Abstract
The relationships between the kinetochore and checkpoint control remain unresolved. Here, we report the characterization of the in vivo behavior of Cdc20 and Mad2 and the relevant spindle assembly checkpoint (SAC) functions in the neuroblasts of a Drosophila Mps1 weak allele (aldB4–2). aldB4–2 third instar larvae brain samples contain only around 16% endogenous Mps1 protein, and the SAC function is abolished. However, this does not lead to rapid anaphase onset and mitotic exit, in contrast to the loss of Mad2 alone in a mad2EY mutant. The level of GFP-Cdc20 recruitment to the kinetochore is unaffected in aldB4–2 neuroblasts, while the level of GFP-Mad2 is reduced to just about 20%. Cdc20 and Mad2 display only monophasic exponential kinetics at the kinetochores. The aldB4–2 heterozygotes expressed approximately 65% of normal Mps1 protein levels, and this is enough to restore the SAC function. The kinetochore recruitment of GFP-Mad2 in response to SAC activation increases by around 80% in heterozygotes, compared with just about 20% in aldB4–2 mutant. This suggests a correlation between Mps1 levels and Mad2 kinetochore localization and perhaps the existence of a threshold level at which Mps1 is fully functional. The failure to arrest the mitotic progression in aldB4–2 neuroblasts in response to colchicine treatment suggests that when Mps1 levels are low, approximately 20% of normal GFP-Mad2, alongside normal levels of GFP-Cdc20 kinetochore recruitments, is insufficient for triggering SAC signal propagation.
Keywords: Drosophila melanogaster, Cdc20, Mad2, Mps1, spindle assembly checkpoint
Introduction
The spindle assembly checkpoint (SAC) is a complex surveillance mechanism that ensures faithful chromosome segregation during mitosis by monitoring the attachment of chromosomes to the spindle microtubules. The SAC negatively regulates the activation of the anaphase-promoting complex or cyclosome (APC/C)-mediated proteolysis pathway to prevent the destruction of two key substrates, cyclin B and securin. Many of the SAC components, such as Mad1, Mad2, Bub1, BubR1, Bub3, Mps1, Zw10 and Rod and Aurora B kinase, have been identified.1 They are all kinetochore dynamic proteins.2 Although several accounts disagree as to the requirement of the kinetochore in cascading the SAC inhibitory signals, the integrity of the kinetochore is essential for SAC function,3,4 and an initial diffusible Mad2-Cdc20 inhibitory signal catalytically produced from unattached kinetochores has been observed in vitro.5 The SAC function might be collectively achieved or synergistically affected by the formation of a number of closely related inhibitory complexes, such as the mitotic checkpoint complex (MCC), including the Mad2, BubR1, Bub3 and Cdc20 proteins as well as other smaller complexes, for example Mad2-Cdc206 and Cdc20-BubR1-Bub3,7-10 in order to prevent Cdc20 from activating the APC/C. It has been suggested that a single unattached kinetochore can produce sufficient signal to arrest mitotic rat kangaroo kidney (PtK2) cells at metaphase,11 and a self-propagation mechanism model has been proposed to explain this phenomenon.12,13 However, in living cells, the exact interaction of Cdc20 and Mad2 in terms of the relationship between the kinetochores and mitotic checkpoint control remains poorly understood.
Mps1 (Monopolar spindle 1) protein kinase has been implicated in various mitotic processes,14-16 and its role in the SAC involves recruitment of the key SAC proteins, such as BubR1, Mad1 and Mad2 as well as Bub1, as shown in human and Xenopus,17-21 and its activity is required for mitotic checkpoint complex formation.22-24 It has also been shown to be required for Cdc20 kinetochore localization in Xenopus egg extracts.17
Here we report the characterization of Cdc20 and Mad2 in vivo behavior in living neuroblasts from the whole-mount third instar larvae brains of a Drosophila Mps1 weak allele (aldB4–2)15 and the related SAC functions. Unlike observations in Xenopus egg extracts, the GFP-Cdc20 kinetochore localization is not affected in the aldB4–2 neuroblast, while the Mad2 kinetochore signal has been significantly reduced. There is a correlation between the Mps1 levels and the levels of the Mad2 protein recruited to the kinetochores. In contrast to the biphasic exponential kinetics observed in the PtK2 cell, Cdc20 only displays monophasic exponential kinetics in Drosophila neuroblasts. Attempted SAC activation by colchicine treatment of the aldB4–2 neuroblasts causes the kinetochore recruitment of the normal levels of Cdc20 and approximately 20% of the normal levels of Mad2, but the cells fail to arrest. This suggests that the kinetochore localization of Cdc20 and Mad2 in Drosophila melanogaster neuroblasts is itself insufficient for triggering and promoting the SAC signal propagation when the Mps1 level is also low.
Results
Third instar larvae brain samples from an aldB4–2 fly line expresses a significantly reduced level of endogenous Mps1
We have previously shown that Mad2 is not required for kinetochore recruitment of Cdc20 both in syncytial embryos and neuroblasts.25 To further characterize Cdc20 behavior in SAC-deficient cells, and to understand the relationship between the kinetochore and checkpoint control, a Drosophila mps1 homolog mutant allele (aldB4–2) was used. The aldB4–2 line was originally anticipated to be a null for Mps1,15 but the endogenous Mps1 levels within this fly line had never before been confirmed by western blot. We have introduced a copy of the GFP-Cdc20 transgenic construct inserted on the second chromosome or a copy of the GFP-Mad2 construct inserted on the X chromosome into this aldB4–2 mutant background. Brain samples prepared from aldB4–2, gfp-cdc20; aldB4–2 or gfp-mad2; aldB4–2 third instar larvae were examined by western blotting.
The western blot results confirmed that the newly established recombinant lines of gfp-cdc20; aldB4–2 and gfp-mad2; aldB4–2 carry the GFP-Cdc20 and GFP-Mad2 fusion proteins, respectively (Fig. 1). However, they reveal that there is approximately 16.4% of detectable endogenous Mps1 protein still present in the homozygous mutant (aldB4–2) third instar brain sample as well as in the newly established recombinants (Fig. 1).
