Abstract
Urinary tract infections primarily caused by uropathogenic strains of Escherichia coli (E. coli) remain a significant public health problem in both developed and developing countries. An important virulence determinant in uropathogenesis is biofilm formation which requires expression of fimbriae, flagella, and other surface components such as lipopolysaccharides. In this study, we explored the regulation of uvrY and csrA genes in biofilm formation, motility and virulence determinants in uropathogenic E. coli. We found that mutation in uvrY suppressed biofilm formation on abiotic surfaces such as polyvinyl chloride, polystyrene and glass, and complementation of uvrY in the mutant restored the biofilm phenotype. We further evaluated the role of uvrY gene in expression of type 1 fimbriae, an important adhesin that facilitates adhesion to various abiotic surfaces. We found that phase variation of type 1 fimbriae between fimbriated and afimbriated mode was modulated by uvrY at its transcriptional level. Deletion mutant of uvrY lowered expression of fimbrial recombinase genes, such as fimB, fimE, and fimA, a gene encoding major fimbrial subunit. Furthermore, transcription of virulence specific genes such as papA, hlyB and galU was also reduced in the deletion mutant. Swarming motility and expression of flhD and flhC was also diminished in the mutant. Taken together, our findings unravel a possible mechanism in which uvrY facilitates biofilm formation, persistence and virulence of uropathogenic E. coli.
Introduction
Uropathogenic Escherichia coli (UPEC) causes urinary tract infections (cystitis, pyelonephritis), and septicemia in humans and animals [1], [2]. Zoonotic transmission of antimicrobial-resistant strains of UPEC from animals to humans was proposed [3]. UPEC adherence to urogenital epithelial cells is the first step in the initiation of infection, which is generally accomplished by type 1 fimbriae [4], [5]. In UPEC, the initial process of attachment is mediated by several adhesins, of which, type 1 and pap fimbriae play a critical role in colonization in urinary bladder and kidneys respectively. Type 1 fimbriae mediate UPEC attachment to the bladder epithelium by binding to mannose-containing glycoproteins and promote the early stages of biofilm formation on both biotic and abiotic surfaces [6]. Furthermore, the fimbriae exhibit phase variation by inversion of a 314-bp DNA element (fim switch), which harbors the promoter for several structural proteins [7], [8]. This allows (“on” orientation) or prevents (“off” orientation) transcription of the structural genes in E. coli.
Bacterial biofilms are structured bacterial communities embedded in a self-produced exopolysaccharide matrix on biotic and abiotic surfaces [9], which are, indeed, the major problem of the interference of bacterial eradication with antibiotics. An important hallmark of UPEC is the formation of biofilm, which facilitates UPEC strains to persist in the urogenital tract and interfere with bacterial eradication [10]. Biofilm formation in E. coli requires a set of gene expressions facilitating its initiation, attachment and subsequent maturation. For example, a variety of virulence factors in E. coli, such as hemolysin, fimbriae, lipopolysaccharide (LPS), secreted proteins, capsules and iron-acquisition systems [11], [12], which allow bacterial colonization in the mucosal epithelial cells lining the urogenital tract, invade, and further form intracellular biofilm-like pods in uroepithelial cells [13]. Remarkably, UPEC is capable of forming biofilms on the abiotic surfaces of indwelling medical devices such as catheters [14], [15], which lead to “Catheter-Associated Urinary Tract Infection” (CAUTI) in clinics [16]. CAUTI is especially fatal in immunocompromised, debilitated and diabetic patients [17], [18]. Therefore, biofilm formation in UPEC is important for causing persistent colonization in bladder, kidneys or urine and in hospital settings.
The two-component regulatory systems (TCSs) are a ubiquitous mechanism for coupling various environmental stimuli with the transcription program of bacteria [19], [20]. A typical TCS consists of a sensory protein kinase and a cognate response regulator [19]. The sensory protein kinase monitors environmental signals and transduces the information, via a phosphorelay system, to the response regulator, which, in turn, responds and modulates gene expression. The BarA-UvrY TCS in E. coli is pleiotropic and have been linked with several metabolic processes including biofilm formation, oxidative stress, sigma S expression, and efficient adaptation in carbon utilization [21], [22]. In this system, BarA is the sensor kinase and UvrY is the cognate response regulator [23]. In addition, a critical downstream effect of the BarA-UvrY TCS in E. coli is its regulation on Carbon Storage Regulatory system (Csr) [22]. Csr has been shown to control the ‘switching’ of E. coli from a colonization state to a persistent state [24]. The Csr system in E. coli is composed of the 61 amino acid CsrA protein and two small, non-coding regulatory RNAs, CsrB and CsrC [25]. Transcription of these two small RNAs is regulated by the BarA/UvrY TCS system in E. coli. In this circuit, UvrY enhances transcriptions of CsrB and CsrC which in turn bind and interfere with the activity of RNA binding function of the CsrA. CsrA also regulates the TCS and subsequently controls its own expression in an auto-regulatory loop. It has been shown CsrB and CsrC RNA's contain several degenerative sequences that serve as multiple binding sites for CsrA protein. In addition, CsrA can directly target several E. coli genes, such as the glg operon which encodes genes in glycogen biosynthesis and the pgaA mRNA which encodes a polysaccharide adhesin involved in biofilm formation [26]–[28].
In this study, we investigated the roles of uvrY and csrA in regulating expression of genes relevant to E. coli biofilm formation. Our previous study has shown that mutants deficient in barA and uvrY genes of avian pathogenic E. coli are attenuated in an avian embryo model, exhibited reduced persistence in liver, spleen and decreased invasiveness in chicken embryo cells (12). This attenuation is correlated with the down regulation of fimbriae, exopolysaccharide accumulation and sensitivity to oxidative stress in vivo [29]. Our recent findings further verify that mutation in either barA or uvrY of UPEC CFT073 results in reduced persistence of mutants in bladder, kidneys or urine in an ascending murine UTI model [30]. Because UPEC share similar genes with APEC [31], [32], both studies lead us to hypothesize that the BarA/UvrY/Csr pathway plays an important role in the biofilm formation of UPEC. To prove this hypothesis in UPEC, the goal of this study is to determine the relationships between the BarA/UvrY/Csr pathway and regulation of genes relevant to in vitro biofilm formation of the CFT073 strain. Since fimbriae is a common adhesin present in both commensal and pathogenic E. coli [5], we also compared the regulation of type 1 fimbriae by the BarA/UvrY/Csr system between E. coli and UPEC CFT073 strains. To our knowledge the relationship of the BarA/UvrY/Csr system with the fimbriae in regulating the biofilm formation has not been previously explored. Our results indicated that the BarA/UvrY/Csr system could play key roles in uropathogenesis of UPEC in regulating the expressions of genes relevant to the biofilm formation at both transcriptional and post-transcriptional levels.
Materials and Methods
Bacterial strains, plasmids and oligonucleotides
Bacterial strains and oligonucleotides used in this study are listed in Table 1 and Table 2, respectively. Sequenced strains of uropathogenic E. coli CFT073 and non-pathogenic E. coli K-12, MG1655 were chosen for genetic manipulation in this study [33], [34]. Precise in-frame deletions were constructed in E. coli K-12 and UPEC CFT073 by λ Red recombinase method [35] as previously described [30]. PCR verification and phenotype characterization were performed to confirm the deletions. TR1-5 MG1655 was a kind gift of T. Romeo [36].
