Abstract
Lack of functional SAP protein, due to gene deletion or mutation, is the cause of X-linked lymphoproliferative disease (XLP), characterized by functionally impaired T and NK cells and a high risk of lymphoma development. We have demonstrated earlier that SAP has a pro-apoptotic function in T and B cells. Deficiency of this function might contribute to the pathogenesis of XLP. We have also shown that SAP is a target of p53 in B cell lines. In the present study, we show that activated primary T cells express p53, which induces SAP expression. p53 is functional as a transcription factor in activated T cells and induces the expression of p21, PUMA and MDM2. PARP cleavage in the late phase of activation indicates that T cells expressing high levels of SAP undergo apoptosis. Modifying p53 levels using Nutlin-3, which specifically dissociates the MDM2-p53 interaction, was sufficient to upregulate SAP expression, indicating that SAP is a target of p53 in T cells. We also demonstrated p53’s role as a transcription factor for SAP in activated T cells by ChIP assays. Our result suggests that p53 contributes to T cell homeostasis through the induction of the pro-apoptotic SAP. A high level of SAP is necessary for the activation-induced cell death that is pivotal in termination of the T cell response.
Keywords: SAP, p53, T cell homeostasis, apoptosis, X-linked lymphoproliferative disease
Introduction
The tumor suppressor p53 has a key role in regulating the delicate balance between survival and death in mammalian cells.1 A tight regulation of cell cycle progression is critical for the maintenance of cellular homeostasis. Cellular stress, like DNA damage, hypoxia, oncogene activation (“oncogenic stress”) and heat shock, triggers p53 stabilization and activation, resulting in transcriptional activation or suppression of specific p53 target genes that decide the fate of the cell. Functional impairment of p53 may occur by mutations that alter its DNA-binding ability and results in failure to regulate p53 target genes.2 p53 inactivation by mutation or other mechanisms, e.g., viral proteins such as HPV E6 that block p53 function, allows evasion from apoptosis or senescence, leading to tumor development/progression.
The X-linked lymphoproliferative disease (XLP) is a rare immunodeficiency caused by mutations or deletion of the SH2D1A gene.3 Affected individuals are extremely vulnerable to EBV, but not to other herpes virus infections. EBV infects B cells preferentially and induces them to proliferation. In normal individuals, this proliferation is controlled by NK cells and specific T cells. However in XLP individuals, EBV infection leads to uncontrolled T and B cell proliferation.
The product of the gene, SLAM-associated protein (SAP), is an SH2 domain containing small proteins expressed in T, NK and B cells. SAP functions as an adaptor, bridging the SLAM family of proteins with the tyrosine kinase FynT, leading to SLAM receptor-associated signaling.4 Among other deficiencies, in the absence of functional SAP, the cytotoxic functions of T and NK cells are impaired, allowing escape and accumulation of the proliferating EBV-infected B cells.
During immune response, the antigen-specific clonal proliferation of T cells leads to antigen clearance. Subsequently, the T cell response is resolved by reducing the number of circulating T cells by apoptosis, a phenomenon called activation-induced cell death (AICD). In an earlier study, we have shown that SAP is upregulated in activated T cells, and it has a pro-apoptotic role,5 thus contributing to T cell homeostasis confirming a previous hypothesis.6 In addition, we have also shown that SAP is a target of wt p53 in DNA-damaged Burkitt lymphoma cell lines.7 In our present work, we demonstrate that SAP is a target of p53 in activated primary T cells, which demonstrates p53’s involvement in T cell homeostasis through induction of SAP.
Results
SAP and p53 expression is upregulated in PHA-stimulated T cells
We have previously reported that SAP is upregulated in activated T cells5 and is a target of p53 in DNA-damaged B cells.7 Putting the two separate results together, we hypothesize that p53 contributes to the expression of SAP in T cells. In order to study this possibility, we first analyzed the levels of SAP and p53 proteins in primary T cells that were cultured with or without PHA for different time periods (Fig. 1A). Both proteins were upregulated in the activated T cells with similar kinetics, and their levels peaked on day 4 of culture. Kinetics of proliferation in activated T cells measured by tritiated (3H) thymidine incorporation showed a maximum on day 2 of culture and dropped at later time points (Fig. 1B). Both SAP and p53 proteins were upregulated at a similar level and kinetics in the two major T cell subsets (CD4+ and CD8+) stimulated with anti CD3/CD28 beads (Fig. 1C). These findings, together with our previous results,7 suggested that p53 contributes to the expression of SAP in activated T cells.