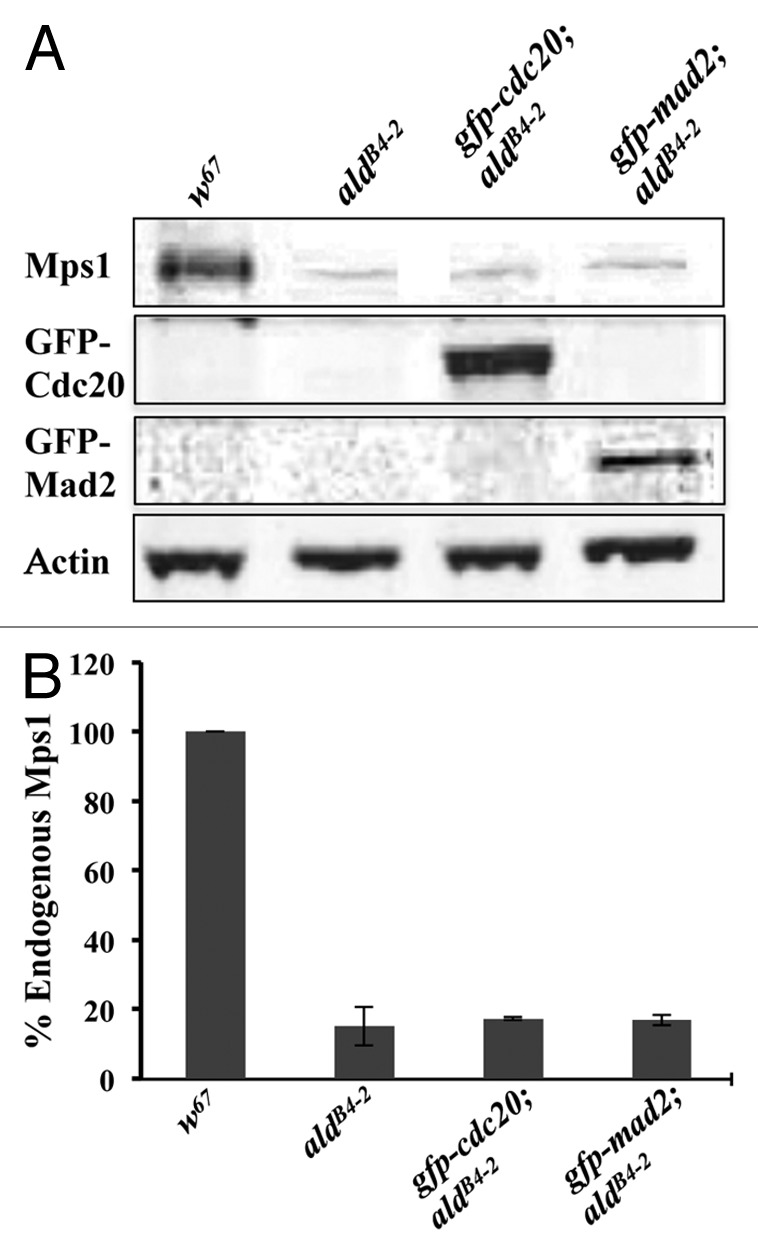
Figure 1. Western blot results showing that the Mps1 weak allele aldB4–2 retains approximately 16.4% of the endogenous Mps1 protein. (A) Western blot results showing samples prepared from the third instar larvae brains for w67, aldB4–2, gfp-cdc20; aldB4–2 and gfp-mad2; aldB4–2 lines. A small amount of the Mps1 endogenous protein in samples from aldB4–2, gfp-cdc20; aldB4–2 and gfp-mad2; aldB4–2 lines can be still detected. Expression of GFP-Cdc20 (panel 2) and GFP-Mad2 (panel 3) fusion proteins has been confirmed in the relevant aldB4–2 genetic background as indicated. The actin bands are used as a loading control. All the antibodies were used in 1:500 dilutions. The western blot bands were imaged using an Odyssey machine. (B) Quantification results showing the western blot band intensities for Mps1 in relation to the above samples after normalization with the intensity of the relevant actin band. The data are presented as a percentage of the Mps1 in the wild-type (w67) sample. The intensity for each sample was quantified from three independent western blots using TINA software.
The SAC in aldB4–2 neuroblasts is defective and mitosis displays increased lengths of transition in different stages
With colchicine treatment of the third instar brains, the neuroblasts from gfp-cdc20; ald+ arrest at metaphase and show an accumulation of kinetochore fusion protein (Fig. 2, top panel); in contrast, gfp-cdc20; aldB4–2 cells failed to arrest at metaphase, and progress through mitosis. This is revealed by the continuous cycling of the GFP-Cdc20 fusion proteins on and off the kinetochores and the reappearance of the dark daughter nucleus as the fusion protein relocated to the cytoplasm (Fig. 2, middle panel arrowhead labeled image at 35:00 time point). This defective SAC function can be rescued by introducing a copy of the gfp-mps1 transgene into the aldB4–2 mutant background (Fig. 2, bottom panel though the GFP-Mps1 fluorescent signals are weaker). Thus, the SAC-defective phenotype of gfp-cdc20; aldB4–2 results from the reduction of Mps1 protein activity.
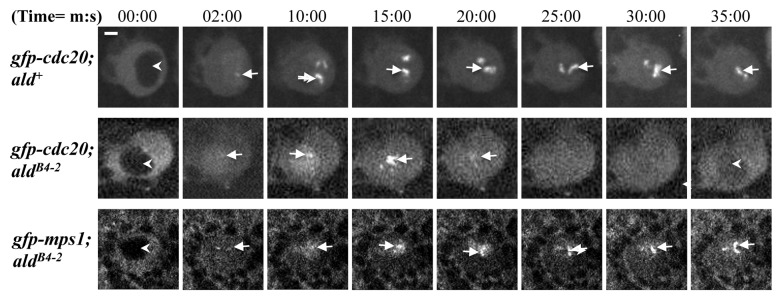
Figure 2. SAC in aldB4–2 neuroblasts is defective, and this can be rescued by introducing an ectopically expressed GFP-Mps1 fusion protein. Third instar larvae prepared from wild-type (gfp-cdc20; ald+, top panel), an Mps1 mutant (gfp-cdc20; aldB4–2, middle panel) and a rescued Mps1 mutant line (gfp-mps1; aldB4–2, bottom panel) were treated with 5 μM colchicine. GFP-Cdc20 behavior was then examined in the neuroblasts. The GFP-Cdc20 fluorescent signal accumulates predominantly in the neuroblast cytoplasm in interphase (arrowheads in all panels at 00:00 time point). It is strong and persistently associated with arrested kinetochores after colchicine treatment (top panel, arrows) in wild-type neuroblasts. In contrast, the GFP-Cdc20 signal continues to oscillate on and off the kinetochores (middle panel, arrows), and the protein was transported to the cytoplasm, when the cell enters interphase so that the daughter nucleus reappears as a dark shape (arrowheads in the middle panel at 00:00 and 35:00 min time points). This suggests that the aldB4–2 neuroblast mitosis has bypassed the SAC arrest. This defective SAC can be restored by introducing a copy of the gfp-mps1 fusion transgene. The kinetochores with strong and persistent GFP-Mps1 fluorescent signals reappear (bottom panel, arrows), suggesting the cell cycle is arrested (bottom panel). The images were scanned using a spinning disk confocal at 22°C with 488 nm laser excitation. Time, minutes, seconds. Bar = 2 μm.
We then compared the mitotic progression in neuroblasts using GFP-Cdc20 as a mitotic progression marker25 (Fig. 3) using lines of gfp-cdc20; aldB4–2 and gfp-cdc20; mad2EY as well as the gfp-cdc20; ald+ (in a wild-type background). GFP-Cdc20 enters the nucleus after nuclear envelope breakdown (NEB), and its entry was clearly visible in all of our experiments in neuroblasts; the nuclear entry occurs just as chromatin assumes a prophase configuration, as illustrated using co-expressed his2BmRFP as a chromatin marker (Fig. 3A, 01, 11 and 21). Metaphase is marked by the stage when the GFP-Cdc20 kinetochore signals begin to decline (Fig. 3A, 4, 14 and 24) a stage that is immediately before condensed chromosome segregation (Fig. 3A, 05, 15 and 25). Telophase is marked by the stage when the mother cell starts to change its shape to form the bud of the daughter cell (Fig. 3A, 07, 17 and 27) where the two separated chromosome masses are farthest apart within the mother cell (Fig. 3A, 08, 18 and 28). Interphase is marked by the time when nuclear GFP-Cdc20 starts to be transported to the cytoplasm and where a dark nucleus could just be visualized, a stage beginning with decondensing of the chromosome (Fig. 3A, 10, 20 and 30).
