Abstract
Recently, EDI3 was identified as a key factor for choline metabolism that controls tumor cell migration and is associated with metastasis in endometrial carcinomas. EDI3 cleaves glycerophosphocholine (GPC) to form choline and glycerol-3-phosphate (G3P). Choline is then further metabolized to phosphatidylcholine (PtdC), the major lipid in membranes and a key player in membrane-mediated cell signaling. The second product, G3P, is a precursor molecule for several lipids with central roles in signaling, for example lysophosphatidic acid (LPA), phosphatidic acid (PA) and diacylglycerol (DAG). LPA activates intracellular signaling pathways by binding to specific LPA receptors, including membrane-bound G protein-coupled receptors and the intracellular nuclear receptor, PPARγ. Conversely, PA and DAG mediate signaling by acting as lipid anchors that bind and activate several signaling proteins. For example, binding of GTPases and PKC to PA and DAG, respectively, increases the activation of signaling networks, mediating processes such as migration, adhesion, proliferation or anti-apoptosis—all relevant for tumor development. We present a concept by which EDI3 either directly generates signaling molecules or provides “membrane anchors” for downstream signaling factors. As a result, EDI3 links choline metabolism to signaling activities resulting in a more malignant phenotype.
Keywords: glycerophosphodiesterase, phosphatidic acid, lysophosphatidic acid, choline metabolism, signaling networks
Key Role of EDI3 in Choline Metabolism: The Missing Link in the Kennedy Pathway
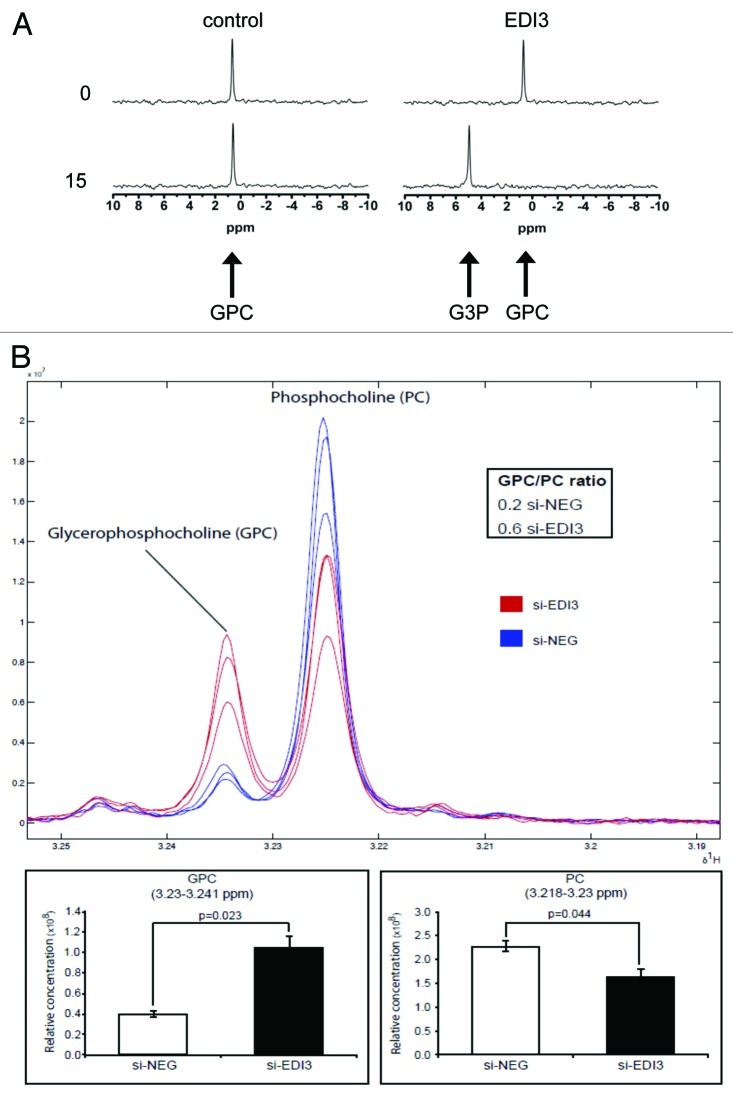
In a screening set of endometrial carcinomas consisting of pairs of tumor tissue with identical histopathological features, which later did or did not form metastasis, we identified three differentially expressed genes. However, only one—EDI3—was confirmed to be positively associated with metastasis in independent sets of carcinomas.1 EDI3 showed a 99% homology to glycerophosphodiesterase 5 (GDE5), a so-far poorly characterized member of the GDE protein family. No other member of the family was, at the time, implicated in tumor development. As an initial step to understand the function of EDI3, we tried to identify EDI3′s preferred substrate. Therefore, possible candidate compounds were incubated with both lysates from cells overexpressing EDI3 and recombinant EDI3 protein, resulting in efficient cleavage of glycerophosphocholine (GPC) to form choline and glycerol-3-phosphate (G3P) (Fig. 1A). To analyze whether EDI3 also influences intracellular concentrations of choline metabolites, we performed siRNA knockdowns in several tumor cell lines (MCF7, MDA-MB-231 and AN3-CA). Knockdown of EDI3 always led to increased intracellular levels of the substrate, GPC and decreased levels of phosphocholine (PC) (Fig. 1B), formed from the phosphorylation of choline by choline kinase and representing the first step of the Kennedy pathway (Fig. 2). Production of choline from the cleavage of GPC provides an important source of choline needed for several downstream signaling pathways, including the synthesis of various phospholipids, such as the Kennedy pathway (Fig. 2). However, the enzyme responsible for the cleavage remained unknown. Our experiments using specific siRNA oligos to knockdown EDI3, together with EDI3 enzymatic assays with purified protein, demonstrated that EDI3 is the responsible enzyme providing choline for the first step of the Kennedy pathway.
