Abstract
Reprogramming somatic cells into a pluripotent state is expected to initiate a new era in medicine. Because the precise underlying mechanism of reprogramming remains unclear, many efforts have been made to optimize induced pluripotent stem cell (iPSC) engineering. However, satisfactory results have not yet been attained. In this review, we focus on recent roadblocks in iPSC reprogramming engineering, such as the inefficiency of the process, tumorigenicity and heterogeneity of the generation. We conclude that cell reprogramming is a naturally occurring phenomenon rather than a biological technique. We will only be able to mimic the natural process of reprogramming when we fully understand its underlying mechanism. Finally, we highlight the alternative method of direct conversion, which avoids the use of iPSCs to generate cell materials for patient-specific cell therapy.
Keywords: iPSC engineering, reprogramming, efficiency, safety, reprogramming factor cocktails, delivery system, identification, natural reprogramming, direct conversion
Introduction
Since 2006, stem cell research has largely focused on reprogramming somatic cells into a pluripotent state to create iPSCs. Like embryonic stem cells, iPSCs can differentiate into the cells of the three germ layers; however, the use of iPSCs bypasses the need for embryonic tissues or oocytes. Therefore, iPSCs were expected to provide cell materials for patient-specific cell therapy while circumventing the ethical and immunologic issues associated with ESCs. Many types of human seed cell types have been successfully reprogrammed into iPSCs, including dermal fibroblasts,1 keratinocytes,2 astrocytes,3 neural progenitor cells,4 neural stem cells,5 hepatocytes,6 pancreatic β cells,7 adipose tissue-derived stem cells (ADSCs),8 peripheral blood cells,9 hematopoietic cells,10 cord blood cells,11 amniotic fluid cells,12 amniotic membrane mesenchyme cells,13 umbilical vein endothelial cells14 and prostate cancer cells.15 Although great efforts have been invested to improve iPSC engineering technology, several issues remain unresolved, including the time that is required to produce these cells and the technique’s low efficiency and safety. Some advanced strategies have slightly improved single issues, but until now, these improvements have been unsatisfactory. In most cases, efficiency and safety cannot be simultaneously guaranteed. Because the field is rapidly advancing, it is important to seriously consider the iPSC generation assessment system. Here, we focus on the recent roadblocks in iPSC engineering and compare them with direct conversion (the conversion of a somatic cell to another somatic cell). We propose that because direct conversion bypasses the pluripotent stage, it requires less time and produces cells with higher efficiency and greater safety. Due to these advantages, direct conversion may be a more suitable clinical technique.
The Issues of Reprogramming Factors in iPSC Engineering
According to proposed transcriptomics, the cell stage is believed to be determined by 10–20 critical TFs. These core factors typically include several key TFs, such as Oct4 (O), Sox2 (S), Klf4 (K), c-Myc (M), Lin28 (L) and Nanog (N).16,17 Additionally, small molecular compounds and microRNAs (miRNAs) are applied during the artificial reprogramming process. Although some cells are easily converted to pluripotent cells, iPSC engineering is plagued by low efficiency11,17-23 and high heterogeneity.17,24 Some core TFs induce tumor formation (oncogenes)25; therefore, alternate TF types have been carefully selected from the pool of factors that are abundant in stem cells but absent in somatic cells, such as Glis1, a Gli-like TF. However, when Glis1 was chosen as a reprogramming factor to replace c-Myc, incompletely reprogrammed human cells were observed.26 To optimize iPSC generation, reprogramming factor cocktails containing fewer TFs (with or without small molecular compounds) or even miRNAs alone have been tested in numerous studies. However, none of these strategies has generated satisfactory results.
First, the reduced number of TFs was thought to improve the ease and safety of the procedure. However, iPSC generation efficiency was reduced with the decreasing number of TFs (Table 1). iPSCs were successfully generated in a less efficient manner when neural stem cells (from mouse and human) and mouse trophoblast stem cells were reprogrammed with a single TF (Oct4); however, terminally differentiated cells did not display this effect. More significantly, by reducing the number of TFs, the reprogramming efficiency and the iPSC quality were reduced. When adult mouse NSCs were reprogrammed into iPSCs using 1 TF (O), 2 TFs (OK) or 4 TFs (OSKM), the neuronal and hematoendothelial differentiation in the 1 TF-derived and 2 TF-derived iPSCs was less effective than that in 4 TF-derived iPSCs.32 A recent study demonstrated that TF stoichiometry influenced the efficiency and quality of iPSC generation33 when all of the TFs (OSKM) were used in equal stoichiometry in most studies. By observing 16 stoichiometric combinations of OSKM transduced by lentiviral vectors in mouse embryonic fibroblast (MEF)-derived iPSCs, the combination of a high Oct4 level and moderate levels of Sox2 and Klf4 reportedly conferred the best efficiency and differentiation capacity. These results suggest that the dosage and proportions of pluripotency factors are critical in iPSC engineering. The overexpression of transcription factors directly or indirectly contributes to incomplete or heterogenetic conversion. The reprogramming network should be balanced as a whole and function similar to the natural reprogramming process; otherwise, the establishment of pluripotency may be inefficient or lead to tumorigenesis. Furthermore, the reprogramming efficiency of adult stem cells has been demonstrated to be significantly higher than that of terminally differentiated cells that originated from the same developmental lineage or were located in the same anatomical site.34-36 Unlike terminally differentiated cells, adult stem cells can be successfully transformed into iPSCs using fewer TFs (Table 1); even a single TF (Oct4) was sufficient. However, these adult stem cells are produced at very low efficiencies and are difficult to obtain from patients in sufficient quantities.
