Abstract
Tissue heat stabilization is a vital component in successful mammalian neuropeptidomic studies. Heat stabilization using focused microwave irradiation, conventional microwave irradiation, boiling, and treatment with the Denator Stabilizor T1 have all proven effective in arresting post-mortem protein degradation. Although research has reported the presence of protein fragments in crustacean hemolymph when protease inhibitors were not added to the sample, the degree to which postmortem protease activity affects neuropeptidomic tissue studies in crustacean species has not been investigated in depth. This work examines the need for Stabilizor T1 or boiling tissue stabilization methods for neuropeptide studies of Callinectes sapidus (blue crab) pericardial organ tissue. Neuropeptides in stabilized and non-stabilized tissue are extracted using acidified methanol or N,N-Dimethylformamide (DMF) and analyzed by MALDI-TOF and nanoLC-ESI-MS/MS platforms. Post-mortem fragments did not significantly affect MALDI analysis in the range m/z 650–1600, but observations in ESI MS/MS experiments suggest that putative post-mortem fragments can mask neuropeptide signal and add spectral complexity to crustacean neuropeptidomic studies. The impact of the added spectral complexity did not dramatically affect the number of detected neuropeptides between stabilized and non-stabilized tissues. However, it is prudent that neuropeptidomic studies of crustacean species include a preliminary experiment using the heat stabilization method to assess the extent of neuropeptide masking by larger, highly charged molecular species.
Keywords: Neuropeptide, crustacean, Callinectes sapidus, heat stabilization, DiLeu, MALDI, ESI, peptidomics, Stabilizor T1, post-mortem
Introduction
Neuropeptides are a class of signaling molecules that act as neurohormones, neuromodulators, or neurotransmitters in the central nervous system. These signaling peptides are involved in a variety of physiological roles including cell-cell communication, feeding, pain, depression, and memory.1–5 The precise regulation of physiological processes that neuropeptides display have made them the focus of many researchers striving to further understand development, homeostasis, animal behavior, and disease progression mechanisms.6–12 Studies of this nature can be divided up into two categories: targeted quantitative neuropeptidomics and discovery neuropeptidomics. In the first category, researchers use a list of interesting peptides for experiments that aim to fully characterize the roles these molecules play in behavior or disease. For these studies, antibodies are commonly used to study peptide distribution and measure peptide location and relative abundance.13–15 In the second category, the focus of the experiment is to identify and characterize all of the peptides in a neural organ and then follow the expression changes these peptides undergo during the progression of a study. The ability to characterize all of the peptides in a neural organ at once resulted from many years of analytical method development.16 An important technology that was advanced during this time was biological mass spectrometry.
The development of matrix-assisted laser desorption/ionization (MALDI)17, 18 and electrospray ionization (ESI)19 techniques and increases in mass analyzer sensitivity and resolving power have allowed mass spectrometry to become a prominent tool in global and targeted studies of neuropeptide expression in neural organs.6, 9, 12, 20–23 Although mass spectrometry is a highly sensitive technique, the investigation of neuropeptides in neural tissue is still challenging, and further analytical method development is warranted.16 Signaling peptides are normally present at low concentrations (μM or nM), often have non-ideal amino acid sequences when it comes to molecular ionization efficiencies, and reside in a mass-to-charge (m/z) range that can become overrun with degradation fragments from abundant proteins. Technological advancements and methodological improvements can help solve the first two challenges, but the latter challenge requires the development of innovative sample collection and preparation techniques.
Researchers have been aware of the post-mortem degradation problems in neuropeptidomic tissue research for years.24–28 Several strategies have been developed to minimize degradation products in the neuropeptide m/z range. Focused microwave irradiation has been used to stabilize rodent brains prior to dissection.26, 29, 30 A drawback of this technique is that it is expensive and specific to stabilizing only one organ of interest, the brain. More commonly, efforts have been directed towards stabilizing tissue after sacrificing the animal. Snap-freezing, heat deactivation by conventional microwave irradiation, and boiling are additional tissue stabilization methods commonly employed.6, 24, 25 An alternative strategy for post-sacrifice tissue stabilization that can be applied to a variety of tissues was developed in Per Andren’s laboratory.27, 31 The technology rapidly and uniformly raises the temperature of tissue to 90 °C, halting proteolytic activity. Reported results using this technology were similar to data gathered using focused microwave sacrifice.27
This technology has demonstrated a clear advantage in mammalian peptidomics, but the extent that post-mortem degradation affects peptidomic studies in the crustacean model species is unknown. Many crustacean species used for neurochemical peptidomic research do not have sequenced genomes. Therefore, peptidomic studies performed in these species rely on the assumption that the presence of proteolytic fragments in the neuropeptide m/z range is minimal and do not suppress neuropeptide signal. The presence of protein fragments in crustacean hemolymph when protease inhibitors were not added to the sample has been reported previously.32 However, the degree to which post-mortem protease activity affects neuropeptidomic tissue studies in crustaceans has not been investigated in depth. The assumption is tested here by comparing the neuropeptide contents in Callinectes sapidus (blue crab) pericardial organs (POs) treated by heat stabilization using Denator Stabilizor T1 or boiling to non-heat treated tissues. Neuropeptides were extracted from treated or non-treated POs using acidified methanol (acMeOH) or N,N-Dimethylformamide (DMF) and mass analyzed using MALDI and ESI mass spectrometers.
Materials and Methods
All chemicals and reagents were purchased from Thermo Fisher Scientific (Waltham, MA) unless otherwise specified.
Animals and Tissue Collection
Blue crabs were obtained from commercial suppliers (The Crab Place, Crisfield, MD; Maryland Blue Crab Express, Darlington, MD) and placed in an artificial seawater tank at 12–13 °C before use. Crabs were allowed to acclimate to the tank conditions for two days prior to dissection. Thirty minutes prior to dissection, crabs were cold anesthetized on ice. Details of animal treatment and dissection were described previously by Kutz et al.33 In this study, both pericardial organs (POs) were isolated in chilled physiological saline (composition: 440 mM NaCl; 11 mM KCl; 13 mM CaCl2; 26 mM MgCl2; 10 mM HEPES acid; pH 7.45). A total of 24 crabs were used in this study.
Sample Preparation
Immediately following dissection, POs were briefly rinsed in water and thermally stabilized using a Denator Stabilizor T1 tissue stabilization device (Gothenburg, Sweden), boiled in water, or placed directly in 20 μL of ice-cold neuropeptide extraction solvent. Extraction solvents were composed of either acidified methanol (acMeOH, 90:9:1 MeOH:H2O:glacial acetic acid, v:v:v) or N,N-Dimethylformamide (DMF). On average, approximately four minutes passed during the PO microdissection of each pericardial ridge. For tissue stabilized with the Stabilizor T1, each PO was placed in a Stabilizor cartridge, inserted into the device, and stabilized using the fresh preserve tissue function. The stabilization process involved uniformly heating the tissue to 95 °C for 30–45 sec (depending on thickness). After stabilization, tissue was removed from the cartridge, placed in the appropriate extraction solvent, and stored at −80 °C until needed. Tissue stabilized by boiling was placed into a 0.6 mL microcentrifuge tube containing 20 μL DMF and then suspended in a beaker of boiling water for 10 min. Stabilized tissue was then stored at −80 °C until needed.
