SUMMARY
How homologous chromosomes (homologs) find their partner, pair and recombine during meiosis constitutes the central phenomenon in eukaryotic genetics. It is widely believed that in most organisms SPO11-mediated DNA double-strand breaks (DSBs) introduced during prophase I precede and are required for efficient homolog pairing. We now show that in the mouse, a significant level of homolog pairing precedes programmed DNA cleavage. Strikingly, this early chromosome pairing still requires SPO11, but is neither dependent on its ability to make DSBs nor homologous recombination proteins. Intriguingly, SUN1, a protein required for telomere attachment to the nuclear envelope and for post-DSB synapsis, is also required for early pre-DSB homolog pairing. Furthermore, pre-DSB pairing at telomeres persists upon entry into prophase I, and is most likely important for initiation of synapsis. Our findings suggest that the DSB-triggered homology search may mainly serve to proofread and stabilize the pre-DSB pairing of homologous chromosomes.
INTRODUCTION
Homolog pairing is the process of alignment and physical juxtaposition of whole or segments of homologous chromosomes. Despite decades of research, how homologous chromosomes find each other in the nucleus in order to initiate the pairing process remains a puzzle. It is believed that in most organisms, repair of the DSBs (Neale and Keeney, 2006; San Filippo et al., 2008) introduced in leptotene – the onset of prophase I – by SPO11, an evolutionally conserved type II topoisomerase-like protein (Keeney, 2001), initiates a genome wide search for homology. This search drives the homolog pairing and alignment, ultimately leading to the lengthwise pairing and synapsis (the stabilization of homolog interactions by the polymerization of a proteinacious structure called the synaptonemal complex) of all homologs achieved by pachytene (Neale and Keeney, 2006). However, DSB-independent pairing has been reported in some organisms (Martínez-Pérez et al., 1999; Prieto et al., 2004) and has been extensively studied and characterized in flies and worms (Dernburg et al., 1998; Gerton and Hawley, 2005; McKim et al., 1998; Phillips et al., 2009; Tsai and McKee, 2011). Although not as widely accepted, the incisive studies in budding yeast on DSB-independent pairing (Burgess et al., 1999; Cha et al., 2000; Weiner and Kleckner, 1994) have provided the impetus to re-examine this issue in mammals, where neither DSB-independent nor pre-DSB pairing has been reported (Scherthan et al., 1996). Here we report that in mouse spermatocytes a significant proportion of homolog pairing is established prior to the introduction of SPO11 mediated DSBs, is maintained and further stabilized by meiotic recombination and is most likely important for initiation of synapsis.
RESULTS
Overview of morphological classification and definition of pairing
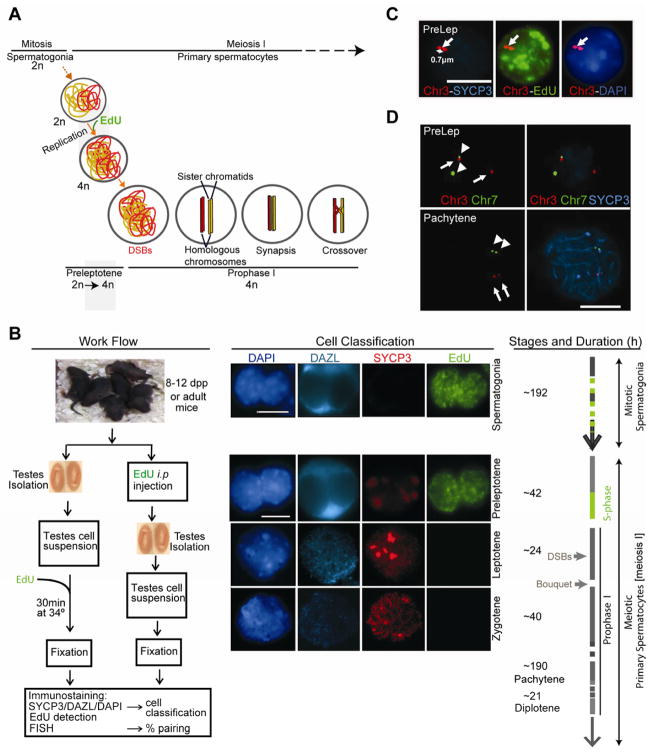
In order to determine whether any degree of homolog pairing was established before DSB formation, we analyzed pairing in preleptotene, the stage preceding entry into prophase I, using prepuberal mice (8–12 or 21 days post partum, dpp) that are enriched for preleptotene spermatocytes (Figures 1A, 1B and 1C). During mouse spermatogenesis, type B spermatogonia divide to form preleptotene primary spermatocytes, which undergo a final round of DNA replication (meiotic S) before entering meiotic prophase I (Bellvé et al., 1977; Scherthan et al., 1996). In order to mark cells preceding the stage during which DSBs are introduced, we labeled replicating cells by either injecting mice intraperitoneally (i.p.) or culturing mouse testes cell suspensions for 30 min (the duration of meiotic replication is estimated to be 12–24 hr) with the thymidine analog 5-ethynyl-2′-deoxyuridine (EdU) (Chehrehasa et al., 2009; Salic and Mitchison, 2008) (Figure 1B). Since there are no stage-specific cell surface markers, we used morphological characteristics to distinguish the different cell types in the testis (Scherthan et al., 2000; Scherthan et al., 1996). Briefly, we differentiated EdU-positive structurally preserved nuclei (SPN) of late preleptotene spermatocytes from spermatogonia based on their more spherical nuclear shape, relatively larger size, distinct peripheral distribution of bright DAPI-stained heterochromatic DNA clusters and weakly-stained intranuclear axial element protein SYCP3 non-linear aggregates (Figure 1B; see Extended Experimental Procedure for a detailed description of the morphological characteristics used to distinguish the different cell types).
Figure 1. Overview of morphological classification of cell types and criteria to define pairing.
(A), Schematic illustration of meiosis I in male mice. Type B spermatogonia divide into preleptotene spermatocytes, which carry out one round of DNA replication before entering prophase I. At the onset of prophase I, SPO11 introduces the DSBs that trigger homolog recombination, synapsis and ultimately, the formation of crossover DNA products (chiasmata) that ensures the accurate reductional segregation of chromosomes in the first meiotic division. We used incorporation of the thymidine analog EdU to mark preleptotene spermatocytes.
(B), Schematic illustration of the experimental set-up, mouse spermatogenesis and the criteria used for cell classification. Immunofluorescence staining of spermatogonia, preleptotene, leptotene and zygotene spermatocytes is shown. Nuclei are stained with a 488-azide (EdU), synaptonemal complex protein SYCP3, a germ cell specific marker (DAZL) and DAPI.
