Abstract
Our previous studies have revealed that the human FOXF1 gene, encoding a transcription factor member of the forkhead box (FOX) family, functions as a tumor suppressor and its expression is frequently silenced in breast cancer via DNA hypermethylation. Moreover, we recently reported that FOXF1 expression is preferentially silenced in colorectal cancer cell lines with inactive p53 and knockdown of FOXF1 caused genomic instability in FOXF1-expressing colorectal cancer cells with a defect in the p53-p21WAF1 checkpoint, suggesting that FOXF1 plays a key role in colorectal tumorigenesis. Given that the in vivo role of FOXF1 in colorectal cancer remains unknown, the study here was aimed at delineating the clinical relevance of FOXF1 in colorectal adenocarcinomas. To characterize FOXF1 protein expression in colorectal cancer, designed tissue microarrays, comprising 50 cases of primary colorectal adenocarcinoma paired with matched adjacent normal tissue, were utilized in the immunohistochemistry (IHC) study. The IHC results showed that for adjacent normal colorectal tissue, the FOXF1 protein was only detected in stroma, not in epithelium, with either cytoplasmic staining (70% of total cases) or a mix of cytoplasmic and nuclear staining (6%). In contrast, for colorectal adenocarcinomas, FOXF1 staining was predominately identified in the cytoplasm of tumor epithelial cells (40% of total cases) and tumor-associated stromal cells of some cases (10%) also exhibited FOXF1 positivity in their cytoplasm. Cytoplasmic FOXF1 protein expression in tumor epithelial cells positively correlated with the histologic grade, depth of invasion, stage and lymphatic metastasis of colorectal adenocarcinomas (p < 0.05). Moreover, in silico meta-analysis of Oncomine’s cancer microarray database indicates that FOXF1 mRNA is overexpressed in a significant subset of colorectal adenocarcinoma tumors compared with normal colorectal tissue and other types of cancers. Our findings for the first time have revealed that the FOXF1 protein is overexpressed as well as mislocalized in cancerous epithelial cells and underexpressed/lost in tumor-associated stromal fibroblasts of colorectal adenocarcinomas, and suggest that FOXF1 is a potential prognostic marker due to its association with the malignancy and metastasis of colorectal cancer.
Keywords: colorectal cancer, FOXF1, tumor suppressor gene, tissue microarrays, immunohistochemistry, cytoplasmic mislocalization
Introduction
As one of the major causes of cancer-related mortality in the world (Jemal et al., 2010), colorectal cancer is surgically curable at early stages, but this advanced disease at the metastatic stage is associated with high mortality rates. In spite of advances in the chemotherapeutic regimens and combined radiotherapy for the treatment of advanced colorectal cancer (Aggarwal and Chu, 2005), surgery is still the only curative form of treatment. Furthermore, the development of the current therapeutic concepts, especially for late-stage colorectal tumors, is immature and far from optimal. Therefore, even though mortality rates have improved, the discovery of new biomarkers for predicting malignancy as well as prognosis and the identification of new therapeutic targets are still urgently needed for colorectal cancer.
Forkhead box (FOX) proteins function as transcription factors with the evolutionarily conserved DNA-binding domain termed forkhead box or the winged helix domain (Hannenhalli and Kaestner, 2009; Katoh and Katoh, 2004; Lai et al., 1993). Through the transcriptional control of gene expression, many FOX protein members have been documented to play imperative roles in embryonic development as well as organogenesis and also in the regulation of a variety of physiological processes, such as cell cycle progression, cell survival, cellular metabolism, life span, and immune responses (Carlsson and Mahlapuu, 2002; Hannenhalli and Kaestner, 2009; van der Horst and Burgering, 2007). Consequently, dysregulating the functions, subcellular localization and expression of FOX transcription factors leads to the development and progression of diseases, in particular cancer (Hannenhalli and Kaestner, 2009; Myatt and Lam, 2007).
