Abstract
Embryonic stem cells (ESCs) depend on extensive regulatory networks to coordinate their self-renewal and differentiation. The polyamine pathway regulator AMD1 was recently implicated in ESC self-renewal and directed differentiation of ESCs to neural precursor cells (NPCs). The polyamines spermine and spermidine are essential for a wide range of biological processes, and their levels are tightly regulated. Here, we review the polyamine pathway and discuss how it can impact polyamine levels, cellular methylation and hypusinated EIF5A levels. We discuss how it could feed into regulation of ESC self-renewal and directed differentiation. We show that in addition to AMD1, a second rate-limiting enzyme in the polyamine pathway, ODC1, can also promote ESC self-renewal, and that both Amd1 and Odc1 can partially substitute for Myc during cellular reprogramming. We propose that both Amd1 and Odc1 are essential regulators of ESCs and function to ensure high polyamine levels to promote ESC self-renewal.
Keywords: polyamines, embryonic stem cells, iPSC, Amd1, Spermine, Odc1
Introduction
Embryonic stem cells are derived from the inner cell mass of the blastocyst embryo and can both self-renew indefinitely and give rise to all the cell types of the body. They hold huge potential in regenerative medicine. ESCs and their differentiated derivatives could be used to model disease progression, screen for cell type-specific drug toxicity and for replacement cell therapy. More recently, the establishment of somatic cell reprogramming to produce induced pluripotent stem cells (iPSCs) has enabled the generation of disease and patient-specific iPSCs for disease modeling and potentially for therapeutic application.1,2 Recently, significant progress has been made in understanding the molecular networks that regulate the decision of ESCs and iPSCs to self-renew or differentiate to specific lineages.
Many regulators of ESC self-renewal have been described, and a complex network of factors has now been shown to work coordinately to regulate the ESC state. Signaling pathways feed into a complex network of transcription factors that function to regulate the balance between self-renewal and differentiation. Differentiation can be triggered through a variety of pathways, ultimately resulting in dramatic changes to the transcriptome that are orchestrated by a selection of transcription factors and chromatin modifiers.3 The core pluripotency transcription factors, OCT4, SOX2 and NANOG, have been well characterized in ESCs, and their target genes have been described in numerous genome-wide studies. Recently, a large number of additional transcription factors have been shown to have a role in regulating self-renewal and pluripotency.3
In ESCs, transcription factors have much greater access to the chromatin than in somatic cells, which is the result of the chromatin being in a more “open” formation. Chromatin structure is regulated by chromatin remodeling factors, DNA methylation, post-translational modifications of histones and the incorporation of histone variants. Chromatin organization is tightly controlled to allow maintenance of the ESC state or to facilitate differentiation toward specific lineages.4 Histone-modifying complexes function to modify the histone proteins predominantly by methylation or acetylation of certain residues. DNA methylation is dramatically altered on differentiation, and the promoters of pluripotency genes OCT4 and NANOG show reduced methylation in ESCs.5 The polycomb repressive complex 2 (PRC2) is a histone-modifying complex that functions to repress transcription through histone methylation. Inactivation of the PRC complex proteins results in an enhanced ESC state and compromised differentiation, highlighting the essential role these complexes play in ESCs.6,7 The dramatic changes in chromatin structure that occur on ESC differentiation result in the regulation of gene transcription, which will ultimately drive the differentiation process.7 A picture has now arisen of a transcription-focused regulatory module directing the fate of ESCs.
Mouse embryonic stem cells require the cytokine LIF when cultured without feeder cells.8 LIF activates a number of signaling pathways, one of which acts through activation and phosphorylation of STAT3 to maintain ESC self-renewal.9,10 pSTAT3 is a transcription factor that, when activated, dimerizes and translocates to the nucleus to activate transcription of its target genes.11 One of the targets of pSTAT3 is the transcription factor Myc, which subsequently activates a core set of pluripotency genes to promote self-renewal.12 On withdrawal of LIF from ESC cultures, STAT3 is dephosphorylated and Myc mRNA levels drop. In addition, LIF withdrawal results in the phosphorylation of MYC protein, mediated by GSK3B, which targets it for degradation.11,13 Confirming its role as a self-renewal factor, MYC was shown to be able to maintain the ESC state in the absence of LIF when a non-degradable form was overexpressed following LIF withdrawal.13 A stabilized form of MYC is one of only a small number of proteins that have been demonstrated to be able to maintain mouse ESC self-renewal in the absence of LIF.11
While transcription factors have received considerable attention as core regulators of ESCs, more recently, a variety of screening methods have identified new stem cell regulators that function outside the nucleus. miRNAs have been shown to be essential for ESC differentiation, and a number have been shown to be directly involved in the directed differentiation of ESCs to specific lineages.14 An RNAi screen set out to identify novel regulators of ESC self-renewal identified a wide selection of mRNAs that when knocked down, induce ESCs to differentiate. Interestingly, a significant number of these were RNA regulators, implicating post-transcriptional control in self-renewal and differentiation.15 We recently performed a screen to identify mRNAs that are translationally regulated on differentiation of mouse ESCs and identified AMD1 as a new regulator of ESCs.16 AMD1 (adenosylmethionine decarboxylase) is a key regulator of the polyamine synthesis pathway and is widely expressed in eukaryotic cells.17 We showed that Amd1 is essential for mouse ESC self-renewal and can promote the ESC state in the absence of self-renewal factor LIF when overexpressed. In this study, AMD1 was shown to promote high levels of MYC, which is an established self-renewal factor and previously demonstrated target of the polyamine pathway.13,18,19 This was the first demonstration of a role for the polyamine pathway in the regulation of ESC self-renewal.