Table 1. TF cocktails in human and mouse models.
| TF | Animal | Donor cells | Delivery system | Efficiency (%) |
|---|---|---|---|---|
| OSKMN24 |
Mouse |
B cells |
Lentivirus |
> 0.5 |
| OSK+L-Myc19 |
MEFs |
Retrovirus |
~0.4 |
|
| OSKM16 |
MEFs |
Retrovirus |
0.1 |
|
| OSK20 |
MEFs |
Retrovirus |
0.01 |
|
| OKM21 |
Dermal papilla |
Retrovirus |
1.4 |
|
| OK21 |
Dermal papilla |
Retrovirus |
0.02 |
|
| OK22 |
Neural stem cells |
Retrovirus |
< 0.1 |
|
| O18 |
Neural stem cells |
Retrovirus |
< 0.01 |
|
| OSKMLN27 |
Human | Newborn foreskin fibroblasts |
Lentivirus |
0.1 |
| OSK+L-Myc19 |
Adult dermal fibroblasts |
Retrovirus |
0.03 |
|
| OSLN17 |
Fetal fibroblasts |
Retrovirus |
0.01 |
|
| OSKM1 |
Dermal fibroblasts |
Retrovirus |
0.01 |
|
| OSKM28 |
CD34+ mobilized peripheral Blood cells |
Retrovirus |
0.01 |
|
| OSLN11 |
Cord blood endothelial cells |
Lentivirus |
< 0.01 |
|
| OSK20 |
Dermal fibroblasts |
Retrovirus |
0.002 |
|
| OSK29 |
Liver progenitor cells |
Retrovirus |
NA |
|
| OSK2 |
Keratinocytes |
Retrovirus |
NA |
|
| OS (+VPA)30 |
Fibroblasts |
Retrovirus |
~1 |
|
| OS31 |
Cord blood stem cells |
Retrovirus |
< 0.01 |
|
| O18 | Neural stem cells | Retrovirus | < 0.004 |
Abbreviations: O, Oct4; S, Sox2; K, Klf4; M, c-Myc; N, Nanog; MEFs, mouse embryonic fibroblasts; L, Lin28; NA, not available;. VPA, valproic acid
Second, several research groups have used small molecular compounds or miRNAs as TF alternatives when engineering iPSCs. The addition of small molecules facilitates reprogramming and, to some extent, lowers the risk of tumorigenesis and improves efficiency by altering epigenetic features or affecting signaling pathways.30,37-44 However, these practices also have several downsides. (1) The addition of small molecular compounds and miRNAs can be beneficial to some extent, but the specificity of these factors in targeting cellular pathways is difficult to assess, and some of the compounds may confer negative effects. For example, 5′-azacytidine, a DNA methyltransferase inhibitor that was shown to improve reprogramming in mice,45 induces DNA damage.46 SV40 large T antigen and telomerase reverse transcriptase (TERT) greatly enhanced the generation of iPSCs that were derived from the somatic cells of large mammals, such as human,47 sheep48 and goat.49 However, these factors are potent viral oncoproteins that can inactivate the p53 and the retinoblastoma pathways.50 (2) The non-specific and broad action of the chemicals may induce the dysregulation of gene expression. For example, although vitamin C is a natural nutrient, it reportedly accelerates reprogramming by reducing p53 levels, which may also affect safety.51 Several studies have demonstrated that epigenomic reprogramming is incomplete, especially at early passages soon after derivation.52-54 Additionally, miRNA expression is difficult to control post-induction. (3) Although the relative reprogramming efficiency rate was increased by approximately 5- to 100-fold,30,37-44,55 the absolute reprogramming efficiency rate was only approximately 1% in humans,43 which is insufficient for subsequent uses. (4) The inhibition of tumor suppressor pathways contributes to reprogramming but also brings serious safety concerns. Many studies have demonstrated that the downregulation of tumor suppressor components, such as p53, p21, p16 and p19, when reprogramming mouse fibroblasts greatly enhances the efficiency and kinetics.56,57 Among these factors, p53, which is known as the “master watchman” to genome mutations, regulates cell growth and apoptosis and is situated at the crossroads of a signaling network. p53 activation has been shown to directly induce miR-145, which then represses the expression of some TFs, including OSK, and induces differentiation.58 miR-34, a p53 target, has also been reported to play an essential role in restraining reprogramming.59 When the p53–21 pathway was inhibited by knocking down p21, the efficiency of iPSC generation from human dermal fibroblasts (HDFs) was increased to approximately 10%.60 On observation in reprogramming iPSCs from MEFs and mouse pre-B cells by lentiviral vectors, transducing OSKM, which increased the cell division rate and cell proliferation by inhibiting the p53-p21 pathway or overexpressing Lin28, resulted in remarkably accelerated kinetics of early iPSC formation rather than enhancing the overall efficiency.24 The authors assumed that a low-frequency epigenetic event was required to initiate the reprogramming event. Therefore, the enhanced cell division rate increased the odds that these events would occur.61 Meanwhile, however, the donor cells were more susceptible to genome damage. The subtle regulation of cell proliferation without introducing genome mutations is a difficult problem.