Figure 1. EDI3 cleaves glycerophosphocholine (GPC) to choline and glycerol-3-phosphate (G3P). (A) Activity of the purified EDI3 protein which metabolizes GPC to form G3P. Additional generation of choline has been demonstrated by an enzyme-coupled spectrophotometric assay. (B) Knockdown of EDI3 in MCF-7 cells increases intracellular concentrations of the substrate GPC and decreases phosphocholine (PC), which is formed from the product, choline (from Stewart et al., 2012).1
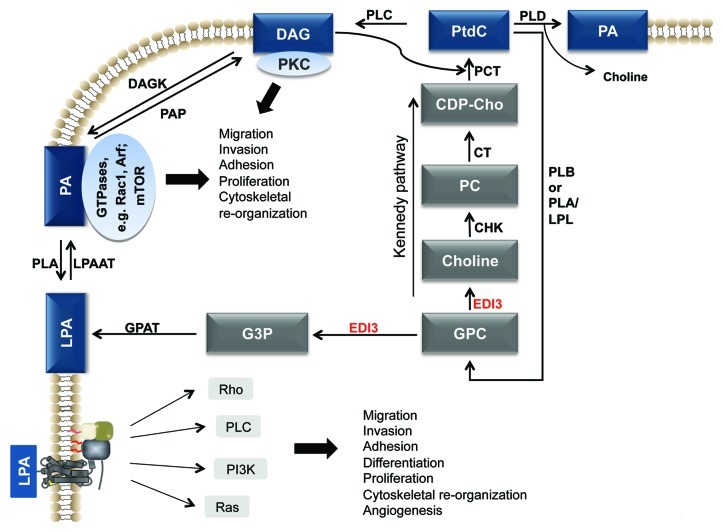
Figure 2. Concept of the link between choline metabolism and intracellular signaling. A proposed concept of how EDI3 links choline metabolism to altered signaling network activities. Choline is further metabolized to the major membrane lipid PtdC. Tumor cells have a higher demand for this lipid to facilitate restructuring of the cell membrane for division and migration. G3P fuels the G3P-LPA-PA-DAG pathway which creates anchoring points for GTPases and PKC and activates intracellular signaling (from Marchan et al., 2012). Abbreviations: CDP-Cho, cytidine 5′-diphosphocholine; CHK, choline kinase; CT, CDP, phosphocholine cytidyltransferase; DAG, diacylglycerol; DAGK, DAG kinase; G3P, glycerol-3-phosphate; LPL, lysophospholipase; GPAT, G3P acyltransferase; LPAAT, LPA acyltransferase; PAP, PA phosphatase; PCT, diacylglycerol choline phosphotransferase; PLA, phospholipase A; PLB, phospholipase B; PLC, phospholipase C; PLD, phospholipase D; PtdC, phosphatidylcholine (from Marchan et al., 2012).22
EDI3 Controls Tumor Cell Migration via PKC Signaling
To identify possible consequences of altered choline metabolism, we tested various cell lines (MCF7, MDA-MB-231 and AN3-CA) after EDI3 siRNA knockdown, using only conditions where EDI3 activity was reduced by approximately 70%. Decreased EDI3 expression and activity did not influence most of the analyzed endpoints, including proliferation and apoptosis. However, in all analyzed cell lines, a decrease in EDI3 activity led to reduced cell migration.1 A general phosphodiesterase inhibitor that was identified after screening with purified protein as capable of inhibiting EDI3 caused a similar effect. Accordingly, overexpression of EDI3 in MCF-7 cells increased migration activity. To identify the mechanism by which EDI3 influences cell migration, we tested numerous possible signaling pathways. Interestingly, only PKC was functionally relevant in both MCF-7 and AN3-CA cells. EDI3 siRNA decreased PKCα protein levels with the opposite effect observed in cells overexpressing EDI3. Activation or inhibition of PKCα caused increased or decreased migration, respectively. Moreover, siRNA knockdown of PKCα in EDI3-overexpressing cells countered the increased migration rate due to EDI3 overexpression.