The human FOXF1 gene, previously named as Forkhead RElated ACtivator (FREAC)-1 (Hellqvist et al., 1996), encodes a homologue of the mouse forkhead box-F1 (Foxf1) transcription factor. Gene knockout studies have shown that the function of mouse Foxf1 is indispensable for organ morphogenesis, including the lung, liver, gallbladder, esophagus, and trachea (Kalinichenko et al., 2003; Kalinichenko et al., 2002; Mahlapuu et al., 2001a). Despite the largely unknown role of FOXF1 in cancer, several lines of evidence have linked human FOXF1 function to tumorigenesis (Lo et al., 2010; Nilsson et al., 2010; Saito et al., 2010). In lung cancer, FOXF1 regulates cancer-associated fibroblasts to stimulate cancer cell migration and xenograft tumor growth (Saito et al., 2010). Besides its oncogenic role in tumor-associated stromal fibroblasts, another evidence has shown that ectopic overexpression of FoxF1 in a mouse mammary epithelial cell line induced epithelial-to-mesenchymal transition (EMT), which in turn increased invasiveness of epithelial cells in vitro, and enhanced growth of breast carcinoma xenograft tumors (Nilsson et al., 2010). In contrast to these oncogenic roles of FOXF1 mentioned above, we recently discovered that human FOXF1 plays a novel tumor-suppressor role in cell-cycle regulation of mammary epithelial cells, which is frequently silenced in breast cancer through epigenetic mechanisms. Analogous to our findings, it has been found that FOXF1 expression is underexpressed in prostate cancer (Watson et al., 2004). Our and other studies, taken together, suggest that FOXF1 may play a dual role in tumorigenesis as an oncogene or a tumor suppressor gene depending on tissue cell types and disease stages. In addition to the relevance of FOXF1 in lung and breast cancers, we recently have revealed that FOXF1 expression is predominantly silenced in colorectal cancer cell lines with the inactive p53 gene (Lo et al., 2012). By using wild-type, p53-null and p21WAF1-null HCT116 colorectal cancer cell line models, we have demonstrated that knockdown of FOXF1 by siRNA led to DNA over-replication (a mechanism causing genomic instability) in colorectal cancer cells with a defect in the p53-p21WAF1 checkpoint (Lo et al., 2012). Hence, our findings imply that FOXF1 might play a key role in colorectal carcinogenesis. To unravel the clinical relevance of FOXF1 in colorectal cancer, this study was aimed at identifying the distribution of expressed FOXF1 proteins in the tumor tissue of colorectal cancer patients and delineating its correlation with clinicopathological data.
Materials and Methods
Transfection
The colorectal carcinoma cell lines, DLD-1 and HCT116, were obtained from ATCC (American Type Culture Collection, Manassas, VA, USA) and cultured in suitable mediums according to the ATCC online instructions. For transfection, 6×105 DLD-1 cells (or 2×105 HCT116 cells) cultured in 6-well cell culture plates were transfected with 4 μg of hemagglutinin (HA)-tagged FOXF1 expression plasmid DNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Twenty four hours after transfection, transfected cells were re-plated back into the wells of 6-well cell culture plates containing coverslips. After 24-hour incubation, transfected cells on coverslips were fixed by 2% formaldehyde for 30 min at 4°C and then permeabilized in ice-cold 100% methanol for 10 min at −20°C. After fixation, the fixed cells were subjected to immunostaining assays.
Immunofluorescence (IF) and immunocytochemistry (ICC) assays
IF assays were performed as previously described (Lo et al., 2010). In brief, methanol-fixed cells were rehydrated in 1×PBS for 10 min and then incubated in the blocking solution (1×PBS containing 10% goat serum) for 30 min. After blocking, cells were subjected to primary (anti-FOXF1 antibody, ARP32296_T100, Aviva System Biology, San Diego, CA, USA) and then secondary (Alexa series, invitrogen) antibody reactions according to manufacturer’s instructions. After immunostaining, nuclear DNA of cells was stained with 10μg/ml 4′6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich, St. Louis, MO) and coverslips were mounted on slides in mounting medium (Vecta Shield, Vector Laboratories, Burlingame, CA, USA). Immunostained cells on slides were examined under a fluorescence microscope (Nikon Eclipse E800). ICC assays were performed in the same way as previously described immunohistochemistry (IHC) assays (Lo et al., 2010) except omitting deparaffinization with xylene and a series of procedures specialized for tissue sections such as the dehydration-rehydration cycle, Epitope Retrieval and microwave heating. Briefly, the endogenous peroxidase of cells was blocked by 0.3% H2O2 in methanol. The non-specific stain was blocked by 10% of normal goat serum for 20 min. Primary antibodies were then applied at the concentration of 1 μg/ml for the anti-FOXF1 antibody (rabbit polyclonal, ARP32296_T100, Aviva System Biology) and 0.5 μg/ml for the anti-HA tag rabbit monoclonal antibody (Cell signaling, Danvers, MA, USA). 1 μg/ml of rabbit IgG was included in assays as a negative control. The avidin-biotin-peroxidase complex (ABC) method (Vector Laboratories) was used in the following secondary antibody detection and color developing according to manufacturer’s instructions. Counterstain was done by staining fixed cells on coverslips with DAKO hematoxylin (Dako, Carpinteria, CA, USA). After counterstaining, stained cells were dehydrated in ethanol, and then mounted on slides with the mounting solution (SecureMount™, Thermo Fisher Scientific Company, Waltham, MA, USA) after the complete dryness of stained cells.
Human colorectal cancer tissue microarrays
A tissue microarray (TMA) containing 50 cases of primary colorectal adenocarcinoma paired with adjacent noncancerous tissue (≤ 1.5 cm away from the tumor) was used in the study. The tissue samples from colorectal cancer patients (43 to 80 years old with a mean age of 65 ± 9 years) who underwent surgery between January 2010 and June 2010 were provided by the Chonnam National University Hwasun Hospital National Biobank of Korea, a member of the National Biobank of Korea, which is supported by the Ministry of Health, Welfare and Family Affairs. Characterization of clinicpathological features of patients was based on the Cancer staging system from the American Joint Committee. All tissue samples were obtained with informed consent from patients under institutional review board-approved protocols. TMA slides were constructed with a custom-built precision instrument (Beecher Instruments Inc., Sun Prairie, WI, USA). Three core tissue biopsies exhibiting carcinoma with a diameter of 2 mm were punched from individual donor paraffin-embedded tissue blocks and precisely arrayed into a new recipient block. Normal colorectal tissue from two representative areas of donor tissue blocks was taken and served as a baseline control. Four-micron sections of TMA blocks were cut and placed on polylysine-coated slides. These slides were used for IHC analysis.