Table 1. Bacterial strains and plasmids used in this study.
| Strains | Genotype | Source |
| CFT073 | Wild-type Uropathogenic E. coli | H. L. Mobley [33] |
| SM3010 | ΔuvrY::cam | CFT073 |
| SM3011 | ΔcsrA::cam | CFT073 |
| SM3013 | ΔuvrY::cam/p-uvrY | SM3010 |
| SM3014 | ΔcsrA::cam/p-csrA | SM3011 |
| MG1655 | Wild-type K-12 λ- rph-1 | D. J. Jin [34] |
| AM1002 | ΔuvrY::cam | MG1655 |
| AM1005 | ΔuvrY::cam/p-uvrY | AM1002 |
| TR1-5MG1655 | ΔcsrA::kan | T. Romeo [36] |
| AM1006 | ΔcsrA::kan/p-csrA | TR1-5 MG1655 |
| Plasmids | ||
| pBR322 | Cloning Vector | Invitrogen |
| pSM2 | pBR322 containing uvrY gene; Apr | [29] |
| pSM7 | pBR322 containing csrA gene; Apr | [29] |
| pBB2-1 | fimA-lacZYA on pPR274 | William R. Schwan [40] |
| pWS124-17 | fimA-lacZYA locked on on pPP2-6 | William R. Schwan [40] |
Table 2. List of oligonucleotides used in the study.
| Primer designation | Sequence (5′-3′) | Gene/target sequence |
| OSM64 | CCCGAATTCATAATTTCATCGTAGGGCTTACTGTGA | uvrY |
| OSM65 | CCCCTGCAGATGCACGCCTGGCTGGGTTAC | |
| OSM250 | AGCGTTCTGTAAGCCTGTGAAGGT | rrnA |
| OSM251 | TAACGTTGGACAGGAACCCTTGGT | |
| OSM260 | ACCGTTCAGTTAGGACAGGTTCGT | fimA |
| OSM261 | TCTGCAGAGCCAGAACGTTGGTAT | |
| OSM271 | GGAATCGGTGTAGATGTAACCCC | Icd |
| OSM272 | CGTCCTGACCATAAACCTGTGTGG | |
| OSM287 | GCTCGTCACGGTCGCAACAA | lrhA |
| OSM288 | ACATCCAGCGCTAATTTCGG | |
| OSM295 | CAGTAATGCTGCTCGTTTTGCCG | fim promoter [39] |
| OSM296 | GACAGAGCCGACAGAACAACG | |
| OSM297 | CGACAGCAGAGCTGGTCGCTC | fim switch orientation [39] |
| OSM298 | GTAAATTATTTCTCTTGTAAAT | |
| TAATTTCACATCACCTCCGC | ||
| OSM299 | GCGGAGGTGATGTGAAATTAA | |
| TTTACAATAGAAATAATTTAC | ||
| OSM309 | ACTCTGCGGACCACTTGGGA | papA |
| OSM310 | CCAACTATTCCTCAGGGGCA | |
| OSM315 | GATGAAACCGCAGAAGGCTT | kpsE |
| OSM316 | GCGATGCGATGTGACATCTC | |
| OSM347 | CAAGGGCGCTGGTGAACAAC | hlyB |
| OSM348 | AACAGGAACTCGCTGAACCC | |
| OSM351 | AGTTCGTCAGGGTCTGGCGA | galU |
| OSM352 | CAACGCCATATGCGGTCACA | |
| OSM357 | CCATGATGCAGGCGGTTTGT | fimE |
| OSM358 | CCACGGCTTCACGCTCATCA | |
| OSM359 | GCCAAAGCAAAACCACACGA | fimB |
| OSM360 | AACGCACCCGCTATTGAACA | |
| OSM363 | TTTCATGGTCTGCGTGTTAGTG | ipuA |
| OSM364 | TTACCCGCAGCAGAAACTATGT |
The uvrY gene was amplified in the divergent yecF promoter using primer pairs OSM 64 and OSM 65 (Table 2). The amplified product was cloned into vector pCR2.1 using TOPO-TA cloning method (Invitrogen, Carlsbad, CA). Plasmids were sequenced to confirm the amplification and cloning. A 700-bp BamH 1-EcoR V DNA fragment was cloned into pBR322; the open reading frame (ORF) of the uvrY gene was placed in the same orientation as the tet gene in the vector.
Assessment of biofilm formation by crystal violet staining
Biofilm formation was evaluated on three abiotic surfaces - PVC, Polystyrene, and borosilicate cover glass as previously described [37]. Strains were grown in Luria broth (LB) supplemented with appropriate antibiotics without shaking and diluted (1∶100) in 50 ml LB broth with necessary antibiotic and grown at 37°C for 1 hour. Approximately equal amount of cells were taken as starting culture as determined by cell density and colony forming units (CFU). To evaluate biofilm formation on abiotic surfaces such as polystyrene or PVC, microtiter plates were used. For evaluation of biofilm on the glass surface, sterile borosilicate cover slips were added in petri plates (BD, Falcon, USA). Cells were added either to sterile petri plates or to wells of polystyrene or PVC microtiter plates. The plates were incubated at room temperature for the duration of the experiment. Media was periodically removed every 24 hours by washing with 20 ml of 1× phosphate buffer saline (PBS) (pH 7.4) and fresh media was added with appropriate antibiotics. Crystal Violet staining was performed from cells grown either in polystyrene or PVC microtiter plates after forty-eight hours incubation at room temperature. Cover slips or wells were thoroughly washed with 1× PBS. After washing, cover slips were dry-fixed for 1 hour at 60°C. 0.1% Crystal Violet (SIGMA Chemicals, MO, USA) dissolved in a mixture (1∶1∶18) of isopropanol: ethyl alcohol: PBS (pH 7.4) was added either to the wells or to the cover slips and allowed to stand at room temperature for 10 min. Excess crystal violet was then completely removed by washing at least twice with PBS. The coverslips were allowed to dry, broken with a glass cutter and taken into 1.5 ml microfuge tubes. To dissolve the crystal violet dye, 33% acetic acid was added and OD570 value was measured with appropriate dilution. Three replicates were used per strain and experiments were repeated three times.
Assays for fim switch orientation
Assays for determining the orientation of fim switch were performed as described earlier [38], [39]. In brief, after isolation of chromosomal DNA, 1 µg genomic DNA was used as a template to determine the “ON” and “OFF” phase respectively by using two sets of primer pairs, respectively (Table 2). In another assay, the fimA promoter element encompassing invertible region was amplified by PCR method. Equal amount of fimA promoter DNA was digested by SnaB1 and resolved in 2% agarose gel. The “ON” and “OFF” populations were determined by different sizes of DNA fragment either in PCR amplification or digestion experiment. These experiments were repeated three times.