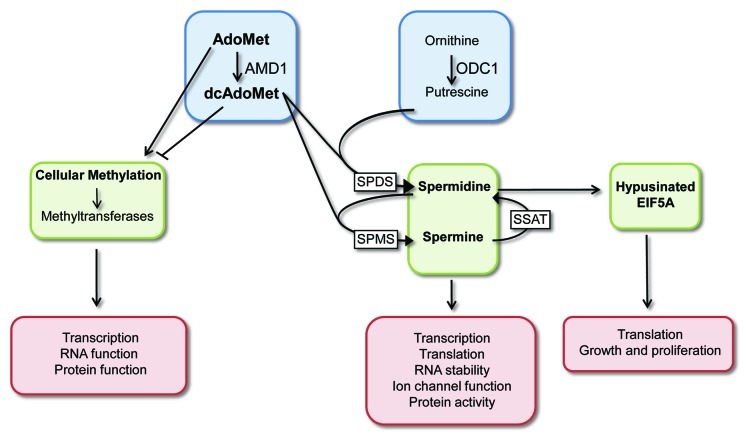
The polyamines, spermine and spermidine, and the diamine putrescine, are organic cations that are found ubiquitously in mammalian cells and have been implicated in a wide range of physiological and pathophysiological processes. Polyamines are essential for growth, and their levels and ratios have been shown to be reduced with age in specific tissues. A large number of diseases and disorders display altered polyamine levels, including Alzheimer, Parkinson, cardiovascular disease and cancer.20-22 In addition, polyamines play essential roles in cellular stress responses.20 Despite their pleiotropic effects, few molecular targets have been defined. It is estimated that the majority of polyamines in the cell are associated with RNA, a smaller portion with DNA, and only a small percentage associates with phospholipids or exists in an unbound state.23 They have been shown to regulate translation of specific mRNAs,24,25 and in intestinal epithelial cells polyamines regulate the RNA binding protein HuR to control target mRNA stability.26,27 Polyamines have also been shown to regulate gene-specific transcription28 and kinase activity.23 The levels of the polyamines are tightly regulated at the level of synthesis, uptake, degradation and secretion.29 The two rate-limiting enzymes in their synthesis are ODC1 and AMD1. ODC1 (ornithine decarboxylase) decarboxylates ornithine to putrescine, which is converted into the higher order polyamines, spermidine and spermine. AMD1 decarboxylates adenosylmethionine (AdoMet) to form dcAdoMet. This functions as the aminopropyl donor for the formation of spermidine and spermine from putrescine (Fig. 1).23 The levels and enzyme activities of AMD1 and ODC1 directly impact the levels of intracellular polyamines, and, as such, they are regulated at the level of transcription, translation, protein degradation and enzyme activity to ensure polyamine levels remain within the required range for a given cell type.30
Figure 1. Overview of the polyamine pathway. AMD1 promotes the conversion of AdoMet to dcAdoMet, and the ratio and levels of these feed into three major downstream pathways: high dcAdoMet results in high spermine and spermidine levels, both of which are implicated in regulation of gene expression and protein function. High spermidine levels result in increased levels of hypusinated EIF5A, which can influence translation, growth and proliferation. AdoMet is the methyl donor for DNA methyltransferases in the cell, and high dcAdoMet functions to inhibit methytransferase activity. Methylation impacts on gene expression control at the RNA and DNA levels and the activity of proteins and phospholipids. ODC1 functions to decarboxylate ornithine to produce putrescine, which is required for the synthesis of spermidine and spermine. Abbreviations: SPMS, spermine synthase; SPDS, spermidine synthase; SSAT, spermidine/spermine N1-acetyltransferase.
