Summary
Background
Circadian (~24hr) rhythms offer one of the best examples of how gene expression is tied to behavior. Circadian pacemaker neurons contain molecular clocks that control ~24hr rhythms in gene expression that in turn regulate electrical activity rhythms to control behavior.
Results
Here we demonstrate the inverse relationship: there are broad transcriptional changes in Drosophila clock neurons (LNvs) in response to altered electrical activity, including a large set of circadian genes. Hyperexciting LNvs creates a morning-like expression profile for many circadian genes while hyperpolarization leads to an evening-like transcriptional state. The electrical effects robustly persist in per0 mutant LNvs but not in cyc0 mutant LNvs suggesting that neuronal activity interacts with the transcriptional activators of the core circadian clock. Bioinformatic and immunocytochemical analyses suggest that CREB family transcription factors link LNv electrical state to circadian gene expression.
Conclusions
The electrical state of a clock neuron can impose time-of-day to its transcriptional program. We propose that this acts as an internal zeitgeber to add robustness and precision to circadian behavioral rhythms.
Introduction
Electrical activity can initiate transcriptional changes in neurons that alter their synaptic strength and this has been extensively studied in the context of learning and memory [1]. Hundreds of activity-dependent genes have been identified as have specific transcription factors that regulate gene expression in response to electrical activity e.g.[2–5].
The interplay between electrical activity and transcription is also relevant to circadian pacemaker neurons where ~24hr rhythms in gene expression and neural activity define the functional state of clock neurons [6]. Rather than simply transmitting the state of the molecular clock to downstream neurons to regulate circadian behavior, an emerging view is that neural activity supports molecular clock oscillations. Preventing mammalian pacemaker neurons in the Suprachiasmatic Nucleus (SCN) from firing action potentials or reducing extracellular K+ dampened mPeriod1 transcriptional rhythms [7, 8]. Furthermore, hyperpolarizing or hyperexciting the master Drosophila circadian pacemaker neurons (LNvs) alters rhythmic clock protein accumulation [9–11], although the effects of hyperpolarization are less severe if LNvs are only hyperpolarized in adulthood [12]. Ca2+ influx has also been implicated in phase-shifting the molluscan retinal clock [13]. While these studies suggest a relationship between membrane excitability and the molecular oscillator in pacemaker neurons, there is relatively little known about the nature of this interaction.
Here, we characterized the transcriptional responses to spatially, temporally and directionally controlled changes to membrane excitability by performing microarrays on circadian pacemaker neurons purified from Drosophila. Strikingly, a large set of circadian genes is bi-directionally regulated with reference to the cell’s electrical state, implying a potent effect of neuronal excitability on circadian gene expression. Many of these changes correlate with the nature of the alterations such that hyper-excitation created a morning-like expression profile and hyperpolarization created an evening-like expression profile. Hyperexcitation induced a morning-like transcriptional program even in a per0 background whereas the effect of hyperpolarization on circadian gene expression was disrupted in cyc0 mutants, implying that neuronal activity interacts with the core clock transcriptional activators. We establish links between activity-dependent pathways and circadian gene expression and provide evidence that the CREB transcription factor family – integrators of neuronal activity for transcription in learning and memory – are part of the activity-dependent regulatory network in circadian pacemaker neurons.
Results
Broad transcriptional response in LNvs to increased electrical excitability
To test how increased electrical excitability affects gene expression in LNvs, brains were dissected from larvae with an LNv-specific Gal4 (Pdf-Gal4)[14] expressing UAS-NaChBac (NCB), a low activation threshold voltage-gated Na+ channel from bacteria, which increases LNv excitability in the evening [15]. These Pdf>NCB larvae also contained a Pdf-RFP transgene to label LNvs for flow cytometry. Larval LNvs were isolated on day 2 in constant darkness (DD) at CT15 (subjective evening) when expression of core clock genes like period (per) and timeless (tim) is high.
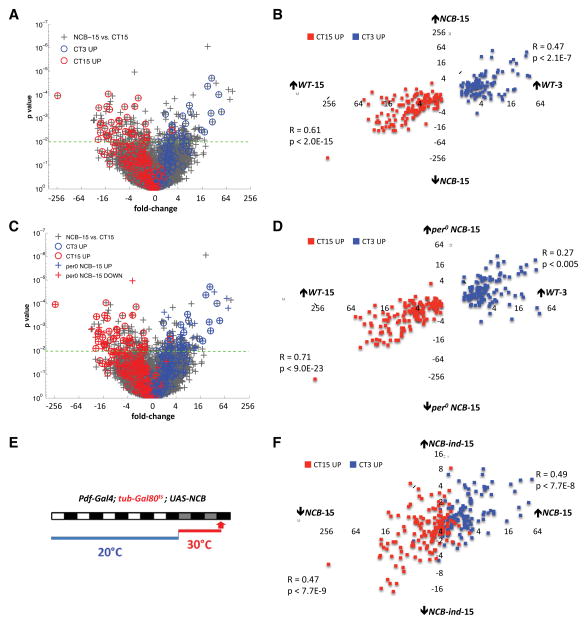
We compared expression profiles from LNvs expressing NCB at CT15 (NCB-15) with those from control LNvs isolated at CT15 (WT-15). We identified 336 mRNAs either up- or down-regulated in NCB-15 (p<.01, False Discovery Rate (FDR) < 8%, fold change (FC) > 1.5, Table S1). In separate LNv GeneChip experiments, we identified 249 “circadian transcripts” with differential expression in wild type larvae between CT3 (subjective morning, WT-3) and CT15 (Table S2). We noticed that 81 of the 336 NCB-regulated transcripts show circadian regulation in wild-type LNvs (Fig. 1A–B and Table S2). Interestingly, circadian transcripts up-regulated in NCB-15 are more highly expressed at CT3 than at CT15 in wild type LNvs. For example, Clock (Clk) and cryptochrome (cry) transcript levels were up-regulated 8-fold (p<.001) and 5-fold (p<.005) respectively in NCB-15. A scatter plot of fold-change versus p-value in the NCB-15 versus WT-15 comparison (Fig. 1A) illustrates circadian genes high at WT-3 (blue circles) or WT-15 (red circles) among the ~19,000 probesets (grey “+”). Of the 110 circadian mRNAs normally high at WT-3, 36 are significantly up-regulated in NCB-15, and conversely, 45 of 139 of circadian mRNAs normally high at WT-15 are significantly down-regulated in NCB-15 (p<0.01, FC>1.5). We also noticed a clear split in how expression of the remaining circadian transcripts is affected by NCB at CT15. Although many fall below the p<.01 significance line, genes normally higher at CT3 than CT15 tend to be on the right of the scatter plot (i.e. up-regulated in NCB-15) and the converse for genes normally higher at CT15 (Fig. 1A).