(C), Illustrative image of a preleptotene spermatocyte analyzed by combined Immunofluorescence-Fluorescent in situ hybridization (IF-FISH) showing a Chr3-INT probe, 488-azide (EdU), SYCP3 and DAPI stained nucleus in a structurally preserved preleptotene (PreLep) spermatocyte.
(D), The frequency of non-homologous pairing between mouse chr3 and chr7 determined by co-FISH is 8% (n = 50). PreLep (top) and pachytene (bottom) spermatocytes are shown. Chr3-INT probes (arrows), Chr7-INT probes (arrowheads).
Scale bar: 10 μm. See also Figure S1.
After isolation and fixation of cells using techniques that preserve the nuclear architecture (Scherthan et al., 2000) (Figures 1C and 1D), we measured the extent of homolog pairing by fluorescent in situ hybridization (FISH), using bacterial artificial chromosome (BAC) probes (Figure S1A and Extended Experimental Procedure) targeting loci on interstitial chromosomal sites (at least 29 Mb from the chromosome ends). In fully synapsed pachytene chromosomes, the FISH foci of two paired homologs are not always fused (Figure 1D). If we used the criteria of scoring only fused foci as paired homologs, the frequency of paired homologs in pachytene cells was spuriously below 60% (Figures S1B–S1D). Hence, we determined the distance between the foci centers of fully paired/synapsed chromosomes in pachytene cells, and established this distance as the threshold to define whether homologs are paired at a certain locus. Thus, two homologs were defined as paired if the distance between their foci centers was equal to or less than 1 μm (Figures 1D, S1B and S1C) in structurally preserved nuclei (SPN), in which the nuclear architecture is well preserved by permeabilizing and fixing the cells without lysing them.
Significant homolog pairing detected prior to programmed DSBs in mice
We began by analyzing pairing in different cell types (Figure 1B), using a chromosome 3 interstitial probe (Chr3-INT, Figure S1A and Extended Experimental Procedure). Surprisingly, we found that homologs were paired in approximately 35% of preleptotene (pre-prophase I) spermatocytes and returned to roughly pre-meiotic levels in leptotene (P < 0.001, n = 478, where n = total number of cells analyzed, Fisher’s exact test; Figures 2A and S2A). As a control, we measured the heterologous association frequency between different interstitial loci located on chromosomes 3 and 7 as 8% (n = 50, Figures 1D and S2A). Consistent with the stabilization of homolog interactions via synapsis occurring later during prophase I, we detected paired homologs in 85% and 95% of spermatocytes in zygotene and pachytene, respectively (Figure 2A). We observed a similar proportion of cells with paired homologs, irrespective of the age of the mice (21 dpp prepuberal or 2-month old, not shown) or the chromosome monitored. This early pairing in meiotic cells was even higher (~45%) in arguably the best structurally preserved sample, frozen tissue sections (Figures 2A and 2B). These findings demonstrate that a significant level of homolog pairing occurs in preleptotene spermatocytes (before DSBs), but declines upon entry into prophase I (in leptotene spermatocytes).
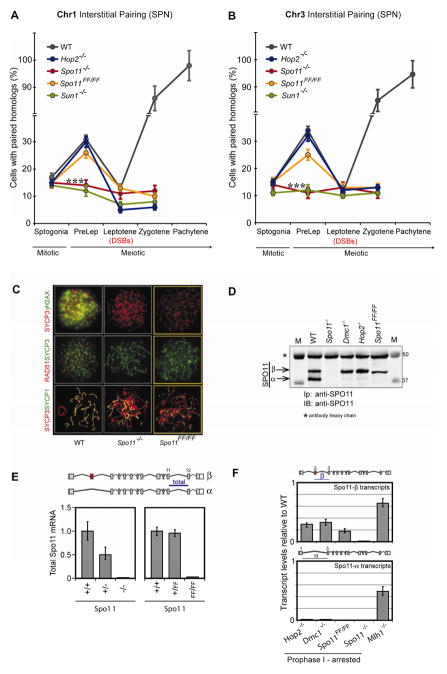
Figure 2. A significant level of homolog pairing is detected in preleptotene spermatocytes prior to programmed DSBs.
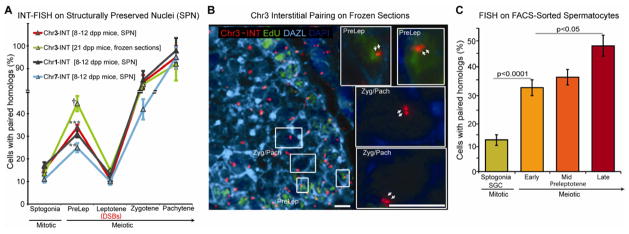
(A), Assessment of pairing during early spermatogenesis, using chr1, chr3 and chr7 interstitial probes in either structurally preserved nuclei (SPN) from prepuberal (8–12 dpp) mice or frozen tissue sections of 21 dpp mice. Statistical significance of the difference between samples was assessed by Fisher’s exact test. The p-values for difference in pairing between preleptotene spermatocytes and spermatogonia are given for each probe († P < 0.0001, *** P < 0.001, ** P < 0.01, n = 321 to 609). The error bar is an estimation of the standard deviation (SD), based on the counting error (square root of n).
(B), IF-FISH on frozen testis tissue sections of 21 dpp EdU-injected mice: Frozen sections were stained with 488-azide (EdU), DAZL (marker for germ cells), and hybridized with a Chr3-INT probe. Shown is a representative image taken with 40X magnification. Images of sections taken with 100X magnification (see inserts) were used for the analysis. Preleptotene (PreLep), and zygotene (Zyg)/pachytene (Pach) spermatocytes. Scale bar, 10 μm.
(C), Four cell populations isolated by FACS (see Figures S2B and S2C) enriched in either spermatogonia (spermatogonia, SPGN + seminal germ cells, SGC) or early, mid and late preleptotene spermatocytes, were analyzed by Chr1-INT FISH. Statistical significance of the difference between samples was assessed by Fisher’s exact test and the p-values indicated. The error bar is an estimation of the standard deviation (SD), based on the counting error. For each data point, 234 to 450 total number of cells were analyzed.
See also Figure S2.
Preleptotene lasts about 42 hours in the mouse. In order to evaluate whether progression through preleptotene correlates with an increase in pairing, we used fluorescence-activated cell sorting (FACS) (Bastos et al., 2005) to isolate early, mid and late preleptotene spermatocytes, based on their increasing DNA content (Figures 2C, S2B and S2C), and determined the percentage of cells with paired homologs in these three cell populations, as well as in spermatogonia. Consistent with previous reports in budding yeast (Cha et al., 2000), we observed that pairing increased significantly towards the end of the preleptotene stage, right before entering prophase, when meiotic DSBs are introduced.