Modulation of the levels of active AMD1 will alter the ratio of AdoMet to dcAdoMet in the cell, and this has important consequences for a number of different pathways (Fig. 1). The levels of dcAdoMet are generally significantly lower than the levels of AdoMet, and, once produced, it is directed to the polyamine synthesis pathway.29 Increased dcAdoMet will enable higher levels of spermine and spermidine to be produced, both of which impact on a variety of biological processes. Increased spermidine also promotes the levels of the modified amino acid hypusine that is present in the translation factor EIF5A and is required for its function.31,32 Hypusine is created through the post-translational modification of a lysine residue in EIF5A (Fig. 1). EIF5A is important for growth and protein synthesis and has recently been shown to function as a tumor suppressor.29,33 AMD1 activity has been shown to impact the levels of hypusinated EIF5A, and the addition of an inhibitor to AMD1 resulted in decreased spermidine levels and a decrease in the levels of hypusine-containing EIF5A.34 In addition to being required for the formation of the polyamines, AdoMet functions as the methyl donor for methylation reactions in the cell (Fig. 1). The methyl group from AdoMet is transferred to DNA via the DNA methyltransferases to create m5CpG. In addition to DNA, RNA, proteins and phospholipids are modified by methylation.35 The importance of the ratio of AdoMet to dcAdoMet is highlighted by reports that dcAdoMet, which cannot function as a methyl donor, acts to inhibit methyltransferase activity.36 Methylation of CpG islands in DNA functions to regulate gene expression, and methylation of RNA and protein can influence their function. As a result, fluctuations in the ratio of the methyl donor AdoMet and dcAdoMet in the cell can have dramatic and varied consequences. The extensive feedback loops that exist in the cell to maintain appropriate polyamine levels make interpretation of data from manipulation of ODC1 or AMD1 protein levels or activities complex. Regulation of AMD1 and ODC1 levels have the potential to affect polyamine levels, methylation status and hypusinated EIF5A levels, all of which can influence different biological processes (Fig. 1).
Results and Discussion
Odc1 promotes ESC self-renewal
We recently showed that Amd1 is essential for ESC self-renewal, and that ESCs start to differentiate when its levels are decreased. We also showed that AMD1 levels are decreased to allow NPC differentiation, and that the addition of spermine was sufficient to inhibit accumulation of SOX1, a marker for NPCs.16 To further explore the role of the polyamine pathway in self-renewal, we addressed the role of ODC1 in ESCs. We used siRNAs to knockdown Odc1 in mouse ESCs and saw no effect on self-renewal after a 60% decrease in Odc1 mRNA levels (unpublished data). ESCs may be less sensitive to changes in ODC1 levels, or this could be the result of feedback mechanisms that enhance ODC1 enzyme activity or promote its translation when the mRNA levels are decreased.
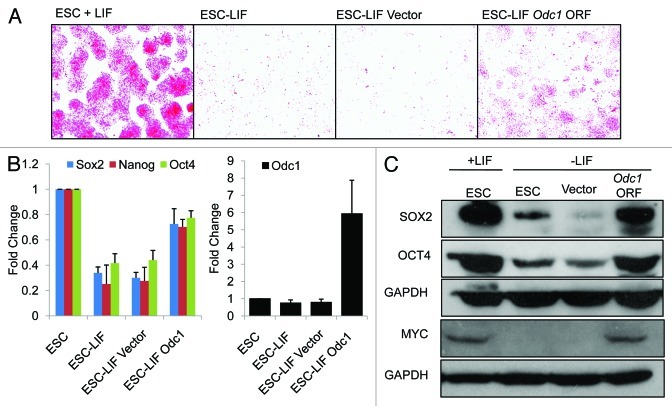
To determine if overexpression of Odc1 can promote self-renewal of ESCs under conditions that promote differentiation, we overexpressed the Odc1 ORF in the pTracer vector in cells that had been induced to differentiate in the absence of LIF. We see that overexpression of Odc1 does promote ESC self-renewal in the absence of LIF as shown by alkaline phosphatase (AP) staining (Fig. 2A). Real-time PCR data shows that self-renewal markers Oct4, Nanog and Sox2 remain elevated when Odc1 is overexpressed in the absence of LIF (Fig. 2B), and western blots show that the protein levels of OCT4 and SOX2 remain high (Fig. 2C). These data demonstrate that overexpression of Odc1 in ESCs can bypass the requirement for LIF in the maintenance of self-renewal. We previously showed that Amd1 overexpression may promote self-renewal in the absence of LIF by promoting high MYC levels.16 Here we show that following withdrawal of LIF, Odc1 overexpression also promoted high levels of MYC protein (Fig. 2C). MYC has been shown to be able to sustain ESC self-renewal in the absence of LIF when a non-degradable form is overexpressed.13 We propose that AMD1 and ODC1 promote ESC self-renewal by enabling high MYC levels in the absence of LIF.
Figure 2. Overexpression of Odc1 can promote the ES cell state in the absence of LIF. (A) Alkaline phosphatase staining of ESCs grown in the absence of LIF with and without Odc1 overexpression for 5 d. ESCs and cells transfected with the empty vector are shown as a control. (B) Real-time PCR showing the maintenance of high levels of Oct4, Sox2 and Klf4 when Odc1 is overexpressed in cells differentiated in the absence of LIF for 5 d. ESCs and cells transfected with the empty vector are shown as a control. (C) Western blot analysis showing high levels of OCT4 and SOX2 on overexpression of Odc1. GAPDH is shown as a loading control.