β-galactosidase assay
The fim-lacZYA constructs were kind gifts from Dr. William R. Schwann [40]. The fim-lacZYA constructs were cloned in single copy plasmid to mimic chromosomal regulation. The plasmids pBB2-1 and pWS124-17 were used to determine the relative level of fimA expression in various mutants and their complements. β-galactosidase assay was performed in accordance with Miller's method [41]. This assay was repeated three times with three replicates per sample.
cDNA synthesis, quantitative RT-PCR, and real time RT-PCR
Quantitative polymerase chain reaction (qPCR) and quantitative real-time polymerase chain reaction (qRT-PCR) were performed according to the manufacturers' instructions. Total RNA was isolated in accordance with RNeasy mini protocol (Qiagen, CA) and eluted in a final 50 µl H2O. In brief, four ml of culture was added to 0.9 ml ice cold stop solution (Phenol∶Ethanol - 1∶19) and centrifuged at 11,000 rpm for 15 min at 4°C. Lyzozyme was used at a final concentration of 1 mg/ml to disrupt the cells. The integrity of RNA was examined by measuring OD260/280 ratio and by denaturing gel electrophoresis. To remove possible DNA contamination, total RNA was further subjected to a rigorous DNase (Turbo DNA-free, Ambion) treatment. For qPCR, the first-strand cDNA was synthesized in a mixture including 5 µg total RNA, Moloney Murine Leukemia Virus reverse transcriptase, Superscript II RnaseH− (Invitrogen, Carlsbad, CA), and 50 ng random hexamers (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. The quality of cDNA synthesis was determined by electrophoresis on 1.2% agarose gels and the concentration was measured by using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE). A 16S ribosomal RNA (rrnA) was used as an internal control. PCR amplification was performed for up to 30 cycles in a 25 µl total volume with Taq polymerase in a Biometra T-Gradient PCR instrument (Biometra, Horsham, PA). At various cycle intervals, a gene-specific and a control reaction tube was removed respectively. Five µl of the reaction products was resolved on a 1.2% agarose gel and the double-stranded DNA (dsDNA) product intensities were quantified using a Bio-Rad Gel Documentation system (Bio-Rad, Hercules, CA). The linear range of amplification for the rrnA gene was from 5 to 15 cycles in all backgrounds. The above set of samples was subjected to the qRT-PCR amplification under the identical condition in a Light Cycler (Roche, Indianapolis, IN) with SYBR Green I PCR Master Mix. The fluorescence signal from SYBR Green intercalation was monitored to quantify dsDNA product formed after each PCR cycle in comparison with a housekeeping gene, rrnA or icd as appropriate. This experiment was repeated three times.
RNA stability assay
Cells were harvested for RNA isolation from late log phase during which CsrA is maximally expressed [42]. Rifampicin (Sigma, Aldrich) was added into the culture medium at a final concentration of 500 µg/ml to inhibit transcription initiation. Samples were harvested just before and after 2.5, 5, 7.5, 10 min after addition of rifampicin. The cells were harvested at 14,000 rpm in a microcentrifuge and frozen in solid CO2-ethanol, within 2 min. Total RNA isolation, cDNA synthesis, and qRT-PCR experiment were performed as described above. The amount of remaining fimA and lrhA mRNA was calculated from the band intensities by normalizing with intensities of housekeeping gene, icd. This experiment was repeated twice.
Swarming assay
Strains were grown 48 hours under static conditions in LB broth with relevant antibiotics for three passages. Swarming assays were performed on LB media containing 0.6% Agar and 0.5% glucose. The cell number was assessed by measuring OD600 and by plating. To initiate swarming, 1 µl of normalized cells were stabbed into the middle of the soft agar plates and the plates were incubated overnight at 37°C. The swarming zone was photographed with an Olympus C765 Ultra zoom camera. This experiment was performed in triplicates.
Image and statistical analysis
DNA quantification from agarose gels was performed by using the software, ImageJ. For fim switch orientation assay, band intensities of various strains were compared with respect to the intensity of the wild-type ON or OFF band, set at 100%. For qRT-PCR, band intensities of strains were compared to that of the wild-type set at one, normalized with a housekeeping gene, rrnA. All statistical analyses were performed by Graphpad Prism. One way analysis of variance followed by Dunnett's test was carried to assess statistical significance. A p value less than 0.05 was considered significant.
Results
Loss of uvrY abolished biofilm production in UPEC
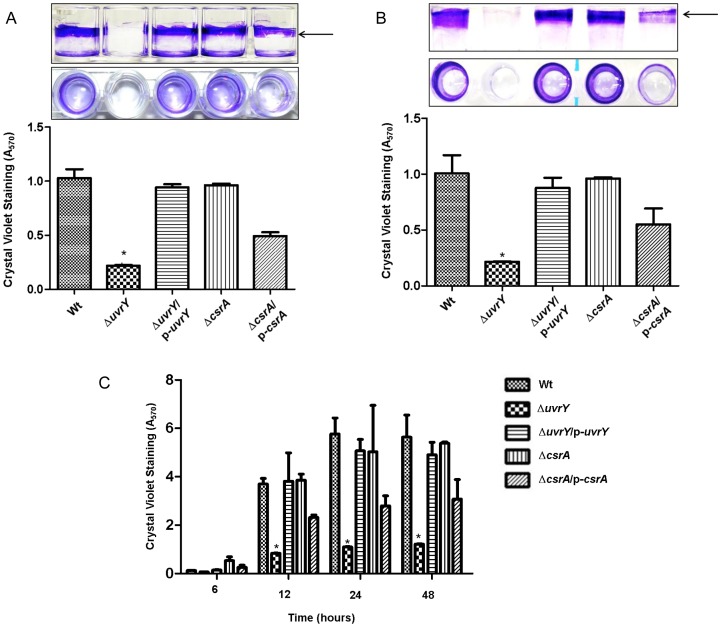
To demonstrate the role of uvrY or csrA in biofilm formation, we deleted the uvrY or csrA gene in UPEC CFT073. To restore the gene function, a complementation experiment was carried out by transforming a plasmid expressing individual gene product. Our results showed that deletion of uvrY in CFT073 abolished biofilm production on abiotic surface such as polystyrene, PVC or borosilicate glass (Fig. 1A, B & C, lane 2), as compared to that of the wild-type (Fig. 1A, B & C, lane 1). Complementation of ΔuvrY in trans restored the biofilm formation to a similar level as that of the wild-type (Fig. 1A, B & C, lane 3). However, the ΔcsrA mutant in CFT073 showed similar levels of the biofilm production as complemented ΔuvrY strain (Fig. 1A, B & C, lane 4). Furthermore, complementation of ΔcsrA reduced biofilm production but not as low as ΔuvrY mutant (Fig. 1A, B & C, lane 5). Loss of uvrY diminished attachment to glass at all-time tested on the glass surface (Fig. 1C, lane 2). These results suggest that the uvrY gene product significantly facilitates the biofilm development, whereas the over-expression of csrA leads to inhibition of biofilm formation in CFT073 strain.
Figure 1. Biofilm formation on various abiotic surfaces in UPEC CFT073.
Crystal violet staining was performed to assess air-liquid biofilm formation on abiotic surfaces such as polystyrene (A) or PVC (B) at 48 hours, whereas crystal violet staining was performed to assess biofilm biomass on glass (C) at 6, 12, 24 and 48 hours. Strains were grown in LB broth with appropriate antibiotics without aeration at room temperature. Bars represent means of three experiments with three replicates per sample. The error bars represent standard errors of three replicates.