Figure 1. Hyperexciting LNvs creates a morning-like transcript profile in the evening.
(A) Scatter-plot of fold change versus p-value for all mRNAs in the NCB-15 vs. WT-15 comparison (grey “+”). We identifed 336 NCB-regulated mRNAs using p<.01, FDR < 8 and FC > 1.5 as cut-offs. The 249 circadian genes are identified with blue (high at CT3) or red (high at CT15) circles.
(B) Scatter-plot of wild type circadian fold differences (WT-3 vs. WT-15) for each of the 249 circadian mRNAs against their NCB-fold differences (NCB-15 vs. WT-15). Blue boxes: mRNAs normally high at WT-3. Red boxes: mRNAs normally high at WT-15. Pearson correlation coefficients (R values) were transformed to test hypotheses of no correlation (p values).
(C) Re-plot of the NCB-15 versus WT-15 comparison from Figure 1A with the addition of 246 mRNAs either up- (blue “+”) or down-regulated (red “+”) in the per0; NCB-15 vs. WT-15 comparison.
(D) Scatter-plot of circadian fold differences (WT-3 vs. WT-15) for all circadian mRNAs against their per0; NCB-fold differences (per0; NCB-15 vs. WT-15).
(E) Pdf-Gal4>NCB/tub-gal80ts larvae were entrained in LD cycles at 20°C, when Gal80t.s represses Gal4 activity to prevent NCB-expression. Larvae were transferred to DD at 30°C to induce NCB-expression (NCB-ind-15) and dissected at CT15 on day 2 in DD. Control larvae without a NCB transgene went through the same temperature shifts (Pdf-Gal4>+/tub-gal80ts, WT-ind-15).
(F) Scatter-plot of fold differences for each of the 249 circadian mRNAs between the NCB-15 vs. WT-15 and NCB-ind-15 vs. WT-ind-15 comparisons.
To quantify the relationship between NCB-regulation and circadian phase for the full set of circadian mRNAs, we plotted wild-type circadian fold differences (WT-3 vs. WT-15) for all 249 circadian mRNAs against their NCB-fold differences (NCB-15 vs. WT-15, Fig. 1B). The linear correlations are highly significant (R=0.47, p<2.1E-7 and R=0.61, p<2.0E-15 for up- and down-regulated mRNAs, respectively), indicating that artificially hyperexciting LNvs via NCB creates a morning-like transcript profile in the evening.
The NCB effect persists in the absence of a functional molecular oscillator
A central notion in circadian biology is that a clock cell’s circadian transcription follows its molecular clock. However the NCB data reveal a potent influence of electrical state on circadian gene expression. The per0 mutation stops circadian rhythms in gene expression since it removes the key transcriptional repressor that inhibits the activity of CLOCK (CLK) and CYCLE (CYC), the transcriptional activators that together activate per and tim expression [16]. To test whether NCB requires an intact molecular oscillator to impose a morning-like transcriptional state in the evening, we performed GeneChips on LNvs expressing NCB in a per0 mutant background isolated at CT15 (per0; NCB-15).
We identified 246 differentially expressed mRNAs when comparing per0; NCB-15 to WT-15 (Table S3). Of these, 64 mRNAs are circadian and 44 overlap with the NCB-15 set (Table S3). Fig. 1C superimposes these 246 mRNAs (colored “+”) on the original NCB-15 versus WT-15 comparison. There is a clear pattern: mRNAs up-regulated in per0; NCB-15 also tend to be up-regulated in NCB-15, and mRNAs down-regulated in per0; NCB-15 tend to be down-regulated in NCB-15.
To quantify this relationship, we compared wild-type circadian fold differences for all 249 circadian mRNAs with their per0; NCB-fold differences. The positive correlation is highly significant (R=0.27, p<0.005 and R=0.71, p<9.0E-23 for up- and down-regulated mRNAs, respectively, Fig. 1D). Thus hyperexciting LNvs induces a morning-like transcriptional program even without per.
NCB-regulated transcripts are temporally inducible
Altered synaptic activity can disrupt neural network development and lead to compensatory molecular changes [17]. Furthermore, some gene expression changes observed in mutant Drosophila with chronic nervous system hyperexcitation were not seen with acute excitation [2]. We tested if inducing NCB expression in larval LNvs after their differentiation recapitulates the effects of constitutive NCB-expression. For this, we added a temperature sensitive tubulin-Gal80ts transgene (tub-Gal80ts, [18]) into the Pdf-Gal4; UAS-NCB background to control NCB expression.
Larvae were raised in Light:Dark (LD) cycles at 20°C so that Gal80ts repressed Gal4 activity to prevent NCB-expression during LNv development and entrainment. Larvae were then shifted to DD at 30°C, inactivating Gal80ts to induce NCB expression and dissected at CT15 on day 2 in DD (Fig. 1E). We generated expression profiles from LNvs from Pdf-RFP/Pdf-Gal4; tub-Gal80ts/UAS-NaChBac larvae (NCB-ind-15) and LNvs from larvae following the same temperature regime without a NCB transgene (WT-ind-15: Pdf-RFP/Pdf-Gal4; tub-Gal80ts/+).
We found 315 mRNAs differentially-expressed between NCB-ind-15 and WT-ind-15 (NCB-ind-15 mRNAs, Table S4). Comparing the overlap between NCB-ind-15, NCB-15, and circadian datasets, 50 of the 315 NCB-ind-15 mRNAs overlap with the NCB-15 dataset, and 20 of these 50 transcripts are normally expressed with a circadian rhythm (Table S4). We also noted significant positive correlations (R=0.49, p<7.7E-8; R=0.47, p<7.7E-9) between levels of the 249 circadian transcripts in the NCB-15 and NCB-ind-15 conditions (Fig. 1F). Thus hyper-excitation affects circadian transcripts even when induced post-differentiation. However, 75% of the 50 mRNAs altered by chronic and induced NCB show reduced fold-change and statistical significance in the induced condition, which likely reflects a cumulative and/or stronger effect of constitutive NCB expression. The mRNAs that only respond to chronic NCB-expression may represent homeostatic responses to persistent hyper-excitation. However, the ability of NCB to alter circadian gene expression even when more acutely induced is consistent with the idea that neuronal activity may normally help generate circadian rhythms in LNv gene expression.
Hyperpolarizing LNvs in the morning induces an evening-like circadian expression pattern
If the changes in gene expression induced by NCB in LNvs are a function of electrical polarity, many could occur with a reciprocal manipulation. To test this, we used the mammalian Inward Rectifier K+ channel Kir2.1 (Kir) since Kir-expressing adult LNvs are hyperpolarized and lose spontaneous action potential firing [10]. Kir-expressing LNvs were isolated at CT3 and CT15 (Kir-3 and Kir-15).