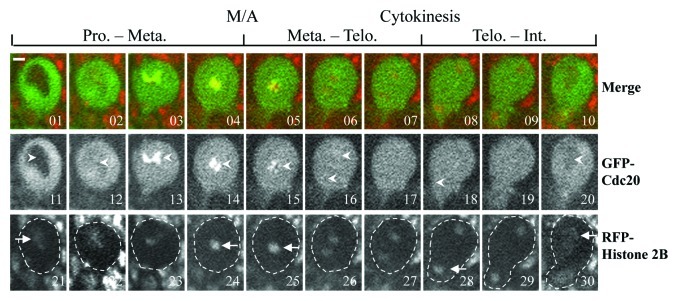
Figure 3. Comparison of the mitotic progression in neuroblasts using GFP-Cdc20 as marker. GFP-Cdc20 was excluded from the interphase (Int.) nucleus (images 1 and 11, arrowheads), entering the nucleus by early prophase (Pro.) (Images 2 and 12, arrowhead). GFP-Cdc20 could be readily observed on prophase and prometaphase kinetochores (Images 3, 4, 13 and14, arrowheads) and persisted on metaphase (Met.) (Images 5 and 15, arrowheads) and greatly declined from anaphase kinetochores (Images 6 and 16, arrowheads). The beginning of cytokinesis immediately after telophase (Telo.) is marked by the arrowhead at image 18, when the cell begins to elongate and bud. Interphase in the daughter cell is marked by the beginning of the reappearance of the dark nucleus as the GFP-Cdc20 relocated to the cytoplasm (Image 20, arrowhead) the chromatin morphologies showing relevant cell cycle stages were determined using coexpressed His2B-mRFP as a marker (Images 21–30, arrows). The entire neuroblast was encompassed in the dashed-line circled region. Time-lapse images were recorded using a spinning disk confocal system at 22°C. Bar = 2 μm. Please refer to Table 1 for the actual timing of mitotic progression.
Table 1 shows the changes to the timings for the various transitions between the stages of the cell cycle. The time to complete prophase to metaphase is significantly shortened in Mad2 mutant neuroblasts but prolonged in Mps1 mutant neuroblasts. The time for progressing through telophase to early interphase is unchanged in Mps1 mutant neuroblasts but shortened in Mad2 mutants compared with wild-type (Table 1).
Table 1. Comparison of the mitotic progressions in Drosophila wild-type, Mad2 and Mps1 mutant neuroblasts.
| Sample | Genotype | Time to complete P*→M* (min) |
Time to complete M*→T* (min) |
Time to complete T→EI* (min) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| |
|
Mean ± SD |
p value |
n |
Mean ± SD |
p value |
n |
Mean ± SD |
p value |
n |
| 1 |
gfp-cdc20;ald+ |
10.92 ± 4.67 |
|
48 |
3.6 ± 1.46 |
|
58 |
11.2 ± 2.16 |
|
50 |
| 2 |
gfp-cdc20;mad2EY |
7.71 ± 2.15 |
0.0031 (1:2)# |
33 |
2.2 ± 0.65 |
0.0001 (1:2)# |
48 |
9.6 ± 1.98 |
0.0006 (1:2)# |
41 |
| 3 | gfp-cdc20;aldB4–2 | 13.76 ± 4.62 | 0.0004 (1:3)# | 50 | 5.0 ± 2.82 | 0.0016 (1:3)# | 69 | 11.6 ± 3.13 | 0.4446 (1:3)# | 60 |
: P, prophase; M, metaphase; T, Telophase; EI, early interphase, #: p value compared between samples in the brackets.
The level of kinetochore recruitment of Cdc20 in neuroblasts is not affected in the aldB4–2 mutant
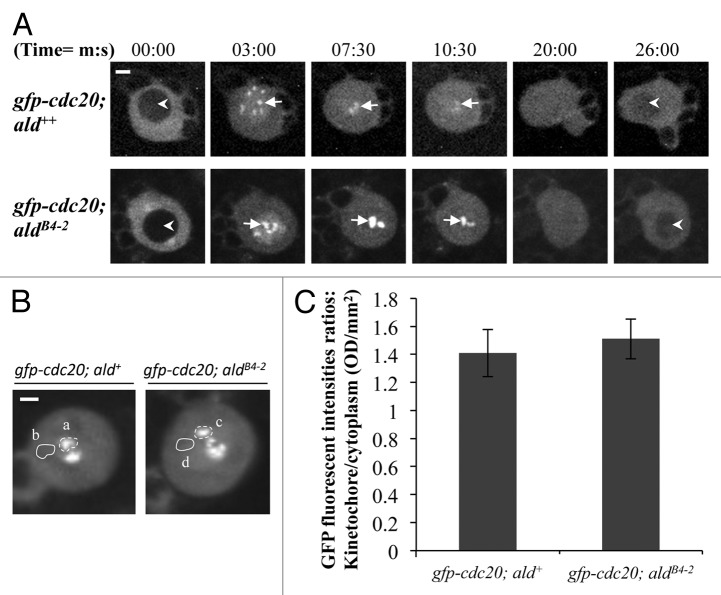
It has been shown that Mps1 acts upstream of Mad1 in the SAC cascade for kinetochore localization of Mad2 both in Xenopus egg extracts and human HeLa cells.17,24,26,27 The depletion of the Mps1 in Xenopus egg extracts also induces a loss of the kinetochore localization of the Cdc20.17 Mps1 might also indirectly affect the BubR1 kinetochore localization in both Xenopus egg extracts and HeLa cells as it is required for ECENPE kinetochore localization and the latter is required for kinetochore recruitment of BubR1.17,27 We have previously shown that Mad2 is not required for Cdc20 kinetochore recruitment but that BubR1 plays a critical role in this process in Drosophila.25 Therefore, we examined how Cdc20 behaves in the Drosophila Mps1 mutant (aldB4–2) neuroblast in order to understand how the SAC cascade on the kinetochore affects the behavior of Cdc20. In the wild-type background, GFP-Cdc20 localizes to the kinetochore during metaphase (arrows, top panel, Fig. 4A). A similar localization pattern can also be observed when GFP-Cdc20 is expressed in the aldB4–2 mutant background (arrows, bottom panel, Fig. 4A). The highest average fluorescent intensities of the GFP-Cdc20 signals at the kinetochores during mitosis from both lines were quantified (Fig. 4B, a and c) and compared with the cytoplasmic level (Fig. 4B, b and d). These results show no significant difference in the overall levels of the localized kinetochore GFP-Cdc20 fluorescent intensities in wild-type and aldB4–2 mutant neuroblasts (Fig. 4C).
Figure 4. The level of kinetochore recruitment of Cdc20 is not affected by reduction of the Mps1 in aldB4–2 mutant. (A) Spinning disk confocal time-lapse images show GFP-Cdc20 localization patterns in third instar larvae neuroblasts from wild-type (gfp-cdc20; ald+, top panel) or Mps1 mutant (gfp-cdc20; aldB4–2, bottom panel). Arrows indicate kinetochore-associated GFP-Cdc20 signals. Arrowheads indicate the prophase nucleus just before NEB (nuclear envelope breakdown, Image at 00:00 time point) or the early interphase nucleus (Image at 26:00 time point) where GFP-Cdc20 was excluded from the nucleus. Bar = 2μm. (B) GFP-Cdc20 fluorescent intensities at prometaphase kinetochores (selected regions a and c) were quantified and compared after subtraction of the relevant cytoplasmic backgrounds (selected regions b and d). Spinning disk confocal images displaying the peak levels of the GFP-Cdc20 at the prometaphase kinetochores were taken from either the wild-type (gfp-cdc20; ald+, left) or the Mps1 mutant (gfp-cdc20; aldB4–2, right). Bar = 3 μm. (C) The graph shows the quantification results. There is no significant different between the highest intensities of the GFP-Cdc20 recruited at prometaphase kinetochores in Mps1 mutant (gfp-cdc20; aldB4–2) and in the wild-type (gfp-cdc20; ald+) neuroblasts. Twenty individual neuroblast images from each fly line were used for this quantification. Time, minutes, seconds.