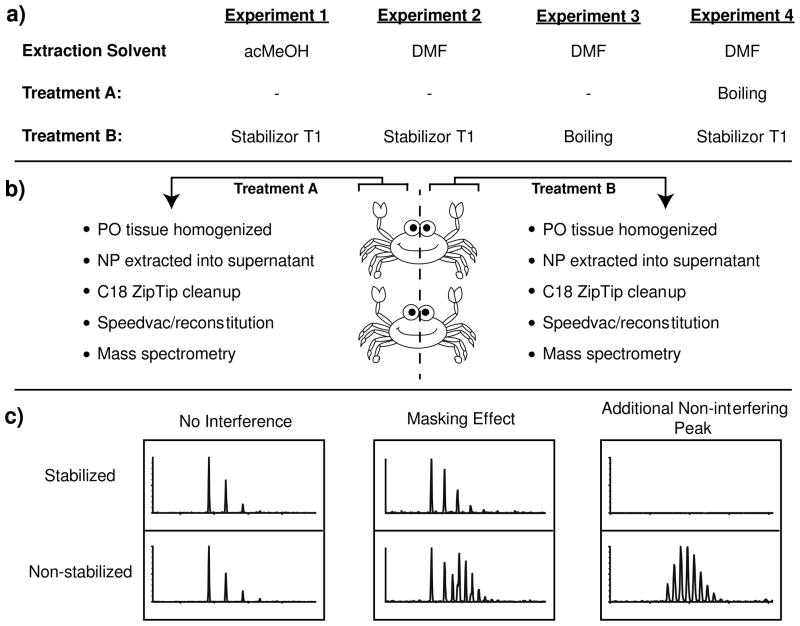
To minimize biological variability between the crabs that could skew the experiments, four independent comparisons of blue crab PO tissue handling/extraction were made in this work. Each comparison required two crabs (4 POs) and was performed in biological triplicate (Figure 1). POs removed from the left side of the crabs were either treated with the Stabilizor T1 (experiments 1, 2, and 4) or boiled (experiment 3). For experiments 1, 2, and 4 the right POs were not stabilized. In experiment 3, the two right POs were boiled in DMF prior to extraction. Stabilized and non-stabilized tissue were placed directly into a minimal volume of extraction solvent and stored at −80 °C. In the first experiment, neuropeptides were extracted with acMeOH from tissue that was either stabilized (Stabilizor T1) or non-stabilized. In the second, third, and fourth experiments, neuropeptides were extracted from stabilized and non-stabilized tissues using DMF. Experiment 2 compared stabilized (Stabilizor T1) to non-stabilized tissue. Experiment 3 compared stabilized (boiling) to non-stabilized tissue. Experiment 4 compared stabilized (Stabilizor T1) to stabilized (boiling) tissue.
Figure 1.
Experimental workflow. (a) Four experiments were carried out in this study. In experiment 1, acMeOH was used for neuropeptide extraction, whereas in experiments 2, 3, and 4, DMF was used to extract neuropeptides. (a, b) Right and left POs were removed from two crabs. Removed right POs were pooled together and processed using the “Treatment A” workflow. Removed left POs were pooled together and processed using the “Treatment B” workflow. POs were either placed directly into an extraction solvent (acMeOH or DMF) or subjected to tissue stabilization (Stabilizer T1 or boiling) and then placed in extraction solvent. Tissue was then homogenized, neuropeptides extracted, concentrated, and subjected to mass spectral analysis by nanoLC-ESI-MS/MS and MALDI-TOF-MS. (c) It was expected to find instances where no neuropeptide interference would be observed, regions where an isotopic envelope from non-stabilized samples would mask neuropeptide signal, and regions where ion peaks present only in non-stabilized tissues would be detected, constituting additional non-interfering peaks.
For each part of a single comparison, stabilized or non-stabilized POs were placed in a 0.1 mL glass tissue grinder (Wheaton), submerged in 50 μL of appropriate ice-cold extraction solvent, and mechanically homogenized. The homogenate was transferred to a clean 0.6 mL low retention microcentrifuge tube and centrifuged at 16000 × g for 10 min. The supernatant was removed from the tissue pellet and placed in a clean vial, and the pellet was re-extracted two additional times. The combined supernatant volumes were concentrated to dryness using a Savant SVC 100 Speedvac (Thermo Fisher Scientific) vacuum concentrator at medium heat. The crude extract was resuspended in 50 μL of 0.1% formic acid(aq) (v/v) and processed by C18 Ziptips (Millipore) according to the manufacturer’s protocol to remove lipids, salts, and large proteins from the sample. Following vacuum concentration of the eluent, peptides were resuspended in 11 μL of 0.1 % formic acid(aq) and subjected to mass spectrometric analysis.
MALDI-TOF/TOF Mass Spectrometry and Data Analysis
Each neuropeptide sample was analyzed with an Applied Biosystems (Framingham, MA) 4800 MALDI TOF/TOF mass spectrometer equipped with a 200 Hz, 355 nm Nd:YAG laser. Sample aliquots of 0.5 μL were spotted onto a metal MALDI target, mixed with 0.5 μL of α-Cyano-4-hydroxycinnamic acid MALDI matrix (Sigma-Aldrich, St. Louis, MO), and allowed to dry at room temperature prior to analysis. A 0.5 μL aliquot of Sigma-Aldrich ProteoMass peptide MALDI-MS calibration standard was also spotted on the plate to be used for instrument calibration. Instrument parameters were optimized using the 4000 Series Explorer Software (Applied Biosystems). All data were acquired over the m/z range 500–2000 in positive reflectron mode. Data was analyzed using Applied Biosystems Explorer software (version 4.9, build 115) and peptides were identified based on accurate mass (50 ppm tolerance) using an in-house database of identified crustacean neuropeptides (Supplemental Database 1)
nanoLC-ESI-MS/MS Mass Spectrometry and Data Analysis
A Waters Synapt G2 QTOF mass spectrometer coupled to a Waters nanoAcquity ultra performance liquid chromatograph system was used for nano-liquid chromatography electrospray ionization tandem mass spectrometry (nanoLC-ESI-MS/MS) analysis. Mobile phase A was water in 0.1% FA(aq) (v/v), and mobile phase B was ACN in 0.1% FA(aq) (v/v). A 2 μL injection of neuropeptide rich PO extract was loaded onto a Symmetry C18 nanoAcquity trap column (180 μm × 20 mm, 5 μm) at a flow rate of 5 μL·min−1 of 97% mobile phase A for 3 min. Peptides were separated using a 1.7 μm BEH C18 75 μm × 100 mm column with a 60 min gradient from 3% to 35% mobile phase B at a flow rate of 300 nL·min−1 and a column temperature of 35 °C. Electrospray emitter tips were prepared in house from 75 μm i.d., 360 μm o.d. capillary tubing (Polymicro Technologies, Phoenix, AZ) using a Sutter P-2000 laser capillary puller (Novator, CA).