Figure 1. p53 and SAP expression is upregulated in PHA activated T cells. (A) T cells were isolated from peripheral blood of healthy donors and cultured in medium with or without PHA (1 µg/ml) for the indicated time periods. p53 and SAP expression was detected by immunoblots. Results from three different donors are shown. (B) 3H thymidine incorporation assay to determine T cell proliferation. At the mentioned time points, 0.1 x 106 viable cells from bulk culture were plated in 200 µl medium per well in a 96-well plate and 1 µCi of 3H thymidine was added to each well and cultured for 16 h at 37°C with 5% CO2 under humidified conditions. Results of three different donors are presented. (C) p53 and SAP expression in CD4+ and CD8+ subsets of T cells upon activation by anti CD3 and CD28 antibodies.
p53 is functional as a transcription factor in activated T cells
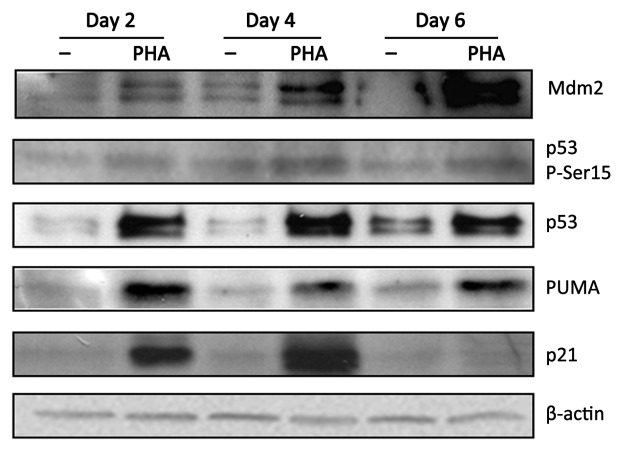
To obtain further support for our hypothesis, we also tested the functionality of p53 in these cells by checking the expression of three well-characterized p53 targets: p21, PUMA and MDM2 by immunoblotting. The three proteins were upregulated in activated T cells after 2 days of culture (Fig. 2). By day 6, p21 expression was undetectable, possibly as a result of ongoing apoptosis. In contrast, MDM2 and PUMA were expressed even at later time points, suggesting that p53 expressed during the later stages of T cell activation might still be functional as a transcription factor.
Figure 2. p53 target genes: p21, PUMA and MDM2 are expressed in PHA-activated T cells. T cells were isolated from peripheral blood of healthy donors and were cultured in medium with or without PHA (1 µg/ml) for the indicated time periods. p21, PUMA, MDM-2, p53 and phosphorylated p53 (Ser-15) expression was detected by immunoblots.
p53 is known to undergo a variety of post-translational modifications in response to different stimuli.8 One of them, phosphorylation of Serine-15 occurs as a response to DNA damage.9 We detected a slight increase of phosphorylated p53 levels in activated T cells (Fig. 2). However, this induction was clearly lower than the induction of total p53 levels, suggesting that p53 induction during T cell activation does not occur as a response to DNA damage.
Activated primary T cells expressing high levels of SAP and p53 undergo apoptosis
T cell homeostasis is maintained by activation-induced cell death (AICD) following restimulation. We have previously reported that SAP has a pro-apoptotic role in T cell lines.5 Based on this finding, we examined PARP cleavage in activated primary T cells as a signature for apoptosis (Fig. 3). In line with the higher levels of SAP, on day 4 and 6 of culture, activated T cells showed increased PARP cleavage. The kinetics of SAP upregulation, the decrease in T cell proliferation (Fig. 1A and B) and PARP cleavage are consistent with the involvement of SAP in apoptosis of activated T cells.
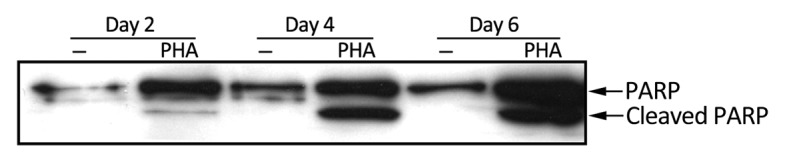
Figure 3. PHA-stimulated T cells in late stage of activation undergo apoptosis. PARP cleavage was determined by immunoblot at the indicated time points in control and PHA (1 µg/ml) activated primary T cells.