The absence of Mad2 only causes slower Cdc20 dynamic at kinetochores
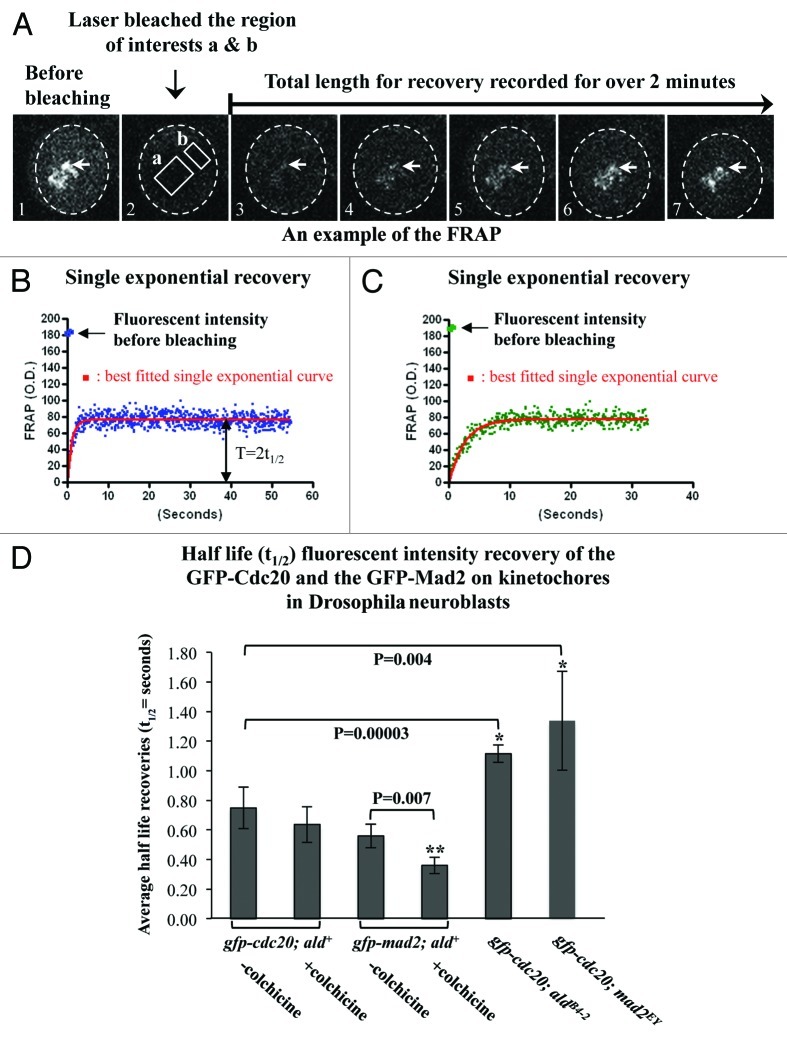
In living mitotic rat kangaroo kidney (PtK2) cells, quantitative results of the fluorescence recovery after photobleaching (FRAP) showed that GFP-Cdc20 possesses biphasic exponential kinetics at unattached kinetochores (fast kinetics t1/2≈1–3 s and slow kinetics t1/2≈21–23 s), while GFP-Mad2 only displays a slow monophasic recovery (t1/2≈19 s).2 The slower phase of GFP-Cdc20 on the kinetochore has been suggested to reflect the complex formation with Mad2, since it was tension-insensitive and disappeared in cells that lacked Mad2 or where Cdc20 carries no major Mad2-binding domain on the kinetochores.2 If this is also the case in Drosophila, we would then expect to see biphasic exponential kinetics for GFP-Cdc20 FRAP and slower phase kinetics that would disappear when GFP-Cdc20 FRAP was measured in a Mad2-null mutant. In fact, the results measured from neuroblasts (Fig. 5A) unequivocally showed monophasic kinetics from the unattached prophase kinetochores (half-life recovery t1/2 = 0.75 ± 0.15 s) (Table 2 and Fig. 5B) and metaphase kinetochores (colchicine arrested kinetochores, half-life recovery t1/2 = 0.64 ± 0.12 s) (Table 2 and Fig. 5C). These rather fast half-life recoveries are similar to the first phase observed for GFP-Cdc20 from PtK2 cells mentioned above. As with the observations in PtK2 cells, there is approximately 50% fluorescent intensity recovery in neuroblasts (Table 2). The value of the half-life recovery was slowed slightly, but it was statistically significant, when it was measured in the neuroblasts prepared from the mad2EY-null mutant (t1/2 = 1.33 ± 0.39 s) or from the aldB4–2 (t1/2 = 1.10 ± 0.07 s) (Table 2 and Fig. 5D). As was observed in PtK2 cells, the GFP-Mad2 kinetics also displayed monophasic kinetics, but they appear to be much faster both at unattached kinetochores (prophase, t1/2 = 0.56 ± 0.08 s) or metaphase kinetochores after colchicine treatment (t1/2 = 0.36 ± 0.06 s) than was observed in PtK2 cells (19 ± 7 s) (Table 2 and Fig. 5D). Approximately 35% of GFP-Mad2 fluorescent intensity recovery occurred (Table 2) in contrast to about 91% recovery in PtK2 cells. Thus, Drosophila Mad2 only affects Cdc20 dynamics at kinetochores, and the Cdc20 and Mad2 dynamic recruitment kinetics at kinetochores are different than those described in PtK2 cells.2
Figure 5. The absence of Mad2 causes slower Cdc20 dynamics at kinetochores. (A) An example of the FRAP experiments performed on the GFP-Cdc20 signals from a group of prometaphase kinetochores (arrows) in neuroblasts from the gfp-cdc20; ald+ wild-type line. Image A1 displays the GFP-Cdc20 fluorescent intensity on the prometaphase kinetochores before photobleaching. Image A2 shows that the GFP-Cdc20 fluorescent intensity at kinetochores in the region of interest “a” was reduced to levels equivalent to that of “b” in cytoplasm after photobleaching. Images A3–7 are selected images from the time-lapse movies showing the GFP-Cdc20 fluorescent intensity recoveries on kinetochores monitored over 2 min post bleach. The cytoplasmic area of the neuroblast is encompassed within the dashed circle in each picture. (B) An example of the normalized GFP-Cdc20 fluorescence intensity recovery time-lapse curve (blue dots) in neuroblasts from the wild-type background (gfp-cdc20; ald+ genotype). The red line represents the best-fitted single exponential curve for the GFP-Cdc20 analyzed by fitting nonlinear regression curves using GraphPad Prism. The original fluorescent intensity level before bleaching has been indicated by an arrow. The value representing the half-life recovery (t1/2) has been indicated by the double arrowheads. (C) An example of the normalized GFP-Cdc20 fluorescent intensity recovery time-lapse curve (green dots) in neuroblasts from Mps1 (aldB4–2) mutant background (gfp-cdc20; aldB4–2). The red line shows the best-fitted single exponential curve. (D) FRAP analysis results representing the half-life fluorescent intensity recoveries at prometaphase or colchicine arrested metaphase kinetochores post-photo bleach. The asterisks indicate the p values showing significant differences between the compared pairs. The numbers of the experiments used in these assays were indicated in Table 2.