It is currently unclear how AMD1 and ODC1 regulate MYC in ESCs. The ribosomal load of Myc mRNA in control and Amd1-overexpressing cells was similar, suggesting translation rate was not affected, and Myc mRNA levels were not consistently increased on Amd1 overexpression (unpublished data). On differentiation of ESCs, MYC protein is rapidly degraded following phosphorylation by GSK3B.13 Given the dramatic effect this has on MYC protein levels, it is unlikely regulation at the RNA level would result in the MYC levels observed on Amd1 and Odc1 overexpression. As such, we suggest AMD1 and ODC1 promote high levels of polyamines, which, in turn, function to stabilize MYC protein to promote ESC self-renewal. Polyamines can regulate signaling pathways through modulation of kinase activity. It is possible, therefore, that the polyamines prevent the phosphorylation of MYC protein that signals its degradation by the proteasome, thus promoting ESC self-renewal.
Amd1 and Odc1 enhance iPSC derivation
Induced pluripotent stem cells (iPSCs) possess the self-renewal and pluripotency phenotypes of ESCs and can be derived by reprogramming somatic cells through the expression of a set of core transcription factors Oct4, Sox2, Klf4 and Myc (OSKM).1,37 The efficiency of iPSC conversion, however, is low. Myc is not essential for reprogramming but dramatically increases its efficiency. A link between the polyamine pathway and Myc has been shown in several other systems. The polyamine pathway is required for high levels of Myc mRNA in human colon carcinoma cells19 and has been shown to promote increased Myc translation in rat intestinal epithelial cells.18 Given that ODC1 and AMD1 can promote high MYC levels and ESC self-renewal, we sought to determine if their expression could enhance reprogramming. We proposed that overexpression of AMD1 and ODC1 could substitute for Myc during cellular reprogramming. Mouse fibroblasts were transfected with the reprogramming factors Oct4, Sox2 and Nanog alone or with Odc1 or Amd1. Inclusion of either Amd1 or Odc1 greatly enhanced the efficiency of iPSC generation in the absence of Myc (Fig. 3). Amd1 and Odc1 had no effect on the efficiency of iPSC generation when Myc was included in the reprogramming cocktail (Fig. 3). Given the established link between polyamines and MYC and our demonstration that Amd1 and Odc1 promote high MYC levels in ESCs, we believe that AMD1 and ODC1 function to promote reprogramming by promoting high MYC levels (Fig. 4). Spermidine has been shown to be able to enhance the efficiency of iPSC derivation, further demonstrating the importance of the polyamine pathway in reprogramming.38
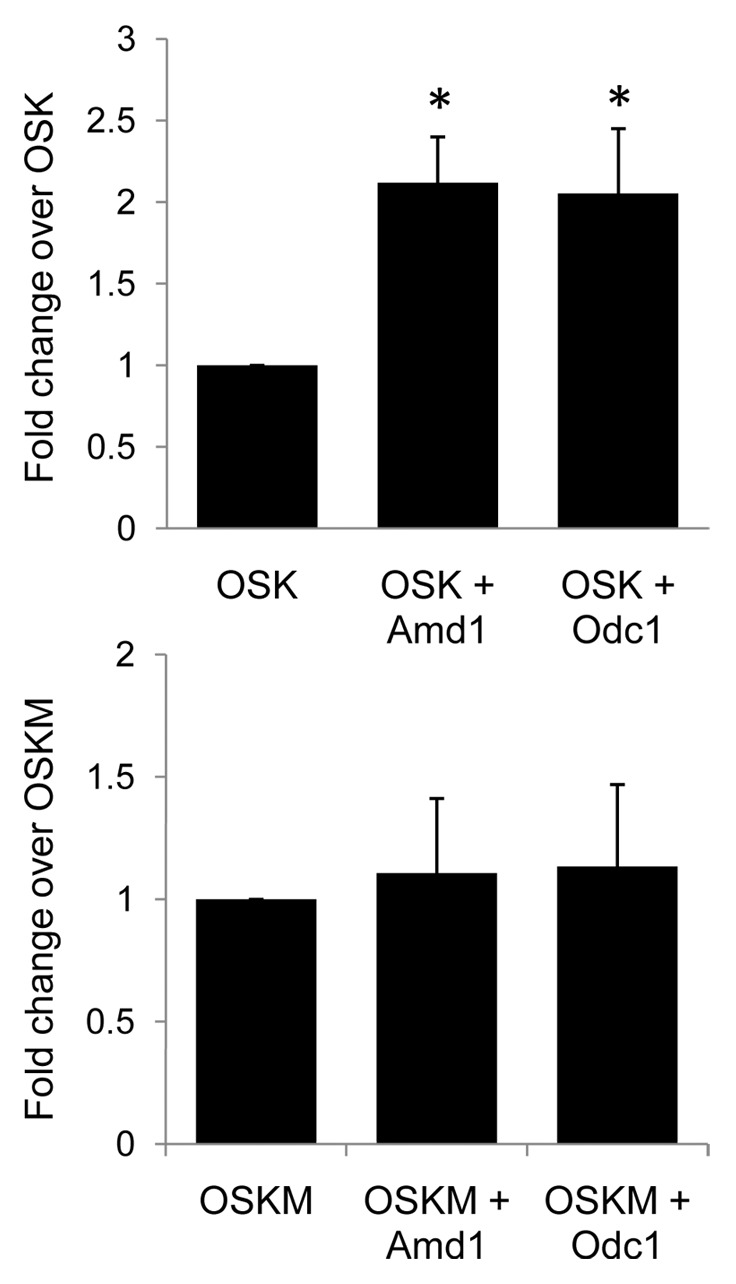
Figure 3.Amd1 and Odc1 enhance the efficiency of iPSC derivation in combination with OSK. (A) iPSCs were generated from mouse fibroblasts using Oct4, Sox2 and Klf4 (OSK) alone or in combination with Odc1 or Amd1. The same was done with Oct4, Sox2, Klf4 and Myc (OSKM). The relative number of Oct4-GFP positive colonies are shown. Values are means +/− SD *p < 0.005.