The Issues of Delivery Systems in iPSC Engineering
Since the discovery of iPSCs, many studies have focused on optimizing TF delivery systems. Here, we summarize the advantages and disadvantages of the current delivery systems (Table 2). To avoid viral vector integration, naked DNA transgenes and non-conservative transposon remobilization, researchers have designed several non-integrative systems, including the integration-defective viral system, the non-integrative DNA-based system, RNA-based delivery and the protein-based system. A dilemma exists when choosing a system, because generally speaking, the higher the transfection rate of the method, the higher the risk of tumorigenesis it runs (Table 2). Although they are generally extremely inefficient, non-integrative systems do not necessarily prevent aberrant epigenetic remodeling or the expression of lineage-specification genes. According to recent research, the non-integrative system based on mRNA is promising due to its increased efficiency and kinetics.72 Nevertheless, the system demands high dosages of the reprogramming factors, which may represent an oncogenic risk, because the increased Myc expression levels may affect genomic stability. It is also difficult to chemically synthesize long mRNAs, and large mRNAs cannot yet be synthesized.
Table 2. Advantages and disadvantages of different delivery system methods.
| Delivery system | Advantages | Disadvantages | ||
|---|---|---|---|---|
| Virus |
Retrovirus16,25 |
Efficient and stable |
Genomic integration; Random insertion of transgenes; Viral transgenes reactivated in the iPSC generation |
|
| Lentivirus17,62 |
Very efficient and stable |
Genomic integration; Random insertion of transgenes; Viral transgenes reactivated in the iPSC generation |
||
| Adenovirus63 |
No genome integration |
Extremely inefficienct; A substantial proportion of tetraploids |
||
| Sendai virus64,65 |
Efficiency; No genome integration |
Very sensitive to the TF transgene sequence content; Difficult to be eliminated from the host cells; Prone to generating aneuploid iPSC lines due to longer exposure to Myc |
||
| DNA- based |
Integrative |
Linear DNA+ polycistronic system66 |
Transgene-free and vector-free |
Inefficient; Requirement of selection for clones free of integration, labor intensive |
| Transposon + helper plasmid67,68 |
Precise deletion of the transgenes |
Inefficienct; Its long-term effect on iPSCs still needs to be evaluated |
||
| Non- integrative |
Non-replicating episome69 |
Relatively simple precedures; |
Inefficient; Requirement of selection for clones free of integration, labor intensive |
|
| Replicating episome70 |
Relatively simple precedures; Can be removed by drug selection system during culturing; |
Extremely inefficient; Requirement of selection for clones free of integration, labor intensive |
||
| Minicircle71 |
Deletion of potentially methylatable prokaryotic backbone sequences |
Inefficient; Requirement of selection for clones free of integration, labor intensive |
||
| Others | mRNAs72 |
Efficient; No need to manually pick clones free of integration |
Many techniques and pieces of equipment required; Labor intensive |
|
| Protein73 | No need to manually pick clones free of integraion |
Extremely inefficient; Hard to be reproducibly purified |
||
In addition to systems based on different transcriptional levels, researchers have designed systems such as the Cre/LoxP system,74 the piggyBac (PB) transposon system,66 a polycistronic system consisting of self-cleavage peptide 2A sequences and an internal ribosome entry site (IRES)75 to remove transgenes. Although they effectively prevent transgenes, none of these systems is completely safe. For example, in the Cre/LoxP system, a portion of the vector backbone remains at the integration site after deletion by Cre, causing insertion mutations. Cre-excisable transgenes also leave a genomic scar that includes the loxP site. Additionally, the PB transposon system theoretically permits a more precise excision when compared with the Cre/LoxP system; however, its safety has not been well-documented, which is likely due to the low efficiency of the PB transposon system. Moreover, the current excision systems may still leave a short sequence of exogenous DNA (mostly viral LTR) in the iPSC genome.
Identification Issues of iPSC Generation
The ultimate goal of iPSC engineering is to generate artificial autologous cells similar to ESCs, the embryo’s natural reprogramming product. The deeper recognition of iPSCs has caused some researchers to question some former studies claiming that they “have high efficiency or free of transgenes or integrations by some novel method.” In addition to tumorigenesis, the concerns regarding incomplete iPSC generation, immune tolerance to autologous iPSCs,76 the different genomic methylation between iPSCs and ESCs77,78 and the epigenetic memory of donor tissue79 have been discussed in an increasing number of studies. This type of iPSC generation may not be qualified in application. Therefore, there is an urgent need to generate accurate, biologically meaningful methods to identify authentic iPS clones during engineering or at least to assess the differences between iPSCs that are derived from a specific strategy and authentic ESCs.