Nutlin-3-mediated upregulation of p53 in T cells induces SAP expression
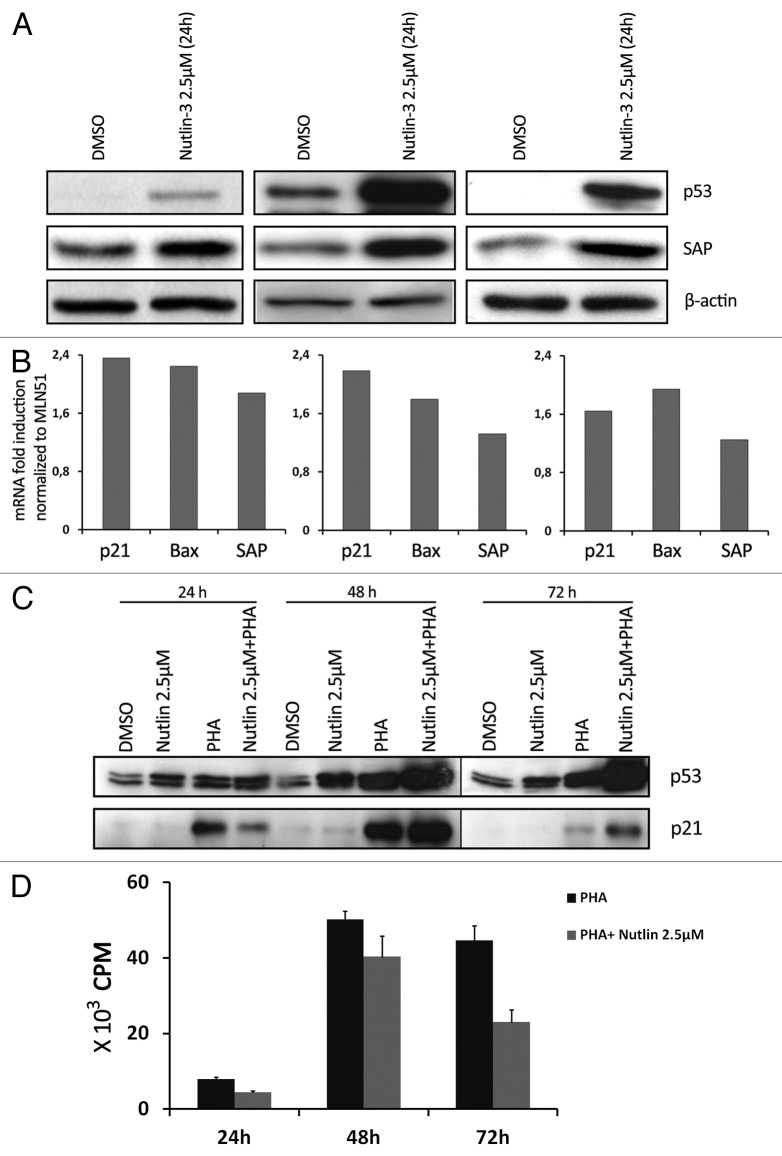
In order to study whether p53 contributes to the upregulation of SAP in T cells, we induced p53 by using the small-molecule Nutlin-3, which selectively interacts with MDM2 and protects p53 from degradation.10 Exposing primary T cells from different donors to 2.5 µM Nutlin-3 for 24 h regularly led to upregulation of both p53 and SAP expression (Fig. 4A). However, the p21 protein levels remained unchanged in response to this dose of Nutlin-3 and at the time points analyzed (Fig. 4C and unpublished data). The induction of SAP protein levels was not merely a consequence of T cell activation brought about by Nutlin-3 treatment, because activation markers (CD69 and CD95) were not induced (not shown). We also measured SAP mRNA by real-time PCR after a 6-h Nutlin-3 (2.5 µM) treatment in T cells. SAP mRNA was induced during this short treatment (Fig. 4B), suggesting that SAP is a direct transcriptional target of p53. Other p53-responsive genes, like p21 and BAX, were also upregulated. Since p21 mRNA but not p21 protein was upregulated, the expression of p21 was determined in T cells treated only with Nutlin-3 in parallel with activated T cells treated with Nutlin-3. p21 expression was upregulated in activated T cells, and the expression was further increased in cells that, in addition to activation, were also treated with Nutlin-3. Again, no p21 protein expression was observed in T cells treated with Nutlin-3 alone (Fig. 4C), showing that this cell cycle inhibitor could only be induced in cycling T cells. 3H-thymidine incorporation showed that activated T cells treated with Nutlin-3 could still proliferate (Fig. 4D), although at a lower rate than the non-treated T cells, which is in agreement with the amount of p21 protein.