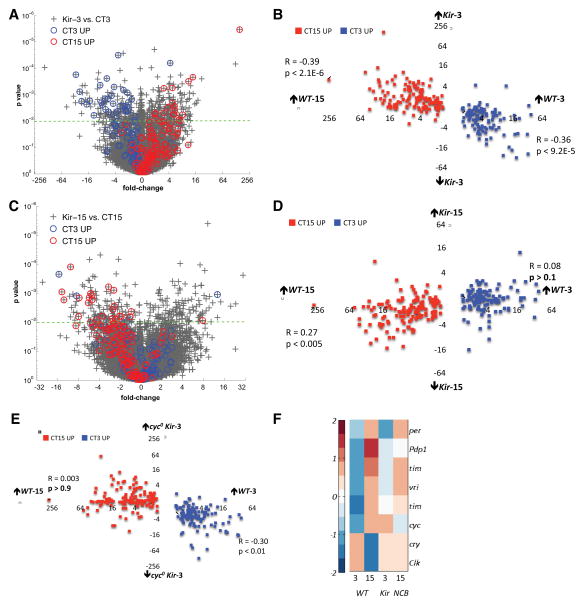
To identify changes in expression caused by decreased LNv excitability, Kir-3 was compared to control WT-3. Applying the same cutoffs for differential regulation as for NCB, we identified 319 Kir-regulated mRNAs (Fig. 2A and Table S5). Many (50/319) Kir-regulated mRNAs show circadian oscillations in wild-type LNvs, with a correlation between their normal phase (high at WT-3 or WT-15) and the direction of Kir-regulation (Fig. 2A and Table S5). To quantify this relationship, we compared wild-type circadian fold differences (WT-3 vs. WT-15) for each of the 249 circadian mRNAs with their Kir-fold differences (Kir-3 vs. WT-3) (Fig. 2B). We observed a significant negative correlation, with mRNAs high at WT-3 down-regulated in Kir-3 (R=−0.36, p<9.2E-5) and mRNAs high at WT-15 up-regulated in Kir-3 (R=−0.39, p<2.1E-6).
Figure 2. Hyperpolarizing LNvs at morning induces an evening-like circadian expression pattern.
(A&C) Scatter-plot of fold change versus p-value for all mRNAs in the Kir-3 vs. WT-3 (A) and Kir-15 vs. WT-15 (C) comparisons. Circadian genes (249) are identified with blue (high at WT-3) and red (high at WT-15) circles.
(B&D) Scatter-plot of wild type circadian fold differences (WT-3 vs. WT-15) for each of the 249 circadian mRNAs against their Kir-3 fold differences (Kir-3 vs. WT-3, B) and Kir-15-fold differences (Kir-15 vs. WT-15, D).
(E) Scatter-plot of wild type circadian fold differences (WT-3 vs. WT-15) for all circadian mRNAs against their cyc0; Kir-3 fold differences (cyc0; Kir-3 vs. WT-3).
(F) Expression values for core clock mRNAs (row) across four genomic conditions (columns) standardized by mean centering (row mean = 0, row standard deviation =1) and assigned color-map values based on their standard deviation from the row mean.
However, there were less dramatic differences in the Kir-15 vs. WT-15 comparison (Fig. 2C–D). Kir does alter expression of some circadian genes normally high at WT-15 (R=0.27, p<0.005), consistent with the documented effects of Kir on some clock protein oscillations [9]. However, Kir does not impose an overall morning-like expression profile in the evening, since expression of most circadian genes normally higher at CT3 than CT15 is relatively unchanged in Kir-15 LNvs (Fig. 2D). Thus Kir specifically induces an evening-like expression profile in the morning rather than reversing the circadian transcriptional profile independent of sampling time.
The resting membrane potential of s-LNvs (the adult version of larval LNvs) shows diurnal variations, with increased excitability around dawn [19]. Strikingly, we found that hyperexciting LNvs via NCB in the evening induced a morning-like transcript profile, while hyperpolarizing LNvs via Kir in the morning created an evening-like profile. The traditional view has been that the molecular clock encodes time-of-day by driving the activity of properly timed neural outputs. The data here indicate the inverse relationship: neural activity can bi-directionally regulate circadian gene expression.
Electrical activity interacts with the core transcriptional activators
NCB up-regulation of morning genes and down-regulation of evening genes persisted in per0 mutant LNvs, indicating that electrical activity can circumvent the repression step in the molecular clock. We next examined if electrical activity interacts with the clock transcriptional activators. For this, we generated expression profiles of LNvs isolated at CT3 expressing Kir in a cyc0 mutant background (cyc0; Kir-3). We chose this genotype because CLK/CYC activates evening genes including per, tim, PAR-domain protein 1 (Pdp1), and vrille (vri), while morning genes such as Clk and cry are highly expressed in cyc0 mutants [20, 21]. However, evening genes are up-regulated while morning genes are down-regulated in Kir-3 LNvs (Fig. 2A–B). Thus it seemed informative to assay circadian gene expression with two conflicting manipulations (Kir and cyc0).
Fig. 2E shows that Kir expression does not up-regulate mRNAs normally high in the evening in a cyc0 mutant background (cyc0; Kir-3 vs. WT-3, R=0.003, p>0.9). However, cyc was not essential for the down-regulation of the mRNAs high at WT-3 (R=−0.30, p<0.01), raising the possibility that some mRNAs high at WT-3 are controlled by activity-dependent transcription factors.
The core molecular clock drives rhythmic electrical activity in clock neurons [19, 22–24]. The data here reveal that electrical activity cooperates with the core clock transcriptional activators to regulate circadian transcription (Fig. 2E). We propose that this mutually dependent interaction helps determine the phase and/or amplitude of circadian oscillations for many mRNAs.
Expression of Kir or NCB in LNvs affects behavioral rhythms and can change molecular clock rhythms [9, 11]. However, this has only been characterized at the protein level for TIM, PER and PDP1. To understand the extent of electrical effects on core clock gene expression, we examined which are regulated by neuronal excitability. A heat map showing expression of the core Drosophila clock genes across genomic experiments (Fig. 2F) revealed that some core clock mRNAs are sensitive to altered neuronal activity (FC>1.5, p<0.01). The normally low Clk and cry RNA levels at CT15 were up-regulated by NCB whereas cyc was up-regulated by Kir at CT3. tim mRNA levels were slightly down-regulated in NCB-15 but not in Kir-3. Pdp1 mRNA levels were not affected by Kir or NCB expression and per mRNA levels are only down-regulated ~3-fold when comparing Kir-15 vs WT-15. These modest effects on per, tim and Pdp1 RNA levels suggest that post-transcriptional regulation also contributes to the effects of electrical activity on clock protein abundance [9, 11]. Overall, Fig. 2F indicates that altering LNv excitability fine-tunes core clock mRNA levels. Fig. 2F is also in line with the modest effects on PER protein rhythms with inducible Kir expression in LNvs [12]. Thus the most dramatic effects of electrical activity are not on core clock genes (see Tables S1 and S4).