Pre-DSB homolog pairing is independent of meiotic homologous recombination
To gain insight into the mechanisms underlining preleptotene (or pre-DSB) pairing, we investigated several genetic factors that could play a role. We found that neither DMC1 (Bishop et al., 1992; Pittman et al., 1998; Shinohara et al., 1992), the meiosis specific homolog of the recombinase RAD51 (Neale and Keeney, 2006; San Filippo et al., 2008) (not shown, see below), nor HOP2 (Petukhova et al., 2003; Pezza et al., 2007), another meiotic recombination protein required for RAD51/DMC1 dependent meiotic DSB repair (Figures 3A and 3B), are required for preleptotene pairing. Note that DSB and recombination markers (γH2AX and RAD51) were not detected in replicating preleptotene spermatocytes (Figure S3B). This indicates that meiotic homologous recombination is most likely not required for this process, despite being required for the later pairing occurring in prophase I (Neale and Keeney, 2006; San Filippo et al., 2008).
Figure 3. A DSB-independent activity of SPO11 and the telomere tethering protein SUN1 are both required for preleptotene pairing but the meiotic homologous recombination machinery is dispensable.
(A, B), Preleptotene pairing is disrupted in Spo11−/− and Sun1−/− but unaffected in Spo11FF/FF or Hop2−/− mice, as determined with a Chr1-INT probe (A) and a Chr3-INT probe (B). *** P < 0.001 (n = 282 to 551, where n = total number of cells analyzed), applies to the difference in pairing between wild-type or Spo11FF/FF and Spo11−/− or Sun1−/− mice. Statistical significance of difference between samples was assessed by Fisher’s Exact Test. The error bar is an estimation of the standard deviation (SD), based on the counting error.
(C), Spo11FF/FF spermatocytes are defective in DSB formation, meiotic recombination and synapsis. IF analysis of surface-spread preparations shows that leptotene spermatocytes of both Spo11FF/FF and Spo11−/− are devoid of γH2AX staining (a marker for DSBs) and RAD51 foci (a marker for homologous recombination intermediates). Zygotene-like spermatocytes only show minimal co-localization of SYCP3 and SYCP1 staining (indicative of defective synapsis). Scale bar, 10 μm.
(D), Spo11FF/FF mice, like other mutants arrested in prophase I, express primarily the SPO11-β polypeptide. Total testis extracts from wild-type, Spo11−/−, Dmc1−/−, Hop2−/−, and Spo11FF/FF mice (5 mg of total protein) were precipitated/blotted with anti-SPO11 antibody.
(E), The mutated Spo11::YY137,138FF allele is transcribed. In order to verify expression of the mutated allele, total Spo11 transcripts were quantified by reverse transcription-qPCR (RT-qPCR) of total RNA from adult mice testes, (see Extended Experimental Procedures). As previously reported, Spo11 heterozygous knockout (+/−) testes carry half the amount of transcripts when compared to WT mice (because only one allele is being transcribed) (Bellani et al., 2010). Given that homozygous Spo11FF/FF mice are arrested in prophase, their total Spo11 transcript levels are very low (because the majority of Spo11 transcripts correspond to Spo11-α, which is expressed after mid prophase) (Bellani et al., 2010). Nevertheless, heterozygous knockin mice (+/FF) showed comparable levels of total transcripts to WT mice, indicating that both the WT and mutant (FF) alleles are being transcribed. Error bars represent the SD.
(F), Homozygous Spo11FF/FF mice synthesize primarily Spo11-β transcripts. Total testis RNA from Spo11FF/FF mice, and from several mutants arrested in either prophase I (Hop2−/−, Dmc1−/−) or metaphase I (Mlh1−/−) were analyzed by RT-qPCR, using Taqman assays targeting isoform-specific exon junctions in order to quantify Spo11-α and Spo11-β transcripts Bars represent the ratios of Spo11-α or -β transcripts in homozygous mutants relative to a wild-type littermate. Thus, Spo11FF/FF mice synthesize primarily Spo11-β transcripts at levels comparable to those of other prophase I arrested mutant mice. In contrast a mutant mouse arrested in metaphase I express both isoforms. Error bars represent the SD. See also Figure S3.
Preleptotene Pairing Requires SPO11 But Not It’s Cleavage Activity
In mice, SPO11 deficiency results in the absence of DSBs, defective recombination and synapsis, ultimately leading to early pachytene arrest (Baudat et al., 2000; Romanienko and Camerini-Otero, 2000). To test whether SPO11 plays a role in homolog pairing in preleptotene, we investigated the level of pairing in Spo11−/− spermatocytes, and found that homolog pairing was completely abolished both in preleptotene spermatocytes and, as previously reported (Baudat et al., 2000; Romanienko and Camerini-Otero, 2000), in prophase arrested cells (zygotene-like, Figures 3A and 3B). Surprisingly, Mei1−/− mice (Libby et al., 2003), which also lack DSBs in prophase I in spite of normal SPO11 expression (data not shown), showed no reduction in pre-prophase I pairing (not shown, see below), suggesting that preleptotene pairing requires SPO11 but not DSBs.
This observation prompted us to investigate whether the DSB catalytic activity of SPO11 was actually required. Wild type mice express two major SPO11 isoforms (both carrying the catalytic tyrosine), α and β, which are expressed with different timing. Specifically, SPO11β is expressed in early spermatocytes, whereas the SPO11a polypeptide is hardly synthesized until past early pachynema (Bellani et al., 2010). We generated a knock-in mouse expressing a catalytic mutant of SPO11 (Spo11YY137,138FF, hereafter referred to as Spo11FF/FF), by replacing the codons for the catalytic tyrosine(s) with those of phenylalanine(s) in the Spo11 locus (Figure S3A; see also Extended Experimental Procedures). Immunostaining of Spo11FF/FF spermatocytes with antibodies against γH2AX (a marker of DSBs), RAD51 (a marker for homologous recombination intermediates) and SYCP1 (a marker of synapsis) revealed that these mice lack meiotic DSBs and consequently are defective in recombination and synapsis (Figure 3C). Thus, Spo11FF/FF mice arrest in early prophase I, just like Spo11−/− mice (Figure 3C), even though they synthesize Spo11-β transcripts and express wild type levels of the SPO11-β protein (the isoform expressed in early prophase I, see (Bellani et al., 2010) (Figures 3D–3F).