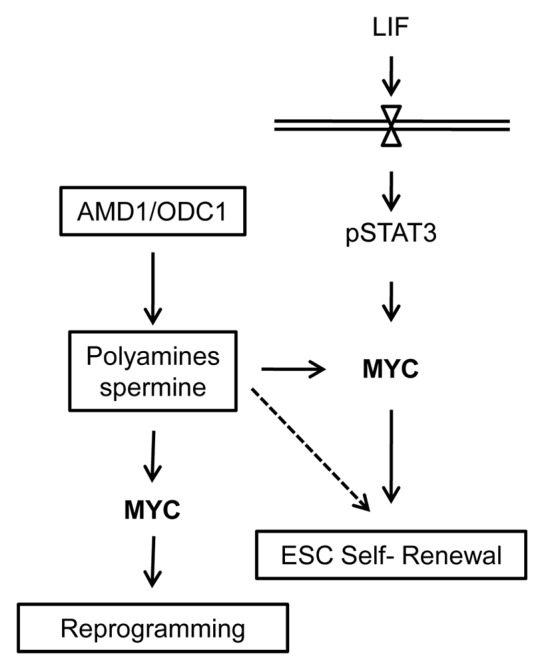
Figure 4. A model showing the role of the polyamine regulators AMD1 and ODC1 in regulating ESC self-renewal and iPSC generation. ODC1 and AMD1 function to regulate the levels of polyamines in ESCs. When expressed at high levels, they promote increased MYC levels to support ESC self-renewal and somatic cell reprogramming. The polyamines likely regulated additional targets in addition to MYC to promote ESC self-renewal (dotted line).
We have demonstrated that both the polyamine regulators AMD1 and ODC1 promote the ESC state in mouse. This confirms earlier observations showing that mice that do not express functional AMD1 cannot survive and embryos die during early gastrulation.39 Embryos without functional ODC1 develop past the blastocyst stage but die just before gastrulation.40 Embryonic stem cells could not be derived from Amd1−/− mice except on the addition of exogenous spermidine.39 Our previous studies showed that the addition of spermine but not spermidine could rescue the Amd1-knockdown phenotype in mouse ESCs.16 This difference is likely the result of the complexity of the polyamine pathway and the ability of cells to readily convert between the polyamines spermine and spermidine (Fig. 1).
We have shown previously that AMD1 can promote ESC self-renewal, and that the addition of spermine can rescue the Amd1-knockdown phenotype.16 We now show that ODC1, when overexpressed, can function in a similar manner, enabling ESCs to grow in the absence of LIF. Increased ODC1 will lead to increased levels of putrescine and the polyamines spermine and spermidine. Together, these data strongly suggest that the essential role of AMD1 and ODC1 in ESCs is to promote high levels of the polyamines, specifically spermine. Our data also suggests that AMD1 and ODC1 mediate their effect on self-renewal and iPSC generation by promoting high levels of MYC protein (Fig. 4).18,19
The polyamines have the potential to affect a wide range of biological processes, and it is very likely that they work on multiple pathways to promote self-renewal. Histone acetylases and deacetylateses are known to be regulated by high polyamine levels, resulting in modification of histone acetylation and altered gene expression. Spermidine can promote longevity in a number of organisms and functions to inhibit histone acetyletransferases, resulting in reduced histone acetylation.20,41 It is possible that in ESCs, high AMD1 and ODC1 promote high polyamine levels that, in turn, modulate histone acetylation, leading to altered gene expression. A link between reprogramming and longevity was proposed following the demonstration that rapamycin, in addition to other longevity factors, including spermidine, could enhance iPSC derivation. While the mechanism by which these factors promote reprogramming is unclear, it was proposed that they could function by preventing cellular senescence, promoting EMT or overcoming additional reprogramming barriers.38,42 The link between longevity and iPSC derivation is interesting, and further investigation is required to understand the mechanistic relationship between these two processes.