Interactions between electrical activity and circadian transcription
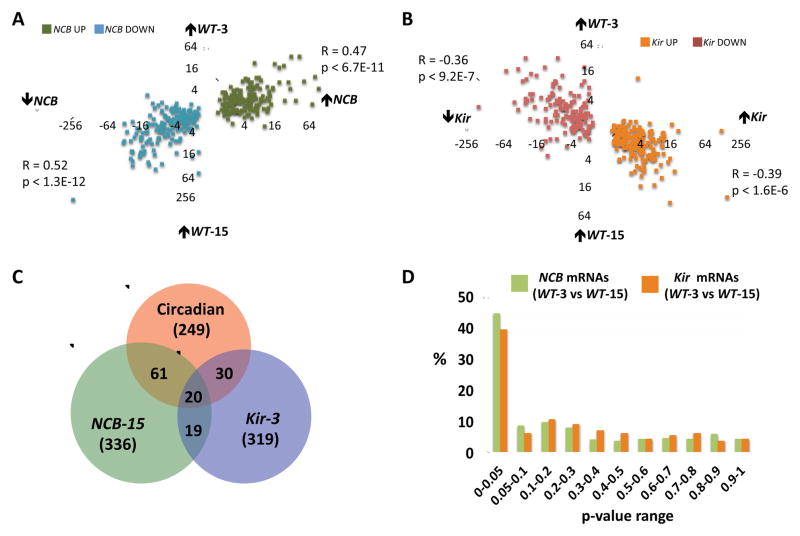
The data so far has demonstrated bi-directional regulation of the circadian transcriptional program with reference to the electrical state of LNvs. Since the electrical activity in wild-type LNvs shows daily changes [19, 24], electrically regulated mRNAs could have circadian expression as a result of rhythmic LNv excitability. To search for evidence of this, we measured the circadian changes in expression for all 336 NCB-regulated mRNAs (the inverse of Fig. 1 & 2). We plotted the NCB-fold differences for these mRNAs (NCB-15 vs. WT-15) against their circadian fold differences (WT-3 vs. WT-15). The linear correlations are highly significant (R=0.47, p<6.7E-11 and R=0.52, p<1.3E-12 for up- and down-regulated mRNAs, respectively, Fig. 3A). Importantly, the mRNAs up-regulated by NCB are normally higher in the morning while the down-regulated NCB transcripts are normally higher in the evening. These data indicate many of the transcripts regulated by NCB likely normally exhibit circadian changes in expression in wild-type LNvs.
Figure 3. A strong relationship between electrical activity and circadian gene expression in LNvs.
(A) Scatter-plot of NCB fold differences (NCB-15 vs. WT-15) for all 336 NCB-regulated mRNAs against their circadian fold differences (WT-3 vs. WT-15). mRNAs up- and down-regulated by NCB are in green and blue respectively.
(B) Scatter-plot of Kir fold differences (Kir-3 vs. WT-3) for all 319 Kir-regulated mRNAs against their circadian fold differences (WT-3 vs. WT-15). mRNAs up- and down-regulated by Kir are in orange red respectively.
(C) Overlap of significantly regulated genes (p<0.01; FC>1.5) from 3 independent genomic experiments: WT-3 vs. WT-15 (red); NCB-15 vs. WT-15 (green); and Kir-3 vs. WT-3 (blue).
(D) p-values of 336 NCB-regulated mRNAs (green) or 319 Kir-regulated mRNAs (orange) from the WT-3 vs. WT-15 comparison.
See also Figure S1.
Next we plotted Kir fold differences for all 319 Kir regulated mRNAs (Kir-3 vs. WT-3) against their circadian fold differences (WT-3 vs. WT-15). Here too, we found a significant negative correlation, with mRNAs up-regulated by Kir normally higher at WT-15 (R=−0.36, p<9.2E-7) and mRNAs down-regulated by Kir normally higher at WT-3 (R=−0.39, p<1.6E-6, Fig. 3B). Together, these data indicate that a significant fraction of electrically regulated mRNAs exhibit circadian changes in LNvs: i.e. genes activated by increased electrical activity tend to be higher in the subjective morning and mRNAs up-regulated by hyper-polarization tend to be higher in the evening.
However, we had found that only 111 of 616 (18%) of the NCB and/or Kir regulated mRNAs are under circadian control with p<0.01 (Fig. 3C). To evaluate the relationship between these high confidence activity-dependent transcripts and circadian phase, we measured the p-values for rhythmic expression (different between WT-3 vs. WT-15) for all 336 NCB- and 319 Kir-regulated mRNAs. We found that 44.3% of NCB-regulated mRNAs (149/336) and 39.2% of Kir regulated mRNAs (125/319) show circadian regulation with p<0.05 (Fig. 3D). Thus a large fraction of electrically regulated mRNAs display circadian expression, but some were filtered out using our original cutoffs (FC>1.5, p<0.01, Fig. 3C). Our analysis may also underestimate the extent of circadian regulation in the NCB and Kir regulated mRNAs since we only sampled two timepoints per day and may have missed genes whose expression peaks with different phases.
We also re-examined how many circadian transcripts are altered by NCB and Kir with p<0.05 by plotting the p-values of all 249 circadian transcripts (WT-3 vs. WT-15, FC>1.5, p<0.01) from NCB-15 vs. WT-15 and Kir-3 vs. WT-3 comparisons (Fig. S1A and S1B). 33% and 20% of the 249 circadian mRNAs are NCB- and Kir-regulated with a cut-off of p<0.01 (Fig. 3C). However, these numbers rise to 52% for NCB and 40% for Kir when the p-value is relaxed to 0.05 (Fig. S1). Overall these analyses indicate a major overlap between activity-dependent and circadian gene expression in LNvs.
To better understand the directionality of electrical effects on circadian transcripts, we plotted the fold change differences of the circadian transcripts in response to NCB or Kir without filtering by p-value or fold-change cutoffs. Kir down-regulates 92% (101/110) of the transcripts normally high at CT3 (Kir-3 vs WT-3) while NCB up-regulates 85% (94/110) of these transcripts (NCB-15 vs WT-15) (Fig. S1C). Conversely, Kir up-regulates 84% (117/139) of the transcripts normally high at CT15 (Kir-3 vs WT-3) while NCB down-regulates 91% (126/139) of these transcripts (NCB-15 vs WT-15) (Fig. S1D). These data further demonstrate the dramatic reorganization of LNv circadian gene expression by Kir and NCB.