The mass spectrometer was calibrated before use with the fragment ions of Glu-fibrinopeptide over the m/z range 50–2000. Data was acquired in resolution mode using either MSE (where the E represents collision energy) or data dependent acquisition (DDA) mode. For MSE acquisitions, low energy and high energy scans were collected at 1 Hz with a high energy collision energy ramp of 10–35 eV. For DDA data, a 1 Hz survey scan was followed by three MS/MS scans if ions in the survey scan had a charge of +2, +3, or +4. Ions subjected to MS/MS were actively excluded from subsequent MS/MS scans for 60 sec. MS/MS spectra were collected using a low mass collision energy ramp of 10–25 eV and a high mass collision energy ramp of 25–45 eV. For both MSE and DDA modes, lockmass correction scans of Glu-fibrinopeptide (m/z 785.8421) were collected every 60 sec during the acquisition. The capillary voltage was set to 2.80 kV, sampling cone voltage was 30 V, extraction cone was 4.0 V, and the source temperature was set to 70 °C.
Acquired data were analyzed using MassLynx 4.1 software. Peptides between [M+H]+ m/z 600 – 2200 were identified by accurate mass (30 ppm), retention times (± 1 min), and tandem mass spectra using an in-house database of identified crustacean neuropeptides and PepSeq de novo sequencing software. For peptides with [M+H]+ > m/z 2200, Mascot was used for identification. MSE files were processed using ProteinLynx Global Server (v.2.5.1) using the Apex 3D algorithm to generate pkl files. The pkl files were searched against an in-house crustacean neuropeptide/neurohormone database (1957 non-redundant entries from reported literature and from Uniprot search of “(neuro)peptide” and arthropod species) using Mascot (ver 2.3.02). For Mascot searches, MS peptide tolerance was set to ± 30 ppm, MS/MS tolerance was 0.8 Da, and variable modifications amidation (C-term), pyro Glu (N-term Q), pyro Glu (N-term E) and oxidation (M) were specified. Peptides with scores > 40 were considered valid.
The identification of putative interfering protein fragments was determined manually by summing up 2.5 min segments of low energy MSE data and comparing between samples in a given experiment using MassLynx 4.1 software.
Results and Discussion
Neuropeptide Extraction Solvent Rationale
Two neuropeptide extraction methods used in crustacean neuropeptidomic research were used in this study. Acidified methanol is commonly used to extract neuropeptides from neural tissues.34–36 However, when relative quantification experiments are performed using a set of in-house developed 4-plex DiLeu isobaric tags, DMF is the preferred extraction solvent.35 Table 1 and Supplemental Table 1 demonstrate that DMF had comparable neuropeptide extraction efficiency to acMeOH in this study.
Table 1.
Neuropeptides observed in ESI mass spectra of each sample preparation experimenta
| Experiment 1 | Experiment 2 | Experiment 3 | Experiment 4 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Family | [M+ H]+ | Sequence | acMeOH | Stabilizor T1 | DMF | Stabilizor T1 | DMF | Boiling | Boiling | Stabilizor T1 |
| AST-A | 781.39 | DPYAFGLa | + | + | ||||||
| 854.40 | DGPYSFGLa | + | + | + | + | |||||
| 883.43 | SNPYSFGLa | + | ||||||||
| b893.42 | SGHYNFGLa | + | + | + | ||||||
| 897.46 | ARGYDFGLa | + | + | + | + | + | + | + | + | |
| 909.49 | ARPYSFGLa | + | + | + | + | + | + | + | + | |
| 911.47 | ARAYDFGLa | + | + | |||||||
| 928.45 | SSGQYAFGLa | + | + | + | ||||||
| 937.53 | PRVYSFGLa | + | + | + | + | + | + | + | + | |
| 939.50 | TRPYSFGLa | + | + | + | + | + | + | + | + | |
| 962.51 | APQPYAFGLa | + | + | + | + | + | + | + | ||
| b986.48 | RNMYSFGLa | + | + | + | + | + | + | + | + | |
| 998.49 | TSPQYSFGLa | + | + | |||||||
| 1004.48 | FSGTYNFGLa | + | + | + | + | + | + | + | + | |
| 1021.51 | THPTYSFGLa | + | + | + | + | + | + | + | + | |
| 1097.60 | AGLSALYSFGLa | + | + | |||||||
| 1167.58 | AAGLQNYDFGLa | + | + | + | + | + | + | + | + | |
| 2051.03 | GSGQYAYGLGKKAGQYSFGLa | + | + | + | + | + | + | |||
|
| ||||||||||
| AST-B | 1031.51 | AWSNLQGAWa | + | + | + | + | + | + | + | |
| 1061.55 | AGWSSLKGAWa | + | + | + | + | + | + | + | + | |
| 1107.52 | AGWSSMRGAWa | + | + | + | + | + | + | + | + | |
| 1123.51 | AGWSSM(O)RGAWa | + | + | + | + | + | + | + | + | |
| b1165.55 | NWNKFQGSWa | + | + | + | + | + | + | |||
| 1182.57 | TSWGKFQGSWa | + | + | + | + | + | + | + | + | |
| 1209.58 | TGWNKFQGSWa | + | + | + | + | + | + | + | + | |
| 1220.58 | SGDWSSLRGAWa | + | + | + | + | + | + | + | + | |
| 1222.58 | GNWNKFQGSWa | + | + | + | + | + | + | + | + | |
| 1252.59 | NNWSKFQGSWa | + | + | + | + | + | + | + | + | |
| 1253.57 | NDWSKFGQSWa | + | + | + | + | + | ||||
| 1293.63 | STNWSSLRSAWa | + | + | + | + | + | + | + | + | |
| 1380.64 | NNNWTKFQGSWa | + | + | + | + | + | + | + | + | |
| 1470.70 | VPNDWAHFRGSWa | + | + | + | + | + | + | + | + | |
| 1586.84 | MFAPLAWPKGGARWa | + | + | + | + | + | + | + | + | |
|
| ||||||||||
| AST-C | b1899.83 | pQIRYHQcYFNPIScF | + | + | + | + | + | + | + | + |
|
| ||||||||||
| CCAP | 956.38 | PFCNAFTGCa | + | + | + | + | + | + | + | + |
|
| ||||||||||
| FLP | b695.40 | NFLRFa | + | + | + | + | + | + | + | + |
| b851.50 | RNFLRFa | + | ||||||||
| b916.52 | THPFLRFa | + | + | + | + | + | + | |||
| b925.45 | DDNFLRFa | + | + | + | + | + | ||||
| 926.52 | SKNYLRFa | + | + | + | + | + | + | + | + | |
| 965.54 | NRNFLRFa | + | + | + | + | + | + | + | + | |
| 966.53 | DRNFLRFa | + | + | + | + | + | + | + | + | |
| 1022.56 | GNRNFLRFa | + | + | + | + | + | + | + | + | |
| 1104.61 | GAHKNYLRFa | + | + | + | + | + | + | |||
| 1124.63 | GLSRNYLRFa | + | + | + | + | + | + | + | ||
| b1132.60 | NVGSHGFLRFa | + | + | + | + | + | ||||