Immunohistochemistry (IHC) analysis of colorectal cancer tissue microarrays
Automated IHC staining of a TMA slide with the anti-FOXF1 antibody (1:50 dilution) or with the anti-p53 antibody (clone DO-7, 1:50 dilution, Dako) was performed by using the Bond-max system (Leica Microsystems, Buffalo Grove, IL, USA) according to manufacturer’s instructions. The anti-p53 antibody is able to detect both the wild-type and mutant-type p53 proteins. The FOXF1 or p53 staining of tissue was interpreted as positive when >10% of tumor or stromal cells exhibited positive staining (Dix et al., 1994). The correlation between FOXF1 protein expression and clinicopathological characteristics of colorectal cancer patients was analyzed by the chi-square test (χ2 test) using the SPSS statistical software (version 11.5 for windows, SPSS Inc., Chicago, IL, USA).
In silico analysis of FOXF1 gene expression in a variety of cancers
We used Oncomine’s Cancer Microarray Database (Rhodes et al., 2004) (http://www.oncomine.org) to perform the in silico analysis of the FOXF1 gene expression in normal colon, colorectal adenoma, colorectal adenocarcinoma and other cancers. The Mann-Whitney test was used to evaluate the statistical differences of FOXF1 expression between normal colon tissue and different groups of colorectal adenocarcinomas.
Results
Validation of the anti-FOXF1 antibody
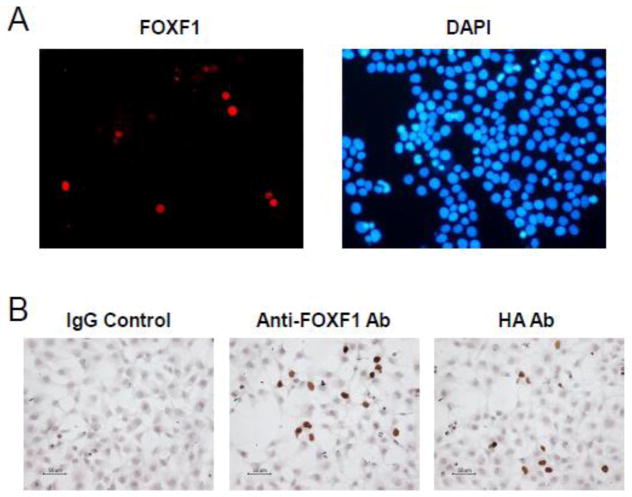
The anti-FOXF1 antibody (Cat. No. ARP32296_T100) used in this study was obtained from the Aviva System Biology and had been characterized with breast cancer cell lines and tumors (Lo et al., 2010). To further validate whether this antibody can specifically detect the FOXF1 protein in colorectal carcinoma cells, we performed IF assays to analyze the protein expression of HA-tagged FOXF1 in colorectal cancer cell lines DLD-1 and HCT116. As shown in Fig. 1A, HA-tagged FOXF1-transfected DLD-1 cells exhibited nuclear FOXF1 staining in IF assays. To further confirm whether this anti-FOXF1 antibody works well in the immunostaining system for IHC assays, HCT116 cells were transfected with the HA-tagged FOXF1 expression plasmid DNA and then subjected to ICC assays based on the traditional IHC protocol with some modifications as described in “Materials and Methods”. The anti-HA antibody and rabbit IgG immunoglobin were included in ICC assays as positive and negative controls. Consistent with the anti-HA staining results, the anti-FOXF1 antibody specifically detected expressed FOXF1 proteins in the nuclei of transfected HCT116 cells (Fig. 1B). These testing results, taken together, have demonstrated that this anti-FOXF1 antibody can unambiguously detect functional FOXF1 proteins in the cell nucleus.
Fig. 1.
The anti-FOXF1 antibody specifically stains nuclear FOXF1 proteins in FOXF1-transfected colorectal cancer cells. (A) Immunofluorescence analysis of ectopic expression of HA-tagged FOXF1 in DLD-1 colorectal cancer cells using the anti-FOXF1 antibody. The left panel is HA-FOXF1-transfected DLD-1 cells stained with the anti-FOXF1 antibody and the goat anti-rabbit IgG Alexa 568 antibody. The right panel is the co-staining of cells with the DNA staining dye DAPI. (B) Immunocytochemistry analysis of HA-tagged FOXF1 expression in HCT116 colorectal cancer cells using either the anti-FOXF1 or anti-HA antibody. HA-FOXF1-transfected HCT116 cells stained with rabbit IgG and the anti-HA antibody served as negative and positive controls, respectively, in comparison with those stained with the anti-FOXF1 antibody. Immunocytochemistry analysis was performed as described in “Materials and Methods”.