Regulation of fimbrial switch by uvrY and csrA in UPEC and E. coli K-12
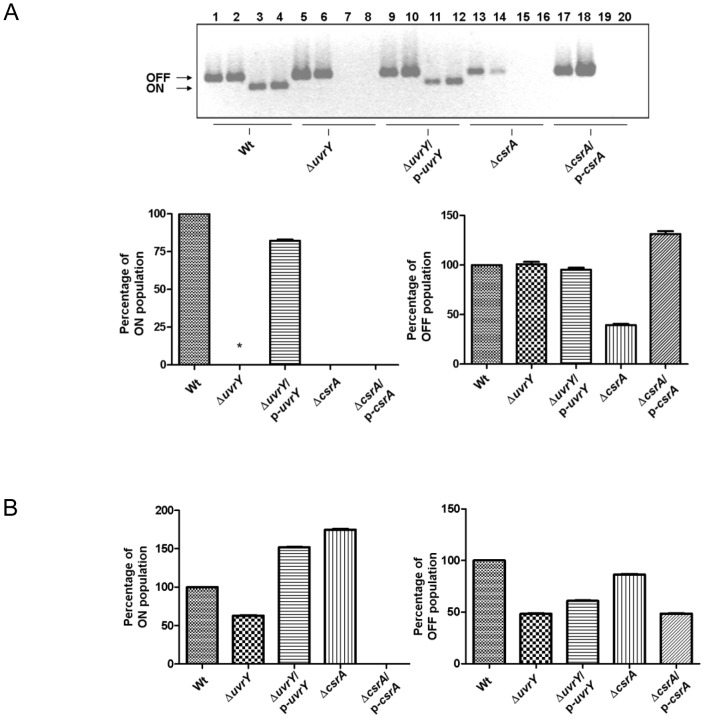
An important attribute of type 1 fimbrial expression is its ability to switch between “ON” and “OFF” phase characterized by fimbriated and afimbriated phase respectively. The switch between phase OFF or ON is mediated by a 314-bp invertible element called fimS, which includes the fimA promoter flanked by 9 bp inverted repeats [43]. Site-specific recombination event alters the orientation of the promoter leading to switch between phase-ON and phase- OFF phase, thus controlling transcription of type 1 fimbriae. Two recombinase, fimB and fimE, and other regulators are involved in the regulation of this genetic switch [44]. FimB mediates interconversion between ON and OFF phase, however, FimE preferentially switches from ON to OFF phase [45]. In addition, environmental signals and DNA topology also play a role in switching the gene orientation [46]. Our data showed that mutation in uvrY reduced the fimbrial population (Fig. 2A & 2B, left panel, lane 2) in UPEC and E. coli K-12. The ON population in uvrY mutant was much lower in UPEC than in K-12. The function of uvrY was further confirmed by complementation whereby the expression of uvrY restored the ON population in both K-12 and CFT073 (Fig. 2A & 2B, left panel, lane 3). Mutation in csrA diminished the ON population in UPEC (Fig. 2A, left panel, lane 4) whereas in K-12, it enhanced the ON population (Fig. 2B, left panel, lane 4). We found that csrA mutant grew slower and exhibited mucoid and smooth colony phenotype. In UPEC, the overexpression of csrA by a plasmid expressing csrA gene did not restore the ON population but in K-12 the ON population was reduced (Fig. 2A & 2B, lane 5). In addition, we used single copy plasmids as well as multi-copy plasmids in our complementation experiments. Most importantly, the mutant strains were sequenced to verify the secondary mutations were around the intended sequences. However, our approach was unable to reveal whether there are mutations elsewhere in the chromosome. Anyway, these results suggest a distinct type 1 fimbrial gene regulation exists between K-12 and UPEC. Loss of uvrY caused a reduction in the OFF population in K-12 but not in UPEC (Fig. 2A & 2B, right panel, lane 2). Complementation of uvrY did not significantly alter OFF population neither in K-12 or CFT073 (Fig. 2A & B, right panel, lane 3). In UPEC, mutation in csrA lowered OFF population while complementation enhanced the OFF population (Fig. 2A, right panel, lanes 4&5) , the effect in K-12 was inverse (Fig. 2A & 2B, right panel, lanes 4&5). Overall, uvrY and csrA affects phase variation of fimbrial switch “ON” or “OFF” although their underlying mechanisms might be different.
Figure 2. Orientation of fim switch in CFT073 (A) and MG1655 (B).
Direction of the fim switch was determined by using a PCR inversion assay. Densitometric analysis was performed to assess relative intensity of bands from agarose gels by using a software, ImageJ. The OFF or ON band intensity of the wild-type was set at 100%. Two independent replicates for each strain were used for this assay. This assay was repeated three times.
Mutation in uvrY influences transcription of adhesins in UPEC or E. coli K-12
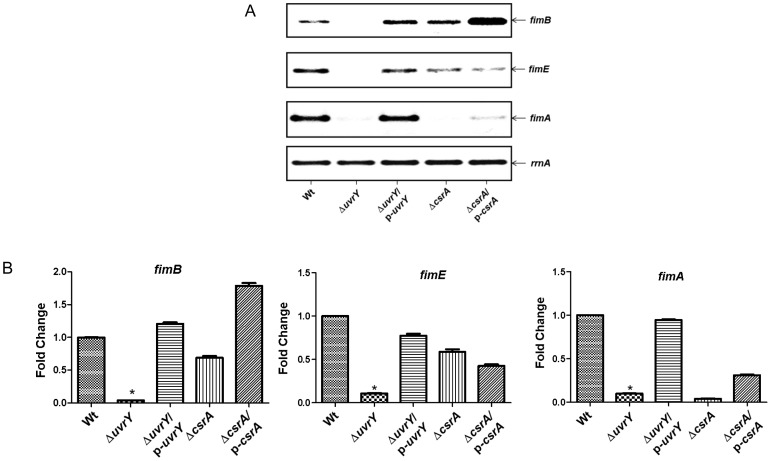
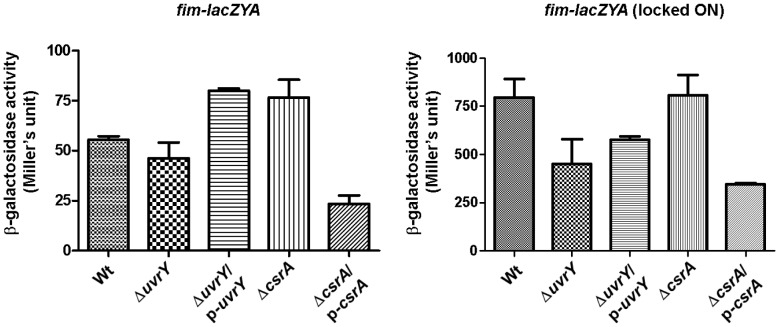
Type 1 fimbriae are crucial for biofilm formation in E. coli. Importantly, type 1 fimbriae are associated with colonization of the bladder [5], [47]. On the other hand, Pap fimbriae (Pyelonephritis associated pilus) are associated with UPEC adherence to epithelial cells and essential for colonization in the kidneys [48]. To determine whether uvrY has an effect on expression of these adhesins, we evaluated the expression of fimA and papA by semi qRT-PCR in UPEC. Mutation in uvrY turned off the transcription of fimA (Fig. 3A & 3B, lane 2) and expression levels of fimA were restored to that of wild-type (Fig. 3A & 3B, lane 1) upon uvrY complementation (Fig. 3A & 3B, lane 3). In addition, the transcription of two recombinase fimB and fimE was undetectable when uvrY was deleted in UPEC (Fig. 3A, lane 2), however, they were restored upon complementation to the level of wild-type (Fig. 3A & B, lane 3). In UPEC, loss of csrA diminished the transcription of fimB and fimA and complementation of the mutant increased the expression level of both transcripts (Fig. 3B). In K-12, a plasmid based lacZ reporter construct fused with the entire fim operon was used to evaluate the effects the expression of fim operon on loss of uvrY or csrA gene. In K-12, uvrY mutation marginally reduced the expression of fimbrial operon reporter construct, while complementation of the mutant increased the expression level higher than that of the wild-type (Fig. 4, lane 2). In addition, deletion of the csrA increased expression of fim-lacZYA in reporter construct and complementation of the mutant reduced the expression (Fig. 4, lane 4 & 5). These data indicate that deletion of the csrA in K-12 exerts a remarkable effect on fim switch orientation.