| 1146.61 | GYSKNYLRFa | + | + | + | + | + | + | + | + | |
| 1147.65 | APQRNFLRFa | + | + | + | + | + | + | + | + | |
| 1158.62 | YGNRSFLRFa | + | + | + | + | + | + | + | + | |
|
| ||||||||||
| Myosuppressin | 1125.57 | pQDLDHVFLR | + | + | + | + | + | |||
| 1271.65 | pQDLDHVFLRFa | + | + | + | + | + | + | + | + | |
| 1288.68 | QDLDHVFLRFa | + | + | + | + | + | + | |||
|
| ||||||||||
| Orcomyotropin | 1186.52 | FDAFTTGFGHS | + | + | + | + | + | + | + | + |
|
| ||||||||||
| Orcokinin | 1228.56 | NFDEIDRSSFa | + | + | # | + | ||||
| 1256.55 | NFDEIDRSGFG | + | + | |||||||
| 1270.57 | NFDEIDRSGFA | + | + | + | + | + | + | + | + | |
| 1403.62 | NFDEIDRSGFGF | + | + | + | + | + | + | + | + | |
| 1433.63 | NFDEIDRSSFGF | # | + | # | + | # | # | + | ||
| 1474.66 | NFDEIDRSGFGFA | + | + | + | + | + | + | + | + | |
| 1502.69 | NFDEIDRSGFGFV | + | + | + | + | + | + | + | + | |
| 1504.67 | NFDEIDRSSFGFA | # | + | # | + | # | # | # | + | |
| 1532.70 | NFDEIDRSSFGFV | # | + | # | + | # | # | # | + | |
| 1547.68 | NFDEIDRSSFGFN | + | + | + | + | + | + | + | + | |
|
| ||||||||||
| Others | 844.48 | HLGSLYRa | + | + | + | + | + | + | + | + |
|
| ||||||||||
| Proctolin | 649.37 | RYLPT | + | + | + | + | + | + | + | + |
|
| ||||||||||
| RYamide | 784.41 | FVGGSRYa | + | + | + | + | + | + | + | + |
| 832.41 | FYANRYa | + | + | + | + | + | + | + | + | |
| 959.47 | SGFYAPRYa | + | + | + | + | + | + | + | + | |
| 976.46 | SGFYANRYa | + | + | + | + | + | + | + | + | |
| 1027.54 | SRFVGGSRYa | + | + | + | + | |||||
| 1030.47 | pEGFYSQRYa | + | + | # | + | + | + | # | + | |
| 1114.58 | SSRFVGGSRYa | + | + | + | + | + | + | + | + | |
|
| ||||||||||
| Total Peptides | 60 | 62 | 59 | 61 | 64 | 61 | 57 | 58 | ||
| Unique Peptides | 2 | 1 | 5 | 2 | 6 | 2 | 6 | 4 | ||
AST, allatostatin; CCAP, crustacean cardioactive peptide; CPRP, crustacean hyperglycemic hormone precursor related peptide; FLP, FMRFamide like peptide. Amidation indicated by lowercase a. Pyroglutamation indicated by lowercase p. Disulfide bond indicated by lowercase c. Methionine oxidation indicated by M(O).
indicates peptide had BPI > 1000, but no MS/MS event triggered
indicates that peptide was observed in ESI spectra
indicates that there are additional peaks at [M+2H]+2 that mask neuropeptide signal
We also tested the assumption that the neuropeptide expression between right and left POs are similar since left POs from two crabs were pooled for one extraction protocol and POs from the right side of two crabs were pooled for a different extraction protocol in each experiment.37, 38 To test this assumption, POs from two crabs were dissected out and the two left and right POs were pooled together separately. Neuropeptides were extracted from both PO pools with DMF and analyzed on the Synapt G2 QTOF mass spectrometer. Supplemental Figure 1 shows that the base peak ion (BPI) chromatograms for left and right POs were similar, suggesting that differences in neuropeptide expression in all experiments should only depend on the extraction protocols used, not the location of the POs used.
Four neuropeptide extraction protocols were compared in this study. Figure 1 shows an overview of the experimental workflow. Experiment 1 compared the conventional acMeOH extraction protocol to an acMeOH extraction protocol incorporating the Stabilizor T1 for tissue stabilization. DMF was used as the extraction solvent in experiments 2, 3, and 4. Experiment 2 compared the normal DMF neuropeptide extraction method to a DMF method that utilized the Denator Stabilizor T1 for tissue stabilization. Experiment 3 compared the normal DMF extraction protocol to a protocol that utilized a 10 min boiling period to stabilize the tissue prior to DMF extraction. Experiment 4 compared the DMF extraction protocol utilizing boiling stabilization to a DMF extraction protocol utilizing Stabilizor T1 stabilization. Experiments 3 and 4 were designed to compare the performance of boiling to Stabilizor T1 stabilization.
MALDI-TOF-MS Analysis
Previous research has shown that mouse brain stabilized with the Stabilizor T1 showed increased detection of endogenous neuropeptides as compared to non-stabilized tissue in MALDI MS experiments.31 Figure 2a and 2b show the MALDI mass spectra of acMeOH versus Stabilizor T1 extracted tissue and DMF versus Stabilizor T1 extracted tissue, respectively. In Figure 2a, maximum intensities of both mass spectra were set to the base peak ion (BPI) of the non-stabilized acMeOH extracted tissue (3.1 × 104). The only molecular ion in the Stabilizor T1 stabilized tissue that exceeded this threshold intensity was RYamide pEGFYSQRYa (m/z 1030.45), which had an intensity of 5.8 × 104. In both mass spectra, the other neuropeptide molecular ions present in the spectra did not exceed 20% relative intensity. Overall, a similar number of neuropeptides were identified in acMeOH extracted non-stabilized (44 peptides, 2 unique) and stabilized (45 peptides, 3 unique) tissue. Figure 2b, shows a similar pattern for DMF versus Stabilizor T1 treated tissue. Again, both mass spectra intensities were set to the BPI of the non-stabilized tissue, with only m/z 1030.45 exceeding the intensity threshold in the stabilized tissue. A total of 40 neuropeptides (zero unique) were identified in DMF non-stabilized tissue extracts and 43 neuropeptides (3 unique) were identified in DMF stabilized tissue extracts. The results of DMF versus boiling stabilization and boiling stabilization versus Stabilizor T1 stabilization experiments were similar to those observed in Figure 2. Supplemental Table 1 summarizes the neuropeptides identified in each of the four MALDI experiments.
Figure 2.