Characterization of FOXF1 staining patterns in colorectal cancer tissue microarrays
Given that in vivo FOXF1 expression in colorectal carcinomas has never been characterized and reported before, we performed IHC analysis of colorectal tumor TMA including 50 cases of colorectal adenocarcinomas as well as their corresponding, adjacent normal colorectal tissue to examine FOXF1 protein expression patterns in colorectal tumors and adjacent normal colonic mucosa. The IHC results of FOXF1 expression patterns in tissue cell types (epithelial and stromal cells) and their subcellular localization (nuclear and cytoplasmic) were summarized in Table 1 and the representative staining images are shown in Figs. 2, 3 and 4. In adjacent normal colonic mucosa specimens, FOXF1 staining was only observed in stromal cells of 76% of examined cases and no staining in intestinal epithelial cells of any examined cases (Table 1). The FOXF1 staining in stromal cells of normal colorectal tissue was predominantly localized in the cytoplasm of examined cases (70%) and few cases (6%) manifested a mosaic of nuclear and cytoplasmic FOXF1 staining in tissue (Table 1 and Fig. 2). For FOXF1-positive colorectal adenocarcinoma specimens (40% of all cases), FOXF1 staining was unexpectedly observed exclusively in the cytoplasm of tumor epithelial cells (Table 1, Figs. 3 and 4), whereas tumor-associated stromal cells of 90% of examined cases were FOXF1-negative and remaining 10% of positive cases showed weak, cytoplasmic FOXF1 staining (Table 1 and Fig. 4). These observations indicate that the FOXF1 protein is overexpressed and mislocalized in the cytoplasm of tumor epithelial cells in a significant subset of examined colorectal adenocarcinoma cases and the majority of tumor tissue samples underexpressed/lost FOXF1 in tumor-associated stromal cells.
Table 1.
Protein expression of FOXF1 in adjacent normal colonic mucosa and colorectal adenocarcinomas.
| Tissue type (n) | Cell type | Subcellular localization | FOXF1 (+) (%) |
|---|---|---|---|
| Normal colon (50) | Epithelial | Nuclear | 0 |
| Cytoplasmic | 0 | ||
| Mixed | 0 | ||
| Stromal | Nuclear | 0 | |
| Cytoplasmic | 35 (70) | ||
| Mixed | 3 (6) | ||
| Adenocarcinomas (50) | Epithelial | Nuclear | 0 |
| Cytoplasmic | 20 (40) | ||
| Mixed | 0 | ||
| Stromal | Nuclear | 0 | |
| Cytoplasmic | 5 (10) | ||
| Mixed | 0 |
Fig. 2.
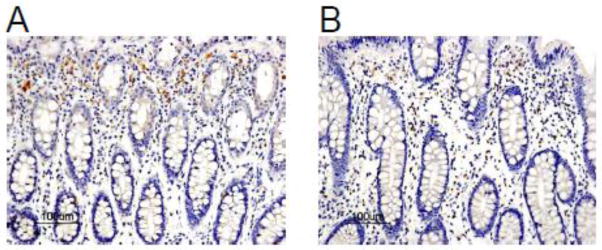
The FOXF1 expression pattern in adjacent normal human colorectal mucosa. (A) A representative case of adjacent normal human colorectal mucosa specimens showed FOXF1 staining predominantly in the cytoplasm of stromal cells. (B) A representative case of adjacent normal human colorectal mucosa specimens showed a heterogeneous pattern of nuclear and cytoplasmic FOXF1 staining in stromal cells.
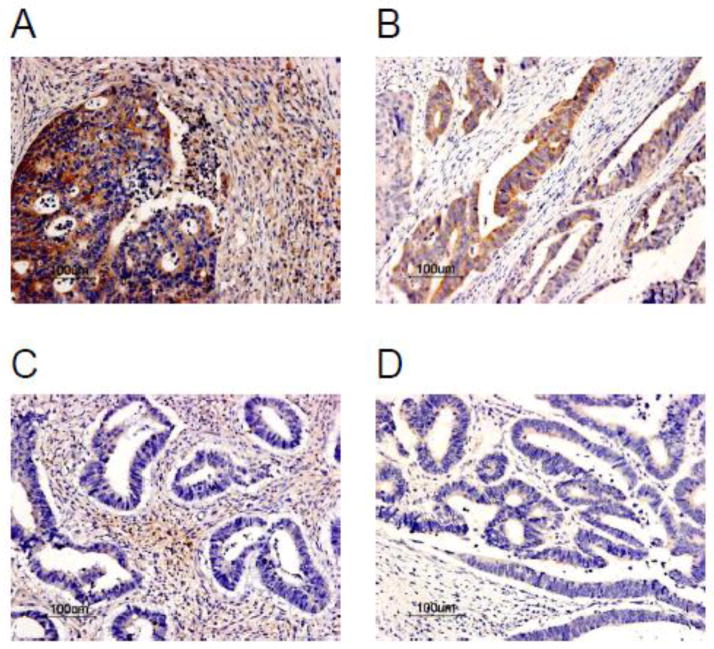
Fig. 3.