Most importantly, pairing was restored to wild type levels in preleptotene spermatocytes of Spo11FF/FF mice (Figures 3A, 3B and S3C). Since Spo11FF/FF mice show wild type levels of pairing during preleptotene but only express the β isoform (Figure 3D), our results imply an early role for SPO11-β in pre-DSB pairing. Consistent with this notion, preleptotene homolog pairing was not rescued in a Spo11−/− mouse complemented with a Spo11-α transgene (F.P., K.A.B. and R.D.C-O, unpublished results). Taken together, these findings demonstrate that a DSB-independent activity of SPO11 is required for preleptotene pairing in mice. This is consistent with a previous report in budding yeast (Cha et al., 2000).
Pre-DSB homolog pairing requires the telomere tethering protein SUN1
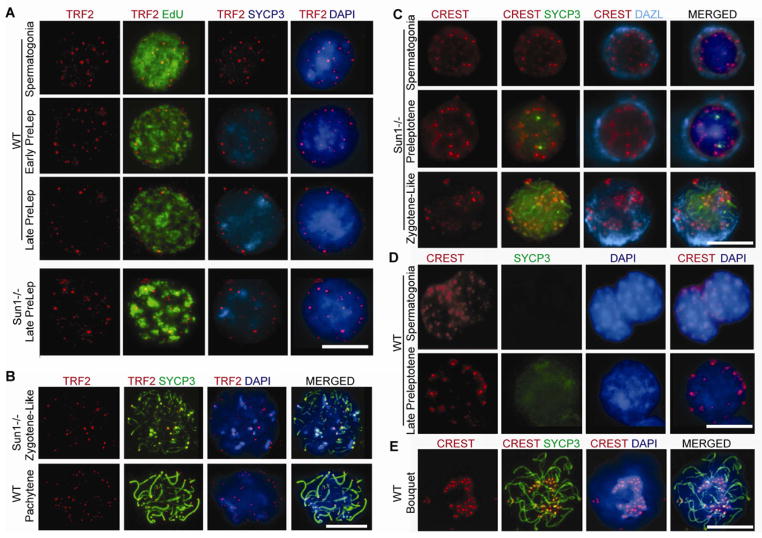
Since chromosome movements in early prophase I promote post-DSB prophase pairing and synapsis (Koszul and Kleckner, 2009; Scherthan et al., 2007), we asked if SUN1, a nuclear membrane protein required to tether the ends of chromosomes to the nuclear envelope (NE) that has been implicated in both chromosome movements and bouquet formation occurring later in prophase I (at the leptotene-zygotene transition) (Ding et al., 2007; Hiraoka and Dernburg, 2009), could also be promoting pre-DSB pairing. Lack of SUN1 leads to asynapsis during meiotic prophase and consequently disrupts gametogenesis in mice (Chi et al., 2009; Ding et al., 2007). We noted that SUN1 showed a significantly high expression during early mouse spermatogenesis (Figures S4A and S4B) and also localized to the NE in preleptotene spermatocytes similar to the telomere repeat binding factor (TRF1) protein (Figures S4C–S4G) (Scherthan et al., 2000), suggesting that it might have an early role prior to prophase I. We found that not only is SUN1 required to anchor chromosomal ends to the NE in prophase (Ding et al., 2007), but it is also required to do so much earlier in preleptotene spermatocytes (Figures 4A–4D). Nevertheless, we could not detect bouquet formation (Figure 4E) (Ding et al., 2007) in either Sun1−/− or WT preleptotene spermatocytes, arguing against the possibility of a general clustering of telomeres to one side of the nuclear periphery (bouquet) in preleptotene. Most importantly, we found that homolog pairing is abolished in Sun1−/− preleptotene spermatocytes (Figures 3A and 3B). Altogether our data indicate that preleptotene pairing requires SUN1 and its associated telomere anchoring function, but not bouquet formation per se.
Figure 4. SUN1 is required for telomere attachment to the nuclear envelope in preleptotene spermatocytes.
(A), Telomere re-localization to the nuclear envelope in late preleptotene spermatocytes requires SUN1: IF with antibodies against telomere repeat binding factor 2 (TRF2), and axial/lateral element protein SYCP3, counter stained with DAPI and 488-azide (EdU, for cell classification) in WT and Sun1−/− mice at indicated stages, showing that telomeres (TRF2) relocalize to the nuclear periphery in late preleptotene spermatocytes of WT mice but not in Sun1−/− mice.
(B), Control showing that TRF2 binding to the telomeres is not impaired in Sun1−/− mice.
(C), IF with CREST antiserum labeling centromeres and antibodies against the axial/lateral element protein SYCP3 and germ cell marker DAZL in Sun1−/− mice at indicated stages, showing the distribution of CREST foci localization in both the nuclear periphery and nuclear lumen at all stages in Sun1−/− mice.
(D), In contrast to (C) the distribution of CREST foci is restricted to nuclear peripheral localization in preleptotene in WT mice.
(E), WT zygotene spermatocyte with the centromeres clustered into a bouquet. 2.4% of 1008 total WT leptotene/zygotene prophase spermatocytes displayed a bouquet configuration, consistent with previous reports (see (Liebe et al., 2006) and references therein). In contrast, bouquets are not observed either in Sun1−/− mice (C) or in preleptotene of WT mice (D) of over 500 nuclei analyzed at each stage per mice.
Scale bar, 10 μm. See also Figure S4.