MALDI-TOF-MS spectra for one biological replicate of a) acMeOH versus Stabilizor T1 and b) DMF versus Stabilizor T1. Mass spectra between the two sample treatments in both experiments are similar. No noticeable interfering, ionization efficiency perturbing molecular ions were observed in m/z 650 to 1600.
In total 13 A-type allatostatins, 14 B-type allatostatins, one C-type allatostatin, a crustacean cardioactive peptide (CCAP), two crustacean hyperglycemic hormone precursor related peptides (CPRP), six FMRFamide like peptides (FLP), one myosuppressin, an orcomyotropin, 10 orcokinins, five RYamides, and one other peptide were observed by MALDI analysis. In each experiment, approximately the same numbers of peptides were observed between sample treatments. In addition, all intense ions observed from m/z 650 – 1600 were identified as neuropeptides. It appears that neuropeptide detection from m/z 650 – 1600 in blue crab POs were not affected by post-mortem protein fragments. Although roughly the same numbers of neuropeptides were identified in the MALDI mass spectra from non-stabilized and stabilized tissue, the extent of post-mortem degradation was also investigated in ESI mass spectra. In MALDI, singly charged ion species are predominantly observed from m/z 650 – 1600. In contrast, ESI produces predominantly multiply charged ions in the m/z range 300 – 1600.
Base Peak Ion Chromatograms in nanoLC-ESI-MS/MS Experiments
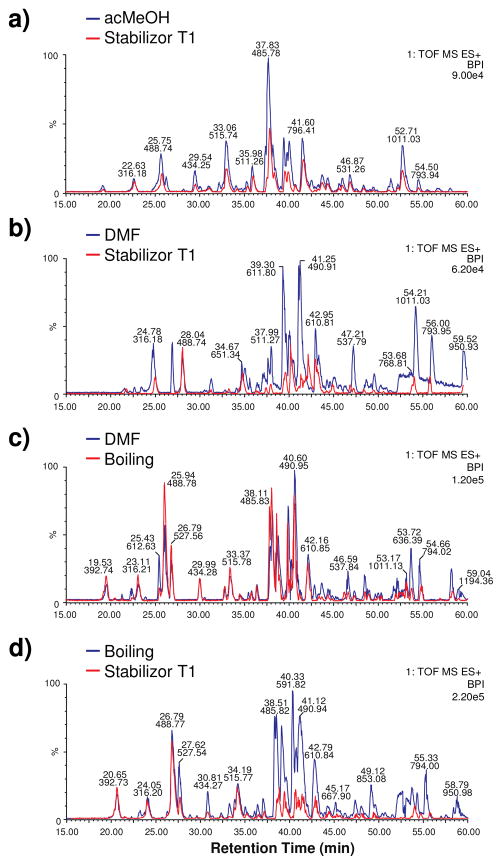
Figure 3 shows the ESI BPI chromatograms for a single biological replicate for each experiment. The same trend shown for each experiment in Figure 3 was observed in each biological replicates. After inspection of the four BPI chromatograms for all four experiments, three observations are readily apparent. First, Figure 3a, 3b, and 3d depict Stabilizor T1 treated tissue versus non-stabilized acMeOH, non-stabilized DMF, and stabilized boiling tissue extract, respectively. In each case, the red BPI chromatogram trace (Stabilizor T1 stabilization) has an observably lower BPI intensity compared to tissue extracts not treated with Stabilizor T1. Second, Figure 3b and 3d show that after 50 min of separation, more intense peaks are observed in DMF tissue extracts not treated with the Stabilizor T1. Third, Figure 3c illustrates that BPI intensity differences were observably minimal between tissues treated with boiling stabilization versus tissue only treated with DMF extraction. In addition to the BPI chromatogram profiles in Figure 3, Supplemental Figure 2 shows 2.5 min summed mass spectra of each experiment across three quarters of the elution gradient (17.5 – 60 min). In all experiments using the Stabilizor T1, it was observed that at each time point the BPI intensities of all mass spectra were greater in tissue not stabilized with the Stabilizor T1. For experiment 3, the mass spectrum BPI intensities appeared similar between tissues stabilized by boiling and tissues only treated with DMF.
Figure 3.
Base peak ion (BPI) chromatograms of each neuropeptide sample preparation experiment. a) acMeOH versus Stabilizor T1. b) DMF versus Stabilizor T1. c) DMF versus boiling. d) Boiling versus Stabilizor T1. In all biological replicates, the BPI of tissue stabilized with Stabilizor T1 prior to neuropeptide extraction was lower than non-stabilized, or boiling stabilized issue.
Molecular ion intensities of tissues treated with the Stabilizor T1 had lower intensities than tissue samples stabilized by boiling or not stabilized at all. We hypothesize that the decreased signal intensities observed in the BPIs were due to the loss associated with an additional step in a sample handling process. In addition, it was difficult to remove all of the stabilized PO tissue from the Denator Stabilizor T1 tissue card used during the stabilization process. The blue crab PO is approximately 100 μm thick and 3 mm long prior to being placed in the Denator card. We believe that this technical difficulty can reconcile the reduced signal observed in all Stabilizor T1 BPI chromatograms. Since the experiments performed in this study use the minimum amount of PO material needed to get good ion signal for three 2 μL injections, the signal loss associated with Stabilizor T1 treatment may be decreased when pooling enough POs for discovery neuropeptidomic experiments (> 10).21, 36
Neuropeptide Signal Masking in Non-Stabilizor T1 Treated Tissues
Table 1 shows the peptides identified in each experimental comparison. In total, 18 AST-A peptides, 15 AST-B peptides, one AST-C, a CCAP, 14 FLPs, three myosuppressins, an orcomyotropin, 10 orcokinins, proctolin, seven RYamide peptides, and one other peptide were identified from all four experiments using only two POs (one per crab). For a peptide to be counted as present in an experiment, it had to be observed in at least 2 of the 3 biological replicates and validated by accurate mass, retention times between experiments, and MS/MS spectra if the peptide was in high enough abundance. Supplemental Figure 3 shows MS/MS de novo sequenced spectra for neuropeptides identified in Table 1. The total number of neuropeptides identified within each experiment was similar between experiments and averaged to be 60. Within each experiment, the number of neuropeptides identified using either stabilized or non-stabilized tissue was similar as well, suggesting that potential postmortem fragments did not affect neuropeptide detection. Upon initial review, there did not seem to be a great advantage to implementing the Stabilizor T1 into crustacean PO neuropeptidomic studies. A closer inspection of the mass spectra data showed that tissue stabilized with the Stabilizor T1 contained endogenous neuropeptide signals that were masked by interfering ions only present in non-Stabilizor T1 treated samples. A total of five neuropeptides were observed to be masked entirely or partially by an isotopic envelope from an ion species unique to non-Stabilizor T1 treated samples. Four of the masking events occurred during orcokinin elution times and one event occurred during the elution time of a RYamide. Figure 4 gives two examples of the spectral masking observed in the experiments affecting orcokinin peptide detection. Figure 4a, b, and c show the interference observed for the doubly charged molecular ion of NFDEIDRSSFGFA (m/z 752.84) in acMeOH, DMF, and boiling extraction experiments, respectively. The top panel of each sub-figure demonstrates that tissue treated with the Stabilizor T1 lacked the interference observed in non-Stabilizor T1 treated tissues (bottom panel). The bottom panel of each sub-figure shows that the mass spectral profile of m/z 752.84+2 was masked by a quadruply charged molecular ion in tissues not treated with Stabilizor T1. Without the Stabilizor T1, orcokinin NFDEIDRSSFGFA would not be identified in an ESI-MS/MS experiment, as was the case in a previous study.36 Fortunately, MALDI-MS experiments were performed in the previous study, and the neuropeptide was identified in the blue crab PO peptidome, highlighting the advantage of combining MALDI and ESI-based approaches. Figure 4d,e, and f show that orcokinin NFDEIDRSSFGF (m/z 717.32+2) is clearly observed in Stabilizor T1 treated tissues (top panel) but masked in non-Stabilizor T1 treated tissues by a triply charged molecular ion (bottom panel).