Figure 4. SAP expression is upregulated in Nutlin-3-treated T cells. (A) p53 and SAP expression was determined in Nutlin-3 (2.5 µM)-treated primary T cells for 24 h. Results from three different donors are shown. (B) mRNA expression of p21, Bax and SAP was determined in primary T cells treated for 6 h with Nutlin-3 (2.5 µM). Results from three different donors are shown. (C) Kinetics of p53 and p21 expression in control, Nutlin-3 (2.5 µM), PHA (1 µg/ml) and Nutlin-3 (2.5 µM) +PHA (1 µg/ml)-treated T cells. (D) 3H thymidine incorporation assay to determine proliferation of PHA (1 µg/ml)-stimulated cells and Nutlin-3 (2.5 µM) + PHA (1 µg/ml)-treated T cells.
p53 binds to response elements upstream of the SAP transcriptional start site in activated primary T cells
p53 protein activates its target genes by binding to specific DNA sequences. The SAP promoter contains two such p53-response elements (RE). One of them is located in the SAP promoter region between positions 37112 and 37161 (p53 RE-1) and the other in the 5′-UTR (p53 RE-2) between positions 39092 and 39136.7 We tested the physical interaction of p53 with the two p53 REs in activated T cells by chromatin immunoprecipitation (ChIP) assay. We found that p53 binds to both p53 response elements, although the fold increase of binding is different between the two sites and in different donors (Fig. 5). p53 also bound to the p53 response element on the MDM2 promoter, which was used as a positive control. The physical interaction between p53 and the p53 REs in the SAP promoter confirms that p53 contributes to the upregulation of SAP expression in activated T cells.
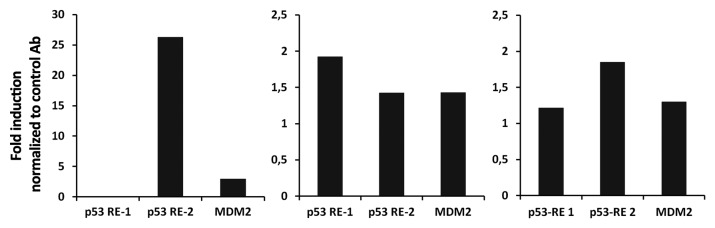
Figure 5. p53 binds to both of the p53 response elements on the SAP promoter. T cells negatively isolated from buffy coat were cultured with PHA (1 µg/ml) for 3 days. Cells were fixed with 1% formaldehyde, sonicated to fragment DNA and immunoprecipitated with antibody-specific against p53 (DO-1) and a non-specific antibody of the same isotype directed against SV40 T antigen. The abundance of precipitated DNA fragments was measured by quantitative PCR using primers specific to p53 RE-1 and p53 RE-2 of the SAP gene. MDM2 served as positive control. Results from three different donors are shown.
Discussion
SAP expression was first identified in T cells, but later it was also found in B cell lymphomas and T and NK cell-derived leukemias.11-13 SAP is involved in cellular signaling in T and NK cells, e.g., by bridging FynT and SLAM, which results in the phosphorylation of tyrosine residues on SLAM, triggering the cellular14 activation cascade. Moreover, two independent reports have shown that SAP has a function in activation-induced cell death of T cells.5,15 Mutation or deletion of the SAP gene causes the XLP immunodeficiency syndrome.16-18 In immunocompetent individuals, primary infection with EBV can lead to a self-limiting lymphoproliferatve disease, infectious mononucleosis. The virally induced B cell proliferation is contained by T cells and NK cells. In XLP patients, however, this surveillance malfunctions, and EBV infection can lead to fulminant, fatal lymphoproliferation. In addition to the functional impairment of the EBV-specific CD8 T and NK cells, the absence of functional SAP in XLP patients disrupts normal T cell homeostasis. Activation-induced cell death is impaired, resulting in a large number of circulating activated T cells, leading to lymphocytic infiltration and tissue damage.
The involvement of p53 in regulating the progression of cell cycle and apoptosis has been intensively investigated. These functions are executed by a large number of p53 transcriptional targets like, p21, Bax, PUMA and PIG3.19-21
We have previously shown that SAP is a transcriptional target of wt p53 in B cell lines.7 The current study demonstrates that p53 also contributes to SAP expression in activated T cells. p53 accumulates in activated T cells and is functional as a transcription factor as monitored by increases in the expression of p53 targets such as MDM2, PUMA and p21. Activated T cells that express high p53 levels also express SAP. The proliferative ability of cells expressing SAP is impaired, as determined by 3H-thymidine uptake. These cells also show an increased apoptotic propensity, as demonstrated by PARP cleavage. This result is in line with our previous findings in a T-ALL cell line, where activation-induced cell death could be related to the high level of SAP.5 The expression level of SAP may be a crucial factor in tilting the balance between T cell proliferation and apoptosis. Higher levels of SAP can interact with Valosin-containing protein (VCP/p97) and may interfere with NFκB signaling.5,22
To test the involvement of p53 in regulating SAP levels in activated T cells, we used Nutlin-3 to specifically induce p53 accumulation and p53 transcription activity. Nutlin-3 treatment of non-activated T cells for 6 h resulted in upregulation of SAP mRNA along with other known p53-responsive genes, i.e., p21 and Bax,23 which is consistent with SAP being a transcription target of p53. Treatment of primary T cells with Nutlin-3 for 24 h also resulted in SAP protein upregulation. However, at this time point, p21 protein was not expressed. Elevation of p21 protein levels by Nutlin-3 treatment could be seen only upon concomitant T cell activation by PHA. This suggests that p21 protein expression is only sustained in proliferating cells, where this cell cycle inhibitor has a clear function.