Pre-DSB subtelomeric pairing is preserved throughout prophase I, is further stabilized by recombination and most likely facilitates initiation of synapsis
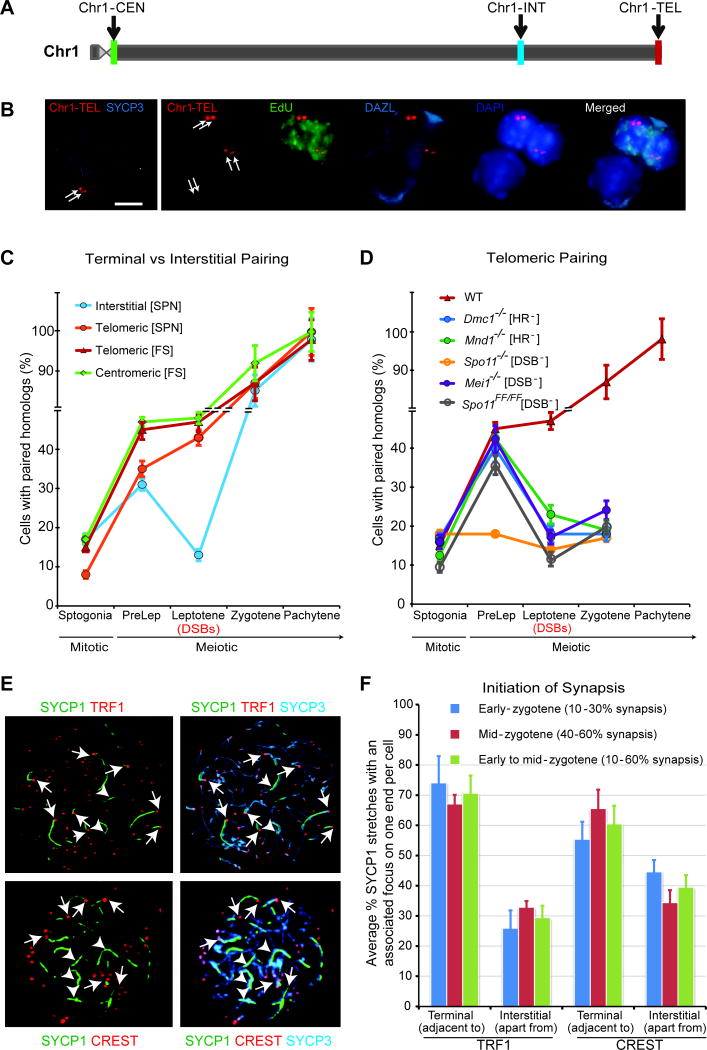
Since the homology search performed by the homologous recombination machinery during leptotene/zygotene is thought to mediate prophase pairing (Neale and Keeney, 2006; San Filippo et al., 2008), we were puzzled by the decline in pairing observed in leptotene spermatocytes when using interstitial probes (Figures 2A, 3A and 3B). Given our data demonstrating that the telomere tethering protein SUN1 is involved in preleptotene pairing, we wondered whether such a decline would be observed with probes targeting the ends of chromosomes. Strikingly, using chromosome 1 subtelomeric (Chr1-TEL) and subcentromeric (Chr1-CEN) probes (and hence close to the other telomere since mouse chromosomes are telocentric) (Figures 5A–5C, Table S2A and Figure S1A), we found that the proportion of cells exhibiting telomeric pairing of homologs in preleptotene was maintained or increased in leptotene. We note that the preleptotene homologous subtelomeric pairing of 35–50% (n = 457 to 495; Figure 5C) is significantly higher than the experimentally determined non homologous subtelomeric pairing of 14% (n = 77, P < 0.0001). Therefore, while a degree of random peripheral telomere/centromere clustering may occur at preleptotene (Scherthan et al., 2000) (Figures 4A, S4C and S4D), we conclude that a highly significant proportion of the subtelomeric interactions we observe are homologous, and that this terminal association does not decline upon progression into leptotene. In agreement with this notion, immunofluorescence staining using a CREST serum (which labels centromeres) revealed that on average 60% of the chromosomes were associated (pair wise but not necessarily fused) at their centromeric ends in preleptotene spermatocytes (well above the 23% association observed for spermatogonia), and that this centromere association is maintained upon progression into leptotene (data not shown).
Figure 5. Pre-DSB pairing at telomeres persists upon entry into prophase I and is most likely important for initiation of synapsis at chromosomal ends.
(A), Chr1 ideogram showing the loci position of the interstitial (Chr1-INT), subtelomeric (Chr1-TEL) and subcentromeric (Chr1-CEN) probes (see Extended Experimental Procedure).
(B), Sample IF-FISH images showing the terminal position of the Chr1-TEL loci in a pachytene surface spread preparation (left panel), and Chr1-TEL foci in preleptotene (top two cells, EdU+, green) and zygotene/pachytene (bottom cell, EdU−) cells in a SPN preparation (right panel). Scale bar, 10 μm.
(C), Terminal pairing achieved in preleptotene spermatocytes is preserved upon entry into leptotene, whereas interstitial pairing is lost. Terminal versus interstitial homolog pairing was assessed on either structurally preserved nuclei (SPN) or frozen tissues sections (FS) of wild type mice, probed with interstitial (Chr1-INT), subtelomeric (Chr1-TEL) or subcentromeric (Chr1-CEN) probes. Notice that the preleptotene homologous telomeric pairing was 35–50% (n = 457 to 495; Table S1A), depending on the probe used, and is significantly higher than the experimentally determined average non homologous telomeric pairing between chr1 and chr3 of 14% in SPN, (n = 77, P < 0.0001) (see Extended Experimental Procedures). Note that the experimentally determined average non homologous telomeric pairing between chr1 and chr3 in frozen sections was 12% (Figure S5). The error bar is an estimation of the standard deviation (SD), based on the counting error.
(D), Maintaining terminal pairing upon entry into leptotene requires DSB formation and meiotic recombination. Frozen tissue sections of wild type (WT), Dmc1−/−, Mnd1−/−, Spo11−/−, Mei1−/− and Spo11FF/FF mice were probed with Chr1-TEL probe. Notice that telomeric pairing is also significantly disrupted during preleptotene in Spo11−/− but not in DSB or HR impaired mutants (Table S1B). HR−/−, homologous recombination impaired; DSB−/−, DSB impaired. The error bar is an estimation of the standard deviation (SD), based on the counting error.
(E), Synapsis initiates from the terminal ends of chromosomes in mice. Sample images of chromosome spreads from WT spermatocytes stained with antibodies against the central element protein SYCP1, the axial/lateral element protein SYCP3 and the telomeric protein TRF1 or centromere marker, CREST are shown, indicating that in early zygotene spermatocytes TRF1/CREST signals lie adjacent or associate with (but not necessarily co-localize with) the ends of most short SYCP1-stretches. Arrows: terminal synapsis, arrowheads: interstitial synapsis. Scale bar, 10 μm.
(F), Quantification of (E), the proportion of synapsed homologs per nuclei in early zygotene (10–30% synapsis) and in mid-zygotene (40–60% synapsis) spermatocytes, as assessed by SYCP1/SYCP3 co-localization, either adjacent to (terminal synapsis) or apart from (interstitial synapsis) TRF1/CREST foci. For each classification, about 100 nuclei were analyzed. Note that the early to mid-zygotene (10–60% synapsis) classification is a pool of the above two classifications. To determine the resolution of this analysis, we first estimated the threshold length of SYCP1 short stretches from 126 measurements to be 2.8 μm (median). Similarly, we determined the mean overall length of all chromosomes to be 13.4 μm and computed the resolution or estimated fraction of chromosome carrying SYCP1 short stretches to be about 20% (2.8 μm/13.4 μm). The error bar is an estimation of the standard deviation (SD), based on the counting error.