Contribution of EDI3′s Downstream Metabolites to Cellular Signaling
Signaling via lysophosphatidic acid
Many open questions remain from our original work with EDI3, including how to resolve EDI3′s activity with its downstream signaling and metastatic phenotype. EDI3 was shown to influence lipids important for signaling, such as LPA, PA and DAG. LPA is a ubiquitous single-chain phospholipid that acts both as an intermediate for the de novo synthesis of glycerolipids and, extracellularly, as a lipid mediator that binds and activates specific G protein-coupled receptors. This powerful signaling molecule mediates downstream signaling pathways via Rho, Cdc42 and Rac GTPases, PI3K-Akt, Ras-MAPK, phospholipase C and PKC to control cellular migration, invasion, proliferation, differentiation, cytoskeletal reorganization, angiogenesis, inflammation and resistance to apoptosis.2,3 LPA arises from several metabolic pathways in the cell. Phospholipids are hydrolyzed by the sequential activity of phospholipase D (PLD) followed by phospholipase A2 (PLA2), to form LPA. Alternatively, lysophosphatidylcholine is cleaved by the ectoenzyme, autotaxin, which has phospholipase D activity to form extracellular LPA. Perhaps most specific for EDI3 is the direct acylation of G3P, EDI3′s enzymatic product, by G3P acyltransferase to form LPA. LPA formed via the latter pathway has recently been shown to activate the peroxisome proliferator activated receptor-γ (PPARγ).4 PPARγ binds to its PPAR response elements (PPRE) found on several genes with diverse cellular functions, such as fatty acid metabolism, adipocyte differentiation, lipid transport and proliferation.5
The concept of membrane anchors resulting from choline metabolism
Numerous signaling proteins require anchoring to membrane lipids in order to achieve full activity. Membrane binding leads to downstream signaling that regulates critical processes for tumor development, such as proliferation, migration, adhesion and apoptosis. Possible membrane anchors relevant for EDI3 include DAG and PA. Like LPA, DAG and PA can be directly formed from the hydrolysis of PtdC molecules (Fig. 2), which, in addition to providing lipid signaling, are also necessary to facilitate restructuring of the cell membrane for division and migration, both critical during tumor development.
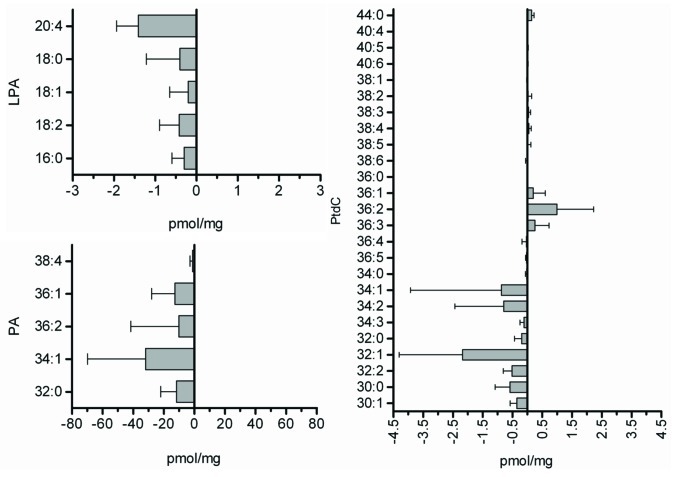
Knocking down EDI3 in several cell lines decreased PKCα, which is activated upon binding to DAG and the anionic phospholipid, phosphatidylserine (PS). The binding of PKC to these lipids frees PKC’s substrate binding site of a pseudosubstrate, facilitating the binding and activation of PKC’s true targets. PKC binds DAG and PS via its N-terminal C1 domain. A ring of positive residues at the middle of the domain facilitates binding to PS and other anionic phospholipids. The interaction between the positive residues of the C1 domain and anionic phospholipids repositions the C1 domain, allowing it to insert itself into the membrane bilayer and bind DAG.6 This binding is facilitated by a groove of hydrophobic residues at the upper third of the C1 domain, and the strength of binding is determined by a conserved residue at position 22.7 Although DAG was not directly measured in our study, it is directly formed from the de-phosphorylation of PA by PA phosphatase. Alternatively, DAG can be produced from the hydrolysis of PtdC, many species of which were altered by EDI3 knockdown (Fig. 3).