Figure 3. Effect of uvrY and csrA on fimB, fimE and fimA transcription in UPEC strain CFT073.
A. Transcript levels of fimB, fimE and fimA were evaluated by quantitative RT-PCR. The rrnA was used as an internal control. B. Relative expression levels of various transcripts were calculated by normalization of the actual intensity of bands with rrnA control and expressed as fold change relative to the wild-type. The wild-type was set at one. Bars represent mean ± S.E.M of three replicates.
Figure 4. Expression of fim operon in K-12 strain MG1655.
β-galalactosidase assay was performed in relevant genetic background harboring a single copy plasmid with fimA-lacZYA transcriptional fusions either at invertible or at locked ON orientation. Bars represent means ± SEM of three experiments.
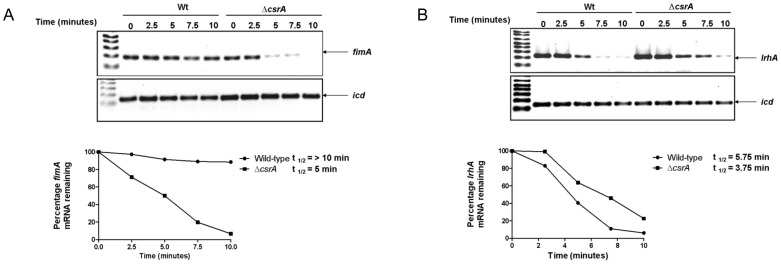
Mutation of csrA decreases mRNA stability of fimA in E. coli K-12
CsrA regulates various target transcripts either positively or negatively by affecting messenger RNA stability [26], [49]. We found that CsrA had a significant effect on the regulation of type 1 fimbriae in E. coli K-12. In contrast, this effect was not significant in CFT073 strain. To understand this effect, we further examined the role of CsrA in affecting mRNA stability of fimA in E. coli K-12. Deletion of csrA destabilized the fimA transcript in E. coli K-12 (Fig. 5A, lanes 6–10). This was evident by fact that the half-life of fimA transcript in wild-type K-12 was greater than 10 min, whereas that of fimA transcript in ΔcsrA mutant was reduced to ∼5 min. LrhA is known to repress type 1 fimbriae expression in E. coli [50]. Under similar conditions, the half-life of lrhA transcript was only mildly increased in the absence of csrA (Fig. 5B, lanes 6–10), but not significantly different from that of wild-type (Fig. 5B, lanes 1–5).
Figure 5. Effect of csrA on message stability of fimA (A) and lrhA (B) in K-12 strain MG1655.
Total RNA was harvested from late log growth phase. The mRNA stability of fimA, lrhA or icd (housekeeping control) was assessed for 10 mins after addition of rifampicin. The relative intensities of the wild-type and the mutant was compared to the intensity of icd mRNA. Normalized transcript levels at time zero prior to rifampicin treatment was set at 100%. This experiment was repeated two times.
Loss of uvrY reduces expression of several virulence specific genes in UPEC
We further evaluated the role of uvrY or csrA in expressing virulence specific genes in UPEC, such as papA encoding the major subunit of P fimbriae, hlyB involved in hemolysin export, and galU, an important factor for biofilm formation. Loss of uvrY completely abolished papA, reduced expression of hlyB and galU genes. The complementation experiment restored their expressions to the levels of wild-type (Fig. 6, lanes 2 & 3). However, loss of csrA did not increase the papA transcript level, whereas complementation of csrA reduced the papA transcript level relative to that of the wild-type. Interestingly, mutation in csrA increased expression of hlyB and galU around 3 and 7 fold respectively and upon complementation, the mutant exhibited a reduced expression of those genes similar to the level of wild-type (Fig. 6, lanes 4 and 5). These results strongly suggest the potential regulatory roles of uvrY and csrA in control of the virulence genes in UPEC.
Figure 6. Relative expression levels of papA, hlyB and galU in UPEC strain CFT073.
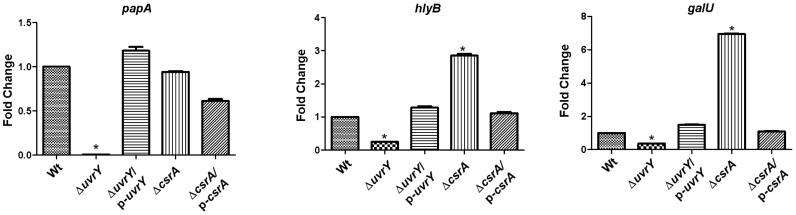
Bacterial strains were grown in LB broth for 48 hours at room temperature. Intensities of bands from agarose gels were subjected to image analysis post semi-quantitative RT-PCR from various strains. Transcript levels of rrnA were used as an internal control. Fold change was represented by the expression levels of genes in various strains relative to the wild-type, set at 100%. Bars represented means ± S.E.M. of three replicates.
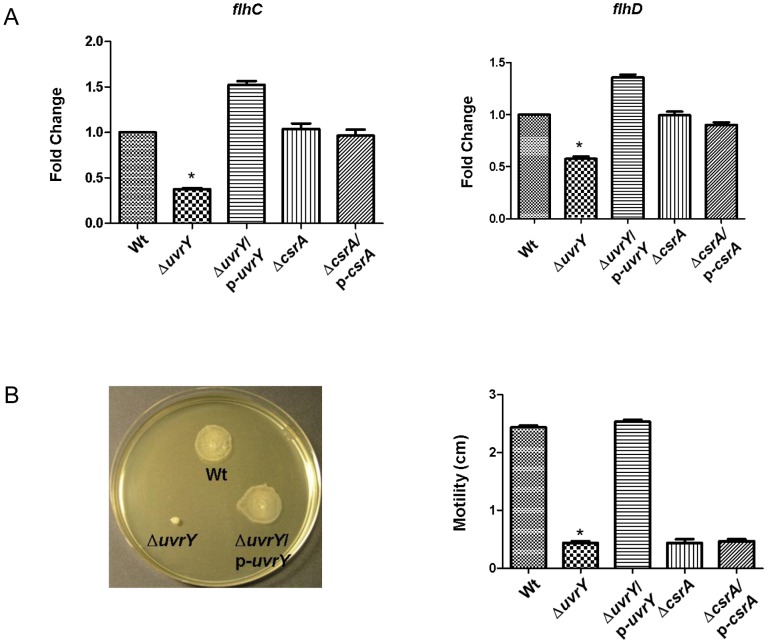
Loss of uvrY affects swarming motility in UPEC
In E. coli, the flagellum is critical for initial attachment and overcoming repulsion between similarly charged bacterial and inert surfaces as well for forming biofilms [51]. Swarming motility is a flagellum-dependent form of bacterial motility that facilitates migration of bacteria on viscous substrates [52]. The swarmer cells undergo differentiation, which is characterized by elongation and an increase in flagellum number and eventual migration of the swarmer cells. In view of swarming motility in facilitating biofilm formation, we further characterized role of uvrY or csrA in regulating the expression of flagellum. In addition, the expression of flagella is dependent upon hierarchy of a cluster of genes, which are controlled by the master regulator, flhDC. Our results showed that loss of uvrY reduced expressions of flhC and flhD in UPEC (Fig. 7A, lane 2); their expression levels were restored to that of the wild-type upon the complementation (Fig. 7A, lane 3) (Fig. 7A, lane 1). Loss of uvrY also reduced the swarming motility in UPEC and swarming was restored to that of wild-type upon complementation (Fig. 7B). These results clearly indicate that uvrY influences swarming motility in UPEC.