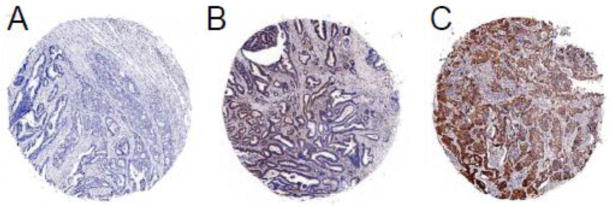
FOXF1 protein expression patterns in colorectal adenocarcinoma tumors. IHC analysis of FOXF1 protein expression in colorectal tumor TMA exhibited negative staining (A) or a homogeneous staining pattern at different intensities in the cytoplasm of tumor epithelial cells (B – weak, C – strong).
Fig. 4.
Detailed FOXF1 protein expression patterns in human colorectal adenocarcinoma tumors. Four types of FOXF1 staining results were found in IHC analysis of colorectal tumors, including FOXF1 positivity in the cytoplasm of both tumor epithelial and stromal cells (A), tumor epithelial cells only (B), tumor stromal cells only (C) and FOXF1 negativity in both tumor epithelial and stromal cells (D).
The correlation between FOXF1 staining and clinicopathological data of colorectal cancers
The results from IHC analysis of colorectal adenocarcinoma TMA were analyzed to reveal the correlation between FOXF1 protein staining patterns and clinicopathological characteristics of colorectal cancer patients. Due to the limited sample size, the correlation between the FOXF1 IHC positivity in adjacent normal colorectal tissue samples and clinicopathological characteristics of colorectal cancer patients is not statistically significant (data not shown). In contrast, the FOXF1 IHC positivity in tumor epithelial cells of colorectal adenocarcinoma tissue samples was statistically associated with higher histologic grades, more invasive status, advanced stages and lymphatic metastasis (Table 2). These results indicate that overexpression of the mislocalized FOXF1 protein in colorectal tumor epithelial cells positively correlates with poor prognostic factors. Given our previous findings indicating the positive correlation between FOXF1 expression and the functional status of p53 in colorectal cancer cell lines (Lo et al., 2012), we examined the relationship between FOXF1 and p53 IHC positivity. As shown in Table 2, this tumor sample cohort did not exhibit the statistically significant correlation between FOXF1 and p53 protein expression.
Table 2.
Correlation between FOXF1 protein expression in colorectal adenocarcinomas and clinicopathological features.
| Clinicopathological Characteristics | n | FOXF1(+) (%) | χ2/P value |
|---|---|---|---|
| Age (years) | |||
| < 65 | 21 | 7 (33.3) | 0.670/0.413 |
| ≥ 65 | 29 | 13 (44.8) | |
| Gender | |||
| Male | 27 | 13 (48.1) | 1.624/0.203 |
| Female | 23 | 7 (30.4) | |
| Grade* | |||
| G1 | 29 | 8 (27.6) | 4.433/0.0352 |
| G2–G3 | 21 | 12 (57.1) | |
| Depth of Invasion | |||
| T1–T2 | 6 | 0 (0.0) | 4.545/0.033 |
| T3–T4 | 44 | 20 (45.5) | |
| Stage | |||
| I | 6 | 0 (0.0) | 7.579/0.0226 |
| II | 30 | 11 (36.7) | |
| III | 14 | 9 (64.3) | |
| Lymphatic Metastasis | |||
| No | 36 | 11 (30.6) | 4.778/0.0288 |
| Yes | 14 | 9 (64.3) | |
| p53 | |||
| Positive | 27 | 13 (48.1) | 1.624/0.203 |
| Negative | 23 | 7 (30.4) | |
G1: well differentiated, G2: moderately differentiated, G3: poorly differentiated.
FOXF1 mRNA expression in normal colon, benign and malignant colorectal tumors
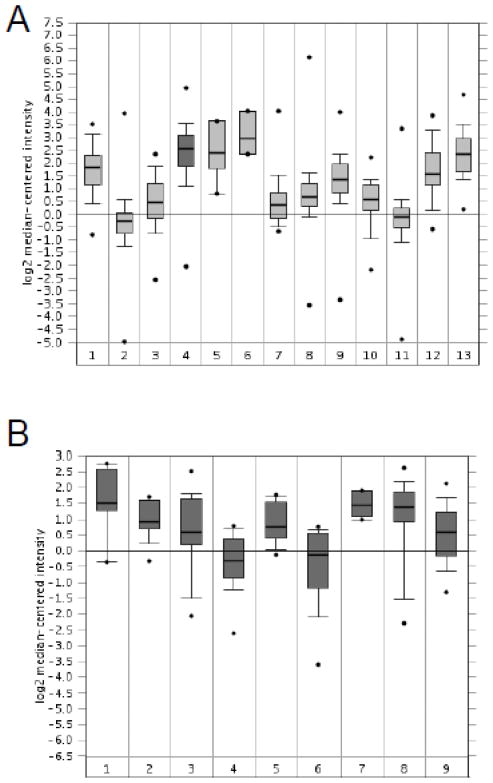
Due to the limited sensitivity of the FOXF1 antibody, the expression range of the FOXF1 gene in colorectal tumors could not be completely elucidated by IHC analysis. To reveal the overall expression status of FOXF1 in colorectal tumors, in silico meta-analysis of Oncomine’s microarray datasets was performed. Skrzypczak’s and Kaiser’s microarray datasets (Kaiser et al., 2007; Skrzypczak et al., 2010) were analyzed to examine FOXF1 mRNA expression in normal colon, colorectal adenoma (benign) and colorectal adenocarcinoma (malignant). As shown in Fig. 5A, meta-analysis of Skrzypczak’s dataset showed that FOXF1 was overexpressed in colorectal adenocarcinomas compared with normal colorectal tissue and adenoma specimens. In Kaiser’s microarray dataset, meta-analysis showed that FOXF1 was expressed in a broad range from underexpression (the low dataset) to overexpression (the high dataset) in colorectal adenocarcinoma specimens compared with normal colon tissue samples (Fig. 5B and 5C). In comparison to Kaiser’s dataset, Skrzypczak’s dataset only displayed FOXF1 overexpression in colorectal adenocarcinoma (Fig. 5). This might be attributable to the small sample size of colorectal adenocarcinomas in Skrzypczak’s microarray dataset. Overall these in silico expression analysis results indicate that FOXF1 is either underexpressed or overexpressed in a significant subset of colorectal adenocarcinoma tumors.