Earlier we have reported that the human SAP promoter has two p53 response elements (p53 RE-1 and 2), and we showed by ChIP assay that p53 binds to p53 RE-1 in Burkitt lymphoma cells upon DNA damage.7 In the present study, we showed that p53 binds to both p53 REs in the SAP promoter in activated T cells, indicating that binding of p53 to the response elements in the SAP promoter is cell type-specific. The extent of p53 binding to the REs seems to be donor-dependent as well.
Ets-1 and Ets-2 have earlier been identified as transcriptional factors regulating SAP expression in T cells.24 Another study on human T cells showed that Ets-1 expression is downregulated while Ets-2 expression is upregulated in activated T cells.25,26 It is possible that SAP expression is under the control of Ets-1 in resting T cells. Based on the report that expression of Ets-1 decreases upon T cell activation and based on our present findings, we suggest that p53 expressed during the course of T cell activation contributes to the upregulation of SAP, possibly together with Ets-2. After the initial phase of T cell activation, when SAP contributes to the activation of T cells through the SLAM family receptors, events like the high levels of ROS may lead to p53 activation, which, in turn, contributes to the increased levels of SAP. After reaching this critical high level, SAP can manifest its pro-apoptotic function.
Upregulation of p53 in activated T cells has been observed in a limited number of studies27,28 but its significance in relation to T cell survival was not emphasized. We propose that p53 contributes to the regulation of T cell homeostasis by inducing the expression of the proapoptotic SAP. In addition, it is possible that other proteins known to regulate T cell survival are also induced by p53 during T cell activation. For example, CD95 is a pivotal molecule responsible for T cell death,29 and it is also a target of p53.20 Moreover, the lifespan of T cells has been shown to be regulated by the expression of hTERT.30,31 p53 represses telomerase activity by downregulating hTERT expression in tumor lines.32,33 Putting together our findings and results from other studies, the regulation of SAP, CD95, PUMA and hTERT by p53 and their role in the T cell survival/death, we hypothesize that p53 contributes to T cell homeostasis in several different ways, within a common network.
It is very interesting to note that p53-knockout mice show normal T cell development and a normal immune response, but are prone to develop T cell lymphomas.34,35 In addition, CTLs generated from p53−/− mice were more readily expanded than those generated from p53+/+ mice,36 suggesting that the role of p53 in regulating T cell survival is more evident after long-term exposure to the antigen. Similarly, a humanized mouse model in which p53 was silenced showed a normal development of T cells and a normal T cell response to antigen stimulation. However, upon prolonged in vitro antigen stimulation, T cells with silenced p53 showed a significant growth advantage over T cells carrying p53.37
In vivo, the role of p53 in T cell homeostasis is probably manifested following all T cell responses, but it might be more pronounced in circumstances where the antigen challenge persists for a prolonged time, for example, in infectious mononucleosis where the activation and proliferation of T cells is extremely strong.
Materials and Methods
Separation, activation and treatment of primary T cell
Peripheral blood mononuclear cells (PBMCs) were separated from buffy coats of healthy donors by Ficoll-Paque (GE Health Care, 17–1440–02). T cells from PBMCs were obtained by negative selection. T cells were cultured in complete RPMI with or without 1 µg/ml PHA (Sigma, L8902). On the indicated time points, 0.1 × 106 viable cells were plated in 200 µl of medium in a 96-well plate. One µCi 3H-thymidine (Perkin Elmer, NET02250UC) was added to each well and incubated at 37°C in 5% CO2 for 16 h. The cells were harvested on a glass fiber filter, and the radioactivity was measured in a liquid scintillation counter (Perkin Elmer, Microbeta 1450). Cells were treated with 2.5 µM Nutlin-3 (Sigma, N6287) and in the control cultures DMSO was added to the medium.
Immunoblotting
The cells were lysed in SDS and 2-mercaptoethanol containing lysis buffer and aliquots corresponding to 2 × 105 cells were loaded in each lane in a SDS-PAGE gel. Immunoblotting was performed as described earlier.11 The following antibodies were used to detect the respective proteins; anti-SAP rabbit serum (kind gift from Dr. Janos Sumegi, Cincinnati Children’s Hospital Medical Center), DO-7, p21,38 Puma (Cell Signaling, 4976), PARP (BD PharMingen, 556362), MDM2 (Santa Cruz, SC-813). As a control for equal amounts of protein loaded β-actin (Sigma, A1978) was detected.