Various criteria have been used to assess successful reprogramming. In most studies, the authors evaluate the degree to which the iPSCs resemble ESCs with respect to traits including cell morphology, the expression of stem cell genes and proteins, karyotype analysis, doubling time, embryoid body formation, teratoma formation, viable chimera formation and specific differentiation ability. However, simple, straightforward assays are also widely used; some of these assays are not considered to be precise or qualified, such as colony appearance, alkaline phosphatase (AP) staining or Nanog expression. The use of such assays leads to the identification of false positives and the incorrect evaluation of reprogramming efficiency. Additional assays for aspects such as the genomic methylation state,80 miRNA profiling,81 histone modification and proteomic profiling82 have demonstrated the difference between iPSCs and ESCs.
More significantly, the identification of patient-specific iPSCs has been considered. Recently, the in vitro and in vivo activities of patient-specific iPSC-derived hepatic cells were evaluated by human-specific albumin staining on the recipient mouse liver and the detection of secreted human-specific liver proteins in the serum or plasma of the recipient mouse.83 Sixteen skin fibroblast-derived iPSC lines from 10 healthy controls and six patients with amyotrophic lateral sclerosis (from individuals of both sexes, various ages and different tissues) were tested for their pluripotency and differentiation capacity to generate neurons for cell therapy.84 All 16 lines passed the test, while 13 of them produced functional motor neurons with a range of efficiencies that were similar to those of human ESCs. Another study demonstrated that low-passage female human iPSCs generated from patients with Lesch-Nyhan syndrome, retained the inactive X chromosome, suggesting that the transcriptional derepression of genes on the inactive X chromosome could not be reversed by differentiation or further reprogramming.85
No reports to date have described iPSCs that are qualified for use in human therapies. We tend to doubt that the iPSCs derived by inefficient systems were too rare to be evaluated properly, rather than be free of transgenes or integration as earlier thought. Additional questions regarding iPSCs still remain. Are iPSCs and ESCs truly equivalent? Will the subtle differences between these two cell types affect their therapeutic potential or research applications? To determine whether this type of iPSC generation is qualified for human therapy, the distinct origins, modes of derivation and roles in research should be reconsidered, and the quality-associated function should be evaluated in vitro.
Future Directions
At this point in iPSC research, we must reconsider whether the original idea of iPSC generation is reasonable. We should alternatively consider direct conversion, which avoids the iPSC step, to generate cell materials for patient-specific cell therapies.
Cell reprogramming is not a novel biological technique
Although reprogramming has only recently been discussed as an attractive scientific technology, it is not a novel biological technique. The first reprogramming study was performed in 1952;86 Briggs and King injected late-blastula-stage leopard frog nuclei into enucleated frog eggs to generate hybrid eggs, which then sometimes divided and developed into complete embryos. This reprogramming procedure was known as somatic cell nuclear transfer (SCNT). Ten years after that first experiment, SCNT was performed with partial blastulae derived from Xenopus intestinal epithelium cells, and the embryos developed normally until the feeding tadpole stage; a portion of these embryos eventually formed adult Xenopus.87 The best example of reprogramming in mammals occurred in 1996. The nuclei of terminally differentiated gland cells from a 6-y-old adult sheep were injected into enucleated mature oocytes and reprogrammed into a totipotent state to produce a mature adult.88
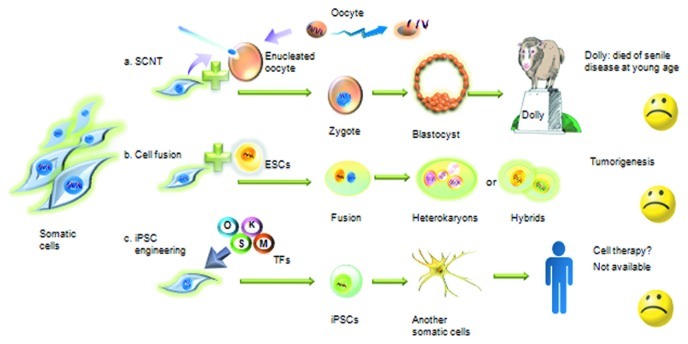
Among the current strategies that are used to reprogram cells to a pluripotent state, SCNT is the best mimic of natural reprogramming, and it significantly differs from the other methods in the two following ways (Fig. 1). First, when the nuclei are transferred to newly enucleated cells, the nuclei that will be reprogrammed are removed from the microenvironment of the primary cytoplasm. Second, the cytoplasm of the enucleated oocyte contains many cytokines that can delicately regulate the introduced nuclei at many levels. Even with these factors, Dolly did not die naturally; she became prematurely senile at 6 y of age, contracted aging-related diseases and died of pneumonia. Dolly died at the same age as the donor sheep that provided the somatic nucleus, which was 6 y old (a sheep’s lifespan is approximately 12 y). Based on these results, it can be concluded that even an ideal reprogramming case cannot radically change a cell’s lifetime. Therefore, it is not surprising that all types of reprogramming methods that utilize TFs and small molecules result in unsatisfactory outcomes, which are primarily related to low efficiency and high heterogeneity. Dolly’s premature senility indicates that the status and function of the current iPSC generation techniques should be thoroughly evaluated. The only way to successfully regulate cell fate and cure clinical refractory diseases is to determine the reprogramming mechanism.