Figure 4.
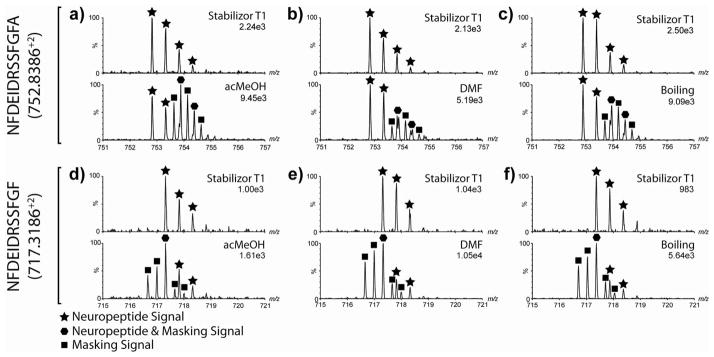
Examples of neuropeptides masked by interfering ion peaks. a), b), and c) show that orcokinin NFDEIDRSSFGFA (m/z 752.84+2) is easily observed in Stabilizor T1 treated tissue extracts but masked by a quadruply charged ion envelope in tissue not treated with the Stabilizor T1. d), e), and f) show that orcokinin NFDEIDRSSFGF (m/z 717.32+2) is easily observed in Stabilizor T1 treated tissue extracts but masked by a triple charged molecular ion in tissue not treated with the Stabilizor T1
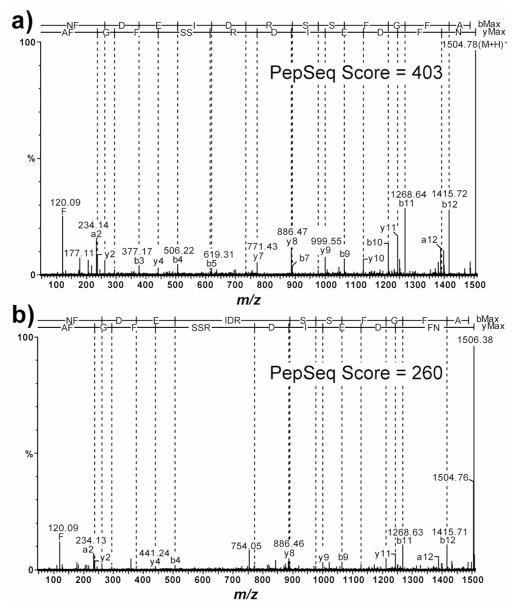
One question resulting from these observations is how MS/MS spectra were affected by the chimeric MS spectra. Figure 5a shows the tandem mass spectrum of NFDEIDRSSFGFA ([M+H]+ = m/z 1504.67) from a Stabilizor T1 treated tissue extract, and Figure 5b shows the MS/MS mass spectrum of the boiling treated tissue extract of the same orcokinin. The MS/MS mass spectrum of the orcokinin in the Stabilizor T1 treated tissue extract received a PepSeq score of 403 whereas the orcokinin in boiled treated tissue extract received a PepSeq score of 203. The disparity in scores can be attributed to the boiled sample’s chimeric MS spectrum resulting in b and y ions from two differently charged molecular ions being present in the same MS/MS spectrum. In this case, the PepSeq algorithm could not determine the [M+H]+ of the orcokinin in the boiled MS/MS spectra because max charge state for fragment ions was selected as +2. The three other orcokinin peptides affected by MS masking interference also showed the same PepSeq score trend. In experiment 2, orcokinin NFDEIDRSSFa ([M+H]+ = m/z 1228.56) was only observed in the Stabilizor T1 treated samples. In the non-stabilized DMF samples, the doubly charged ion (m/z 614.78) was masked by a +6 charged ion envelope. In the case of orcokinins NFDEIDRSSFGF ([M+H]+ = m/z 1433.63) and NFDEIDRSSFGFV ([M+H]+ = m/z 1532.70), the Stabilizor T1 treated tissues had PepSeq scores of 354 and 313, and the non-Stabilizor T1 treated tissues had PepSeq scores of 333 and 113, respectively. In the case of RYamide pEGFYSQRYa, its doubly charged molecular ion m/z 515.73+2 is co-isolated with a much lower abundance quadruply charged ion envelope. The PepSeq scores for the Stabilizor T1 treated tissue versus the scores for the boiling treated tissues were 24 and 32, respectively. Here, the score for the non-Stabilizor T1 treated tissues is slightly higher than the Stabilizor T1 treated tissue.
Figure 5.
MS/MS fragmentation spectra of masked orcokinin NFDEIDRSSFGFA ([M+H]+ = m/z 1504.67) identified using Waters PepSeq algorithm. a) MS/MS of PO tissue extract treated with Stabilizor T1. b) MS/MS of PO tissue extract treated with boiling. Both tissue samples were extracted with DMF. The [M+H]+ ion is correctly identified in the Stabilizor T1 treated sample with a PepSeq score of 403 whereas the [M+H]+ ion is incorrectly identified in the boiled tissue extract resulting in a PepSeq score of 260.