Figure 3. Knockdown of EDI3 in MCF7 cells decreases the concentrations of lysophosphatidic acid (LPA) and phosphatidic acid (PA) species. Moreover, the levels of most phosphatidylcholine (PtdC) species are reduced (from Stewart et al., 2012).1
PA is a backbone lipid for many membrane phospholipids. In addition to its importance as a precursor molecule in phospholipid synthesis and membrane curvature, PA is increasingly recognized for its role as a potent regulator of intracellular signaling, influencing cell growth and proliferation, cytoskeletal reorganization, migration and invasion and membrane trafficking.8,9 Knocking down EDI3 in our study decreased all species of PA measured. PA is generated via various reactions in the cell, and perhaps most relevant for EDI3 is the acylation of LPA, a direct product of EDI3′s G3P product. Alternative pathways include the hydrolysis of PtdC by phospholipase D to form PA and choline and the phosphorylation of DAG by DAG kinase. PLD activity is upregulated in many cancers;10-12 therefore, PLD is intensely studied for its potential as a therapeutic target.12
PA’s activity and importance in signaling is centered on its ability to bind and activate several signaling proteins, including members of the Ras superfamily of GTPases, such as the small GTPase, Rac1,13 the guanine nucleotide exchange factor, Epac114 and the Ras exchange factor, Sos.15 PA has been also shown to bind mTOR8,16 and Raf,17,18 both activators of cellular survival pathways. Many of PA’s targets are mutated or disregulated in cancer, making it necessary to understand the mechanisms underlying the interaction between this lipid anchor and the protein targets. To facilitate anchoring, fatty acids, such as palmitate or myristate, become covalently attached to the protein. Such binding helps the protein insert into the membrane alongside the fatty acid tails of the lipid bilayer, thereby also drastically increasing its effective concentration. Interestingly, the degree, kinetics and strength of membrane anchoring depends both on the “fatty acid anchor,” i.e., its hydrophobic length and degree of unsaturation, and the property of the membrane lipid bilayer, i.e., its charge density, composition and lateral organization.19 For example, PA was previously shown to activate the small GTPase Rac113 via interaction with Rac1’s isoprenylated C-terminal polybasic motif. The concentration of PA in the membrane was also shown to be important for Rac1-binding, where increasing the fraction of PA between 0.3 and 3% in artificially made membranes (liposomes) improved liposome binding to a purified isoprenylated Flag-V12-Rac1.
Interestingly, the existence of a polybasic stretch, such as that present in the hypervariable region of the small GTPase KRas4B, together with a single lipid anchor, such as a farnesyl residue, induces effective lipid sorting. Such interactions can form membrane-associated nanodomains that could potentially operate as effective, high-fidelity signaling platforms. Conversely, palmitoylated and farnesylated N-Ras proteins partition into the disordered regions of the lipid membrane, concentrating more to the domain boundaries of heterogeneous membranes. Moreover, the G-domain mediates the Ras-membrane interaction by inducing different sets of preferred orientations in the active and inactive state with largely parallel orientation of the majority of the helices with respect to the membrane. Therefore, the distinct localization for the different isoforms, exposing them eventually to different pools of effectors and regulators, coupled with a differential G-domain-membrane orientation, suggests a synergy between this type of recognition motif and the specificity conferred by the anchor region, thereby validating the concept of isoform specificity in Ras.19-21
The role of PA as a “membrane anchor” is of high relevance for the recently discovered role of EDI3 in choline metabolism. EDI3 not only fuels the Kennedy pathway with choline to produce PtdC, but also provides G3P, a precursor of the signaling lipid, LPA, and the membrane anchor, PA (Fig. 2; ref. 22). PA, in turn, is metabolized to DAG—an anchor for PKC.23 Therefore, EDI3 occupies an important position at a metabolic crossroad, where downstream pathways lead to production of the major membrane lipid, PtdC, signaling lipids such as LPA24 and, last but not least, the membrane anchor lipids PA and DAG, required for activation of GTPases and PKC, respectively (Fig. 2). Further work is needed to determine how EDI3′s function as a glycerophosphodiesterase alters downstream signaling and cellular processes.