Figure 7. Relative expression levels of flhC and flhD transcripts (A) and swarming motility in UPEC strain CFT073 (B).
Relative transcript levels of flhC and flhD was assessed by qRT-PCR with rrnA as an internal control and represented as fold change relative to the wild-type, set at 100%. Swarming motility was assessed on 0.6% LB agar supplemented with 0.5% glucose. The zone of swarming was measured after 16-hour incubation at 37°C. Bars indicate means ± S.E.M. of three replicates.
Furthermore, the csrA deletion mutant in UPEC exhibited marginal effect on transcriptions of flhC and flhD genes. Over-expression of the csrA in the csrA mutant did not exert an effect on the expression levels of both flhC and flhD transcripts. We also noted that UPEC with a csrA deletion mutant were not as motile as the wild-type; the over-expression of the csrA mutant did not repair the defect in swarming motility, probably due to lack of selective pressure for plasmid maintenance in agar plates. However, the effect of CsrA could be examined by over-production in a wild-type strain as well, especially considering that p-csrA failed to complement the motility defect of the csrA mutant.
Discussion
Biofilm formation in E. coli requires a set of gene expressions for facilitating its initiation, attachment and subsequent maturation. The initial attachment is mediated by several adhesins, of which, type 1 and pap fimbriae play a critical role in colonization in urinary bladder and kidneys respectively. We tested the association of the BarA-UvrY TCS in uropathogenesis of UPEC. UPEC rely on its ability to form intrabacterial biofilms in the bladder and in the pods in a polysaccharide based matrix. Such intrabacterial communities have been demonstrated to express type 1 fimbriae, antigen 43 and exopolysaccharides [53]–[56]. Our previous study has demonstrated that uvrY mutation in CFT073 resulted reduced colonization of the bacteria in the bladder, kidneys or in urine as compared to that of wild type [30]. In this study, we further investigated the roles of uvrY and csrA in regulating expression of genes relevant to the biofilm formation in E. coli.
First, we tested the association of the uvrY and csrA genes in forming biofilms of UPEC. Our findings indicated that loss of uvrY in UPEC significantly reduced the bacterial attachment to various abiotic surfaces. We reasoned that the lowered expression of few adhesins might be responsible for this adhesion defect based on our earlier studies with APEC & UPEC [29], [57]. We investigated the role of uvrY and csrA on type 1 fimbriae switch since type 1 fimbriae is an important factor for biofilm formation on various abiotic surfaces such as PVC, polypropylene or borosilicate glass [51]. As type 1 fimbriae is commonly present in both pathogenic and non-pathogenic strains, a comparative study in E. coli K-12 was also performed to further address whether such regulation holds true in both non-pathogenic and pathogenic strains. Our results indicated differential regulation between E. coli K-12 and CFT073 in type 1 fimbriae regulation as mediated by uvrY or csrA.
It becomes clear that uvrY controls transcriptions of fimA and papA which encode the major fimbrial subunit of type 1 and pap fimbriae in UPEC. Both adhesins are important for persistence of UPEC in bladder and kidneys. Furthermore, deletion of uvrY in UPEC also reduces expression of the master regulator, flhC and flhD. Thus mutation in uvrY affects the bacterial persistence in the bladder or kidneys by several possible ways. Down-regulation of both type 1 and pap fimbriae has been shown to affect adhesion and ability to persist in bladder and kidneys, respectively. The uroplakin particles of the apical cell membrane can mediate interaction with type 1 fimbriae for internalization of UPEC. Thus, mutation in uvrY might also compromise the uptake of UPEC into the bladder epithelial cells. Loss of uvrY also turns OFF the type 1 fimbriae and reduced transcription of fimA, suggesting that high percentage of cells in uvrY deletion mutant probably exists in afimbriated phase. These indicate that a successful colonization in the bladder for initiating the biofilm formation might be impaired in an uvrY mutant. Secondly, a mutation of uvrY in UPEC results in hypersensitivity to hydrogen peroxide [23]. Biofilms formed from an uvrY mutant might subsequently be cleared due to the phagocytosis and oxidative burst by the polymorphonuclear leukocytes. In contrast, biofilms from wild-type strain would be difficult to penetrate due to polysaccharide based matrix and protective uroplakin. Finally, flagellar motility might also play a subtle role in the fitness of UPEC, even though flagellar motility may not be absolutely critical for the virulence [58], [59]. In fact, studies have shown that the flagella is down-regulated during UPEC infection, most likely resulting from avoiding activation of toll like receptor-5 (TLR-5) mediated stimulation and release of proinflammatory cytokines such as IL-8. Flagella also promote ascendance of UPEC from the bladder and initiate upper urinary tract infections, particularly in the kidneys [60], [61]. However, transient expression of flagellar motility is thought to be important for initial colonization of UPEC in the urinary tract. Co-challenge experiments with UPEC and flagellar mutants have demonstrated that flagellar motility is important for colonization of UPEC against a flagellar mutant strain and thereby contribute to fitness of UPEC. Thus, mutation in uvrY might affect fitness or persistence of UPEC in bladder, kidneys, urine or formation of intracellular biofilm in the urinary tract. Mutation in uvrY might also compromise the attachment to such abiotic surfaces, leading to the reduced persistence of UPEC in devices such as catheters. Hence uvrY might be a key regulator in the pathogenesis of UPEC in the bladder or kidneys.
CsrA is a global RNA binding protein which regulates several virulence genes in E. coli. The CsrA protein acts on the translation of target genes and influences stability of target mRNA transcripts in positive and negative regulatory manners. Specifically, CsrA could bind at or near shine-dalgarno sequences, thus blocking ribosome loading and facilitating mRNA decay. In contrast, CsrA could also stabilize mRNA by increasing target transcript levels and subsequently increase the translation of the mRNA. Our results indicate a potential novel role of CsrA in regulation of type 1 fimbriae in E. coli. The regulation of type 1 fimbriae by csrA might be different between K-12 and UPEC. The effect of csrA is clearly pronounced in K-12 background as evident by impact of fimA transcript stability in a csrA deletion mutant. Earlier studies have shown that CsrA is a positive regulator of flhDC in E. coli K-12 at different growth phases under aerobic conditions [49]. Interestingly, CsrA has also been shown to negatively regulate flhDC in Erwinia carotovaora [62]. In this study, we focused on the effect of deleting uvrY and csrA on the expression of flhDC in UPEC CFT073 under biofilm inducing conditions. It is possible that the expression pattern of flhDC could be different due to differences in genetic background, growth conditions and composition of LB media, which in our experiments did not contain glucose.