Table 2. Comparison the dynamic kinetics of the SAC components at kinetochores in living PtK2 cells and Drosophila neuroblasts.
| Proteins | Cells | Phase | - Colchicine, prometaphase unattached KTs3 | Colchicine# or taxol● arrested metaphase KTs | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|
| |
|
|
t1/2 (s) |
Recovery (%) |
n |
t1/2 (s) |
Recovery (%) |
n |
|
|
GFP-Mad2 |
PtK2 |
|
19 ± 7 |
91 ± 17 |
13 |
|
|
|
Howell et al., 2004 |
| Dm nb1(wt2) |
|
0.56 ± 0.08 (a) |
32.8 ± 5.29 |
17 |
0.36 ± 0.06# (b) (b to a: P=0.007) 4 |
34.6 ± 4.73# |
15 |
This study |
|
|
GFP-Cdc20 |
PtK2 cells |
Fast |
1 ± 1 |
53 ± 17 |
10 |
2 ± 1● |
88 ± 12● |
8 |
Howell et al., 2004 |
|
Slow |
23 ± 10 |
47 ± 17 |
10 |
|
|
|
Howell et al., 2004 |
||
| Dm nb (wt) |
|
0.75 ± 0.14 (c) |
51.2 ± 3.85 |
20 |
0.64 ± 0.12# |
52.7 ± 6.39# |
17 |
This study |
|
| Dm nb (mad2-/-) |
|
1.33 ± 0.34 (d) (d to c: P=0.004)4 |
51.1 ± 7.94 |
15 |
|
|
|
This study |
|
| Dm nb (aldB4-2) |
|
1.12 ± 0.06 (e) (e to c: P=0.00003)4 |
39.0 ± 5.69 |
20 |
|
|
|
This study |
|
|
GFP- Cdc20∆1-167 |
PtK2 | 2 ± 1 | 93 ± 11 | 12 | Howell et al., 2004 |
||||
1, Dm nb, Drosophila neuroblast; 2, wt, wild-type; 3, KTs, kinetochores; 4, p values compared between the pairs indicated by the letters in the brackets. a, b, c, d and e, the letters represent the values in the cells used for p value comparison. #, data collected from colchicine treatment; ●, data collected from taxol treatment.
Kinetochore recruitment of Mad2 is insufficient for SAC signal propagation in Mps1 mutant neuroblasts
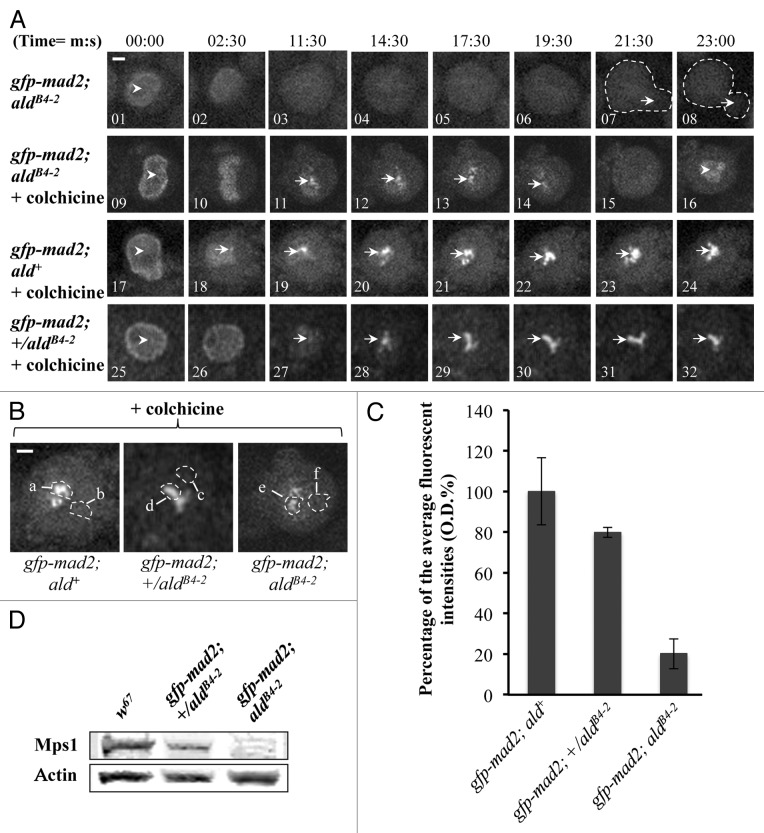
Mad2 is predominantly a nuclear protein (Fig. 6A, 01, 09, 17 and 25) and associates with unattached prophase kinetochores during mitosis in wild-type Drosophila neuroblasts.28 When the third instar larvae brains from the gfp-mad2; ald+ wild-type line were treated with 5 µM colchicine, all the neuroblasts in the brains arrested at metaphase, displaying strong and persistent GFP-Mad2 fluorescent signals at kinetochores over a 30 min observation time (Fig. 6A, bottom panel 19‒24). In contrast, with aldB4–2 mutant neuroblasts from the gfp-mad2; aldB4–2 line, GFP-Mad2 cannot be detected on the kinetochore in normal mitotic progression (Fig. 6A, top panel 03‒06), and the mitotic progression has bypassed the SAC arrest (Fig. 6A, middle panel 15 and 16). However, approximately 20% of the GFP-Mad2 can be recruited to the kinetochore in response to colchicine treatment in these mutant neuroblasts (Fig. 6A, middle panel 11–14, and Fig. 6B and C). This impaired SAC phenomenon is rather surprising and intriguing, as apparently the kinetochore recruitment of Mad2 in the aldB4–2 mutant is still attempted in response to SAC activation, though the level of Mad2 recruitment is significantly reduced in Mps1 mutant neuroblasts, where only around 16% of the normal level of Mps1 is present. Thus, in the presence of this low level of Mps1 kinase activity, kinetochore recruitment of the normal levels of Cdc20 and around 20% of the normal level of Mad2 (compared with the wild-type) is itself insufficient for triggering SAC inhibitory signal propagation. Is there a threshold level of Mps1 protein that would still allow normal mitotic arrest? An intermediate genotype, the heterozygotes of the aldB4–2 line, expressing the GFP-Mad2 fusion protein (gfp-mad2; +/aldB4–2) were created and examined. The actual expression of the Mps1 protein level in the neuroblast samples from the aldB4–2 heterozygotes is approximately 65.1 ± 6.52% of the normal protein level in the wild-type sample (Fig. 6D). All the heterozygous embryos are healthy, viable and developed to be fertile adults (data not shown). The mitotic progression in the neuroblasts derived from this heterozygote is also arrested by 5 µM colchicine treatment (Fig. 6A, bottom panel 25–32). The fluorescent intensity quantification indicates that about 80% of the normal levels of GFP-Mad2 were recruited on to these arrested kinetochores (Fig. 6C). Therefore, there is a dose-dependent correlation between Mps1 and Mad2 for kinetochore localization. The fact that 65% of the normal level of Mps1 is sufficient to support cell cycle progression and overall embryonic development suggests the existence of a threshold level of Mps1 that is required to maintain its SAC functions.
Figure 6. Kinetochore recruitment of Mad2 is insufficient for SAC signal propagation in Mps1 mutant neuroblasts. (A) Top panel: Images showing the GFP-Mad2 fluorescent signals in Mps1 (aldB4–2) mutant neuroblasts under normal mitotic progression. The arrowhead in image 1 indicates nuclear localized GFP-Mad2 at prophase. The arrows in the dashed-line circled regions in images 7 and 8 show the beginning of daughter cell formation. No kinetochore GFP-Mad2 fluorescent signals could be detected throughout the cycle. Middle panel: Images showing the GFP-Mad2 fluorescent signals in Mps1 (aldB4–2) mutant neuroblasts treated with 5 μM colchicine. The arrowhead in image 9 shows the nuclear localized GFP-Mad2 at prophase. The arrows in images 11–14 show that a certain amount of the GFP-Mad2 protein can still be recruited to the kinetochores during the time course. The arrowhead in image 16 shows that the GFP-Mad2 fluorescent signal reappears in the newly formed nucleus to indicate the mitotic progression has bypassed the SAC arrest. Bottom panel: Images showing the GFP-Mad2 fluorescent signals in wild-type neuroblasts treated with 5 μM colchicine. Cells are arrested with strong and persistent kinetochore accumulated GFP-Mad2 fluorescent signals (arrows) during the time course. The arrowhead in image 17 indicates the nuclear localized GFP-Mad2 in prophase. Bar = 2 μm. Time, minutes, seconds. (B) Spinning disk confocal images displaying the region of interests for the peak levels of the GFP-Cdc20 on the kinetochores in neuroblasts after treatment with 5 μM colchicine (region a, d and e) and the relevant cytoplasmic regions (b, c and f) used as the background subtraction. The images were taken from either the wild-type (gfp-mad2; ald+, left); the aldB4–2 heterozygotes (gfp-mad2; +/aldB4–2, middle) or the Mps1 mutant (gfp-mad2; aldB4–2, right). Bar = 3 μm. (C) The graph shows the quantified results measured from “b.” Approximately 20.2 ± 7.4% and 79.8 ± 2.42% of the wild-type levels of GFP-Mad2 can still be recruited to the kinetochores in the neuroblasts of the Mps1 mutant (aldB4–2) and heterozygous background. Twenty individual images from each cell line were used for this quantification. (D) Western blot results showing relevant Mps1 protein expression levels in wild-type, heterozygous and homozygous of the aldB4–2 third instar larvae brain samples. Actin bands act as the loading control.