Choline Metabolism in Relation to Carcinogenesis and Tumor Progression
Although EDI3 was originally discovered in metastasizing endometrial carcinomas,1 our current knowledge on EDI3′s role in tumors in vivo still remains limited. Nevertheless, an overview of the available publications on choline metabolism and tumor development all support an overall increase in choline metabolism, including changes in GPC and PC levels (Table 1 and ref. 22). Controversy still surrounds the role of GPC as a clinical marker.25 Moreover, the role of the absolute levels of GPC, PC and other choline metabolites and their relation to one another remain unclear.22 Nevertheless, strong evidence exists that tumors have increased flux via the Kennedy pathway (Table 1), as shown by preclinical and clinical trials for inhibitors against choline kinase.26 Furthermore, elevated levels of EDI3 in tumors that went on to metastasize further supports an elevated metabolic capacity in cancer cells. However, compared with choline kinase, antagonists of EDI3 would not only block the Kennedy pathway, but also antagonize the pathways leading to the “membrane anchors” PA and DAG, as well as to the signaling lipid LPA. Unfortunately, EDI3 inhibitors with sufficient specificity are not yet available. Development of antagonists with adequate pharmacokinetic properties would certainly support the importance of EDI3 as a target in some cancers. Such antagonists would also allow us to study the consequences of exclusively inhibiting the Kennedy pathway (by choline kinase inhibitors) in comparison to the effect of additional blocking of the G3P-LPA-PA pathway.
Table 1. Overview of studies that analyze GPC, PC and Cho in relation to malignant transformation.
| Key message | Reference |
|---|---|
| Epithelial ovarian cancer cell lines and normal ovary epithelial cells were compared. The cancer cells had higher PC concentrations and a lower GPC/PC ratio compared with the normal cells. |
Iorio et al., 200527 |
| Human liver tumor biopsies showed significant elevation of PC and reduction of GPC compared with histologically normal liver tissue. |
Bell et al., 199328 |
| Choline phospholipid metabolism was compared in cultivated normal human mammary epithelial cells, immortalized, as well as oncogene transformed cells. A “glycerophosphocholine to phosphocholine switch” was observed with immortalization and oncogene transformation. PC and total Cho levels increased with progression from normal to immortalized to oncogene-transformed cells. |
Aboagye and Bhujwalla, 199929 |
| Compared with normal human mammary epithelial cells, breast cancer cell lines show increased PC, increased choline kinase activity and increased choline transport rates. |
Eliyahu et al., 200730 |
| The absolute concentration of all Cho containing compounds (GPC+PC+Cho) was higher in high-grade than low grade gliomas. In low grade gliomas, the signal was largely due to GPC. In high grade gliomas GPC, PC, and Cho contributed similarly. |
Sabatier et al., 199931 |
| Liver tumors and healthy liver tissues were compared. Liver tumors showed increased PC signals and decreased GPC signals. |
Cox et al., 199232 |
| Breast cancer patients (n = 89) received neoadjuvant chemotherapy with epirubicin or paclitaxel. Tumor biopsies were analyzed before and after treatment. Survivors experienced a decrease in choline containing compounds, including GPC and PC. |
Cao et al., 2012b33 |
| Biopsies from 160 breast cancer patients were examined. MR based metabolomics showed differences between hormone receptor positive and negative carcinomas, whereby hormone receptor negative carcinomas had higher levels of GPC, Cho and PC than hormone receptor positive carcinomas. |
Giskeodegard et al., 201034 |
| Altered phospholipid metabolism is observed in tumors, whereby the malignant choline metabolite profile is characterized by low GPC and high PC. The GPC metabolizing enzyme GDPD5 shows higher expression levels in estrogen receptor negative compared with estrogen receptor positive breast carcinomas. |
Cao et al., 2012a35 |
| Mouse tumor model (SCID mice) was established with biopsy tissue from one primary luminal- and one basal-like mammary carcinoma. The tumor tissue of the luminal-like tumor had higher PC but lower GPC compared with the basal-like tumor. |
Moestue et al., 201036 |
| Comparing paired samples of human lung cancer tissue and noninvolved adjacent tissue showed increased PC and GPC in tumor tissue. |
Rocha et al., 201037 |