Additional Non-Interfering Ion Peaks in Non-Stabilizor T1 Treated Tissues
Although only five out of 72 neuropeptides were found to be masked by other ion species in non-Stabilizor T1 treated tissues, many additional highly charged peaks were observed in DMF extracted non-Stabilizor T1 treated tissues versus their Stabilizor T1 treated counterparts. Supplemental Table 2 lists the additional ion peaks observed in non-Stabilizor T1 treated tissues that were not observed in Stabilizor T1 treated samples. In experiment 1, only 15 extra non-interfering ion envelopes were observed in the non-Stabilizor T1 treated acMeOH extracted tissues. Experiments 2 and 3, which use DMF as the extraction solvent have many more non-interfering ion peaks observed in non-Stabilizor T1 treated tissues than were observed in experiment 1. In experiment 2, 97 non-interfering extra ion peaks were observed in the non-Stabilizor T1 tissue and experiment 3 showed 59 non-interfering extra ion peaks in non-Stabilizor T1 tissue. A total of 171 extra ion peaks were observed in experiments 1, 2, and 4. Of those 171 ion peaks, only seven (4 %) had charges less than +3, meaning that the majority of extra ion peaks have [M+H]+ greater than m/z 3000. This suggests that the majority of putative post-mortem protein fragments (extra ion peaks in non-Stabilizor T1 treated tissues) have a molecular weight greater than 3000 Da. It is possible that some of these non-interfering ion peaks could be putative large neurohormones like CPRP, crustacean hyperglycemic hormone, or other neuropeptide prohormone proteolytic fragments. Mascot and accurate mass matching were used to determine if any of the large [M+H]+ ions were known neurohormones or neurohormone fragments. Only one of the [M+H]+ (m/z 3837) was identified as a CPRP peptide (Supplemental Figure 3). The results indicate that the majority of extra peaks observed between Stabilizor T1 treated tissues and non-Stabilizor T1 treated tissues can be classified as putative postmortem protein fragments.
Conclusions
The observations reported in this article suggest that post-mortem protein fragments do not affect crustacean neuropeptidomic studies to the same degree as mammalian studies using MALDI or ESI MS techniques. In MALDI experiments, Stabilizor T1 treated samples produce MS spectra containing peak profiles similar to those observed in samples not treated with heat stabilization methods. In ESI experiments, tissues not treated with the Stabilizor T1 produced more intense and more complex mass spectra over the course of the elution gradient. The added complexity in the non-Stabilizor T1 treated tissue did not dramatically affect the identification of endogenous crustacean neuropeptides through masking effects. The degree of added complexity was more prominent in DMF extracted tissues than in acMeOH extracted tissues. This suggests that acMeOH is a better protease inhibitor extraction solvent than DMF, possibly due to its low pH. Neuropeptides that were affected by coeluting chimeric species were still successfully de novo sequenced in four of five instances. Only one peptide (NFDEIDRSSFa) was completely masked by a chimeric species in non-treated DMF extracted tissues and not clearly observed. Although the majority of extra ion peaks observed in non-Stabilizor T1 treated samples did not mask identified neuropeptides in blue crab PO extracts in this study, neuropeptidomic studies performed in other species and neural organs may be more affected by the high abundant, highly charged ions. We suggest that a preliminary experiment utilizing the Stabilizor T1 in the sample processing workflow should be performed to test its necessity in fully characterizing the neuropeptidome of another neural organ or another crustacean species. The benefits of decreased spectral complexity need to be balanced with the slightly reduced neuropeptide signal to determine if the Stabilizor T1 or other heat deactivation step would be necessary and beneficial for the outcome of the experiment.
Supplementary Material
Acknowledgments
This work is supported in part by the National Institutes of Health grants (1R01DK071801, 1R56DK071801, and 1S10RR029531) and the National Science Foundation grant (CHE-0957784). L. Li acknowledges an H.I. Romnes Faculty Research Fellowship. R.M.S. acknowledges the NIH-supported Clinical Neuroengineering Training Program Predoctoral Fellowship (T32 EB011434).
Footnotes
Supporting Information. Additional information as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hökfelt T, Broberger C, Xu ZQ, Sergeyev V, Ubink R, Diez M. Neuropeptides--an overview. Neuropharmacology. 2000;39(8):1337–1356. doi: 10.1016/s0028-3908(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 2.Jensen J. Regulatory peptides and control of food intake in non-mammalian vertebrates. Comp Biochem Physiol, Part A: Mol Integr Physiol. 2001;128 (3):469–477. doi: 10.1016/s1095-6433(00)00329-9. [DOI] [PubMed] [Google Scholar]
- 3.Kieffer B, Gaveriaux-Ruff C. Exploring the opioid system by gene knockout. Prog Neurobiol. 2002;66 (5):285–306. doi: 10.1016/s0301-0082(02)00008-4. [DOI] [PubMed] [Google Scholar]
- 4.Okada Y, Tsuda Y, Bryant S, Lazarus L. Endomorphins and related opioid peptides. Vitam Horm. 2002;65:257–279. doi: 10.1016/s0083-6729(02)65067-8. [DOI] [PubMed] [Google Scholar]
- 5.Strand F. Neuropeptides: general characteristics and neuropharmaceutical potential in treating CNS disorders. Prog Drug Res. 2003;61:1–37. doi: 10.1007/978-3-0348-8049-7_1. [DOI] [PubMed] [Google Scholar]
- 6.Bora A, Annangudi SP, Millet LJ, Rubakhin SS, Forbes AJ, Kelleher NL, Gillette MU, Sweedler JV. Neuropeptidomics of the supraoptic rat nucleus. J Proteome Res. 2008;7 (11):4992–5003. doi: 10.1021/pr800394e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brockmann A, Annangudi SP, Richmond TA, Ament SA, Xie F, Southey BR, Rodriguez-Zas SR, Robinson GE, Sweedler JV. Quantitative peptidomics reveal brain peptide signatures of behavior. Proc Natl Acad Sci US A. 2009;106 (7):2383–2388. doi: 10.1073/pnas.0813021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen R, Hui L, Cape SS, Wang J, Li L. Comparative Neuropeptidomic Analysis of Food Intake via a Multi-faceted Mass Spectrometric Approach. ACS Chem Neurosci. 2010;1 (3):204–214. doi: 10.1021/cn900028s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fricker LD, Lim J, Pan H, Che FY. Peptidomics: identification and quantification of endogenous peptides in neuroendocrine tissues. Mass Spectrom Rev. 2006;25 (2):327–344. doi: 10.1002/mas.20079. [DOI] [PubMed] [Google Scholar]
- 10.Jiang X, Chen R, Wang J, Metzler A, Tlusty M, Li L. Mass spectral charting of neuropeptidomic expression in the stomatogastric ganglion at multiple developmental stages of the lobster Homarus americanus. ACS Chem Neurosci. 2012;6 (3):439–450. doi: 10.1021/cn200107v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nilsson A, Stroth N, Zhang X, Qi H, Fälth M, Sköld K, Hoyer D, Andrén PE, Svenningsson P. Neuropeptidomics of mouse hypothalamus after imipramine treatment reveal somatostatin as a potential mediator of antidepressant effects. Neuropharmacology. 2012;62 (1):347–357. doi: 10.1016/j.neuropharm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Svensson M, Sköld K, Svenningsson P, Andrén PE. Peptidomics-based discovery of novel neuropeptides. J Proteome Res. 2003;2(2):213–219. doi: 10.1021/pr020010u. [DOI] [PubMed] [Google Scholar]