RNA isolation, cDNA synthesis and real-time PCR
RNA was extracted from cells using the Quick RNA mini prep (Zymo, R1054) according to the manufacturer’s instructions. This was subjected to DNase (Ambion Applied Biosystems, AM2238) treatment, then the RNA was re-purified (Zymo, R1015). One µg of RNA was used for cDNA synthesis (Invitrogen, 11754–050). SAP, p21 and Bax mRNA expression was quantified by real-time PCR. The relative level of each transcript was determined with the LC FastStartDNA Master SYBR green (Roche, 12239264001) I kit in a LightCycler 1.2 instrument (Roche) using the standard curve method. Each PCR mixture was initially denatured at 95°C for 10 min and then cycled 40 times at 95°C for 8 sec, 60°C for 5 sec and 72°C for 8 sec. Target genes were measured simultaneously with the endogenous control MLN51, which was used for normalization. The following are the primer sequences: BAX forward primer: 5′-ACTCCCCCCGAGAGGTCTT-3′, reverse primer: 5′-GCAAAGTAGAAAAGGGCGACAA-3′; SAP forward primer: 5′-TGTACTGCCTATGTGTGCTGTATC-3′, reverse primer: 5′-TCTCAGCACTCCAAGAACCTGT-3′; p21 forward primer: 5′-GCAGACCAGCATGACAGATTT-3′, reverse primer: 5′-GGATTAGGGCTTCCTCTTGGA-3′ and MLN51 forward primer: 5′-CAAGGAAGGTCGTGCTGGTT-3′, reverse primer: 5′-ACCAGACCGGCCACCAT-3′.
Chromatin immunoprecipitation assay
T cells were activated with PHA for 3 d. Twenty × 106 activated cells were cross-linked with 1% formaldehyde (added drop wise) on ice for 20 min; the fixation was stopped by adding glycine to a final concentration of 2.5 µM and incubated for 20 min on ice. Cells were pelleted and washed in ice-cold PBS and resuspended in 1 ml of RIPA buffer. Aliquots of 200 µl were sonicated (Diagenode, BioRupter UCD-200) to yield 200–1,000 bp DNA fragments. Following centrifugation immunoprecipitation was performed. For 1 ml of pooled sonicated lysate, 2 µg of p53-specific antibody, DO-1 (Santa Cruz, SC-136) or 2 µg of a non-specific antibody directed against SV40 T antigen (Oncogen, Pab 419) was added and incubated overnight at 4°C. The immune-complexes were collected by incubating with Protein G Sepharose 50% slurry (GE Health Care, 17–06–18–01) for 2 h. Beads were washed several times, and the bound DNA-p53 complex was eluted. The eluate was treated with RNase, and reversal of the formaldehyde crosslink was performed by protease treatment for 6 h followed by heating at 65°C for overnight. DNA was recovered by phenol/choloroform extraction and ethanol precipitation in the presence of glycogen. p53 binding to the two response elements on the SAP promoter and to MDM2 promoter was determined by quantitative PCR with the LC FastStartDNA Master SYBR green I kit in a LightCycler 1.2 instrument (Roche) using the standard curve method. Target sequences were measured simultaneously with the 3′ untranslated region of dihydrofolate reductase (Dhfr) as endogenous control, which was used for normalization. The following are the primer sequences: SAP p53 RE-1 forward primer: 5′-TTTTGAATTTGCTAAAGTCAGGTTT-3′, reverse primer: 5′-AATAGCACACCTGTCAAAGCAGC-3′; SAP p53 RE-2 forward primer: 5′-CTCTCTGTATGAACCCTGTGTTG-3′, reverse primer: 5′-GCCTTAAACCTCCTTCTCACAC-3′; MDM2 p53 RE forward primer: 5′-CCTTGTAGGCAAATGTGCAA-3′, reverse primer, 5′-GGTCTCTTGTTCCGAAGCTG-3′: Dhfr forward primer 5′-CTTCTCCAAGACCCCAACTGAG-3′, reverse primer: 5′-CAATGT CAAGGACTGGCAAGAG -3′.
Acknowledgments
We thank Prof. Peter H Krammer for critical reading of the manuscript. This work was supported by grants from the Swedish Cancer Society (Cancerfonden) and Karolinska Institutet. H.M., D.S. and N.N. are recipients of cancer research fellowships from the Cancer Research Institute (New York)/Concern Foundation (Los Angeles).