See also Figure S5.
Further analysis of subtelomeric homolog pairing in preleptotene spermatocytes corroborated the finding that neither DMC1 (Bishop et al., 1992; Neale and Keeney, 2006; Pittman et al., 1998) nor HOP2/MND1 (Petukhova et al., 2003; Pezza et al., 2007) homologous recombination proteins (Figure 5D and Table S1B) are required for pre-DSB pairing. In addition, pairing at chromosome ends in preleptotene also required SPO11 but not its DSB catalytic activity (Figure 5D, Table S1B and data not shown). Importantly, mutants deficient for either introducing DSBs (Spo11FF/FF and Mei1−/−) or processing them via homologous recombination (Dmc1−/− and Mnd1−/−) (Figures 5D and data not shown) fail to preserve pre-DSB pairing achieved in preleptotene into leptotene. Thus, DSB repair through homologous recombination stabilizes interactions at telomeric/centromeric (Figures 5D) sites upon entry into prophase I. Taken altogether, these results suggest that preleptotene pairing at the ends of chromosomes is preserved upon entry into leptotene.
These observations are consistent with the notion that prophase I pairing (which drives homolog synapsis) initiates mostly at chromosome ends in human spermatocytes (Barlow and Hultén, 1996; Brown et al., 2005) and verified here in mouse spermatocytes (Figures 5E, 5F and data not shown). Since in mice, in a given cell, initiation of synapsis is asynchronous, we assessed whether initiation of synapsis is biased towards chromosome ends, by identifying spermatocytes displaying 10–30% (early zygotene) or 40–60% (mid-zygotene) partial synapsis as judged by SYCP1/SYCP3 co-localization, and measured the proportion of short SYCP1 stretches lying adjacent (not necessary colocalized) to a TRF1 or CREST focus (Figures 5E and 5F). The resolution of these short SYCP1 stretches is estimated to be about 20% of the length of each chromosome. Hence the probability of randomly finding a CREST or TRF1 foci at the end of a SYCP1 stretch is 20% (one end) or 40% (two ends) respectively. Therefore, the ~60% or ~70% frequency of finding short SYCP1 stretches with an associated CREST or TRF1 focus respectively is highly significant. In conclusion, our finding that in early-mid zygotene spermatocytes, TRF1/CREST signals lie adjacent or associate with the ends of most short SYCP1-stretches strongly suggest that in mouse spermatocytes synapsis initiates mostly at chromosome ends.
DISCUSSION
Our results unambiguously establish that a significant level of pairing occurs in preleptotene spermatocytes entering meiosis, and suggest that the 35 – 50% pre-DSB pairing of homologs at chromosomal ends facilitates pairing and subsequent synapsis of homologs upon entry into prophase I. Although we find a significant degree of homolog pairing (~35%) at interstitial sites in preleptotene spermatocytes, these interactions are unstable and homolog pairing declines upon entry into prophase. We cannot rule out the possibility that such interstitial pairing is only a consequence of the homologous telomeric pairing at this stage and might not be essential for prophase interstitial pairing/synapsis.
We find that while SPO11 is involved in preleptotene pairing, its DSB catalytic activity is dispensable (Figures 3A, 3B, 5D and data not shown). Furthermore, based on our results that SPO11 mediates telomere pairing and reports that SPO11 localizes to telomeres (Zalzman et al., 2010), we posit that SPO11 plays a structural, non-catalytic role perhaps directly or indirectly interacting with SUN1. Given that SUN1 is essential for homolog pairing –and the tethering of chromosomes to the NE– in preleptotene spermatocytes (Figures 3A, 3B, 4, S4C–S4F and data not shown), we interpret this as evidence that re-localizing chromosomes termini to the NE plays a role in pre-DSB preleptotene pairing. We propose a mechanism by which tethering of chromosomes ends to the NE via SUN1 (Chi et al., 2009; Ding et al., 2007) (Figures 4, S4C and S4D) facilitates finding the right partner by confining the search to sequences adjacent to the chromosome ends localized in a volume near the nuclear periphery.
We also find that while DSB-independent pairing at interstitial (non-telomeric) sites is unstable upon entry into prophase, telomeric pairing is maintained, as long as spermatocytes are proficient for DSB formation and recombination (Figures 5C and 5D). Given the evidence that synapsis often initiates from at least one end of the chromosomes in mammals (Barlow and Hultén, 1996; Brown et al., 2005) (Figures 5E, 5F and data not shown), one could invoke a temporal distinction between terminal and interstitial sites with respect to DSB formation or initiation of repair. Subtelomeric sites would be repaired earlier and thus pairing at these terminal sites would be stabilized by recombination per se or through the incipient synapsis triggered as a consequence. On the other hand, interstitial associations that have not yet been stabilized by recombination/synapsis would be more prone to disruption, either by the onset of the extensive chromatin condensation which occurs during leptotene (Zickler and Kleckner, 1999), or by vigorous chromosome movements akin to those observed in yeast (Koszul and Kleckner, 2009; Scherthan et al., 2007) and rats (Parvinen and Söderström, 1976). The fact that these movements were shown to be DSB independent in yeast (Conrad et al., 2008), suggests that they might be responsible as well for the disruption of pairing observed at telomeric sites in the homologous recombination (HR) and DSB mutants (Figure 5D). Thus, only as the breaks are repaired by recombination would these preleptotene interactions be stabilized or restored.
As for the mechanism by which the initial homology can be sensed to effect pre-DSB pairing we can imagine at least two, not mutually exclusive, possibilities. These associations may be promoted by intact DNA-DNA duplexes (Danilowicz et al., 2009; Kleckner and Weiner, 1993) or other nonduplex DNAs, such as single-stranded DNA generated independent of DSBs, although we cannot exclude the possibility that these DNA-mediated events may be facilitated by proteins. An alternative scenario is that protein-DNA and protein-protein interactions play a key role in a mechanism similar to those described for “pairing centers” (Dernburg et al., 1998; Gerton and Hawley, 2005; McKim et al., 1998; Phillips et al., 2009; Tsai and McKee, 2011) in worms and flies. Irrespective of the mechanism used to initiate pre-prophase I pairing, the interstitial interactions are transient and reversible and this feature may be required as a way to either disrupt unwanted associations (Kleckner and Weiner, 1993; Koszul and Kleckner, 2009) and/or to allow for the HR machinery to mediate the strand invasion that is the hallmark of the more precise and intimate alignment of the chromosomal DNA down to the nucleotide level (Figure 6).
Figure 6. Preleptotene DSB-independent pairing in mice.