Fig. 5.
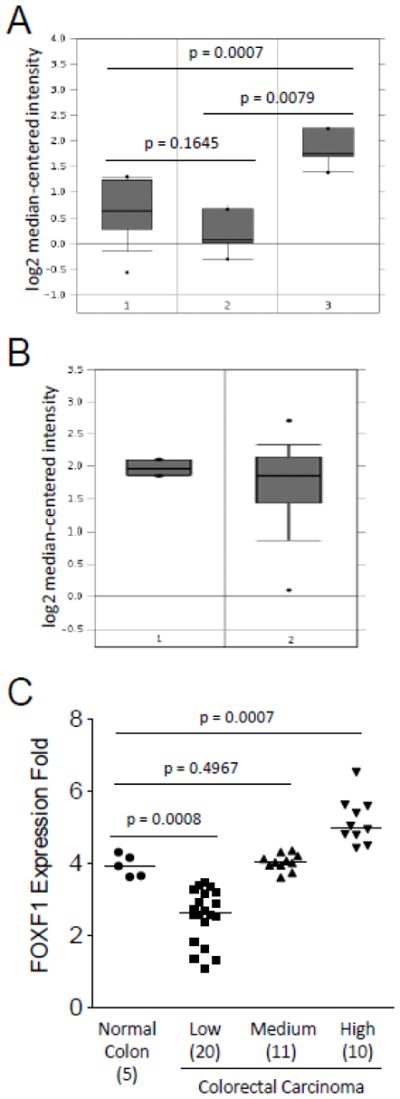
In silico analysis of FOXF1 mRNA expression in normal colon, benign and malignant colorectal tumors. The in silico expression data of the FOXF1 gene in normal colon, benign and malignant colorectal tumors were plotted based on Oncomine’s meta-analysis of previously published Skrzypczak’s (A) and Kaiser’s (B) microarray datasets. Y-axis units are normalized expression values on a log2 scale and the expression level of the FOXF1 gene was normalized across all microarray studies. The 25th and 75th percentiles are indicated by a vertical box and the 10th and 90th percentiles are depicted by error bars. The median of a dataset is shown as a horizontal line within a box. Outliers (indicated by dots) are also shown. The statistical analyses of differences between normal colon tissue, colorectal adenomas and carcinomas are indicated in the plot. The tissue samples in (A) are the following: 1-normal colon (n=10); 2-colorectal adenoma (n=5); 3-colorectal carcinoma (n=5). The tissue samples in (B) are the following: 1-normal colon (n=5); 2-colorectal adenocarcinoma (n=41). (C) FOXF1 expression data from Kaiser’s microarray dataset were plotted as a dot plot based on the expression classification of colorectal tumors (classified as low, medium and high). The statistical analyses of differences between normal colon tissue and different groups of colorectal tumors are indicated in the plot. The number of cases in each data group is indicated in parentheses. The median of a dataset is depicted by a horizontal line.
FOXF1 expression in colorectal cancer compared with other cancer types
Our previous studies have provided comprehensive results to show that FOXF1 expression is frequently silenced in breast cancer via epigenetic mechanisms. However, in this study we found that the FOXF1 protein was overexpressed and functionally inactivated by cytoplasmic sequestration in a significant portion of analyzed colorectal cancer cases. Our studies of breast and colorectal cancers raise a possibility that FOXF1 can be functionally inactivated by either epigenetic silencing or cytoplasmic mislocalization; therefore FOXF1 expression levels in different types of cancers may exhibit either underexpression or overexpression. To demonstrate this possibility, in silico analysis of FOXF1 expression in a variety of cancers was performed using Oncomine’s Cancer Microarray Database. Indeed, meta-analysis of two different multi-cancer microarray dataset sources (Su et al., 2001; Bittner’s microarray dataset was not yet published) consistently showed that overall FOXF1 was underexpressed in breast and ovarian cancers and overexpressed in bladder, colorectal, pancreatic and prostate cancers (Fig. 6).