As discussed earlier, changes in the level or enzyme activity of AMD1 will result in a change in the ratio of AdoMet to dcAdoMet. This can influence cellular methylation, polyamine levels and the levels of hypusinated EIF5A. We showed that the addition of spermine can rescue the differentiation phenotype on knockdown of Amd1, but importantly, spermidine could not.16 As spermidine is essential for the hypusination of EIF5A, this suggests the requirement for AMD1 is not to promote high levels of hypusinated EIF5A. Though our data strongly point to an essential role for AMD1 and ODC1 in regulating polyamine levels, it remains possible that there could be additional effects on methylation that function to promote self-renewal. Further investigation is required to address this.
While the data presented here demonstrate the role of AMD1 and ODC1 in self-renewal, there are also reports that they are important in directed differentiation of ESCs. We previously showed that AMD1 levels have to be downregulated to enable differentiation of mouse ESCs to neural precursor cells. The overexpression of Amd1 resulted in inhibition of NPC differentiation, and this was mimicked by the addition of spermine. This suggests that AMD1 levels have to be downregulated in order to decrease the levels of spermine, which are inhibitory to NPC differentiation. This was shown to be mediated by an NPC-enriched miRNA that targets Amd1 for translational repression.16 Spermine has also been shown to promote the differentiation of mouse ESCs to multi-layer muscle fibers when added during a specific time window of differentiation.43 Polyamines have been implicated in keratinocyte differentiation, kidney organogenesis and neuronal differentiation.44,45 Given the requirement for their enzyme regulators during development, polyamines at different ratios likely play a significant role in directing differentiation to all lineages.
In conclusion, the importance of the polyamine pathway in ESC self-renewal and differentiation is clear. What remains is to define the molecular targets of the pathway both in ESCs and during directed differentiation. Given the wide range of potential molecular targets for this pathway, it is likely that it feeds into the self-renewal and differentiation networks on multiple levels. A detailed understanding of how this pathway functions could aid in realizing the potential of stem cells in disease modeling and cellular therapy.
Methods
Cell culture
Sox1-GFP mouse ESCs46 were cultured on gelatinized dishes in DMEM (GIBCO) supplemented with 15% FBS (GIBCO), 0.2 mM b-Mercaptoethanol, 1X MEM nonessential amino acids (GIBCO), 2 mM L-glutamine (GIBCO) and LIF. Embryoid bodies were formed by culturing ESCs in the absence of LIF.47 ESCs were differentiated to NPCs in N2B27 medium as described.48 BMP4 (R&D Cat No. 5020) was used as described at 10 ng/mL.48 iPSCs were generated as previously described.49 Briefly, MEFs were infected with the viruses pMX-Oct4, Sox2, Klf4 and Amd1 or Odc1 in polybrene (Invitrogen) supplemented 15% FBS-DMEM medium. Forty-eight hours after infection, cells were cultured in mouse ESC medium. GFP-positive colonies were visualized on a fluorescence microscope and quantified on 14 dpi.
Plasmid construction and transfections
The Odc1 ORF was cloned into the PTracer vector (Invitrogen). One ug of empty vector or p-Tracer-Odc1 was transfected into ESCs on day 1 and day 3 of LIF withdrawal. Cells were harvested on day 5 for AP staining, real-time PCR and western blot.
Real-time PCR and western blots
cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen) according to the manufacturer’s instruction. SYBR Green was used with gene-specific primers for qRT-PCR on an ABI PRISM 7900 Sequence Detection Systems. All values were normalized to GAPDH. For western blot, 20 ug of protein was separated on a NuPAGE 4–12% Bis-Tris Gel and transferred to nitrocellulose membrane. ODC1 (Santa Cruz Cat No.sc-21515), OCT4 (Santa Cruz Cat No.sc-5279), GAPDH (Abcam Cat No.ab-9484) and SOX2 (R&D Systems Cat No.MAb2018) antibodies were used at 1:1,000 dilution. For AP staining, cells were stained with the Alkaline Phosphatase Detection Kit (Millipore SCR004) according to the manufacturer’s instructions.
Acknowledgments
We thank the Agency for Science Technology and Research (A*STAR) for funding and Drs. Chin Yan Lim and Queenie Wong Wing Lei for critical reading of the manuscript.