Figure 1. Artificial reprogramming: SCNT, cell fusion and iPSC engineering failed to mimic the natural process. (A) SCNT is the best mimic of natural reprogramming among the current strategies of reprogramming the cell to a pluripotent state. When nuclei are transferred to an enucleated cell, the nuclei that are to be reprogrammed are removed from the primary cytoplasmic microenvironment. The cytoplasm of the enucleated oocyte contains many cytokines that can delicately regulate the introduced nuclei at many levels. (B) Cell fusion of somatic cells and ESCs often leads to heterokaryons or hybrids, which may have considerable potential for tumor formation. (C) Although many researchers have attempted to optimize the process, iPSCs generated by transducing a few TFs into somatic cells have several downsides, such as low efficiency and tumorigenesis.
Somatic cell reprogramming is a common natural phenomenon rather than a biotechnological technique
Nuclear reprogramming to a pluripotent state is a common natural phenomenon. The development of fertilized eggs is an ideal model of this process. By cell-cell fusion, the sperm and oocyte, which harbor distinctive epigenetic modifications, are reprogrammed to generate a totipotent zygote that can differentiate into every specialized cell type in the organism. However, the multiple mechanisms of fertilization and early embryonic development, including the rapid acquisition of specific histone modifications on paternal chromatin, the rapid epigenetic remodeling and the relatively stability of maternal chromatin have remained mysteries.89
Another example of physiological reprogramming was demonstrated by studying the embryonic development of zebrafish.90 Hematopoietic stem cells (HSCs) emerged directly and routinely from endothelial cells lining the aortic floor through a strong bending and subsequent egress of single endothelial cells from the aortic ventral wall into the subaortic space; their reprogramming into floating hematopoietic cells was termed the endothelial-hematopoietic transition.
Notably, cancer studies have revealed additional reprogramming evidence. By observing cancers induced by carcinogens, such as asbestos and arsenic, researchers suggested that aneuploid lesions, rather than gene mutations, appeared in the early stages of cancer development.91,92 Cell fusion is a common physiological phenomenon that is highly regulated and required for development and homeostasis.93 Therefore, it was assumed that abnormal cell fusions between stem cells and somatic cells may have produced tumor stem cells. The mutations were thought to occur in the stem cells, the somatic cells or the fused cells.94 Therefore, mutated or normal somatic cells may be reprogrammed to an immature state via cell-fusion with adult stem cells, eventually leading to tumor formation. All of these findings have demonstrated that reprogramming is an important natural process.
To mimic the natural procedures, embryo-specific modifications must be acquired; the somatic epigenetic features must be totally erased and reprogrammed with precise timing and location. It will only be possible to mimic natural procedures and generate true iPSCs if we fully understand the underlying reprogramming mechanism.
The study of the alternative approaches of “direct conversion/transdifferentiation” has made striking progress
Compared with the process of “one somatic cell type → iPSC → another somatic cell type,” the process of “one somatic cell type → another somatic cell type” bypasses the stem cell stage, generating fewer intermediate byproducts, better downstream products, greater safety and reduced time requirements; therefore, this process is better suited to clinical applications.
Recently, differentiated pancreatic exocrine cells in adult mice were directly converted into cells that closely resembled β-cells that could secrete insulin.95 MEFs and mouse tail-tip fibroblasts (TTFs) were efficiently transdifferentiated into functional neurons.96 After they were transduced with a set of neuronal master regulators, the vast majority of the fibroblasts became postmitotic after 24 h, and immature neuron-like cells were found only 3 d later. Within an additional 2 d, mature neuronal cells with long, branching processes were readily detected, and electrophysiological parameters, such as action potential height and resting membrane potential, also showed signs of maturation over time. The efficiency ranged from 1.8–7.7% in the MEFs and TTFs on day 12. In contrast, iPSC generation and the cells’ subsequent differentiation into a neuronal phenotype can require 1–2 mo each. Based on the availability of donor cells for future applications, a study used skin fibroblasts derived from an Alzheimer disease patient to generate functional neurons through direct conversion; this process had an efficiency of 7.1–8.9% and required less time.97 Using similar strategies, cardiomyocytes were generated with dramatically increased efficiency.98 The use of mouse dermal fibroblasts produced an efficiency of 20% in only 3 d. The MEF kinetics were accelerated 3-fold (11 d compared with 4–5 wk using the indirect approach), and the efficiency was increased more than 10-fold compared with that of the indirect method using iPSCs.
These results are so encouraging that an increasing number of studies have focused on the feasibility of this strategy in clinical applications. More significantly, transplantation therapy with functional-induced hepatocyte-like (iHep) cells directly converted from TTFs resulted in the survival of five of 12 fumarylacetoacetate hydrolase-deficient (Fah−/−) mice; in contrast, all of the mice in the control group died.99 To obtain blood cells for cell therapy, Szabo et al. directly converted human dermal fibroblasts into multipotent hematopoietic progenitors and then into mature blood cells in much less time than required by the iPSC method.100 Compared with the 4 wk required to simply reprogram iPSCs, the entire direct conversion method was completed in 37 d.