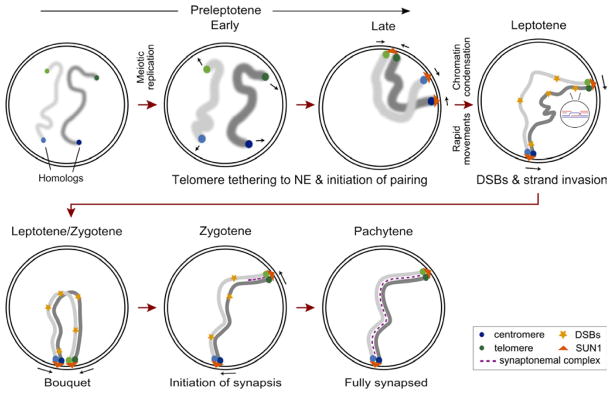
Our data argues for a progressive increase in telomeric homolog pairing which serves to promote interstitial pairing at multiple loci along the whole chromosome, prior to DSB formation occurring at the onset of meiotic prophase I (leptotene). Preleptotene pairing is DSB-independent but requires the topoisomerase II-like protein SPO11, and the protein anchoring telomeres to the nuclear envelope (NE), SUN1. We propose that the tethering of telomeres to the nuclear envelope in late preleptotene (Chehrehasa et al., 2009; Scherthan et al., 1996) (Figures 4 and S4), facilitates the initiation of homolog pairing at subtelomeric regions by simplifying the search for the cognate partner. Upon entry into prophase, DSB-independent pairing at interstitial (non-telomeric) sites is lost, presumably to allow for the removal of unwanted associations and entanglements. However, telomeric pairing is maintained at least at one end, as long as the homologous recombination (HR) machinery is functional. While interstitial interactions are lost, we cannot rule out the possibility that the homologs stay in close proximity. Also, this reversibility in interstitial pairing may permit or promote strand invasion mediated by the HR machinery. Thus the HR may only serve to proofread the initial pairing established prior to DSB formation and as a checkpoint to ensure that ectopic associations are disrupted. Subsequently, “validated” interactions would be stabilized via the polymerization of the synaptonemal complex (synapsis). Furthermore, DSB repair and synapsis, initiating at the preserved preleptotene homologously paired telomeric sites, extends into the chromosome to restore pairing at interstitial sites, ultimately leading to a progressive synapsis (almost zipper-like) of homologs later in prophase I.
While the detailed mechanism of DSB-independent pairing remains to be understood, our data has robustly demonstrated the existence of this phenomenon in mice and has provided insights into some of the key factors involved, namely, a DSB-independent structural role of SPO11 and the telomere anchoring activity of SUN1. Overall, our observations can be summarized in the following model (Figure 6). In preleptotene spermatocytes, centromeres and telomeres associate with the nuclear envelope (Scherthan et al., 1996) (Figures 4, and S4), presumably facilitating homolog associations at the chromosome termini, as well as progressive alignment of interstitial sites. Upon entry into prophase, subtelomeric homolog associations are stabilized in a DSB/HR dependent manner, suggesting that the repair of meiotic DSB on chromosomal termini stabilizes these interactions. Indeed, we observed that most of the initial synapsis occurs at chromosome ends in mouse spermatocytes. Once homologs have initiated synapsis with the right partner at the chromosome end(s), this would facilitate DSB repair and synapsis at interstitial sites within a topologically constrained territory (Mirny, 2011). Thus, DSB-dependent homology search may mainly act as a prophase checkpoint that proofreads the initial pairing to mediate the final stages of proper chromosome synapsis.
EXPERIMENTAL PROCEDURES
Combined immunofluorescence (IF) staining and fluorescent in situ hybridization (FISH)
We used a combination of immunofluorescence (IF) staining and fluorescent in situ hybridization (FISH) to assess pairing at different stages of mouse spermatogenesis. To mark replicating cells, we either injected mice intraperitoneally (i.p.) or cultured mouse testes cell suspension with 5-ethynyl-2′-deoxyuridine (EdU) (Chehrehasa et al., 2009; Salic and Mitchison, 2008). After the cells were isolated and fixed using techniques that preserve the nuclear architecture (Scherthan et al., 2000) followed by IF staining and click chemistry based EdU detection, the extent of homolog pairing was measured by FISH using standard protocols. Chromosome specific probes were prepared to monitor interstitial and subtelomeric/subcentromeric pairing. Two homologs were defined as paired if the distance between their focus centers was ≤ 1.0 μm.
Fluorescence-Activated Cell Sorting (FACS)
We used FACS (Bastos et al., 2005) to obtain purified populations of spermatogonia, and early, mid and late preleptotene spermatocytes.
Fluorescent Microscopy Imaging
All images were acquired with an upright epi-fluorescence microscope, Leica DM6000 B (Leica Microsystems Inc) with OpenLab image capturing software (PerkinElmer), analyzed with OpenLab image analysis software or Volocity software (PerkinElmer) and processed with Photoshop (Adobe). Immuno-fluorescence (IF) images of spread preparations represent single optical sections, while IF-FISH/co-FISH images of structurally preserved nuclei/frozen sections represent projections of 0.2 μm optical sections. All FISH measurements were performed on maximum projections or two-dimensional (2D) projections of three-dimensional (3D) image stacks covering the entire nucleus and when paired, the two foci were verified to be in the same Z plane (Takizawa et al., 2008).
Statistical Analysis
Statistical significance of the difference between pairs of samples was assessed by Fisher’s exact test for count data. The error bar is an estimation of the standard deviation (SD), based on the counting error (Taylor, 1997; a measure of the uncertainties in the estimation of the extent of pairing) and is calculated as the square root of the number of cells with paired signals or homologs.
Mouse strains and constructs
Full details of mouse strains and constructs are described in the Extended Experimental Procedures.
Supplementary Material
Highlights.
Homolog pairing precedes SPO11 mediated programmed DSBs during meiosis in the mouse
A DSB-independent activity of SPO11 is required for this preleptotene pairing
SUN1 anchors telomeres to the nuclear periphery to facilitate the preDSB pairing
PreDSB subtelomeric pairing is more stable and most likely aids synapsis initiation
Acknowledgments
We thank M. Lichten (NCI, NIH) and P. Hsieh (NIDDK, NIH) for comments and discussion. We are also grateful to O. Voloshin, P. Khil and G. Margolin (all of NIDDK, NIH) for comments and help with data analysis. This work was supported by the NIDDK (NIH) Intramural Research Program (R.D.C-O.).