Fig. 6.
In silico analysis of FOXF1 mRNA expression in a variety of cancers. Meta-analysis of FOXF1 gene expression using Bittner’s and Su’s multi-cancer expression microarray datasets from Oncomine’s microarray database are shown in (A) and (B), respectively. The FOXF1 expression level was normalized across all cancer microarray datasets. Y-axis units, percentiles, medians and outliers are indicated as described in Fig. 5. Cancer types in (A) are the following: 1-bladder cancer (n=32); 2-breast cancer (n=328); 3-cervical cancer (n=35); 4-colorectal cancer (n=330); 5-esophageal cancer (n=7); 6-gastric cancer (n=7); 7-head and neck cancer (n=41); 8-kidney cancer (n=254); 9-lung cancer (n=107); 10-lymphoma (n=19); 11-ovarian cancer (n=166); 12-pancreatic cancer (n=19); 13-prostate cancer (n=59). Cancer types in (B) are the following: 1-bladder cancer (n=8); 2-clear cell renal cell carcinoma (n=11); 3-colorectal adenocarcinoma (n=23); 4-ductal breast carcinoma (n=26); 5-lung adenocarcinoma (n=14); 6-ovarian serous surface papillary carcinoma (n=27); 7-pancreatic adenocarcinoma (n=6); 8-prostate adenocarcinoma (n=26); 9-squamous cell lung carcinoma (n=14).
Discussion
It is known that mammalian Foxf1 is predominately expressed in mesenchymal cell types and its functions are crucial for generation of mesoderm-derived tissues (Mahlapuu et al., 2001b). Moreover, animal genetic knockout studies have shown that Foxf factors play key roles in controlling gut development by enhancing the production of extracellular matrices from intestinal fibroblasts and negatively regulating mesenchymal Wnt expression that in turn weakens Wnt-β-catenin signaling in intestinal epithelium (Ormestad et al., 2006). These Foxf-regulated events are indispensable for maintaining the normal functionality of intestinal fibroblasts and the polarization as well as proliferation rate of intestinal epithelial cells, which are essential for the homeostasis and integrity of gut tissue (Ormestad et al., 2006). In the light of these critical roles of Foxf proteins in gut development, alterations in their expression and subcellular localization might be engaged in promoting intestinal tumorigenesis. In our studies, we for the first time found that tumor-associated stromal fibroblasts of most examined colorectal adenocarcinoma cases lost or underexpressed FOXF1 compared with normal stromal fibroblasts of their corresponding adjacent normal colorectal tissue. Therefore, these observations are in line with our hypothesis that loss of (or a reduction in) FOXF1 expression in stromal fibroblasts may participate in colorectal oncogenesis. Although the mechanism leading to silencing of FOXF1 expression in stromal cells is currently unknown and needs to be further investigated, aberrations in stroma-epithelium interactions may be a potential mechanism accounting for this phenomenon as it has been known that epithelium can regulate expression of Foxf factors in stromal fibroblasts via its secreted hedgehog ligands (Mahlapuu et al., 2001a; Ormestad et al., 2006). Identification of alterations in the interactions between stromal and epithelial cells will be a promising direction to address underlying mechanisms giving rise to silencing of FOXF1 expression in tumor-associated stromal cells.
Intriguingly, the FOXF1 IHC staining in the majority of adjacent normal colorectal tissue cases (70%) is cytoplasmic only and few tissue cases (6%) showed a mosaic of nuclear and cytoplasmic FOXF1 staining. Given that this observation is not consistent with the fact that functional FOXF1 is localized in the nucleus, we reexamined the FOXF1 IHC staining in 5 normal colorectal tissue specimens from patients without colorectal cancer. The results manifested a mix of nuclear and cytoplasmic FOXF1 staining patterns in all of these normal colonic mucosa specimens (data not shown). These findings suggest that the subcellular localization of FOXF1 in normal stromal fibroblasts of colorectal mucosa is dynamically regulated, which is more likely through paracrine effects (e.g., from epithelium). Based on these studies, we posited that the dynamic regulation of nucleo-cytoplasmic shuttling of the FOXF1 protein is disturbed in stroma of normal colorectal tissue adjacent to colorectal tumor tissue. Whether this abnormal phenomenon is caused by the paracrine effects from colorectal tumor cells will be an interesting issue and needs further studies to address it.