In the BarA-UvrY-CsrA pathway, a deletion of uvrY or csrA would be expected to show opposite phenotypes, as UvrY stimulates the transcription of csrB and csrC non-coding RNA which sequesters free CsrA in the cell. However, in our experiments, we have observed that the deletion mutants of uvrY and csrA in UPEC are both impaired in motility. These results suggest the BarA-UvrY-CsrA pathway might be regulated differently in UPEC. However, our results are similar to a previous study on uvrY orthologue in Salmonella, sirA on motility and virulence [63]. We have also noted that mutation of csrA in UPEC showed growth defect and formed mucoid colonies. The mucoid phenotype could be due to the over-expression of the exopolysaccharides, such as PGA (poly-β-1,6-N-acetyl-D-glucosamine) and others. These phenotypes have been previously reported [64]. However, since the UvrY and CsrA are part of the same pathway, the effect of uvrY on biofilm formation could be affected either directly or indirectly due a downstream effect on CsrA. It is necessary to conduct experiments characterizing the phenotype of uvrY/csrA double mutant or overproduction of uvrY, csrA or both in the different mutant strains in the future.
Our results further indicate that in UPEC, CsrA represses a set of virulence genes such as hemolysin and LPS biosynthetic genes such as galU. This suggests that a csrA deletion mutant might be highly virulent in UPEC. However, CsrA may not play a crucial role in some other UPEC specific fimbrial gene regulation, such as papA, indicating that under certain niches such as kidneys, a csrA deletion mutant might not be as invasive as the wild-type.
Overall there are numerous differences between K-12 and UPEC which may explain differential regulation between these distinct strains. The genome of UPEC is relatively larger than K-12 and numerous UPEC specific genes might influence gene regulation differentially as compared to non-pathogenic strain. E. coli K-12 is a laboratory adapted strain which lost some important virulence genes over several passages such as O-antigen of LPS. The LPS biosynthesis requires several genes; mutations in rfa and galU genes make E. coli K-12 non-pathogenic. The unique virulence, such as hemolysin secretion, presence of a full-length LPS, and ability to facilitate interaction with kidney cells by p fimbriae, enables UPEC to develop biofilm. Importantly, our findings indicate that UPEC specific genes such as papA, galU and hlyB are also modulated by uvrY at the transcriptional level or by csrA at the post-transcriptional level. Therefore, this work highlights the importance of uvrY in the persistence, motility and virulence, all of which contributes to the uropathogenesis. Importantly, we can predict such regulatory role of uvrY in several gram-negative human pathogens, as this gene is conserved.
Acknowledgments
We thank Dr. Adam Coleman for excellent technical assistance in this study.
Funding Statement
This work was in part supported by USDA-NRI-CSREES Competitive Grant 2004-35204-14749, USDA-Animal Health 2002-1106-0195318 and Maryland Agriculture Experimental Station grant from the University of Maryland (to S.M.) and a National Institutes of Health Grant R01 AI65892 (to X.Z.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Katouli M (2010) Population structure of gut Escherichia coli and its role in development of extra-intestinal infections. Iran J Microbiol 2: 59–72. [PMC free article] [PubMed] [Google Scholar]
- 2. Wiles TJ, Kulesus RR, Mulvey MA (2008) Origins and virulence mechanisms of uropathogenic Escherichia coli . Exp Mol Pathol 85: 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belanger L, Garenaux A, Harel J, Boulianne M, Nadeau E, et al. (2011) Escherichia coli from animal reservoirs as a potential source of human extraintestinal pathogenic E. coli . FEMS Immunol Med Microbiol 62: 1–10. [DOI] [PubMed] [Google Scholar]
- 4. Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ (2000) Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. Embo J 19: 2803–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bahrani-Mougeot FK, Buckles EL, Lockatell CV, Hebel JR, Johnson DE, et al. (2002) Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol Microbiol 45: 1079–1093. [DOI] [PubMed] [Google Scholar]
- 6. Melican K, Sandoval RM, Kader A, Josefsson L, Tanner GA, et al. (2011) Uropathogenic Escherichia coli P and Type 1 fimbriae act in synergy in a living host to facilitate renal colonization leading to nephron obstruction. PLoS Pathog 7: e1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eisenstein BI (1981) Phase variation of type 1 fimbriae in Escherichia coli is under transcriptional control. Science 214: 337–339. [DOI] [PubMed] [Google Scholar]
- 8. Eisenstein BI (1988) Type 1 fimbriae of Escherichia coli: genetic regulation, morphogenesis, and role in pathogenesis. Rev Infect Dis 10 Suppl 2: S341–344. [DOI] [PubMed] [Google Scholar]
- 9. Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284: 1318–1322. [DOI] [PubMed] [Google Scholar]
- 10. Hatt JK, Rather PN (2008) Role of bacterial biofilms in urinary tract infections. Curr Top Microbiol Immunol 322: 163–192. [DOI] [PubMed] [Google Scholar]
- 11. Bower JM, Eto DS, Mulvey MA (2005) Covert operations of uropathogenic Escherichia coli within the urinary tract. Traffic 6: 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wright KJ, Hultgren SJ (2006) Sticky fibers and uropathogenesis: bacterial adhesins in the urinary tract. Future Microbiol 1: 75–87. [DOI] [PubMed] [Google Scholar]
- 13. Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, et al. (2003) Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301: 105–107. [DOI] [PubMed] [Google Scholar]
- 14. Ferrieres L, Hancock V, Klemm P (2007) Specific selection for virulent urinary tract infectious Escherichia coli strains during catheter-associated biofilm formation. FEMS Immunol Med Microbiol 51: 212–219. [DOI] [PubMed] [Google Scholar]
- 15. Harkes G, Dankert J, Feijen J (1992) Growth of uropathogenic Escherichia coli strains at solid surfaces. J Biomater Sci Polym Ed 3: 403–418. [DOI] [PubMed] [Google Scholar]
- 16. Stamm WE (1991) Catheter-associated urinary tract infections: epidemiology, pathogenesis, and prevention. Am J Med 91: 65S–71S. [DOI] [PubMed] [Google Scholar]
- 17. Chenoweth CE, Saint S (2011) Urinary tract infections. Infect Dis Clin North Am 25: 103–115. [DOI] [PubMed] [Google Scholar]
- 18. Foxman B (2002) Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am J Med 113 Suppl 1A: 5S–13S. [DOI] [PubMed] [Google Scholar]
- 19. Mizuno T (1998) His-Asp phosphotransfer signal transduction. J Biochem 123: 555–563. [DOI] [PubMed] [Google Scholar]
- 20. Jung K, Fried L, Behr S, Heermann R (2012) Histidine kinases and response regulators in networks. Curr Opin Microbiol 15: 118–124. [DOI] [PubMed] [Google Scholar]
- 21. Pernestig AK, Georgellis D, Romeo T, Suzuki K, Tomenius H, et al. (2003) The Escherichia coli BarA-UvrY two-component system is needed for efficient switching between glycolytic and gluconeogenic carbon sources. J Bacteriol 185: 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suzuki K, Wang X, Weilbacher T, Pernestig AK, Melefors O, et al. (2002) Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli . J Bacteriol 184: 5130–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pernestig AK, Melefors O, Georgellis D (2001) Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli . J Biol Chem 276: 225–231. [DOI] [PubMed] [Google Scholar]
- 24. Lucchetti-Miganeh C, Burrowes E, Baysse C, Ermel G (2008) The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology 154: 16–29. [DOI] [PubMed] [Google Scholar]
- 25. Romeo T (1998) Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol 29: 1321–1330. [DOI] [PubMed] [Google Scholar]