Footnotes
Supplemental Information includes five Figures and Legends, one Table and Extended Experimental Procedures, and can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barlow AL, Hultén MA. Combined immunocytogenetic and molecular cytogenetic analysis of meiosis I human spermatocytes. Chromosome Res. 1996;4:562–573. doi: 10.1007/BF02261719. [DOI] [PubMed] [Google Scholar]
- Bastos H, Lassalle B, Chicheportiche A, Riou L, Testart J, Allemand I, Fouchet P. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A. 2005;65:40–49. doi: 10.1002/cyto.a.20129. [DOI] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- Bellani MA, Boateng KA, McLeod D, Camerini-Otero RD. The expression profile of the major mouse SPO11 isoforms indicates that SPO11beta introduces double strand breaks and suggests that SPO11alpha has an additional role in prophase in both spermatocytes and oocytes. Mol Cell Biol. 2010;30:4391–4403. doi: 10.1128/MCB.00002-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellvé AR, Cavicchia JC, Millette CF, O’Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Brown PW, Judis L, Chan ER, Schwartz S, Seftel A, Thomas A, Hassold TJ. Meiotic synapsis proceeds from a limited number of subtelomeric sites in the human male. Am J Hum Genet. 2005;77:556–566. doi: 10.1086/468188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SM, Kleckner N, Weiner BM. Somatic pairing of homologs in budding yeast: existence and modulation. Genes Dev. 1999;13:1627–1641. doi: 10.1101/gad.13.12.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha RS, Weiner BM, Keeney S, Dekker J, Kleckner N. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 2000;14:493–503. [PMC free article] [PubMed] [Google Scholar]
- Chehrehasa F, Meedeniya AC, Dwyer P, Abrahamsen G, Mackay-Sim A. EdU, a new thymidine analogue for labelling proliferating cells in the nervous system. J Neurosci Methods. 2009;177:122–130. doi: 10.1016/j.jneumeth.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Chi YH, Cheng LI, Myers T, Ward JM, Williams E, Su Q, Faucette L, Wang JY, Jeang KT. Requirement for Sun1 in the expression of meiotic reproductive genes and piRNA. Development. 2009;136:965–973. doi: 10.1242/dev.029868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 2008;133:1175–1187. doi: 10.1016/j.cell.2008.04.047. [DOI] [PubMed] [Google Scholar]
- Danilowicz C, Lee CH, Kim K, Hatch K, Coljee VW, Kleckner N, Prentiss M. Single molecule detection of direct, homologous, DNA/DNA pairing. Proc Natl Acad Sci U S A. 2009;106:19824–19829. doi: 10.1073/pnas.0911214106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell. 2007;12:863–872. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Gerton JL, Hawley RS. Homologous chromosome interactions in meiosis: diversity amidst conservation. Nat Rev Genet. 2005;6:477–487. doi: 10.1038/nrg1614. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Dev Cell. 2009;17:598–605. doi: 10.1016/j.devcel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Weiner BM. Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harb Symp Quant Biol. 1993;58:553–565. doi: 10.1101/sqb.1993.058.01.062. [DOI] [PubMed] [Google Scholar]
- Koszul R, Kleckner N. Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends Cell Biol. 2009;19:716–724. doi: 10.1016/j.tcb.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby BJ, Reinholdt LG, Schimenti JC. Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. Proc Natl Acad Sci U S A. 2003;100:15706–15711. doi: 10.1073/pnas.2432067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebe B, Petukhova G, Barchi M, Bellani M, Braselmann H, Nakano T, Pandita TK, Jasin M, Fornace A, Meistrich ML, et al. Mutations that affect meiosis in male mice influence the dynamics of the mid-preleptotene and bouquet stages. Exp Cell Res. 2006;312:3768–3781. doi: 10.1016/j.yexcr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Martínez-Pérez E, Shaw P, Reader S, Aragón-Alcaide L, Miller T, Moore G. Homologous chromosome pairing in wheat. J Cell Sci. 1999;112(Pt 11):1761–1769. doi: 10.1242/jcs.112.11.1761. [DOI] [PubMed] [Google Scholar]
- McKim KS, Green-Marroquin BL, Sekelsky JJ, Chin G, Steinberg C, Khodosh R, Hawley RS. Meiotic synapsis in the absence of recombination. Science. 1998;279:876–878. doi: 10.1126/science.279.5352.876. [DOI] [PubMed] [Google Scholar]
- Mirny LA. The fractal globule as a model of chromatin architecture in the cell. Chromosome Res. 2011;19:37–51. doi: 10.1007/s10577-010-9177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvinen M, Söderström KO. Chromosome rotation and formation of synapsis. Nature. 1976;260:534–535. doi: 10.1038/260534a0. [DOI] [PubMed] [Google Scholar]
- Petukhova GV, Romanienko PJ, Camerini-Otero RD. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev Cell. 2003;5:927–936. doi: 10.1016/s1534-5807(03)00369-1. [DOI] [PubMed] [Google Scholar]
- Pezza RJ, Voloshin ON, Vanevski F, Camerini-Otero RD. Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev. 2007;21:1758–1766. doi: 10.1101/gad.1562907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Meng X, Zhang L, Chretien JH, Urnov FD, Dernburg AF. Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nat Cell Biol. 2009;11:934–942. doi: 10.1038/ncb1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, Handel MA, Schimenti JC. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- Prieto P, Santos AP, Moore G, Shaw P. Chromosomes associate premeiotically and in xylem vessel cells via their telomeres and centromeres in diploid rice (Oryza sativa) Chromosoma. 2004;112:300–307. doi: 10.1007/s00412-004-0274-8. [DOI] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Jerratsch M, Li B, Smith S, Hultén M, Lock T, de Lange T. Mammalian meiotic telomeres: protein composition and redistribution in relation to nuclear pores. Mol Biol Cell. 2000;11:4189–4203. doi: 10.1091/mbc.11.12.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, Kaback DB. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104:16934–16939. doi: 10.1073/pnas.0704860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H, Weich S, Schwegler H, Heyting C, Härle M, Cremer T. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol. 1996;134:1109–1125. doi: 10.1083/jcb.134.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Gudla PR, Guo L, Lockett S, Misteli T. Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev. 2008;22:489–498. doi: 10.1101/gad.1634608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JR. An introduction to error analysis: the study of uncertainties in physical measurements. 2. Sausalito, Calif: University Science; 1997. [Google Scholar]
- Tsai JH, McKee BD. Homologous pairing and the role of pairing centers in meiosis. J Cell Sci. 2011;124:1955–1963. doi: 10.1242/jcs.006387. [DOI] [PubMed] [Google Scholar]
- Weiner BM, Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.