Validating the GeneChip data
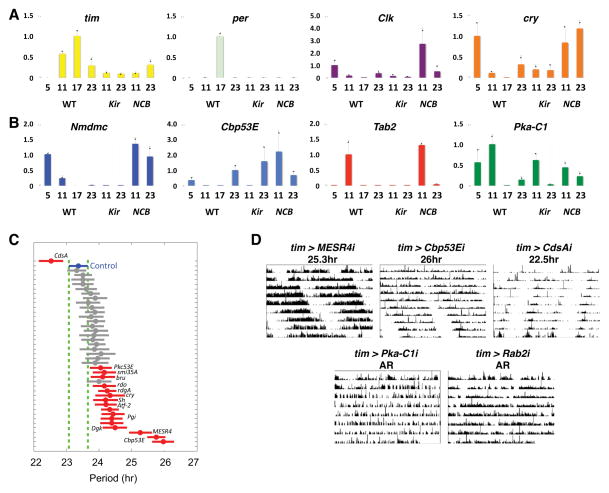
We used qPCR to validate rhythmic expression of several transcripts and the effects of neuronal activity. To provide higher resolution, larval LNvs were isolated at four time-points on day 2 in DD. Initially, we measured the levels of 4 core clock transcripts (per, tim, Clk and cry) and confirmed circadian regulation (Fig. 4A, p<0.01, one-way ANOVA for per and tim, p<0.05 for cry, and p=0.05, t-test for Clk, CT5 vs CT11).
Figure 4. Molecular and circadian behavioral validation of electrically sensitive genes.
(A) Wild-type larval LNvs were isolated at CT5, CT11, CT17 and CT23 and Kir or NCB expressing LNvs at CT11 and CT23 on day 2 in DD. RNA amplification and qPCR are described in Experimental Procedures, with 3 replicates for each condition except WT-17 (2 replicates) to measure expression of tim, per, Clk and cry.
(B) qPCR as in A to analyze Nmdmc, Cbp53E, Tab2 and Pka-C1 expression.
(C) tim(UAS)-Gal4 flies were crossed to 41 UAS-RNAi transformants with UAS-dcr-2 co-expressed to boost RNAi effectiveness. Group mean activity period (circle) with confidence interval are plotted. Non-overlapping intervals (red) are significantly different than control (blue), p<.05 (one-way ANOVA with post-hoc Tukey-Kramer multiple-comparison).
(D) Representative actograms from single RNAi-expressing flies whose activity periods were significantly lengthened (MESR4 and Cbp53E), shortened (CdsA), or arrhythmic (AR, Pka-C1 and Rab2). Period shown is the group mean.
According to our GeneChip experiments, NAD-dependent methylenetetrahydrofolate dehydrogenase (Nmdmc), Calbindin 53E (Cbp53E), TAK1-associated binding protein 2 (Tab2) and Pka-C1 mRNAs exhibited strong circadian changes with higher levels in the subjective morning (WT-3) than evening (WT-15) and were electrically regulated. The additional time-points for qPCR in Fig. 4B give more accurate circadian expression profiles: Nmdmc peaked at CT5 with a robust oscillation (p<0.0001, one-way ANOVA); Tab2 had a sharp peak at CT11 (p<0.05); Cbp53E showed circadian variation with higher levels at CT23 than CT11 and CT17 (p<0.05); and Pka-C1 had higher levels during the subjective day than at CT17 (p<0.05, t-test). In spite of some differences between their profiles, all these transcripts exhibit circadian profiles with higher levels in the subjective day then early evening (CT17), coinciding with their GeneChip values (WT-3 > WT-15).
We also measured levels of these 8 transcripts in Kir or NCB expressing LNvs isolated at two time-points (CT11 and CT23) on day 2 in DD. The GeneChip data showed that, of the core clock regulators, expression of Clk, cry, cyc, tim and per is altered by neuronal excitability with FC>1.5, p<0.01. qPCR revealed that per is Kir and NCB sensitive at both time-points (t-test, p<0.05) and that tim abundance at CT11 was decreased by both Kir and NCB (p<0.01). Clk and cry expression was up-regulated by NCB expression (p<0.05). Of the electrically-sensitive transcripts, Nmdmc was bi-directionally regulated by Kir and NCB (t-test, p<0.05) and Cbp53E and Pka-C1 lost their time-dependent regulation in NCB-expressing LNvs (p>0.1). Finally, Tab2 levels were reduced by Kir expression (p<0.01) and up-regulated by NCB at CT23 (p<0.05).
These findings validate that electrical activity regulates the levels of some circadian transcripts, indicating a central role for neuronal excitability in regulating LNv gene expression. However, changes in LNv electrical state do not completely activate or shutdown core clock transcripts, but rather fine-tune their levels.
A role for activity-dependent genes in circadian behavior
Activity-dependent genes regulate cellular processes that alter neuronal function and behavior e.g. [5, 25]. To test if electrically regulated transcripts affect circadian behavior, we used RNAi to knockdown their expression in clock neurons in vivo and measured adult locomotor rhythms.
Focusing on genes with known functions, we screened 41 transgenic lines that target 31 genes. We identified 19 RNAi lines (14 genes) whose expression in clock neurons altered the period of locomotor rhythms (p<0.05, one-way ANOVA with post-hoc Tukey-Kramer multiple-comparison, Fig. 4C–D; Table S6). RNAi transgenes that target 4 genes in the inositol phosphate/diacyglycerol pathway [CDP diglyceride synthetase (CdsA), Protein C kinase 53E (Pkc53E), Diacyl glycerol kinase (Dgk) and retinal degeneration A (rdgA)] altered circadian period. The most dramatic period-altering phenotype (26hr) came from targeting Cbp53E, a Ca2+-buffering protein, highly related to mammalian Calbindin, implicated in circadian rhythms and light entrainment of the SCN [26]. Finally, an RNAi targeting a regulator of the CREB pathway, Pka-C1, made flies arrhythmic. The phenotypes results from this screen indicate that electrically regulated genes are important in generating 24hr rhythms and provide insight into electrically sensitive signaling pathways that regulate circadian behavior.