- 26. Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, et al. (2005) CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli . Mol Microbiol 56: 1648–1663. [DOI] [PubMed] [Google Scholar]
- 27. Timmermans J, Van Melderen L (2010) Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci 67: 2897–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang H, Liu MY, Romeo T (1996) Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J Bacteriol 178: 1012–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Herren CD, Mitra A, Palaniyandi SK, Coleman A, Elankumaran S, et al. (2006) The BarA-UvrY two-component system regulates virulence in avian pathogenic Escherichia coli O78:K80:H9. Infect Immun 74: 4900–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Palaniyandi S, Mitra A, Herren CD, Lockatell CV, Johnson DE, et al. (2012) BarA-UvrY two-component system regulates virulence of uropathogenic E. coli CFT073. PloS one 7: e31348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Skyberg JA, Johnson TJ, Johnson JR, Clabots C, Logue CM, et al. (2006) Acquisition of avian pathogenic Escherichia coli plasmids by a commensal E. coli isolate enhances its abilities to kill chicken embryos, grow in human urine, and colonize the murine kidney. Infect Immun 74: 6287–6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kariyawasam S, Scaccianoce JA, Nolan LK (2007) Common and specific genomic sequences of avian and human extraintestinal pathogenic Escherichia coli as determined by genomic subtractive hybridization. BMC Microbiol 7: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, et al. (1990) Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 58: 1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guyer MS, Reed RR, Steitz JA, Low KB (1981) Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harbor symposia on quantitative biology 45 Pt 1: 135–140. [DOI] [PubMed] [Google Scholar]
- 35. Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wei B, Shin S, LaPorte D, Wolfe AJ, Romeo T (2000) Global regulatory mutations in csrA and rpoS cause severe central carbon stress in Escherichia coli in the presence of acetate. J Bacteriol 182: 1632–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prouty AM, Schwesinger WH, Gunn JS (2002) Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect Immun 70: 2640–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lim JK, Gunther NWt, Zhao H, Johnson DE, Keay SK, et al. (1998) In vivo phase variation of Escherichia coli type 1 fimbrial genes in women with urinary tract infection. Infect Immun 66: 3303–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bryan A, Roesch P, Davis L, Moritz R, Pellett S, et al. (2006) Regulation of type 1 fimbriae by unlinked FimB- and FimE-like recombinases in uropathogenic Escherichia coli strain CFT073. Infect Immun 74: 1072–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schwan WR, Lee JL, Lenard FA, Matthews BT, Beck MT (2002) Osmolarity and pH growth conditions regulate fim gene transcription and type 1 pilus expression in uropathogenic Escherichia coli . Infect Immun 70: 1391–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller JH (1992) A short course in bacterial genetics: A laboratory manual and handbook for Escherichia coli and related Bacteria: Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42. Gudapaty S, Suzuki K, Wang X, Babitzke P, Romeo T (2001) Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli . J Bacteriol 183: 6017–6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Abraham JM, Freitag CS, Clements JR, Eisenstein BI (1985) An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli . Proc Natl Acad Sci U S A 82: 5724–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klemm P (1986) Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli . Embo J 5: 1389–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McClain MS, Blomfield IC, Eisenstein BI (1991) Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli . J Bacteriol 173: 5308–5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gally DL, Bogan JA, Eisenstein BI, Blomfield IC (1993) Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J Bacteriol 175: 6186–6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Antao EM, Wieler LH, Ewers C (2009) Adhesive threads of extraintestinal pathogenic Escherichia coli . Gut Pathog 1: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lane MC, Mobley HL (2007) Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int 72: 19–25. [DOI] [PubMed] [Google Scholar]
- 49. Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, et al. (2001) Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli . Mol Microbiol 40: 245–256. [DOI] [PubMed] [Google Scholar]
- 50. Blumer C, Kleefeld A, Lehnen D, Heintz M, Dobrindt U, et al. (2005) Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli . Microbiology 151: 3287–3298. [DOI] [PubMed] [Google Scholar]
- 51. Pratt LA, Kolter R (1998) Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30: 285–293. [DOI] [PubMed] [Google Scholar]
- 52. Harshey RM (1994) Bees aren't the only ones: swarming in gram-negative bacteria. Mol Microbiol 13: 389–394. [DOI] [PubMed] [Google Scholar]
- 53. Goller CC, Seed PC (2010) Revisiting the Escherichia coli polysaccharide capsule as a virulence factor during urinary tract infection: contribution to intracellular biofilm development. Virulence 1: 333–337. [DOI] [PubMed] [Google Scholar]
- 54. Anderson GG, Goller CC, Justice S, Hultgren SJ, Seed PC (2010) Polysaccharide capsule and sialic acid-mediated regulation promote biofilm-like intracellular bacterial communities during cystitis. Infect Immun 78: 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wright KJ, Seed PC, Hultgren SJ (2007) Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol 9: 2230–2241. [DOI] [PubMed] [Google Scholar]
- 56. Justice SS, Hung C, Theriot JA, Fletcher DA, Anderson GG, et al. (2004) Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc Natl Acad Sci U S A 101: 1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitra A, Chakraborti N, Mukhopadhyay S (2008) Escherichia coli stress response as a tool for detection of toxicity. In: Sahu SC, editor. Toxicogenomics: A powerful tool for toxicity assessment. Chichester, UK: John Wiley & Sons, Ltd. pp. 199–210.
- 58. Lane MC, Lockatell V, Monterosso G, Lamphier D, Weinert J, et al. (2005) Role of motility in the colonization of uropathogenic Escherichia coli in the urinary tract. Infect Immun 73: 7644–7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wright KJ, Seed PC, Hultgren SJ (2005) Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect Immun 73: 7657–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schwan WR (2008) Flagella allow uropathogenic Escherichia coli ascension into murine kidneys. Int J Med Microbiol 298: 441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lane MC, Alteri CJ, Smith SN, Mobley HL (2007) Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A 104: 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chatterjee A, Cui Y, Chakrabarty P, Chatterjee AK (2010) Regulation of motility in Erwinia carotovora subsp. carotovora: quorum-sensing signal controls FlhDC, the global regulator of flagellar and exoprotein genes, by modulating the production of RsmA, an RNA-binding protein. Mol Plant Microbe Interact 23: 1316–1323. [DOI] [PubMed] [Google Scholar]
- 63. Teplitski M, Al-Agely A, Ahmer BM (2006) Contribution of the SirA regulon to biofilm formation in Salmonella enterica serovar Typhimurium. Microbiology 152: 3411–3424. [DOI] [PubMed] [Google Scholar]
- 64. Jonas K, Edwards AN, Ahmad I, Romeo T, Romling U, et al. (2010) Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium. Environ Microbiol 12: 524–540. [DOI] [PMC free article] [PubMed] [Google Scholar]