Discussion
It is interesting to find that approximately 16% of endogenous Mps1 protein still remains in the aldB4–2 third instar larvae brain samples. This protein is most likely to be left over from maternal inheritance, as the mutant was previously anticipated to be null, since a T to C nonsense mutation has created a stop codon “TAA” at position 48 of the protein N terminus.15 Although we cannot completely rule out the possibility that the low level of detected protein is caused by stop codon read-through, which is often used by viral genomes and has been observed more recently in Drosophila and other metazoa.29 However, analysis of the stop codon context (stop codon and subsequent base) suggests that the high frequency of translational leakage, which is associated with the inefficient stop codon context “TGA-C,” rarely happens with the more effective termination codons of TAA-A or TAA-G.29 Thus, it is less likely that the detected low level of the Mps1 proteins resulted from translational leakage as the stop codon context created by the point mutation in the aldB4–2 is “TAA-G.” Thus, the aldB4–2 mutant has provided a unique opportunity to study the SAC mechanism, in that the SAC still attempts to recruit Mad2 to the kinetochore in its cascading pathway but fails to arrest the cells at metaphase.
In eukaryotes, the SAC is conserved throughout, but at a molecular level its components and how they assemble to form the signal cascade pathways are very divergent. For instance, unlike BubR1 in higher eukaryotes, the yeast BubR1 homolog Mad3 lacks a kinase domain.1,30 The depletion of Mps1 in human culture cells results in the selective loss of Mad2 from the kinetochore.21,26,31,32 In contrast, genetic analyses in yeast and immunodepletion experiments in Xenopus egg extracts show Mps1 affecting not only Mad2, but also Bub1, BubR1/Mad3 and Mad1 kinetochore localization.17,20,23,33 The sustained kinetochore localization of Cdc20 in vivo in aldB4–2 neuroblasts observed in this study, either in normal mitotic progression or when the cells respond to the attempted SAC activation, is different from the observation made in Xenopus egg extracts, where the depletion of the Mps1 is connected with the loss of kinetochore localized Cdc20 in vitro.17 This is yet another example in which the subtle mitotic regulation in Drosophila is different to that found in the vertebrate. The observation of the loss of Mps1 being coincidentally related to the loss of kinetochore Mad2 is, however, consistent with the observations made in yeast, HeLa cells and Xenopus as mentioned as above.
It has been previously reported that there are dosage effects on metaphase arrest in Mps1 mutants.34 It would be of interest to identify if there is a connection between the arrest and how the kinetochore localization of Mad2 responds to different levels of Mps1. Here we show an inability of in vivo aldB4 neuroblasts to retain a functional spindle assembly checkpoint when around 20% of Mad2 and normal levels of Cdc20 can still be recruited to the kinetochore in response to attempted SAC activation. It suggests that these conditions are insufficient to trigger the self-propagation of the SAC signal and so are unable to retain a fully functional SAC in aldB4–2 mutant neuroblasts, even with 20% normal levels of Mad2 present at the kinetochores. Our observations may provide a new angle on reviewing the potential SAC self-propagation mechanism vs. an overall threshold SAC signal requirement for metaphase/anaphase transition. As the results suggest, one possibility is that the decreased Mad2 kinetochore localization observed in the aldB4–2 line in response to the SAC activation is not enough to allow sufficient MCC formation. Otherwise a delayed arrest should occur, if the signal self-propagation mechanism existed.
When the endogenous Mps1 level increased to about 65% of the normal level in the aldB4–2 heterozygous neuroblasts, around 80% of the normal levels of Mad2 protein were recruited onto the kinetochores in contrast to approximately 20% of the normal levels of Mad2 observed in the aldB4–2 mutant. The SAC function in this heterozygous neuroblast has been restored, as mitotic progression is shown to arrest in response to SAC activation with 5 µM colchicine treatment (Fig. 6). This suggests that although there is a correlation between the Mps1 levels and the kinetochore recruitment of Mad2 under the provoked SAC condition, it is not a clear linear correspondence. It appears that this heterozygous level of approximately 65% endogenous Mps1 protein is sufficient to support cell cycle progression as well as embryonic development, as all of the embryos successfully developed to fertile adulthoods. Therefore, there likely exists a threshold level of Mps1 that is needed to maintain its functional activities.
It is surprising to find significant variation in mitotic timing between different fly cell types. It takes, on average, 34 min for cultured wild-type S2 cells to complete mitotic progression from prophase to anaphase onset, and this reduced significantly to about 11 min in the absence of Mad2.35 For the in vivo neuroblasts in third instar larvae brains, there is only a 2 min difference on average between wild-type lines and lines where Mad2 is absent.36 It is even less, only 1 min, in early embryos.14 Comparing the anaphase onsets in Mad2 and Mps1 mutants, our results are consistent with previous reports, in that with the loss of Mad2 alone, mitotic progression from prophase to anaphase onset is shortened by approximately 2 min. However, the overall timing of the progression from metaphase/anaphase onset to early interphase is also reduced by around 3 min (Table 1). This suggests that Mad2 plays a role not only in the timing of mitotic maturation, but also in that of mitotic exit. Surprisingly, the reduction of Mad2 caused by the loss of Mps1 in aldB4–2 mutant is enough to remove the spindle checkpoint in response to colchicine, but it does not lead to rapid anaphase onset and mitotic exit, in contrast to the loss of Mad2 alone in the mad2EY mutant, instead it shows significantly delayed progression between prophase to metaphase and metaphase to telophase by about 3 and 1.5 min, respectively (Table 1). This is unlike the situation in human cells, in which the loss of Mps1 causes a sharp early anaphase similar to the loss of Mad2 and other checkpoint proteins,23 and, as yet, we do not fully understand the exact reasons causing these differences. The use in this study of whole-mount brain samples derived from aldB4–2 mutant third instar larvae, where the cells continue to develop for many generations under low levels of Mps1 activity, may be different to the system in human culture cells, where the phenotypes were examined when the Mps1 is instantly knocked down.23 The low level of Mps1 in the neuroblasts can apparently support the survival of the cell and cell cycle progression, but with overall slower pace. This suggests that Drosophila Mps1 kinase activity may also be required for controlling mitotic entrance as well as the spindle assembly checkpoint, perhaps by regulating the Cdk1 kinase activity in early mitosis either directly or indirectly. This remains to be tested.