These current studies demonstrate that the direct conversion of somatic cells by defined factors can provide a novel strategy to generate patient-specific cells with much higher efficiency, less time and improved quality. In addition, cell division is critical in iPSC formation,24 which may relate to tumorigenesis as described above; however, this process is not apparently required for direct conversion.96,101 The use of a reprogramming method that bypasses the stem cell status is limited with respect to the availability of cellular materials derived from individuals, and the tumorigenicity of these cells is still unclear; however, progress in this area has generated new ideas regarding the use of reprogramming methods with improved efficiencies in clinical applications. In the meantime, these cells should be thoroughly tested to determine whether they have clinical therapeutic uses.
Conclusion
During the past 5 y, the attractive clinical potential of iPSC technology has generated many studies aimed to optimize its performance. The generation of human iPSCs was even praised as “the biological equivalent of the Wright brothers’ first airplane.” However, based on the evidence and discussion provided above, we can safely determine that studies involving iPSCs have hit a roadblock. If our purpose is to advance the iPSC method, we should first clearly determine the precise mechanism underlying reprogramming. If our purpose is clinical therapy, direct conversion would avoid the roadblock. Throughout the history of science, change in perspective has often been the most important. The direct conversion method that bypasses pluripotency represents an emerging technology that will be highlighted in future research studies.
Acknowledgments
We would like to thank Anfei Liu, Yijun Hu for critical reading. This work was financially sponsored by the Shanghai Pujiang Program (07pj14001), the National Natural Science Foundation of China (30771144) and supported by Program for New Century Excellent Talents in University and specially-appointed Professor of Shanghai.
Glossary
Abbreviations:
- iPSC
induced pluripotent stem cell
- ADSCs
adipose tissue-derived stem cells
- O
Oct4
- S
Sox2
- K
Klf4
- M
c-Myc
- L
Lin28
- N
Nanog
- miRNAs
microRNAs
- MEFs
mouse embryonic fibroblasts
- NA
not available
- VPA
valproic acid
- TERT
telomerase reverse transcriptase
- HDFs
human dermal fibroblasts
- PB
piggyBac
- IRES
internal ribosome entry site
- AP
alkaline phosphatase
- SCNT
somatic cell nuclear transfer
- HSCs
hematopoietic stem cells
- TTFs
tail-tip fibroblasts
- iHep
induced hepatocyte-like
- Fah−/−
fumarylacetoacetate hydrolase-deficient
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/22575
Reference
- 1.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–84. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 3.Ruiz S, Brennand K, Panopoulos AD, Herrerías A, Gage FH, Izpisua-Belmonte JC. High-efficient generation of induced pluripotent stem cells from human astrocytes. PLoS One. 2010;5:e15526. doi: 10.1371/journal.pone.0015526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eminli S, Utikal J, Arnold K, Jaenisch R, Hochedlinger K. Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem Cells. 2008;26:2467–74. doi: 10.1634/stemcells.2008-0317. [DOI] [PubMed] [Google Scholar]
- 5.Kim JB, Greber B, Araúzo-Bravo MJ, Meyer J, Park KI, Zaehres H, et al. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649–3. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Ye Z, Kim Y, Sharkis S, Jang YY. Generation of endoderm-derived human induced pluripotent stem cells from primary hepatocytes. Hepatology. 2010;51:1810–9. doi: 10.1002/hep.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stadtfeld M, Brennand K, Hochedlinger K. Reprogramming of pancreatic beta cells into induced pluripotent stem cells. Curr Biol. 2008;18:890–4. doi: 10.1016/j.cub.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aoki T, Ohnishi H, Oda Y, Tadokoro M, Sasao M, Kato H, et al. Generation of induced pluripotent stem cells from human adipose-derived stem cells without c-MYC. Tissue Eng Part A. 2010;16:2197–206. doi: 10.1089/ten.tea.2009.0747. [DOI] [PubMed] [Google Scholar]
- 9.Staerk J, Dawlaty MM, Gao Q, Maetzel D, Hanna J, Sommer CA, et al. Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell. 2010;7:20–4. doi: 10.1016/j.stem.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eminli S, Foudi A, Stadtfeld M, Maherali N, Ahfeldt T, Mostoslavsky G, et al. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat Genet. 2009;41:968–76. doi: 10.1038/ng.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haase A, Olmer R, Schwanke K, Wunderlich S, Merkert S, Hess C, et al. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell. 2009;5:434–41. doi: 10.1016/j.stem.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 12.Li C, Zhou J, Shi G, Ma Y, Yang Y, Gu J, et al. Pluripotency can be rapidly and efficiently induced in human amniotic fluid-derived cells. Hum Mol Genet. 2009;18:4340–9. doi: 10.1093/hmg/ddp386. [DOI] [PubMed] [Google Scholar]
- 13.Cai J, Li W, Su H, Qin D, Yang J, Zhu F, et al. Generation of human induced pluripotent stem cells from umbilical cord matrix and amniotic membrane mesenchymal cells. J Biol Chem. 2010;285:11227–34. doi: 10.1074/jbc.M109.086389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panopoulos AD, Ruiz S, Yi F, Herrerías A, Batchelder EM, Izpisua Belmonte JC. Rapid and highly efficient generation of induced pluripotent stem cells from human umbilical vein endothelial cells. PLoS One. 2011;6:e19743. doi: 10.1371/journal.pone.0019743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, et al. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–24. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 18.Kim JB, Sebastiano V, Wu G, Araúzo-Bravo MJ, Sasse P, Gentile L, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–9. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 19.Nakagawa M, Takizawa N, Narita M, Ichisaka T, Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proc Natl Acad Sci USA. 2010;107:14152–7. doi: 10.1073/pnas.1009374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–6. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 21.Tsai SY, Clavel C, Kim S, Ang YS, Grisanti L, Lee DF, et al. Oct4 and klf4 reprogram dermal papilla cells into induced pluripotent stem cells. Stem Cells. 2010;28:221–8. doi: 10.1002/stem.281. [DOI] [PubMed] [Google Scholar]
