-exome analysis. Eur J Hum Genet. doi: 10.1038/ejhg.2012.42. e-pub ahead of print 29 February 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snider L, Asawachaicharn A, Tyler AE, Geng LN, Petek LM, Maves L, et al. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum Mol Genet. 2009;18:2414–2430. doi: 10.1093/hmg/ddp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15 (Spec1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 44.Mosch K, Franz H, Soeroes S, Singh PB, Fischle W. HP1 recruits activity-dependent neuroprotective protein to H3K9me3 marked pericentromeric heterochromatin for silencing of major satellite repeats. PLoS One. 2011;6:e15894. doi: 10.1371/journal.pone.0015894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szpakowski S, Sun X, Lage JM, Dyer A, Rubinstein J, Kowalski D, et al. Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene. 2009;448:151–167. doi: 10.1016/j.gene.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng W, de Greef JC, Chen YY, Chien R, Kong X, Gregson HC, et al. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsien F, Sun B, Hopkins NE, Vedanarayanan V, Figlewicz D, Winokur S, et al. Hypermethylation of the FSHD syndrome-linked subtelomeric repeat in normal and FSHD cells but not in ICF syndrome cells. Molec Gen Metab. 2001;74:322–331. doi: 10.1006/mgme.2001.3219. [DOI] [PubMed] [Google Scholar]
- 48.de Greef JC, Wohlgemuth M, Chan OA, Hansson KB, Smeets D, Frants RR, et al. Hypomethylation is restricted to the D4Z4 repeat array in phenotypic FSHD. Neurology. 2007;69:1018–1026. doi: 10.1212/01.wnl.0000271391.44352.fe. [DOI] [PubMed] [Google Scholar]
- 49.Lyle R, Wright TJ, Clark LN, Hewitt JE. The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics. 1995;28:389–397. doi: 10.1006/geno.1995.1166. [DOI] [PubMed] [Google Scholar]
- 50.Winokur ST, Bengtsson U, Vargas JC, Wasmuth JJ, Altherr MR, Weiffenbach B, et al. The evolutionary distribution and structural organization of the homeobox-containing repeat D4Z4 indicates a functional role for the ancestral copy in the FSHD region. Hum Mol Genet. 1996;5:1567–1575. doi: 10.1093/hmg/5.10.1567. [DOI] [PubMed] [Google Scholar]
- 51.Foronda D, de Navas LF, Garaulet DL, Sanchez-Herrero E. Function and specificity of Hox genes. Int J Dev Biol. 2009;53:1404–1419. doi: 10.1387/ijdb.072462df. [DOI] [PubMed] [Google Scholar]
- 52.Richards M, Coppee F, Thomas N, Belayew A, Upadhyaya M. Facioscapulohumeral muscular dystrophy (FSHD): an enigma unravelled? Hum Genet. 2011;131:325–340. doi: 10.1007/s00439-011-1100-z. [DOI] [PubMed] [Google Scholar]
- 53.Tsumagari K, Chang S-C, Lacey M, Baribault C, Chittur SV, Sowden J, et al. Gene expression during normal and FSHD myogenesis. BMC Med Genomics. 2011;4:67. doi: 10.1186/1755-8794-4-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood-Allum C, Brennan P, Hewitt M, Lowe J, Tyfield L, Wills A. Clinical and histopathological heterogeneity in patients with 4q35 facioscapulohumeral muscular dystrophy (FSHD) Neuropathol Appl Neurobiol. 2004;30:188–191. doi: 10.1046/j.0305-1846.2003.00520.x. [DOI] [PubMed] [Google Scholar]
- 55.Reed P, Porter NC, Strong J, Pumplin DW, Corse AM, Luther PW, et al. Sarcolemmal reorganization in facioscapulohumeral muscular dystrophy. Ann Neurol. 2006;59:289–297. doi: 10.1002/ana.20750. [DOI] [PubMed] [Google Scholar]
- 56.Carosio S, Berardinelli MG, Aucello M, Musaro A. Impact of ageing on muscle cell regeneration. Ageing Res Rev. 2011;10:35–42. doi: 10.1016/j.arr.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 58.Scharner J, Zammit PS. The muscle satellite cell at 50: the formative years. Skelet Muscle. 2011;1:28. doi: 10.1186/2044-5040-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bosnakovski D, Xu Z, Gang EJ, Galindo CL, Liu M, Simsek T, et al. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 2008;27:2766–2779. doi: 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Homma S, Chen JC, Rahimov F, Beermann ML, Hanger K, Bibat GM, et al. A unique library of myogenic cells from facioscapulohumeral muscular dystrophy subjects and unaffected relatives: family, disease and cell function. Eur J Hum Genet. 2011;20:204–210. doi: 10.1038/ejhg.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, et al. DUX4 activates germline genes, retroelements, and immune mediators: Implications for facioscapulohumeral dystrophy. Dev Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wallace LM, Garwick SE, Mei W, Belayew A, Coppee F, Ladner KJ, et al. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53- dependent myopathy in vivo. Ann Neurol. 2011;69:540–552. doi: 10.1002/ana.22275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vanderplanck C, Ansseau E, Charron S, Stricwant N, Tassin A, Laoudj- Chenivesse D, et al. The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS One. 2011;6:e26820. doi: 10.1371/journal.pone.0026820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carter MG, Stagg CA, Falco G, Yoshikawa T, Bassey UC, Aiba K, et al. An in situ hybridization-based screen for heterogeneously expressed genes in mouse ES cells. Gene Expr Patterns. 2008;8:181–198. doi: 10.1016/j.gep.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kobayashi T, Kageyama R. Hes1 regulates embryonic stem cell differentiation by suppressing Notch signaling. Genes Cells. 2010;15:689–698. doi: 10.1111/j.1365-2443.2010.01413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suzuki N, Furusawa C, Kaneko K. Oscillatory protein expression dynamics endows stem cells with robust differentiation potential. PLoS One. 2011;6:e27232. doi: 10.1371/journal.pone.0027232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MS. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanaka TS. Transcriptional heterogeneity in mouse embryonic stem cells. Reprod Fertil Dev. 2009;21:67–75. doi: 10.1071/rd08219. [DOI] [PubMed] [Google Scholar]
- 69.Tupler R, Perini G, Pellegrino MA, Green MR. Profound misregulation of muscle-specific gene expression in facioscapulohumeral muscular dystrophy. Proc Natl Acad Sci USA. 1999;96:12650–12654. doi: 10.1073/pnas.96.22.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Celegato B, Capitanio D, Pescatori M, Romualdi C, Pacchioni B, Cagnin S, et al. Parallel protein and transcript profiles of FSHD patient muscles correlate to the D4Z4 arrangement and reveal a common impairment of slow to fast fibre differentiation and a general deregulation of MyoD-dependent genes. Proteomics. 2006;6:5303–5321. doi: 10.1002/pmic.200600056. [DOI] [PubMed] [Google Scholar]
- 71.Arashiro P, Eisenberg I, Kho AT, Cerqueira AM, Canovas M, Silva HC, et al. Transcriptional regulation differs in affected facioscapulohumeral muscular dystrophy patients compared to asymptomatic related carriers. Proc Natl Acad Sci USA. 2009;106:6220–6225. doi: 10.1073/pnas.0901573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheli S, Francois S, Bodega B, Ferrari F, Tenedini E, Roncaglia E, et al. Expression profiling of FSHD-1 and FSHD-2 cells during myogenic differentiation evidences common and distinctive gene dysregulation patterns. PLoS One. 2011;6:e20966. doi: 10.1371/journal.pone.0020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fitzsimons RB. Facioscapulohumeral dystrophy: the role of inflammation. Lancet. 1994;344:902–903. doi: 10.1016/s0140-6736(94)92263-2. [DOI] [PubMed] [Google Scholar]
- 74.Arahata K, Ishihara T, Fukunaga H, Orimo S, Lee JH, Goto K, et al. Inflammatory response in facioscapulohumeral muscular dystrophy (FSHD): immunocytochemical and genetic analyses. Muscle Nerve. 1995;2:S56–S66. [PubMed] [Google Scholar]
- 75.Kan HE, Scheenen TW, Wohlgemuth M, Klomp DW, van Loosbroek-Wagenmans I, Padberg GW, et al. Quantitative MR imaging of individual muscle involvement in facioscapulohumeral muscular dystrophy. Neuromuscul Disord. 2009;19:357–362. doi: 10.1016/j.nmd.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 76.Frisullo G, Frusciante R, Nociti V, Tasca G, Renna R, Iorio R, et al. CD8(+) T Cells infacioscapulohumeral muscular dystrophy patients with inflammatory features at muscle MRI. J Clin Immunol. 2011;31:155–166. doi: 10.1007/s10875-010-9474-6. [DOI] [PubMed] [Google Scholar]
- 77.Sun G, Haginoya K, Wu Y, Chiba Y, Nakanishi T, Onuma A, et al. Connective tissue growth factor is overexpressed in muscles of human muscular dystrophy. J Neurol Sci. 2008;267:48–56. doi: 10.1016/j.jns.2007.09.043. [DOI] [PubMed] [Google Scholar]
- 78.Laoudj-Chenivesse D, Carnac G, Bisbal C, Hugon G, Bouillot S, Desnuelle C, et al. Increased levels of adenine nucleotide translocator 1 protein and response to oxidative stress are early events in facioscapulohumeral muscular dystrophy muscle. J Mol Med. 2005;83:216–224. doi: 10.1007/s00109-004-0583-7. [DOI] [PubMed] [Google Scholar]