In PtK2 cells, Cdc20 exhibits biphasic exponential kinetics, while Mad2 only displays monophasic kinetics. The slower phase of the Cdc20 kinetics is similar to that of the Mad2 kinetics and disappears when Mad2 is absent or a Cdc20 mutant, which lacks the major Mad2 binding domain is used.2 We have shown by FRAP analysis using rapid post-bleach data acquisition, that Cdc20 and Mad2 display only monophasic exponential kinetics at the kinetochores in Drosophila neuroblasts (Table 2 and Fig. 5D). The FRAP kinetics half-life (~0.8 s) and the overall rate of recovery of Cdc20 (~51%) are similar to the first phase kinetics and recovery that was observed in PtK2 cells (t1/2 = ~1 s and 50% total recovery).2 However, we have not detected a slower phase, as described in cultured PtK2 cells.2 Meanwhile, Mad2 in Drosophila neuroblast cells in vivo shows faster dynamic kinetics (t1/2 = 0.56 s for prometaphase and 0.36 s for colchicine arrested metaphase, respectively) and a lower level of recovery (~35%) when compared with that observed in PtK2 cells (t1/2 = ~19 s for prometaphase) and 91% overall recovery.2 These results suggest that more than 60% of Drosophila Mad2 is stable on unattached kinetochores in contrast to the 91% that is cycling on and off the PtK2 kinetochores. We have shown previously that it is BubR1 and not Mad2 that is required for the kinetochore localization of Cdc20, and speculate that Mad2 may act to retain Cdc20 localization on kinetochores for dynamic turnover rather than be required for its initial physical localization.25 We show here that in the absence of Mad2, Cdc20 kinetics at kinetochores in mad2EY or aldB4–2 neuroblasts is significantly slowed (t1/2 = approximately 1.3 and 1.1 s, respectively) (Table 2 and Fig. 5D). The slowed kinetics in aldB4–2 neuroblasts is likely to result from the significantly reduced kinetochore Mad2 rather than depletion of Mps1, as the overall level of the kinetochore localized Cdc20 has not been perturbed in these cells. This was also noted in PtK2 cells, when GFP-Cdc20∆1–167 fusion protein, which lacks the major Mad2 binding domain but retains kinetochore localization capability, was FRAP analyzed, showing slowed kinetics from 1‒2 s, at prometaphase.2 Our observations clearly indicate that kinetochore-bound Mad2 influences the Cdc20 kinetochore kinetics, but there is a lack of strong evidence to suggest physical interaction in terms of the Cdc20-Mad2 complex formation. We do not, however, understand the significance of this slowed Cdc20 kinetochore kinetics. Our observations may provide a new insight into the debate as to whether Mad2 is actually part of the MCC complex37 and whether the MCC can form in the absence of direct kinetochore interaction of all components.7 Our observations also suggest that the Drosophila SAC may undergo slightly different pathways at the distal end of the cascade when compared with that observed in Xenopus egg extracts.17 It would be interesting and important to compare and understand the differences between the SAC mechanisms from different species to provide an insight into the complexity of the SAC mechanism in human cells.
Materials and Methods
Mutant fly stocks
A mad2EY-null mutant (EY21687, CG17498, stock #22495) stock was previously purchased from the BDGP/Baylor Gene Disruption Project and characterized.25 aldB4–2 is a Drosophila mps1 homolog mutant semi-lethal allele and is a kind gift from Dr. Gilliland at the College of Science and Health, DePaul University. It was discovered by a germline clone screen for meiotic mutants in Drosophila. Sequencing analysis revealed that aldB4–2 contains an early T to C nonsense mutation, which changes glutamine at position 48 to a stop codon (TAA). This mutant failed to complement the meiotic phenotype of the ald1 allele and could be rescued by introducing a wild-type copy of mps1 (ald+). Therefore, the aldB4–2 line was originally regarded as null for Mps1.15
Transgenic flies
The transgenic Drosophila fly lines that express GFP-Cdc20 fusion protein in a w67 genetic background used in this study have been described previously.25,38 The His2BmRFP transgenic fly line driven by an ubiquitin promoter is a kind gift from Dr. Yohanns Bellaïche at UMR 144 CNRS/Institut Curie. A GFP-Mad2 transgene under mad2 native promoter control inserted on the X chromosome was generated from an original homozygous lethal line with the transgene insertion on the third chromosome obtained from Dr. Karess (CNRS, Gif-sur-Yvette)28 using a (Δ2–3)96B (an immobilisable transposase C transgene construct) based genetic hopping technique. A GFP-Mps1 transgenic line was a gift from Prof. Lehner (Institute of Molecular Life Sciences, UZH). The lines used in this study: gfp-cdc20; mad2EY, gfp-cdc20; aldB4–2 and gfp-mad2; aldB4–2 as well as the gfp-mps1; aldB4–2 were created by genetic recombination crosses.
Dissecting and mounting the third instar larvae brains on coverslips for imaging
Homozygous gfp-cdc20; mad2EY, Cdc20; aldB4–2 or gfp-mad2EY; aldB4–2 or aldB4–2 third instar larvae (non-tubby phenotype) were isolated from the population mixed with the heterozygous (tubby phenotype). The tubby phenotype results from the heterozygotes carrying a TM6 Tub balancer chromosome. The larvae were washed, and the whole brains were dissected in Drosophila Ringer’s solution. They were then transferred into an appropriate amount of fresh Ringer’s solution with or without colchicine (see below) on a coverslip. The samples were then covered with a square piece of YSI standard membrane (YSI Inc., LN#:09K 100561), approximately 1.5 cm2 large. A thin strip of grease (Prod. 33135 3N from BDH VWR) was applied to the edge of the membrane to seal it to the coverslip. This prevents the evaporation of the liquid and allows oxygen to penetrate through the membrane to avoid the brains becoming hypoxic during the experiment.
Antibodies
The affinity purified Cdc20,38 Mad2 and BubR1 rabbit polyclonal antibodies used are described in previous reports.25,39 An anti-Mps1 antibody was raised in rabbit against a region of the Drosophila Mps1. The Mps1 sequence of interest was generated by PCR using the JY412 5′ end forward primer: GAA TTC GAT AGC ATA AGC TTC TCC with EcoR1 onto the 745 bp position of the Mps1 cDNA, and the JY413 3′ end reverse primer: GTC GAC AGG AAG GTT GGT GGT GTA with Sal1 onto the 1,302 bp of the Mps1 cDNA. The PCR product was subcloned into pMal-c2x expression vector at EcoR1 and Sal1 restriction sites and verified by sequencing. The plasmid DNA construct was then expressed in BL21 cells. The MBP-fusion protein was induced by IPTG and purified using an MBP affinity column as described previously.25 The immunization was performed under contract with Cambridge Bioscience, and the anti-serum was affinity purified. The resulting antibody was tested for band specificity on western blot using Drosophila S2 culture cell samples and detected as a specific band at 71 KD that relates to the endogenous Mps1 (Fig. 1). Unfortunately, it is not suitable for immunofluorescence staining with fixed samples. Mouse monoclonal (AC-15) to β actin (Abcam) was used as a loading control.
SDS-PAGE and western blot were performed as described previously.25
Colchicine treatment
Isolated third instar larvae brains were incubated with 5 µM colchicine in Drosophila Ringer’s solution for imaging. This is to depolymerise the microtubules causing the activation of the SAC to arrest cells at metaphase.
Confocal microscopy and time-lapse imaging
A spinning disk confocal system using a VisiTech QLC100 spinning disk head mounted on a Nikon DIAPHOT 300 inverted microscope provided with solid laser sources (488, 561 and 642 nm), was used for all the time-lapse movie collections. A 40x oil or 60x water immersion objective lens was used where appropriate. The images were captured using a photometrics CoolSNAP HQ camera. The laser power input and the exposure time settings were set to the lowest possible levels to reduce photo damage to the cell. The conditions were first set up using third instar larvae brains prepared from the gfp-cdc20; aldB4–2 line for live imaging of the cell cycle progression in neuroblasts and then applied to other samples to keep the conditions consistent. The specifications are 19% of 27.0 mW original input of the 488 nm laser power for the excitation of the GFP fusion protein and 15% of 18 mW original input of the 561 nm laser power for the excitation of the RFP fusion protein. An exposure time of 200 ms was used throughout the experiments. Each experiment lasted for 1 h 40 min with images taken at 30 s intervals. This covers approximately two cell cycles of neuroblast development in the observed brain samples. The images were quantified using either MetaMorph or ImageJ software, giving values for fluorescent intensity or cell cycle length. Experiments were performed at a constant 22°C.