Glossary
Abbreviations:
- ESC
embryonic stem cells
- NPC
neural precursor cells
- iPSC
induced pluripotent stem cell
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/22772
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Yabut O, Bernstein HS. The promise of human embryonic stem cells in aging-associated diseases. Aging (Albany NY) 2011;3:494–508. doi: 10.18632/aging.100328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xue K, Ng JH, Ng HH. Mapping the networks for pluripotency. Philos Trans R Soc Lond B Biol Sci. 2011;366:2238–46. doi: 10.1098/rstb.2011.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tollervey JR, Lunyak VV. Epigenetics: judge, jury and executioner of stem cell fate. Epigenetics. 2012;7:823–40. doi: 10.4161/epi.21141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lagarkova MA, Volchkov PY, Lyakisheva AV, Philonenko ES, Kiselev SL. Diverse epigenetic profile of novel human embryonic stem cell lines. Cell Cycle. 2006;5:416–20. doi: 10.4161/cc.5.4.2440. [DOI] [PubMed] [Google Scholar]
- 6.Walker E, Manias JL, Chang WY, Stanford WL. PCL2 modulates gene regulatory networks controlling self-renewal and commitment in embryonic stem cells. Cell Cycle. 2011;10:45–51. doi: 10.4161/cc.10.1.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ang YS, Gaspar-Maia A, Lemischka IR, Bernstein E. Stem cells and reprogramming: breaking the epigenetic barrier? Trends Pharmacol Sci. 2011;32:394–401. doi: 10.1016/j.tips.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, et al. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–7. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 9.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–60. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, et al. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–9. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirai H, Karian P, Kikyo N. Regulation of embryonic stem cell self-renewal and pluripotency by leukaemia inhibitory factor. Biochem J. 2011;438:11–23. doi: 10.1042/BJ20102152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith KN, Lim JM, Wells L, Dalton S. Myc orchestrates a regulatory network required for the establishment and maintenance of pluripotency. Cell Cycle. 2011;10:592–7. doi: 10.4161/cc.10.4.14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–96. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 14.Berardi E, Pues M, Thorrez L, Sampaolesi M. miRNAs in ESC differentiation. Am J Physiol Heart Circ Physiol. 2012;303:H931–9. doi: 10.1152/ajpheart.00338.2012. [DOI] [PubMed] [Google Scholar]
- 15.Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–20. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- 16.Zhang D, Zhao T, Ang HS, Chong P, Saiki R, Igarashi K, et al. AMD1 is essential for ESC self-renewal and is translationally down-regulated on differentiation to neural precursor cells. Genes Dev. 2012;26:461–73. doi: 10.1101/gad.182998.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pegg AE, Xiong H, Feith DJ, Shantz LM. S-adenosylmethionine decarboxylase: structure, function and regulation by polyamines. Biochem Soc Trans. 1998;26:580–6. doi: 10.1042/bst0260580. [DOI] [PubMed] [Google Scholar]
- 18.Liu L, Rao JN, Zou T, Xiao L, Wang PY, Turner DJ, et al. Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol Biol Cell. 2009;20:4885–98. doi: 10.1091/mbc.E09-07-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Celano P, Baylin SB, Giardiello FM, Nelkin BD, Casero RA., Jr. Effect of polyamine depletion on c-myc expression in human colon carcinoma cells. J Biol Chem. 1988;263:5491–4. [PubMed] [Google Scholar]
- 20.Minois N, Carmona-Gutierrez D, Madeo F. Polyamines in aging and disease. Aging (Albany NY) 2011;3:716–32. doi: 10.18632/aging.100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giordano E, Flamigni F, Guarnieri C, Muscari C, Pignatti C, Stefanelli C, et al. Polyamines in Cardiac Physiology and Disease. Open Heart Failure Journal. 2010;3:25–30. doi: 10.2174/1876535101003020025. [DOI] [Google Scholar]
- 22.Casero RA, Jr., Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov. 2007;6:373–90. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- 23.Igarashi K, Kashiwagi K. Modulation of cellular function by polyamines. Int J Biochem Cell Biol. 2010;42:39–51. doi: 10.1016/j.biocel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Nishimura K, Okudaira H, Ochiai E, Higashi K, Kaneko M, Ishii I, et al. Identification of proteins whose synthesis is preferentially enhanced by polyamines at the level of translation in mammalian cells. Int J Biochem Cell Biol. 2009;41:2251–61. doi: 10.1016/j.biocel.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 25.Ivanov IP, Atkins JF, Michael AJ. A profusion of upstream open reading frame mechanisms in polyamine-responsive translational regulation. Nucleic Acids Res. 2010;38:353–9. doi: 10.1093/nar/gkp1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou T, Mazan-Mamczarz K, Rao JN, Liu L, Marasa BS, Zhang AH, et al. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J Biol Chem. 2006;281:19387–94. doi: 10.1074/jbc.M602344200. [DOI] [PubMed] [Google Scholar]