CrebA and CrebB protein levels are electrically sensitive and are predicted to bind many NCB and Kir-targets
To identify transcription factors (TFs) that bind sequences common to co-regulated genes, we used REDUCE to search for novel n-mers over-represented among genes sharing similar fold-change responses within each condition [27]. We then used TESS to search TF binding site databases to identify factors known to interact with these motifs [28]. REDUCE found several motifs (Table S7), including an 8-mer perfect match to the mammalian CRE-motif (TGACGTCA), which is bound by the ATF/CREB family of TFs. Occurrence of this motif correlates with a set of 31 mRNAs down-regulated in Kir-3 compared to WT-3 (p<5.0E-5, Table S7). Using mammalian CREB positional weight matrices (PWMs) as inputs, we also tested if CRE-containing genes are enriched among the NCB-15 and Kir-3 datasets. We scanned the promoters (3kb) and first introns of all genes in the genome, and identified significant enrichment of putative CRE-targets among the differentially expressed genes for both NCB-15 vs. WT-15 and Kir-3 vs. WT-3 comparisons (Hypergeometric test, p<.005, Table S7).
These data were striking because our cluster analysis (Fig. S2) had revealed that cAMP response element binding protein A (CrebA) and Activating transcription factor-2 (Atf-2), two members of the ATF/CREB-family of transcription factors, exhibited circadian regulation (higher in WT-3 than WT-15) and were bi-directionally regulated by electrical activity (high in NCB-15 and low in Kir-3). Expression of CrebB, a cAMP-responsive member of this family, is also up-regulated in NCB-15 (~8-fold, p<0.05). Expression of Epac and cAMP-dependent protein kinase 1 (Pka-C1), key components of cAMP signaling, also cluster with CrebA and Atf-2 respectively (Fig. S2). The coherent expression pattern of cAMP- and CREB-related genes suggests the importance of transcriptional regulation of this pathway in LNvs in response to NCB and Kir.
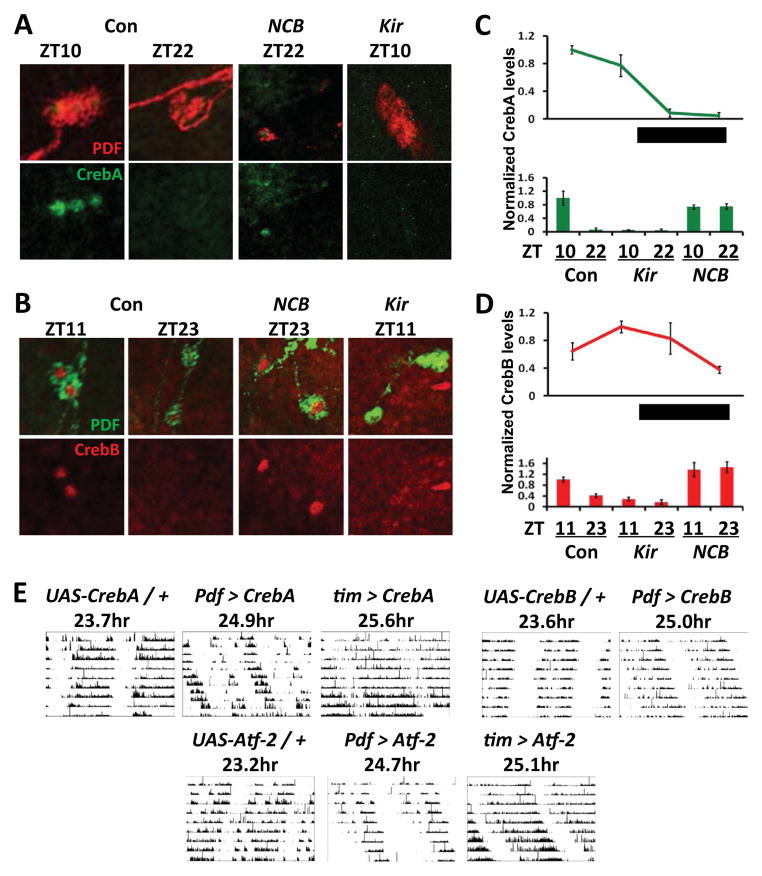
Based on these bioinformatic and expression data, we tested if Creb protein levels follow transcript levels in larval LNvs. We measured CrebA and CrebB levels in LD cycles and detected rhythms in their abundance: CrebA levels were higher during the day (ZT4 and ZT10) than at night (ZT16 and ZT22) and CrebB levels were highest at ZT11 (Fig. 5A–D, p<0.01, one way ANOVA followed by Tukey comparison). Next, we tested whether altering LNv neuronal activity changes CrebA and CrebB protein levels. NCB increased CrebA and CrebB levels at the end of the night, when their levels are normally low, while Kir decreased CrebA and CrebB abundance at the end of the day, when their levels are normally high (p<0.01, Fig. 5A–D). Together, these data further validate the GeneChip expression data by showing that electrical alteration mis-regulates CrebA and CrebB protein levels.
Figure 5. Creb family member protein levels are electrically sensitive and their over-expression alters circadian behavior.
(A) Top: Control y w (Con) larval brains were dissected at ZT10 or ZT22. Pdf > NCB brains were dissected at ZT22 (NCB ZT22) and Pdf > Kir at ZT10 (Kir ZT10) and stained using antibodies to CrebA (green) and PDF (to mark LNvs, red). Bottom: red channel (PDF) removed from images in top panel.
(B) As in A except that brains were stained with antibodies to CrebB (red) and PDF (green) at ZT11 and ZT23.
(C) Top: quantification of CrebA immuno-staining in wildtype (y w) larval brains at four time points in LD. Each data point represents the average CrebA intensity (background corrected) from 5 brain hemispheres, normalized to peak. Bottom: quantification of CrebA in y w (Con), Pdf>Kir (Kir) and Pdf>NCB (NCB) larval brains at ZT10 and ZT22.
(D) As in C except that CrebB immuno-staining was quantified.
(E) Top left: Representative actograms of UAS-CrebA flies crossed to (from left to right): control y w flies (UAS-CrebA/+), flies with two copies of Pdf0.5-Gal4 (Pdf > CrebA), or tim(UAS)-Gal4 flies (tim > CrebA). Top right: Representative actograms of UAS-CrebB flies crossed to either y w flies (UAS-CrebB/+) or flies with two copies of Pdf0.5-Gal4 (Pdf > CrebB). Bottom: Representative actograms of UAS-Atf-2 flies crossed to either control y w flies (UAS- Atf-2/+), flies with two copies of Pdf0.5-Gal4 (Pdf > Atf-2) or tim(UAS)-Gal4 flies (tim > Atf-2). Periods shown are the group mean.
See also Figure S2.