FRAP (fluorescent recovery after photo-bleach) analysis
FRAP experiments were performed using a spinning disk confocal microscopy system (Andor Revolution XD with FRAPPA Device). The isolated whole third instar larvae brains, which were mounted on a coverslip, were placed in a temperature-controlled incubator (Okolab) at 22°C on the stage of an Olympus IX81 microscope and viewed using a 60x water-immersion objective. Appropriate neuroblasts at the prophase stage, when the GFP-Cdc20 has already associated with kinetochores, or colchicine-arrested kinetochores were identified from fluorescent images acquired at 488 nm excitation (15% laser power) 525/50 nm emission. Two regions of interest to photobleach were defined on the image, one on the cytoplasm, the other encompassing the target (either selecting a single kinetochore or a part of the kinetochore cluster, Fig. 5). Photobleaching of the fluorophore to levels equivalent to background (non-cellular area) was achieved using pulses of 488 nm laser light at 15% (dwell time 20 × 60 µs). Images (~13 frames/s) were acquired for 4.2 s pre-bleach, photobleaching of samples took 1.2 ms and then recovery was continuously monitored for up to 2 min post-bleach, using IQ2 software (Andor). Fluorescence intensity over time in regions of interest was calculated using ImageJ. The exponential kinetics of fluorescence recovery after photobleaching was analyzed by fitting nonlinear regression curves using GraphPad Prism.
Acknowledgments
This work was supported by a Wellcome Trust project grant to J.-Y.H. and a Wellcome Trust Equipment grant 087961. We thank Mr. Michael Aitchison and Ms. Maureen Sinclair for technical support, Dr. Roger Karess (CNRS) for supplying a GFP-Mad2 transgenic line driven by the intrinsic Mad2 promoter for comparison. Dr. Yohanns Bellaïche (UMR 144 CNRS/Institut Curie, Paris) for his 2 BmRFP transgenic lines. A GFP-Mps1 transgenic line was a gift from Prof. Lehner (Institute of Molecular Life Sciences). aldB4–2 original semi-lethal line from Dr. Bill Gilliland (College of Science and Health, DePaul University).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/22916
References
- 1.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–93. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 2.Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, Salmon ED. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol. 2004;14:953–64. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 3.Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 4.McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, et al. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003;17:101–14. doi: 10.1101/gad.1040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell. 2009;16:105–17. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poddar A, Stukenberg PT, Burke DJ. Two complexes of spindle checkpoint proteins containing Cdc20 and Mad2 assemble during mitosis independently of the kinetochore in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:867–78. doi: 10.1128/EC.4.5.867-878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–36. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Millband DN, Hardwick KG. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol Cell Biol. 2002;22:2728–42. doi: 10.1128/MCB.22.8.2728-2742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen RH. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J Cell Biol. 2002;158:487–96. doi: 10.1083/jcb.200204048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol Biol Cell. 2002;13:755–66. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol. 1995;130:941–8. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardwick KG. Checkpoint signalling: Mad2 conformers and signal propagation. Curr Biol. 2005;15:R122–4. doi: 10.1016/j.cub.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Peters JM. Checkpoint control: the journey continues. Curr Biol. 2008;18:R170–2. doi: 10.1016/j.cub.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 14.Fischer MG, Heeger S, Häcker U, Lehner CF. The mitotic arrest in response to hypoxia and of polar bodies during early embryogenesis requires Drosophila Mps1. Curr Biol. 2004;14:2019–24. doi: 10.1016/j.cub.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Page SL, Nielsen RJ, Teeter K, Lake CM, Ong S, Wright KR, et al. A germline clone screen for meiotic mutants in Drosophila melanogaster. Fly (Austin) 2007;1:172–81. doi: 10.4161/fly.4720. [DOI] [PubMed] [Google Scholar]
- 16.Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–23. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vigneron S, Prieto S, Bernis C, Labbé JC, Castro A, Lorca T. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol Biol Cell. 2004;15:4584–96. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santaguida S, Tighe A, D’Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, Chen RH. Mps1 phosphorylation by MAP kinase is required for kinetochore localization of spindle-checkpoint proteins. Curr Biol. 2006;16:1764–9. doi: 10.1016/j.cub.2006.07.058. [DOI] [PubMed] [Google Scholar]
- 20.Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, et al. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106:83–93. doi: 10.1016/S0092-8674(01)00410-X. [DOI] [PubMed] [Google Scholar]
- 21.Liu ST, Chan GK, Hittle JC, Fujii G, Lees E, Yen TJ. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol Biol Cell. 2003;14:1638–51. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zich J, Sochaj AM, Syred HM, Milne L, Cook AG, Ohkura H, et al. Kinase activity of fission yeast Mph1 is required for Mad2 and Mad3 to stably bind the anaphase promoting complex. Curr Biol. 2012;22:296–301. doi: 10.1016/j.cub.2011.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maciejowski J, George KA, Terret ME, Zhang C, Shokat KM, Jallepalli PV. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J Cell Biol. 2010;190:89–100. doi: 10.1083/jcb.201001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hewitt L, Tighe A, Santaguida S, White AM, Jones CD, Musacchio A, et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li D, Morley G, Whitaker M, Huang JY. Recruitment of Cdc20 to the kinetochore requires BubR1 but not Mad2 in Drosophila melanogaster. Mol Cell Biol. 2010;30:3384–95. doi: 10.1128/MCB.00258-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tighe A, Staples O, Taylor S. Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores. J Cell Biol. 2008;181:893–901. doi: 10.1083/jcb.200712028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weaver BA, Bonday ZQ, Putkey FR, Kops GJ, Silk AD, Cleveland DW. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162:551–63. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buffin E, Lefebvre C, Huang J, Gagou ME, Karess RE. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol. 2005;15:856–61. doi: 10.1016/j.cub.2005.03.052. [DOI] [PubMed] [Google Scholar]
- 29.Jungreis I, Lin MF, Spokony R, Chan CS, Negre N, Victorsen A, et al. Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res. 2011;21:2096–113. doi: 10.1101/gr.119974.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray AW, Marks D. Can sequencing shed light on cell cycling? Nature. 2001;409:844–6. doi: 10.1038/35057033. [DOI] [PubMed] [Google Scholar]
- 31.Stucke VM, Silljé HH, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 2002;21:1723–32. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jelluma N, Brenkman AB, van den Broek NJ, Cruijsen CW, van Osch MH, Lens SM, et al. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 2008;132:233–46. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 33.Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–6. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 34.Gilliland WD, Wayson SM, Hawley RS. The meiotic defects of mutants in the Drosophila mps1 gene reveal a critical role of Mps1 in the segregation of achiasmate homologs. Curr Biol. 2005;15:672–7. doi: 10.1016/j.cub.2005.02.062. [DOI] [PubMed] [Google Scholar]
- 35.Orr B, Bousbaa H, Sunkel CE. Mad2-independent spindle assembly checkpoint activation and controlled metaphase-anaphase transition in Drosophila S2 cells. Mol Biol Cell. 2007;18:850–63. doi: 10.1091/mbc.E06-07-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buffin E, Emre D, Karess RE. Flies without a spindle checkpoint. Nat Cell Biol. 2007;9:565–72. doi: 10.1038/ncb1570. [DOI] [PubMed] [Google Scholar]
- 37.Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10:1411–20. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raff JW, Jeffers K, Huang JY. The roles of Fzy/Cdc20 and Fzr/Cdh1 in regulating the destruction of cyclin B in space and time. J Cell Biol. 2002;157:1139–49. doi: 10.1083/jcb.200203035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J, Raff JW. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 1999;18:2184–95. doi: 10.1093/emboj/18.8.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]