Glossary
Abbreviations:
- SAP
SLAM-associated protein
- XLP
X-linked lymphoproliferative disease
- ChIP
chromatin immunoprecipitation
- AICD
activation-induced cell death
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/22810
References
- 1.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 2.Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–46. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 3.Purtilo DT. X-linked lymphoproliferative syndrome. An immunodeficiency disorder with acquired agammaglobulinemia, fatal infectious mononucleosis, or malignant lymphoma. Arch Pathol Lab Med. 1981;105:119–21. [PubMed] [Google Scholar]
- 4.Veillette A. Immune regulation by SLAM family receptors and SAP-related adaptors. Nat Rev Immunol. 2006;6:56–66. doi: 10.1038/nri1761. [DOI] [PubMed] [Google Scholar]
- 5.Nagy N, Matskova L, Kis LL, Hellman U, Klein G, Klein E. The proapoptotic function of SAP provides a clue to the clinical picture of X-linked lymphoproliferative disease. Proc Natl Acad Sci USA. 2009;106:11966–71. doi: 10.1073/pnas.0905691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sullivan JL. The abnormal gene in X-linked lymphoproliferative syndrome. Curr Opin Immunol. 1999;11:431–4. doi: 10.1016/S0952-7915(99)80072-7. [DOI] [PubMed] [Google Scholar]
- 7.Nagy N, Takahara M, Nishikawa J, Bourdon JC, Kis LL, Klein G, et al. Wild-type p53 activates SAP expression in lymphoid cells. Oncogene. 2004;23:8563–70. doi: 10.1038/sj.onc.1207908. [DOI] [PubMed] [Google Scholar]
- 8.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 9.Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–7. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 10.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 11.Nagy N, Cerboni C, Mattsson K, Maeda A, Gogolák P, Sümegi J, et al. SH2D1A and SLAM protein expression in human lymphocytes and derived cell lines. Int J Cancer. 2000;88:439–47. doi: 10.1002/1097-0215(20001101)88:3<439::AID-IJC17>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 12.Nagy N, Maeda A, Bandobashi K, Kis LL, Nishikawa J, Trivedi P, et al. SH2D1A expression in Burkitt lymphoma cells is restricted to EBV positive group I lines and is downregulated in parallel with immunoblastic transformation. Int J Cancer. 2002;100:433–40. doi: 10.1002/ijc.10498. [DOI] [PubMed] [Google Scholar]
- 13.Parolini S, Bottino C, Falco M, Augugliaro R, Giliani S, Franceschini R, et al. X-linked lymphoproliferative disease. 2B4 molecules displaying inhibitory rather than activating function are responsible for the inability of natural killer cells to kill Epstein-Barr virus-infected cells. J Exp Med. 2000;192:337–46. doi: 10.1084/jem.192.3.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma CS, Deenick EK. The role of SAP and SLAM family molecules in the humoral immune response. Ann NY Acad Sci. 2011;1217:32–44. doi: 10.1111/j.1749-6632.2010.05824.x. [DOI] [PubMed] [Google Scholar]
- 15.Snow AL, Marsh RA, Krummey SM, Roehrs P, Young LR, Zhang K, et al. Restimulation-induced apoptosis of T cells is impaired in patients with X-linked lymphoproliferative disease caused by SAP deficiency. J Clin Invest. 2009;119:2976–89. doi: 10.1172/JCI39518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coffey AJ, Brooksbank RA, Brandau O, Oohashi T, Howell GR, Bye JM, et al. Host response to EBV infection in X-linked lymphoproliferative disease results from mutations in an SH2-domain encoding gene. Nat Genet. 1998;20:129–35. doi: 10.1038/2424. [DOI] [PubMed] [Google Scholar]
- 17.Nichols KE, Harkin DP, Levitz S, Krainer M, Kolquist KA, Genovese C, et al. Inactivating mutations in an SH2 domain-encoding gene in X-linked lymphoproliferative syndrome. Proc Natl Acad Sci USA. 1998;95:13765–70. doi: 10.1073/pnas.95.23.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D, et al. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature. 1998;395:462–9. doi: 10.1038/26683. [DOI] [PubMed] [Google Scholar]
- 19.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–9. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 20.Owen-Schaub LB, Zhang W, Cusack JC, Angelo LS, Santee SM, Fujiwara T, et al. Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expression. Mol Cell Biol. 1995;15:3032–40. doi: 10.1128/mcb.15.6.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–5. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 22.Nagy N, Matskova L, Hellman U, Klein G, Klein E. The apoptosis modulating role of SAP (SLAM associated protein) contributes to the symptomatology of the X linked lymphoproliferative disease. Cell Cycle. 2009;8:3086–90. doi: 10.4161/cc.8.19.9636. [DOI] [PubMed] [Google Scholar]