- 27.Xiao L, Rao JN, Zou T, Liu L, Marasa BS, Chen J, et al. Polyamines regulate the stability of activating transcription factor-2 mRNA through RNA-binding protein HuR in intestinal epithelial cells. Mol Biol Cell. 2007;18:4579–90. doi: 10.1091/mbc.E07-07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu L, Santora R, Rao JN, Guo X, Zou T, Zhang HM, et al. Activation of TGF-beta-Smad signaling pathway following polyamine depletion in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2003;285:G1056–67. doi: 10.1152/ajpgi.00151.2003. [DOI] [PubMed] [Google Scholar]
- 29.Pegg AE. Mammalian polyamine metabolism and function. IUBMB Life. 2009;61:880–94. doi: 10.1002/iub.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pegg AE, Casero RA., Jr. Current status of the polyamine research field. Methods Mol Biol. 2011;720:3–35. doi: 10.1007/978-1-61779-034-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper HL, Park MH, Folk JE. Posttranslational formation of hypusine in a single major protein occurs generally in growing cells and is associated with activation of lymphocyte growth. Cell. 1982;29:791–7. doi: 10.1016/0092-8674(82)90441-X. [DOI] [PubMed] [Google Scholar]
- 32.Park MH. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A) J Biochem. 2006;139:161–9. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scuoppo C, Miething C, Lindqvist L, Reyes J, Ruse C, Appelmann I, et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature. 2012;487:244–8. doi: 10.1038/nature11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Byers TL, Lakanen JR, Coward JK, Pegg AE. The role of hypusine depletion in cytostasis induced by S-adenosyl-L-methionine decarboxylase inhibition: new evidence provided by 1-methylspermidine and 1,12-dimethylspermine. Biochem J. 1994;303:363–8. doi: 10.1042/bj3030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, et al. S-Adenosylmethionine and methylation. FASEB J. 1996;10:471–80. [PubMed] [Google Scholar]
- 36.Heby O, Persson L, Smith SS. Polyamines, DNA methylation and cell differentiation. Adv Exp Med Biol. 1988;250:291–9. doi: 10.1007/978-1-4684-5637-0_26. [DOI] [PubMed] [Google Scholar]
- 37.Wilmut I, Sullivan G, Chambers I. The evolving biology of cell reprogramming. Philos Trans R Soc Lond B Biol Sci. 2011;366:2183–97. doi: 10.1098/rstb.2011.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen T, Shen L, Yu J, Wan H, Guo A, Chen J, et al. Rapamycin and other longevity-promoting compounds enhance the generation of mouse induced pluripotent stem cells. Aging Cell. 2011;10:908–11. doi: 10.1111/j.1474-9726.2011.00722.x. [DOI] [PubMed] [Google Scholar]
- 39.Nishimura K, Nakatsu F, Kashiwagi K, Ohno H, Saito T, Igarashi K. Essential role of S-adenosylmethionine decarboxylase in mouse embryonic development. Genes Cells. 2002;7:41–7. doi: 10.1046/j.1356-9597.2001.00494.x. [DOI] [PubMed] [Google Scholar]
- 40.Pendeville H, Carpino N, Marine JC, Takahashi Y, Muller M, Martial JA, et al. The ornithine decarboxylase gene is essential for cell survival during early murine development. Mol Cell Biol. 2001;21:6549–58. doi: 10.1128/MCB.21.19.6549-6558.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–14. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 42.Menendez JA, Vellon L, Oliveras-Ferraros C, Cufí S, Vazquez-Martin A. mTOR-regulated senescence and autophagy during reprogramming of somatic cells to pluripotency: a roadmap from energy metabolism to stem cell renewal and aging. Cell Cycle. 2011;10:3658–77. doi: 10.4161/cc.10.21.18128. [DOI] [PubMed] [Google Scholar]
- 43.Sasaki T, Matsuoka H, Saito M. Generation of a multi-layer muscle fiber sheet from mouse ES cells by the spermine action at specific timing and concentration. Differentiation. 2008;76:1023–30. doi: 10.1111/j.1432-0436.2008.00294.x. [DOI] [PubMed] [Google Scholar]
- 44.Pietilä M, Pirinen E, Keskitalo S, Juutinen S, Pasonen-Seppänen S, Keinänen T, et al. Disturbed keratinocyte differentiation in transgenic mice and organotypic keratinocyte cultures as a result of spermidine/spermine N-acetyltransferase overexpression. J Invest Dermatol. 2005;124:596–601. doi: 10.1111/j.0022-202X.2005.23636.x. [DOI] [PubMed] [Google Scholar]
- 45.Loikkanen I, Lin Y, Railo A, Pajunen A, Vainio S. Polyamines are involved in murine kidney development controlling expression of c-ret, E-cadherin, and Pax2/8 genes. Differentiation. 2005;73:303–12. doi: 10.1111/j.1432-0436.2005.00036.x. [DOI] [PubMed] [Google Scholar]
- 46.Aubert J, Stavridis MP, Tweedie S, O’Reilly M, Vierlinger K, Li M, et al. Screening for mammalian neural genes via fluorescence-activated cell sorter purification of neural precursors from Sox1-gfp knock-in mice. Proc Natl Acad Sci USA. 2003;100(Suppl 1):11836–41. doi: 10.1073/pnas.1734197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, et al. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2:448–60. doi: 10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 48.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–6. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 49.Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, et al. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–74. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]