Mammalian CREB plays a role in responding to neuronal activity e.g. [4] and in light-resetting of SCN pacemaker neurons [29]. Furthermore, CrebB mutant flies have either ~1hr shorter periods than wild type flies or are arrhythmic [30]. We tested the behavioral role of CrebA and CrebB in LNvs by over-expression and found that this lengthened the circadian period to 24.9hr and 25.0hr respectively (Fig. 5E, Table S8). UAS-CrebA resulted in even longer rhythms (25.6hr) when expressed with tim-Gal4, a stronger and broader clock neuron driver [31]. Flies had normal period rhythms (23.8hr) when UAS-CrebA was expressed in all clock neurons except LNvs (using tim-Gal4 and Pdf-Gal80), indicating that the long period rhythm requires CrebA over-expression in LNvs (Table S8). Shimizu et al. [32] detected ATF-in adult LNvs and suggested that Atf-2 regulates sleep. We found that over-expressing Atf-2 either in LNvs or in all clock neurons gave a period-lengthening similar to CrebA and CrebB over-expression (Table S8).
Starting by identifying a set of co-regulated genes, our analyses revealed that members of the Drosophila Creb/ATF family likely regulate a set of electrically sensitive circadian genes. Since CrebA, CrebB and Atf-2 RNA and/or protein levels are regulated by neuronal activity in LNvs, and since normal levels of these proteins are required for 24hr rhythms, they are strong candidates for mediating the effect of electrical activity on circadian gene expression.
Discussion
Pacemaker neuron excitability can regulate circadian gene expression
By analyzing the transcriptome of a single pacemaker cell type to spatially, temporally, and directionally controlled changes to its excitability, we identified large sets of mRNAs whose levels can be regulated by the electrical state of LNvs. Strikingly, expression of circadian genes in LNvs correlate with the nature of the alterations: hyperexcitation triggers a morning-like expression profile and hyperpolarization an evening-like profile. These results demonstrate the ability of neural activity to reprogram the transcriptional oscillations in LNvs. Some of this reprogramming occurs through an interaction with the core clock activators but many mRNAs normally high in the morning likely use additional transcriptional mechanism(s) for rhythmic expression. Although constitutively manipulating the electrical state of a neuron is not physiological, the potent transgenes used here allowed us to identify how strongly and broadly electrical activity can affect LNv gene expression. Whether the physiological changes in LNv electrical activity between morning and evening influence LNv gene expression so profoundly remains to be tested. Zeitgebers, such as light:dark cycles, can stabilize the phase and strengthen the amplitude of molecular oscillations [33, 34]. Our data suggest that electrical activity acts as an endogenous zeitgeber to help drive robust molecular oscillations in LNvs.
Given that many synchronizing factors affect the electrical properties of clock neurons [35], a circadian oscillator that incorporates electrical state by adjusting its circadian transcriptome is a potential mechanism for intercellular coupling in circadian networks. Coupling between pacemaker neurons in the mammalian SCN can rescue circadian oscillations in core clock gene mutants [36], although the molecular mechanism is unclear. Our data showing that hyperexcitation of LNvs imposes a morning-like transcriptional program even in per0 mutants, can be viewed as an extreme example of how electrical state can impose time-of-day onto an oscillator.
Activity-dependent expression of a signal transduction pathway and its implications for circadian network synchrony
In the SCN, the circadian transmitter vasointestinal peptide (VIP) activates the CREB-pathway which may help synchronize individual pacemakers [37]. Bi-directionally regulated cAMP/CREB-related genes and the behavioral phenotypes of mis-expressing Creb family members indicate their importance in LNvs. Circadian variations in the availability of CREB-pathway components suggests a signaling pathway with time-of-day representation which could rhythmically regulate LNvs’ response to synchronizing factors in the clock network and/or to entrainment cues. Thus this pathway could admit correctly timed circadian cues and exclude mistimed inputs. The response of the CREB pathway to electrical activity suggests a positive feedback loop where membrane excitability increases pathway activity, which in turn sustains the pathway’s responsiveness to membrane excitability. An autoregulatory role for CREB has been documented in other systems [38] and we noticed the presence of CREs within the CrebA regulatory region (Table S8). Thus we suggest that the CREB-pathway not only relays LNv membrane activity, but also gates LNv responsiveness to membrane activity as a function of time.
Activity-dependent gene expression is critical in synaptic plasticity [1] and even in Alzheimer’s disease [39]. However, activity-dependent transcription in circadian rhythms has been largely unexplored outside of clock resetting, arguably because the CLK/CYC/PER intracellular feedback loop has been viewed as the primary determinant of rhythmic transcription in Drosophila pacemaker neurons. The work here leads us to propose that neural activity acts as an internal zeitgeber and that activity-dependent transcription is a core feature of the multi-oscillator circadian network.
Experimental Procedures
LNv isolation, RNA amplification and analysis
3rd instar larvae were kept in standard LD cycles, transferred to DD and 50 brains dissected centered at either CT3 or CT15, which took ~30 minutes. Dissected brains were dissociated and sorted by flow cytometry as described [40]. mRNA from 150–300 RFP+ cells was amplified using the NuGen WT-Ovation™ Pico System. The resulting single-stranded DNA was either labeled and hybridized to Affymetrix Drosophila 2.0 GeneChips, with 3 biological replicates for each timepoint, or qPCR was performed on 30ng amplified LNv samples with 2–3 biological replicates per timepoint.
Additional experimental procedures including fly stocks, GeneChip data analysis, BIoInformatic analyses, qPCR primer sequences, behavioral analyses are described in the Supplementary information online.
Supplementary Material
Highlights.