- 13.Sithigorngul P, Jarecki JL, Stretton AOW. A specific antibody to neuropeptide AF1 (KNEFIRFamide) recognizes a small subset of neurons in Ascaris suum: differences from Caenorhabditis elegans. J Comp Neurol. 2011;519 (8):1546–1561. doi: 10.1002/cne.22584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Bao L, Ma GQ. Sorting of neuropeptides and neuropeptide receptors into secretory pathways. Prog Neurobiol. 2010;90 (2):276–283. doi: 10.1016/j.pneurobio.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 15.Hsu YWA, Messinger DI, Chung JS, Webster SG, de la Iglesia HO, Christie AE. Members of the crustacean hyperglycemic hormone (CHH) peptide family are differentially distributed both between and within the neuroendocrine organs of Cancer crabs: implications for differential release and pleiotropic function. J Exp Biol. 2006;209 (16):3241–3256. doi: 10.1242/jeb.02372. [DOI] [PubMed] [Google Scholar]
- 16.Li L, Sweedler JV. Peptides in the brain: mass spectrometry-based measurement approaches and challenges. Annu Rev Anal Chem. 2008;1:451–483. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- 17.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem. 1988;60 (20):2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yoshida T, Matsuo T. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1988;2 (8):151–153. [Google Scholar]
- 19.Fenn J, Mann M, Meng C, Wong S, Whitehouse C. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246 (4926):64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 20.Ma M, Sturm RM, Kutz-Naber KK, Fu Q, Li L. Immunoaffinity-based mass spectrometric characterization of the FMRFamide-related peptide family in the pericardial organ of Cancer borealis. Biochem Biophys Res Commun. 2009;390 (2):325–330. doi: 10.1016/j.bbrc.2009.09.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma M, Wang J, Chen R, Li L. Expanding the crustacean neropeptidome using a multifaceted mass spectrometric approach. J Proteome Res. 2009;8 (5):2426–2437. doi: 10.1021/pr801047v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei H, Nolkrantz K, Parkin MC, Chisolm CN, O’callaghan JP, Kennedy RT. Identification and quantification of neuropeptides in brain tissue by capillary liquid chromatography coupled off-line to MALDI-TOF and MALDI-TOF/TOF-MS. Anal Chem. 2006;78 (13):4342–4351. doi: 10.1021/ac052196x. [DOI] [PubMed] [Google Scholar]
- 23.Altelaar AFM, Mohammed S, Brans MAD, Adan RAH, Heck AJR. Improved identification of endogenous peptides from murine nervous tissue by multiplexed peptide extraction methods and multiplexed mass spectrometric analysis. J Proteome Res. 2009;8 (2):870–876. doi: 10.1021/pr800449n. [DOI] [PubMed] [Google Scholar]
- 24.Che FY, Lim J, Pan H, Biswas R, Fricker LD. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol Cell Proteomics. 2005;4 (9):1391–1405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Dowell JA, Heyden WV, Li L. Rat neuropeptidomics by LC-MS/MS and MALDI-FTMS: Enhanced dissection and extraction techniques coupled with 2D RP-RP HPLC. J Proteome Res. 2006;5 (12):3368–3375. doi: 10.1021/pr0603452. [DOI] [PubMed] [Google Scholar]
- 26.Parkin MC, Wei H, O'callaghan JP, Kennedy RT. Sample-dependent effects on the neuropeptidome detected in rat brain tissue preparations by capillary liquid chromatography with tandem mass spectrometry. Anal Chem. 2005;77 (19):6331–6338. doi: 10.1021/ac050712d. [DOI] [PubMed] [Google Scholar]
- 27.Sköld K, Svensson M, Norrman M, Sjögren B, Svenningsson P, Andrén PE. The significance of biochemical and molecular sample integrity in brain proteomics and peptidomics: stathmin 2-20 and peptides as sample quality indicators. Proteomics. 2007;7 (24):4445–4456. doi: 10.1002/pmic.200700142. [DOI] [PubMed] [Google Scholar]
- 28.Colgrave ML, Xi L, Lehnert SA, Flatscher-Bader T, Wadensten H, Nilsson A, Andrén PE, Wijffels G. Neuropeptide profiling of the bovine hypothalamus: thermal stabilization is an effective tool in inhibiting post-mortem degradation. Proteomics. 2011;11 (7):1264–1276. doi: 10.1002/pmic.201000423. [DOI] [PubMed] [Google Scholar]
- 29.Nylander I, Stenfors C, Tan-No K, Mathé AA, Terenius L. A comparison between microwave irradiation and decapitation: basal levels of dynorphin and enkephalin and the effect of chronic morphine treatment on dynorphin peptides. Neuropeptides. 1997;31(4):357–365. doi: 10.1016/s0143-4179(97)90072-x. [DOI] [PubMed] [Google Scholar]
- 30.Theodorsson E, Stenfors C, Mathé AA. Microwave irradiation increases recovery of neuropeptides from brain tissues. Peptides. 1990;11(6):1191–1197. doi: 10.1016/0196-9781(90)90151-t. [DOI] [PubMed] [Google Scholar]
- 31.Svensson M, Boren M, Sköld K, Fälth M, Sjögren B, Andersson M, Svenningsson P, Andrén PE. Heat stabilization of the tissue proteome: a new technology for improved proteomics. J Proteome Res. 2009;8(2):974–981. doi: 10.1021/pr8006446. [DOI] [PubMed] [Google Scholar]
- 32.Chen R, Ma M, Hui L, Zhang J, Li L. Measurement of neuropeptides in crustacean hemolymph via MALDI mass spectrometry. J Am Soc Mass Spectrom. 2009;20 (4):708–718. doi: 10.1016/j.jasms.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kutz KK, Schmidt JJ, Li L. In Situ Tissue Analysis of Neuropeptides by MALDI FTMS In-Cell Accumulation. Anal Chem. 2004;76 (19):5630–5640. doi: 10.1021/ac049255b. [DOI] [PubMed] [Google Scholar]
- 34.Ma M, Gard AL, Xiang F, Wang J, Davoodian N, Lenz PH, Malecha SR, Christie AE, Li L. Combining in silico transcriptome mining and biological mass spectrometry for neuropeptide discovery in the Pacific white shrimp Litopenaeus vannamei. Peptides. 2010;31 (1):27–43. doi: 10.1016/j.peptides.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang F, Ye H, Chen R, Fu Q, Li L. N, N-Dimethyl Leucines as Novel Isobaric Tandem Mass Tags for Quantitative Proteomics and Peptidomics. Anal Chem. 2010;82 (7):2817–2825. doi: 10.1021/ac902778d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hui L, Xiang F, Zhang Y, Li L. Mass spectrometric elucidation of the neuropeptidome of a crustacean neuroendocrine organ. Peptides. 2012;36 (2):230–239. doi: 10.1016/j.peptides.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeKeyser SS, Kutz-Naber KK, Schmidt JJ, Barrett-Wilt GA, Li L. Imaging mass spectrometry of neuropeptides in decapod crustacean neuronal tissues. J Proteome Res. 2007;6 (5):1782–1791. doi: 10.1021/pr060603v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pulver SR, Marder E. Neuromodulatory complement of the pericardial organs in the embryonic lobster, Homarus americanus. J Comp Neurol. 2002;451 (1):79–90. doi: 10.1002/cne.10331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.