| Breast cancer tissue and non-involved breast tissue of 16 patients were compared. Increased levels of Cho, PC and GPC were observed in tumor tissue. |
Gribbestad et al., 199938 |
| Prostate cancer has higher levels of PC and GPC compared with normal prostate. |
Ackerstaff et al., 200139 |
| Chronic lymphocytic leukemia lymphocytes (CLL) and normal lymphocytes were compared. CLL had higher PC and higher GPC concentrations. |
Franks et al., 200240 |
| High-grade prostate carcinomas have higher concentrations of PC and GPC than low-grade prostate carcinomas. |
Keshari et al., 201141 |
| Grade 3 astrocytomas had higher concentrations of PC than grade 2 astrocytomas. GPC increased with the proliferation marker Ki-67. |
McKnight et al., 201142 |
| Low and high grade gliomas were compared for GPC and PC concentrations. GPC was the dominant metabolite in high grade gliomas. |
Righi et al., 200943 |
| PC and GPC were higher in prostate cancer compared with benign prostate tissues. |
Swanson et al., 200844 |
| Total choline concentrations were elevated in untreated pediatric brain tumors compared with controls. Moreover, the GPC to PC ratio decreased in the tumors. | Albers et al., 200545 |
The studies support an overall increase in choline metabolism including metabolites of the Kennedy pathway, such as PC. However, depending on the tumor type, there were variable results with respect to the GPC/PC ratio. To understand this discrepancy, it may be important to differentiate between the metabolic flux through the Kennedy pathway and the concentration of GPC. Increased metabolic flux mediated by for example, EDI3 and CHK may decrease GPC concentrations. However, a tumor may evolve further mechanisms to enhance GPC concentrations. This will, in turn, further increase the metabolic flux through the Kennedy pathway, supporting the need to consider the change to choline metabolism as a whole rather than a single metabolite (from Marchan et al., 2012).22
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/22544
References
- 1.Stewart JD, Marchan R, Lesjak MS, Lambert J, Hergenroeder R, Ellis JK, et al. Choline-releasing glycerophosphodiesterase EDI3 drives tumor cell migration and metastasis. Proc Natl Acad Sci USA. 2012;109:8155–60. doi: 10.1073/pnas.1117654109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi JW, Herr DR, Noguchi K, Yung YC, Lee CW, Mutoh T, et al. LPA receptors: subtypes and biological actions. Annu Rev Pharmacol Toxicol. 2010;50:157–86. doi: 10.1146/annurev.pharmtox.010909.105753. [DOI] [PubMed] [Google Scholar]
- 3.Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer. 2003;3:582–91. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- 4.Stapleton CM, Mashek DG, Wang S, Nagle CA, Cline GW, Thuillier P, et al. Lysophosphatidic acid activates peroxisome proliferator activated receptor-γ in CHO cells that over-express glycerol 3-phosphate acyltransferase-1. PLoS One. 2011;6:e18932. doi: 10.1371/journal.pone.0018932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robbins GT, Nie D. PPAR gamma, bioactive lipids, and cancer progression. Front Biosci. 2012;17:1816–34. doi: 10.2741/4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinberg SF. Structural basis of protein kinase C isoform function. Physiol Rev. 2008;88:1341–78. doi: 10.1152/physrev.00034.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dries DR, Gallegos LL, Newton AC. A single residue in the C1 domain sensitizes novel protein kinase C isoforms to cellular diacylglycerol production. J Biol Chem. 2007;282:826–30. doi: 10.1074/jbc.C600268200. [DOI] [PubMed] [Google Scholar]
- 8.Foster DA. Phosphatidic acid signaling to mTOR: signals for the survival of human cancer cells. Biochim Biophys Acta. 2009;1791:949–55. doi: 10.1016/j.bbalip.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez-Cambronero J. New concepts in phospholipase D signaling in inflammation and cancer. ScientificWorldJournal. 2010;10:1356–69. doi: 10.1100/tsw.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang DW, Choi KY, Min S. Phospholipase D meets Wnt signaling: a new target for cancer therapy. Cancer Res. 2011;71:293–7. doi: 10.1158/0008-5472.CAN-10-2463. [DOI] [PubMed] [Google Scholar]
- 11.Noh DY, Ahn SJ, Lee RA, Park IA, Kim JH, Suh PG, et al. Overexpression of phospholipase D1 in human breast cancer tissues. Cancer Lett. 2000;161:207–14. doi: 10.1016/S0304-3835(00)00612-1. [DOI] [PubMed] [Google Scholar]