- 79.Hirata T, Amano T, Nakatake Y, Amano M, Piao Y, Hoang HG, et al. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci Rep. 2012;2:208. doi: 10.1038/srep00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hryniuk A, Grainger S, Savory JG, Lohnes D. Cdx function is required for maintenance of intestinal identity in the adult. Dev Biol. 2012;363:426–437. doi: 10.1016/j.ydbio.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 81.Gundersen K. Excitation-transcription coupling in skeletal muscle: the molecular pathways of exercise. Biol Rev Camb Philos Soc. 2011;86:564–600. doi: 10.1111/j.1469-185X.2010.00161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barber BA, Rastegar M. Epigenetic control of Hox genes during neurogenesis, development, and disease. Ann Anat. 2010;192:261–274. doi: 10.1016/j.aanat.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 83.Soshnikova N, Duboule D. Epigenetic regulation of vertebrate Hox genes: a dynamic equilibrium. Epigenetics. 2009;4:537–540. doi: 10.4161/epi.4.8.10132. [DOI] [PubMed] [Google Scholar]
- 84.MacLean JA, 2nd, Wilkinson MF. The Rhox genes. Reproduction. 2010;140:195–213. doi: 10.1530/REP-10-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li J, Dani JA, Le W. The role of transcription factor Pitx3 in dopamine neuron development and Parkinson’s disease. Curr Top Med Chem. 2009;9:855–859. [PMC free article] [PubMed] [Google Scholar]
- 86.Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tschopp P, Duboule D. A genetic approach to the transcriptional regulation of Hox gene clusters. Annu Rev Genet. 2011;45:145–166. doi: 10.1146/annurev-genet-102209-163429. [DOI] [PubMed] [Google Scholar]
- 88.Boutet SC, Biressi S, Iori K, Natu V, Rando TA. Taf1 regulates Pax3 protein by monoubiquitination in skeletal muscle progenitors. Mol Cell. 2010;40:749–761. doi: 10.1016/j.molcel.2010.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X, et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190:867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu DX, Lobie PE. Transcriptional activation of p53 by Pitx1. Cell Death Differ. 2007;14:1893–1907. doi: 10.1038/sj.cdd.4402209. [DOI] [PubMed] [Google Scholar]
- 91.Daftary GS, Taylor HS. Endocrine regulation of HOX genes. Endocr Rev. 2006;27:331–355. doi: 10.1210/er.2005-0018. [DOI] [PubMed] [Google Scholar]
- 92.Mann RS, Lelli KM, Joshi R. Hox specificity unique roles for cofactors and collaborators. Curr Top Dev Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mallo M, Wellik DM, Deschamps J. Hox genes and regional patterning of the vertebrate body plan. Dev Biol. 2010;344:7–15. doi: 10.1016/j.ydbio.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Clapp J, Mitchell LM, Bolland DJ, Fantes J, Corcoran AE, Scotting PJ, et al. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2007;81:264–279. doi: 10.1086/519311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu SL, Tsai MS, Wong SH, Hsieh-Li HM, Tsai TS, Chang WT, et al. Characterization of genomic structures and expression profiles of three tandem repeats of a mouse double homeobox gene: Duxbl. Dev Dyn. 2010;239:927–940. doi: 10.1002/dvdy.22210. [DOI] [PubMed] [Google Scholar]
- 96.Kawazu M, Yamamoto G, Yoshimi M, Yamamoto K, Asai T, Ichikawa M, et al. Expression profiling of immature thymocytes revealed a novel homeobox gene that regulates double-negative thymocyte development. J Immunol. 2007;179:5335–5345. doi: 10.4049/jimmunol.179.8.5335. [DOI] [PubMed] [Google Scholar]
- 97.Barro M, Carnac G, Flavier S, Mercier J, Vassetzky Y, Laoudj-Chenivesse D. Myoblasts from affected and non affected FSHD muscles exhibit morphological differentiation defects. J Cell Mol Med. 2010:275–289. doi: 10.1111/j.1582-4934.2008.00368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baumer N, Sandstede ML, Diederichs S, Kohler G, Readhead C, Ji P, et al. Analysis of the genetic interactions between Cyclin A1, Atm and p53 during spermatogenesis. Asian J Androl. 2007;9:739–750. doi: 10.1111/j.1745-7262.2007.00339.x. [DOI] [PubMed] [Google Scholar]
- 99.Bentzinger CF, von Maltzahn J, Rudnicki MA. Extrinsic regulation of satellite cell specification. Stem Cell Res Ther. 2011;1:27. doi: 10.1186/scrt27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kirkpatrick LJ, Yablonka-Reuveni Z, Rosser BW. Retention of Pax3 expression in satellite cells of muscle spindles. J Histochem Cytochem. 2010;58:317–327. doi: 10.1369/jhc.2009.954792. [DOI] [PMC free article] [PubMed] [Google Scholar]