Pacemaker neuron electrical activity can switch circadian gene expression
Many genes expressed with a circadian rhythm are activity-dependent
Creb family members may link pacemaker neuron activity to circadian gene expression
Acknowledgments
We thank Deborah Andrew, Todd Holmes, Jae Park, Michael Rosbash, Jerry Yin, the DSHB, the VDRC and the Bloomington Stock Center for antibodies and fly lines. We thank Peter Lopez and Gelo Victoriano de la Cruz at NYU Medical Center for FACS, and Matthieu Cavey, Ben Collins and Dan Tranchina for comments on the manuscript. We also thank Waverly Diner. This investigation was conducted in a facility constructed with support from NIH Research Facilities Improvement Grant Number C06 RR-15518-01 from NCRR. The NYUCI flow cytometry core is supported by NIH/NCI grant P30CA16087. DM was partly supported by an NYU Dean’s Dissertation Fellowship and GNM by an NYU Dean’s Undergraduate Research Fellowship. The work was supported by NIH grants HD046236, GM085503 (KCG) and GM063911 (JB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan Z, Saraswati S, Adolfsen B, Littleton JT. Genome-wide transcriptional changes associated with enhanced activity in the Drosophila nervous system. Neuron. 2005;48:91–107. doi: 10.1016/j.neuron.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 3.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deisseroth K, Bito H, Tsien RW. Signaling from synapse to nucleus: postsynaptic CREB phosphorylation during multiple forms of hippocampal synaptic plasticity. Neuron. 1996;16:89–101. doi: 10.1016/s0896-6273(00)80026-4. [DOI] [PubMed] [Google Scholar]
- 5.Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- 6.Colwell CS. Linking neural activity and molecular oscillations in the SCN. Nature reviews Neuroscience. 2011;12:553–569. doi: 10.1038/nrn3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- 8.Lundkvist GB, Kwak Y, Davis EK, Tei H, Block GD. A calcium flux is required for circadian rhythm generation in mammalian pacemaker neurons. J Neuroscience. 2005;25:7682–7686. doi: 10.1523/JNEUROSCI.2211-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- 10.Wu Y, Cao G, Nitabach MN. Electrical silencing of PDF neurons advances the phase of non-PDF clock neurons in Drosophila. J Biol Rhythms. 2008;23:117–128. doi: 10.1177/0748730407312984. [DOI] [PubMed] [Google Scholar]
- 11.Nitabach MN, Wu Y, Sheeba V, Lemon WC, Strumbos J, Zelensky PK, White BH, Holmes TC. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neuroscience. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Depetris-Chauvin A, Berni J, Aranovich EJ, Muraro NI, Beckwith EJ, Ceriani MF. Adult-specific electrical silencing of pacemaker neurons uncouples molecular clock from circadian outputs. Current Biology. 2011;21:1783–1793. doi: 10.1016/j.cub.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Block G, Geusz M, Khalsa S, Michel S, Whitmore D. Cellular analysis of a molluscan retinal biological clock. Ciba Foundation Symposium. 1995;183:51–60. doi: 10.1002/9780470514597.ch4. [DOI] [PubMed] [Google Scholar]
- 14.Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. PNAS USA. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, Holmes TC. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Current Biology. 2008;18:1537–1545. doi: 10.1016/j.cub.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardin PE. Molecular genetic analysis of circadian timekeeping in Drosophila. Advances in Genetics. 2011;74:141–173. doi: 10.1016/B978-0-12-387690-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pozo K, Goda Y. Unraveling mechanisms of homeostatic synaptic plasticity. Neuron. 2010;66:337–351. doi: 10.1016/j.neuron.2010.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 19.Cao G, Nitabach MN. Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neuroscience. 2008;28:6493–6501. doi: 10.1523/JNEUROSCI.1503-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glossop NR, Lyons LC, Hardin PE. Interlocked feedback loops within the Drosophila circadian oscillator. Science. 1999;286:766–768. doi: 10.1126/science.286.5440.766. [DOI] [PubMed] [Google Scholar]
- 21.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 22.Herzog ED, Takahashi JS, Block GD. Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nature Neuroscience. 1998;1:708–713. doi: 10.1038/3708. [DOI] [PubMed] [Google Scholar]
- 23.Liu C, Weaver DR, Strogatz SH, Reppert SM. Cellular construction of a circadian clock: period determination in the suprachiasmatic nuclei. Cell. 1997;91:855–860. doi: 10.1016/s0092-8674(00)80473-0. [DOI] [PubMed] [Google Scholar]
- 24.Sheeba V, Gu H, Sharma VK, O’Dowd DK, Holmes TC. Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiology. 2008;99:976–988. doi: 10.1152/jn.00930.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demarque M, Spitzer NC. Activity-dependent expression of Lmx1b regulates specification of serotonergic neurons modulating swimming behavior. Neuron. 2010;67:321–334. doi: 10.1016/j.neuron.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kriegsfeld LJ, Mei DF, Yan L, Witkovsky P, Lesauter J, Hamada T, Silver R. Targeted mutation of the calbindin D28K gene disrupts circadian rhythmicity and entrainment. Eur J Neuroscience. 2008;27:2907–2921. doi: 10.1111/j.1460-9568.2008.06239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bussemaker HJ, Li H, Siggia ED. Regulatory element detection using correlation with expression. Nature Genetics. 2001;27:167–171. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- 28.Schug J. Unit 2.6: Using TESS to Predict Transcription Factor Binding Sites in DNA Sequence. In: Baxevanis AD, editor. Current Protocols in Bioinformatics. J. Wiley and Sons; 2003. [DOI] [PubMed] [Google Scholar]
- 29.Gau D, Lemberger T, von Gall C, Kretz O, Le Minh N, Gass P, Schmid W, Schibler U, Korf HW, Schutz G. Phosphorylation of CREB Ser142 regulates light-induced phase shifts of the circadian clock. Neuron. 2002;34:245–253. doi: 10.1016/s0896-6273(02)00656-6. [DOI] [PubMed] [Google Scholar]
- 30.Belvin MP, Zhou H, Yin JC. The Drosophila dCREB2 gene affects the circadian clock. Neuron. 1999;22:777–787. doi: 10.1016/s0896-6273(00)80736-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu H, Shimoda M, Yamaguchi T, Seong KH, Okamura T, Ishii S. Drosophila ATF-2 regulates sleep and locomotor activity in pacemaker neurons. Mol Cell Biol. 2008;28:6278–6289. doi: 10.1128/MCB.02242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonze D, Halloy J, Goldbeter A. Robustness of circadian rhythms with respect to molecular noise. PNAS USA. 2002;99:673–678. doi: 10.1073/pnas.022628299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wijnen H, Naef F, Boothroyd C, Claridge-Chang A, Young MW. Control of daily transcript oscillations in Drosophila by light and the circadian clock. PLoS Genetics. 2006;2:e39. doi: 10.1371/journal.pgen.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aton SJ, Herzog ED. Come together, right...now: synchronization of rhythms in a mammalian circadian clock. Neuron. 2005;48:531–534. doi: 10.1016/j.neuron.2005.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. PNAS USA. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohamed HA, Yao W, Fioravante D, Smolen PD, Byrne JH. cAMP-response elements in Aplysia creb1, creb2, and Ap-uch promoters: implications for feedback loops modulating long term memory. J Biol Chem. 2005;280:27035–27043. doi: 10.1074/jbc.M502541200. [DOI] [PubMed] [Google Scholar]
- 39.Espana J, Valero J, Minano-Molina AJ, Masgrau R, Martin E, Guardia-Laguarta C, Lleo A, Gimenez-Llort L, Rodriguez-Alvarez J, Saura CA. β-Amyloid disrupts activity-dependent gene transcription required for memory through the CREB coactivator CRTC1. J Nueroscience. 2010;30:9402–9410. doi: 10.1523/JNEUROSCI.2154-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruben M, Drapeau MD, Mizrak D, Blau J. A mechanism for circadian control of pacemaker neuron excitability. J Biol Rhythms. 2012 doi: 10.1177/0748730412455918. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.