- 12.Su W, Chen Q, Frohman MA. Targeting phospholipase D with small-molecule inhibitors as a potential therapeutic approach for cancer metastasis. Future Oncol. 2009;5:1477–86. doi: 10.2217/fon.09.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chae YC, Kim JH, Kim KL, Kim HW, Lee HY, Heo WD, et al. Phospholipase D activity regulates integrin-mediated cell spreading and migration by inducing GTP-Rac translocation to the plasma membrane. Mol Biol Cell. 2008;19:3111–23. doi: 10.1091/mbc.E07-04-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Consonni SV, Gloerich M, Spanjaard E, Bos JL. cAMP regulates DEP domain-mediated binding of the guanine nucleotide exchange factor Epac1 to phosphatidic acid at the plasma membrane. Proc Natl Acad Sci USA. 2012;109:3814–9. doi: 10.1073/pnas.1117599109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao C, Du G, Skowronek K, Frohman MA, Bar-Sagi D. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat Cell Biol. 2007;9:706–12. doi: 10.1038/ncb1594. [DOI] [PubMed] [Google Scholar]
- 16.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–5. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh S, Strum JC, Sciorra VA, Daniel L, Bell RM. Raf-1 kinase possesses distinct binding domains for phosphatidylserine and phosphatidic acid. Phosphatidic acid regulates the translocation of Raf-1 in 12-O-tetradecanoylphorbol-13-acetate-stimulated Madin-Darby canine kidney cells. J Biol Chem. 1996;271:8472–80. doi: 10.1074/jbc.271.14.8472. [DOI] [PubMed] [Google Scholar]
- 18.Rizzo MA, Shome K, Vasudevan C, Stolz DB, Sung TC, Frohman MA, et al. Phospholipase D and its product, phosphatidic acid, mediate agonist-dependent raf-1 translocation to the plasma membrane and the activation of the mitogen-activated protein kinase pathway. J Biol Chem. 1999;274:1131–9. doi: 10.1074/jbc.274.2.1131. [DOI] [PubMed] [Google Scholar]
- 19.Weise K, Kapoor S, Denter C, Nikolaus J, Opitz N, Koch S, et al. Membrane-mediated induction and sorting of K-Ras microdomain signaling platforms. J Am Chem Soc. 2011;133:880–7. doi: 10.1021/ja107532q. [DOI] [PubMed] [Google Scholar]
- 20.Kapoor S, Triola G, Vetter IR, Erlkamp M, Waldmann H, Winter R. Revealing conformational substates of lipidated N-Ras protein by pressure modulation. Proc Natl Acad Sci USA. 2012;109:460–5. doi: 10.1073/pnas.1110553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weise K, Triola G, Brunsveld L, Waldmann H, Winter R. Influence of the lipidation motif on the partitioning and association of N-Ras in model membrane subdomains. J Am Chem Soc. 2009;131:1557–64. doi: 10.1021/ja808691r. [DOI] [PubMed] [Google Scholar]
- 22.Marchan R, Stewart JD, Lesjak MS, Keun HC, Schmitz G, Hengstler JG. Untangling the contribution of choline metabolism to the metastatic process. Proc Natl Acad Sci USA. 2012;109 doi: 10.1073/pnas.1209917109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newton AC. Protein kinase C: poised to signal. Am J Physiol Endocrinol Metab. 2010;298:E395–402. doi: 10.1152/ajpendo.00477.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panupinthu N, Lee HY, Mills GB. Lysophosphatidic acid production and action: critical new players in breast cancer initiation and progression. Br J Cancer. 2010;102:941–6. doi: 10.1038/sj.bjc.6605588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moestue SA, Giskeødegård GF, Cao MD, Bathen TF, Gribbestad IS. Glycerophosphocholine (GPC) is a poorly understood biomarker in breast cancer. Proc Natl Acad Sci USA. 2012;109:E2506. doi: 10.1073/pnas.1208226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallego-Ortega D, Gómez del Pulgar T, Valdés-Mora F, Cebrián A, Lacal JC. Involvement of human choline kinase alpha and beta in carcinogenesis: a different role in lipid metabolism and biological functions. Adv Enzyme Regul. 2011;51:183–94. doi: 10.1016/j.advenzreg.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 27.Iorio E, Mezzanzanica D, Alberti P, Spadaro F, Ramoni C, D’Ascenzo S, et al. Alterations of choline phospholipid metabolism in ovarian tumor progression. Cancer Res. 2005;65:9369–76. doi: 10.1158/0008-5472.CAN-05-1146. [DOI] [PubMed] [Google Scholar]
- 28.Bell JD, Cox IJ, Sargentoni J, Peden CJ, Menon DK, Foster CS, et al. A 31P and 1H-NMR investigation in vitro of normal and abnormal human liver. Biochim Biophys Acta. 1993;1225:71–7. doi: 10.1016/0925-4439(93)90124-J. [DOI] [PubMed] [Google Scholar]
- 29.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999;59:80–4. [PubMed] [Google Scholar]