- 22.Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–50. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 23.Ohmine S, Squillace KA, Hartjes KA, Deeds MC, Armstrong AS, Thatava T, et al. Reprogrammed keratinocytes from elderly type 2 diabetes patients suppress senescence genes to acquire induced pluripotency. Aging (Albany NY) 2012;4:60–73. doi: 10.18632/aging.100428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–7. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 26.Maekawa M, Yamaguchi K, Nakamura T, Shibukawa R, Kodanaka I, Ichisaka T, et al. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474:225–9. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- 27.Liao J, Wu Z, Wang Y, Cheng L, Cui C, Gao Y, et al. Enhanced efficiency of generating induced pluripotent stem (iPS) cells from human somatic cells by a combination of six transcription factors. Cell Res. 2008;18:600–3. doi: 10.1038/cr.2008.51. [DOI] [PubMed] [Google Scholar]
- 28.Loh YH, Agarwal S, Park IH, Urbach A, Huo H, Heffner GC, et al. Generation of induced pluripotent stem cells from human blood. Blood. 2009;113:5476–9. doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Zhou H, Abujarour R, Zhu S, Young Joo J, Lin T, et al. Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells. 2009;27:2992–3000. doi: 10.1002/stem.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–75. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 31.Giorgetti A, Montserrat N, Aasen T, Gonzalez F, Rodríguez-Pizà I, Vassena R, et al. Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell. 2009;5:353–7. doi: 10.1016/j.stem.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Löhle M, Hermann A, Glass H, Kempe A, Schwarz SC, Kim JB, et al. Differentiation efficiency of induced pluripotent stem cells depends on the number of reprogramming factors. Stem Cells. 2012;30:570–9. doi: 10.1002/stem.1016. [DOI] [PubMed] [Google Scholar]
- 33.Tiemann U, Sgodda M, Warlich E, Ballmaier M, Schöler HR, Schambach A, et al. Optimal reprogramming factor stoichiometry increases colony numbers and affects molecular characteristics of murine induced pluripotent stem cells. Cytometry A. 2011;79:426–35. doi: 10.1002/cyto.a.21072. [DOI] [PubMed] [Google Scholar]
- 34.Sun N, Panetta NJ, Gupta DM, Wilson KD, Lee A, Jia F, et al. Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc Natl Acad Sci USA. 2009;106:15720–5. doi: 10.1073/pnas.0908450106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aasen T, Izpisúa Belmonte JC. Isolation and cultivation of human keratinocytes from skin or plucked hair for the generation of induced pluripotent stem cells. Nat Protoc. 2010;5:371–82. doi: 10.1038/nprot.2009.241. [DOI] [PubMed] [Google Scholar]
- 36.Tan KY, Eminli S, Hettmer S, Hochedlinger K, Wagers AJ. Efficient generation of iPS cells from skeletal muscle stem cells. PLoS One. 2011;6:e26406. doi: 10.1371/journal.pone.0026406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gai H, Leung EL, Costantino PD, Aguila JR, Nguyen DM, Fink LM, et al. Generation and characterization of functional cardiomyocytes using induced pluripotent stem cells derived from human fibroblasts. Cell Biol Int. 2009;33:1184–93. doi: 10.1016/j.cellbi.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 38.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–7. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28:713–20. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang T, Chen K, Zeng X, Yang J, Wu Y, Shi X, et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9:575–87. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, et al. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–5. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–9. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Wang W, Yang J, Liu H, Lu D, Chen X, Zenonos Z, et al. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc Natl Acad Sci USA. 2011;108:18283–8. doi: 10.1073/pnas.1100893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q, Xu X, Li J, Liu J, Gu H, Zhang R, et al. Lithium, an anti-psychotic drug, greatly enhances the generation of induced pluripotent stem cells. Cell Res. 2011;21:1424–35. doi: 10.1038/cr.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-Aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol Cell Biol. 2008;28:752–71. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karran P, Offman J, Bignami M. Human mismatch repair, drug-induced DNA damage, and secondary cancer. Biochimie. 2003;85:1149–60. doi: 10.1016/j.biochi.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 47.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–6. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 48.Bao L, He L, Chen J, Wu Z, Liao J, Rao L, et al. Reprogramming of ovine adult fibroblasts to pluripotency via drug-inducible expression of defined factors. Cell Res. 2011;21:600–8. doi: 10.1038/cr.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren J, Pak Y, He L, Qian L, Gu Y, Li H, et al. Generation of hircine-induced pluripotent stem cells by somatic cell reprogramming. Cell Res. 2011;21:849–53. doi: 10.1038/cr.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahuja D, Sáenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene. 2005;24:7729–45. doi: 10.1038/sj.onc.1209046. [DOI] [PubMed] [Google Scholar]
- 51.Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–9. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 52.Pick M, Stelzer Y, Bar-Nur O, Mayshar Y, Eden A, Benvenisty N. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells. 2009;27:2686–90. doi: 10.1002/stem.205. [DOI] [PubMed] [Google Scholar]