- 23.van Leeuwen IM, Higgins M, Campbell J, Brown CJ, McCarthy AR, Pirrie L, et al. Mechanism-specific signatures for small-molecule p53 activators. Cell Cycle. 2011;10:1590–8. doi: 10.4161/cc.10.10.15519. [DOI] [PubMed] [Google Scholar]
- 24.Okamoto S, Ji H, Howie D, Clarke K, Gullo C, Manning S, et al. Expression of the SH2D1A gene is regulated by a combination of transcriptional and post-transcriptional mechanisms. Eur J Immunol. 2004;34:3176–86. doi: 10.1002/eji.200324755. [DOI] [PubMed] [Google Scholar]
- 25.Bhat NK, Thompson CB, Lindsten T, June CH, Fujiwara S, Koizumi S, et al. Reciprocal expression of human ETS1 and ETS2 genes during T-cell activation: regulatory role for the protooncogene ETS1. Proc Natl Acad Sci USA. 1990;87:3723–7. doi: 10.1073/pnas.87.10.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bulanova EG, Budagyan VM, Yarilin AA, Mazurenko NN. Expression of protooncogenes during lymphocyte activation by growth factors. Biochemistry (Mosc) 1997;62:1021–5. [PubMed] [Google Scholar]
- 27.Terada N, Lucas JJ, Gelfand EW. Differential regulation of the tumor suppressor molecules, retinoblastoma susceptibility gene product (Rb) and p53, during cell cycle progression of normal human T cells. J Immunol. 1991;147:698–704. [PubMed] [Google Scholar]
- 28.VanAman SE, Whisler RL. Differential expression of p53 tumor suppressor protein and IL-2 in activated T cells from elderly humans. J Interferon Cytokine Res. 1998;18:315–20. doi: 10.1089/jir.1998.18.315. [DOI] [PubMed] [Google Scholar]
- 29.Dhein J, Walczak H, Bäumler C, Debatin KM, Krammer PH. Autocrine T-cell suicide mediated by APO-1/(Fas/CD95) Nature. 1995;373:438–41. doi: 10.1038/373438a0. [DOI] [PubMed] [Google Scholar]
- 30.Luiten RM, Péne J, Yssel H, Spits H. Ectopic hTERT expression extends the life span of human CD4+ helper and regulatory T-cell clones and confers resistance to oxidative stress-induced apoptosis. Blood. 2003;101:4512–9. doi: 10.1182/blood-2002-07-2018. [DOI] [PubMed] [Google Scholar]
- 31.Roth A, Yssel H, Pene J, Chavez EA, Schertzer M, Lansdorp PM, et al. Telomerase levels control the lifespan of human T lymphocytes. Blood. 2003;102:849–57. doi: 10.1182/blood-2002-07-2015. [DOI] [PubMed] [Google Scholar]
- 32.Kanaya T, Kyo S, Hamada K, Takakura M, Kitagawa Y, Harada H, et al. Adenoviral expression of p53 represses telomerase activity through down-regulation of human telomerase reverse transcriptase transcription. Clin Cancer Res. 2000;6:1239–47. [PubMed] [Google Scholar]
- 33.Xu D, Wang Q, Gruber A, Björkholm M, Chen Z, Zaid A, et al. Downregulation of telomerase reverse transcriptase mRNA expression by wild type p53 in human tumor cells. Oncogene. 2000;19:5123–33. doi: 10.1038/sj.onc.1203890. [DOI] [PubMed] [Google Scholar]
- 34.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr., Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–21. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 35.Donehower LA, Harvey M, Vogel H, McArthur MJ, Montgomery CA, Jr., Park SH, et al. Effects of genetic background on tumorigenesis in p53-deficient mice. Mol Carcinog. 1995;14:16–22. doi: 10.1002/mc.2940140105. [DOI] [PubMed] [Google Scholar]
- 36.Zhou X, Wong S, Walter J, Jacks T, Eisen HN. Increased generation of CD8+ T cell clones in p53 mutant mice. J Immunol. 1999;162:3957–60. [PubMed] [Google Scholar]
- 37.Gimeno R, Weijer K, Voordouw A, Uittenbogaart CH, Legrand N, Alves NL, et al. Monitoring the effect of gene silencing by RNA interference in human CD34+ cells injected into newborn RAG2-/- gammac-/- mice: functional inactivation of p53 in developing T cells. Blood. 2004;104:3886–93. doi: 10.1182/blood-2004-02-0656. [DOI] [PubMed] [Google Scholar]
- 38.Fredersdorf S, Milne AW, Hall PA, Lu X. Characterization of a panel of novel anti-p21Waf1/Cip1 monoclonal antibodies and immunochemical analysis of p21Waf1/Cip1 expression in normal human tissues. Am J Pathol. 1996;148:825–35. [PMC free article] [PubMed] [Google Scholar]