- 30.Eliyahu G, Kreizman T, Degani H. Phosphocholine as a biomarker of breast cancer: molecular and biochemical studies. Int J Cancer. 2007;120:1721–30. doi: 10.1002/ijc.22293. [DOI] [PubMed] [Google Scholar]
- 31.Sabatier J, Gilard V, Malet-Martino M, Ranjeva JP, Terral C, Breil S, et al. Characterization of choline compounds with in vitro 1H magnetic resonance spectroscopy for the discrimination of primary brain tumors. Invest Radiol. 1999;34:230–5. doi: 10.1097/00004424-199903000-00013. [DOI] [PubMed] [Google Scholar]
- 32.Cox IJ, Bell JD, Peden CJ, Iles RA, Foster CS, Watanapa P, et al. In vivo and in vitro 31P magnetic resonance spectroscopy of focal hepatic malignancies. NMR Biomed. 1992;5:114–20. doi: 10.1002/nbm.1940050303. [DOI] [PubMed] [Google Scholar]
- 33.Cao MD, Giskeødegård GF, Bathen TF, Sitter B, Bofin A, Lønning PE, et al. Prognostic value of metabolic response in breast cancer patients receiving neoadjuvant chemotherapy. BMC Cancer. 2012;12:39. doi: 10.1186/1471-2407-12-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giskeødegård GF, Grinde MT, Sitter B, Axelson DE, Lundgren S, Fjøsne HE, et al. Multivariate modeling and prediction of breast cancer prognostic factors using MR metabolomics. J Proteome Res. 2010;9:972–9. doi: 10.1021/pr9008783. [DOI] [PubMed] [Google Scholar]
- 35.Cao MD, Döpkens M, Krishnamachary B, Vesuna F, Gadiya MM, Lønning PE, et al. Glycerophosphodiester phosphodiesterase domain containing 5 (GDPD5) expression correlates with malignant choline phospholipid metabolite profiles in human breast cancer. NMR Biomed. 2012;25:1033–42. doi: 10.1002/nbm.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moestue SA, Borgan E, Huuse EM, Lindholm EM, Sitter B, Børresen-Dale AL, et al. Distinct choline metabolic profiles are associated with differences in gene expression for basal-like and luminal-like breast cancer xenograft models. BMC Cancer. 2010;10:433. doi: 10.1186/1471-2407-10-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rocha CM, Barros AS, Gil AM, Goodfellow BJ, Humpfer E, Spraul M, et al. Metabolic profiling of human lung cancer tissue by 1H high resolution magic angle spinning (HRMAS) NMR spectroscopy. J Proteome Res. 2010;9:319–32. doi: 10.1021/pr9006574. [DOI] [PubMed] [Google Scholar]
- 38.Gribbestad IS, Sitter B, Lundgren S, Krane J, Axelson D. Metabolite composition in breast tumors examined by proton nuclear magnetic resonance spectroscopy. Anticancer Res. 1999;19(3A):1737–46. [PubMed] [Google Scholar]
- 39.Ackerstaff E, Pflug BR, Nelson JB, Bhujwalla ZM. Detection of increased choline compounds with proton nuclear magnetic resonance spectroscopy subsequent to malignant transformation of human prostatic epithelial cells. Cancer Res. 2001;61:3599–603. [PubMed] [Google Scholar]
- 40.Franks SE, Smith MR, Arias-Mendoza F, Shaller C, Padavic-Shaller K, Kappler F, et al. Phosphomonoester concentrations differ between chronic lymphocytic leukemia cells and normal human lymphocytes. Leuk Res. 2002;26:919–26. doi: 10.1016/S0145-2126(02)00035-8. [DOI] [PubMed] [Google Scholar]
- 41.Keshari KR, Tsachres H, Iman R, Delos Santos L, Tabatabai ZL, Shinohara K, et al. Correlation of phospholipid metabolites with prostate cancer pathologic grade, proliferative status and surgical stage - impact of tissue environment. NMR Biomed. 2011;24:691–9. doi: 10.1002/nbm.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKnight TR, Smith KJ, Chu PW, Chiu KS, Cloyd CP, Chang SM, et al. Choline metabolism, proliferation, and angiogenesis in nonenhancing grades 2 and 3 astrocytoma. J Magn Reson Imaging. 2011;33:808–16. doi: 10.1002/jmri.22517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Righi V, Roda JM, Paz J, Mucci A, Tugnoli V, Rodriguez-Tarduchy G, et al. 1H HR-MAS and genomic analysis of human tumor biopsies discriminate between high and low grade astrocytomas. NMR Biomed. 2009;22:629–37. doi: 10.1002/nbm.1377. [DOI] [PubMed] [Google Scholar]
- 44.Swanson MG, Keshari KR, Tabatabai ZL, Simko JP, Shinohara K, Carroll PR, et al. Quantification of choline- and ethanolamine-containing metabolites in human prostate tissues using 1H HR-MAS total correlation spectroscopy. Magn Reson Med. 2008;60:33–40. doi: 10.1002/mrm.21647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Albers MJ, Krieger MD, Gonzalez-Gomez I, Gilles FH, McComb JG, Nelson MD, Jr., et al. Proton-decoupled 31P MRS in untreated pediatric brain tumors. Magn Reson Med. 2005;53:22–9. doi: 10.1002/mrm.20312. [DOI] [PubMed] [Google Scholar]