- 53.Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods. 2010;7:53–5. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Menendez JA, Vellon L, Oliveras-Ferraros C, Cufí S, Vazquez-Martin A. mTOR-regulated senescence and autophagy during reprogramming of somatic cells to pluripotency: a roadmap from energy metabolism to stem cell renewal and aging. Cell Cycle. 2011;10:3658–77. doi: 10.4161/cc.10.21.18128. [DOI] [PubMed] [Google Scholar]
- 56.Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–9. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–4. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–58. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 59.Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol. 2011;13:1353–60. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–5. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao J, Pei G. Why cell reprogramming is functionally linked to aging? Aging (Albany NY) 2011;3:700. doi: 10.18632/aging.100364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–5. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–9. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seki T, Yuasa S, Fukuda K. Derivation of induced pluripotent stem cells from human peripheral circulating T cells. Curr Protoc Stem Cell Biol 2011; Chapter 4:Unit4A 3. [DOI] [PubMed] [Google Scholar]
- 65.Ban H, Nishishita N, Fusaki N, Tabata T, Saeki K, Shikamura M, et al. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci USA. 2011;108:14234–9. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–5. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yusa K, Rad R, Takeda J, Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods. 2009;6:363–9. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–70. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–53. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 70.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197–9. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–30. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–4. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–77. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–9. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao T, Zhang ZN, Rong Z, Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474:212–5. doi: 10.1038/nature10135. [DOI] [PubMed] [Google Scholar]
- 77.Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–23. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang A, Huang K, Shen Y, Xue Z, Cai C, Horvath S, et al. Functional modules distinguish human induced pluripotent stem cells from embryonic stem cells. Stem Cells Dev. 2011;20:1937–50. doi: 10.1089/scd.2010.0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–90. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–3. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilson KD, Venkatasubrahmanyam S, Jia F, Sun N, Butte AJ, Wu JC. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18:749–58. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pewsey E, Bruce C, Georgiou AS, Jones M, Baker D, Ow SY, et al. Proteomics analysis of epithelial cells reprogrammed in cell-free extract. Mol Cell Proteomics. 2009;8:1401–12. doi: 10.1074/mcp.M800478-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Choi SM, Kim Y, Liu H, Chaudhari P, Ye Z, Jang YY. Liver engraftment potential of hepatic cells derived from patient-specific induced pluripotent stem cells. Cell Cycle. 2011;10:2423–7. doi: 10.4161/cc.10.15.16869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, et al. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:279–86. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A, Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Briggs R, King TJ. Transplantation of Living Nuclei From Blastula Cells into Enucleated Frogs’ Eggs. Proc Natl Acad Sci USA. 1952;38:455–63. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol. 1962;10:622–40. [PubMed] [Google Scholar]
- 88.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–3. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 89.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–8. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 90.Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–5. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- 91.Duesberg P, Fabarius A, Hehlmann R. Aneuploidy, the primary cause of the multilateral genomic instability of neoplastic and preneoplastic cells. IUBMB Life. 2004;56:65–81. doi: 10.1080/15216540410001667902. [DOI] [PubMed] [Google Scholar]
- 92.Duesberg P. Does aneuploidy or mutation start cancer? Science. 2005;307:41. doi: 10.1126/science.307.5706.41d. [DOI] [PubMed] [Google Scholar]
- 93.Ogle BM, Cascalho M, Platt JL. Biological implications of cell fusion. Nat Rev Mol Cell Biol. 2005;6:567–75. doi: 10.1038/nrm1678. [DOI] [PubMed] [Google Scholar]
- 94.Bjerkvig R, Tysnes BB, Aboody KS, Najbauer J, Terzis AJ. Opinion: the origin of the cancer stem cell: current controversies and new insights. Nat Rev Cancer. 2005;5:899–904. doi: 10.1038/nrc1740. [DOI] [PubMed] [Google Scholar]
- 95.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–32. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Qiang L, Fujita R, Yamashita T, Angulo S, Rhinn H, Rhee D, et al. Directed conversion of Alzheimer’s disease patient skin fibroblasts into functional neurons. Cell. 2011;146:359–71. doi: 10.1016/j.cell.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 98.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–86. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–9. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 100.Szabo E, Rampalli S, Risueño RM, Schnerch A, Mitchell R, Fiebig-Comyn A, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–6. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 101.Bussmann LH, Schubert A, Vu Manh TP, De Andres L, Desbordes SC, Parra M, et al. A robust and highly efficient immune cell reprogramming system. Cell Stem Cell. 2009;5:554–66. doi: 10.1016/j.stem.2009.10.004. [DOI] [PubMed] [Google Scholar]