Unexpectedly, our novel findings from the IHC study of FOXF1 protein expression showed cytoplasmic mislocalization of overexpressed FOXF1 proteins in a significant subset of colorectal adenocaricinomas (40%), indicating that besides epigenetic silencing (Lo et al., 2010), aberrant mislocalization of FOXF1 to cytoplasm is another pathological mechanism to inactivate nuclear FOXF1 functions. Consistently, in silico meta-analysis of expression microarray datasets of colorectal tumors from Oncomine’s microarray database also indicates that FOXF1 mRNA expression was either underexpressed or overexpressed in a significant subset of colorectal adenocarcinomas compared with normal counterparts. According to IHC analysis of the FOXF1 protein expression pattern in colorectal tumors, FOXF1 mRNA must be overexpressed in colorectal tumor epithelial cells, not in tumor-associated stromal cells. In contrast, FOXF1-expressing breast carcinomas exhibit both nuclear and cytoplasmic FOXF1 staining in tumor epithelial cells, similar to IHC staining patterns of normal mammary ducts (Lo et al., 2010). Furthermore, ductal breast carcinomas only manifest underexpressed FOXF1 profiles compared with normal breast tissue (Lo et al., 2010). Meta-analysis of multi-cancer expression microarray datasets also has revealed that overall colorectal cancers expressed much higher levels of FOXF1 mRNA than breast cancers. Therefore, carcinomas derived from different tissue origins may have their favorable mechanisms to inactivate nuclear functions of FOXF1, either gene silencing via epigenetic mechanisms or protein mislocalization via unknown mechanisms. In breast cancer, the epigenetic mechanism is preferentially engaged in silencing of FOXF1 expression in breast cancer epithelial cells, whereas the cytoplasmic mislocalization mechanism is found to actively participate in inactivating nuclear FOXF1 functions in colorectal tumor epithelial cells. Further studies are required to reveal the mechanisms leading to overexpression as well as cytoplasmic mislocalization of FOXF1 in colorectal cancer epithelial cells and whether cytoplasmic FOXF1 plays a functional role in colorectal carcinogenesis.
Our clinical association studies indicate that cytoplasmic FOXF1 protein expression in tumor epithelial cells positively correlated with poor prognostic factor of colorectal adenocarcinomas such as the higher histologic grades, more invasive status, advanced stages and lymphatic metastasis. These findings suggest that cytoplasmic FOXF1 is a potential indicator to predict the prognosis of colorectal adenocarcinomas. Given that there are no available survival data for colorectal tumor tissue microarrays established in 2010, we are unable to evaluate whether FOXF1 can serve as a prognostic biomarker to predict the outcomes of colorectal cancer patients. Therefore, a study of a large number of colorectal cancer cases with the long-term survival information is mandatory to evaluate the prognostic value of FOXF1 protein expression. In our studies, we did not find any significant correlation between FOXF1 and p53 protein expression. Our previously published studies indicate that FOXF1 expression is preferentially silenced in colorectal cancer cell lines with inactive p53 (Lo et al., 2012). It is well accepted that nuclear p53 IHC positivity in tumor tissue usually indicates the overexpressed p53 mutant protein. We assumed that p53-positive tumor cases express lower levels of FOXF1 compared with normal counterparts. However, the anti-FOXF1 antibody used in this study was unable to detect FOXF1 in adjacent normal colonic epithelium, similar to the situation that endogenous p53 levels were undetectable in epithelium of all examined normal tissue cases as the DO-7 antibody was used in IHC. Therefore, quantitative RT-PCR analysis of FOXF1 expression in conjunction with single-strand conformation polymorphism (SSCP) and IHC analyses of the p53 gene is a better strategy to address this connection. The FOXF1 IHC positivity identified in p53-positive tumor tissue cases is not against our hypothesis due to the reason that overexpressed FOXF1 proteins lose their nuclear transcriptional function by being sequestered in the cytoplasm of tumor epithelial cells. Collectively, our studies indicate that FOXF1 overexpression in colorectal tumor epithelial cells was not related to the p53 IHC positivity.
In summary, this is the first study to unravel that the FOXF1 protein is frequently overexpressed in colorectal tumor epithelial cells and underexpressed/lost in tumor-associated stromal fibroblasts. Importantly, we identified that overexpressed FOXF1 proteins are mislocalized in the cytoplasm of tumor epithelial cells, which positively correlates with poor prognostic factors of colorectal cancer. Therefore, these lines of evidence suggest that cytoplasmic mislocalization of overexpressed FOXF1 is a novel biomarker potentially used for the prognosis of colorectal adenocarcinomas.
Acknowledgments
This work was supported by the Advanced Support Program for Innovative Research Excellence-I (ASPIRE-I) Grants 13010-A026 to P. K. Lo and 13010-A007 to H. Chen from the Office of the Vice President for Research at the University of South Carolina, the American Cancer Society Research Award (RSG-10-067-01-TBE) to H. Chen, the US National Institutes of Health (NIH) Grant (3P20RR017698-08) to F.G. Berger and H. Chen, and the Leading Foreign Research Institute Recruitment Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (MEST) (2011-0030034) to J.S. Lee. The results, views and conclusions reported in this paper are those of the authors and are independent from the funding sources.
Abbreviations
- FOXF1
forkhead box-F1
- IF
immunofluorescence
- ICC
immunocytochemistry
- IHC
immunohistochemistry
- TMA
tissue microarrays
- HA
hemagglutinin
- DAPI
4′6-diamidino-2-phenylindole
Footnotes
Conflict of Interest Statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggarwal S, Chu E. Oncology. Vol. 19. Williston Park, N.Y: 2005. Current therapies for advanced colorectal cancer; pp. 589–595. [PubMed] [Google Scholar]
- Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Developmental Biology. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- Dix B, Robbins P, Carrello S, House A, Iacopetta B. Comparison of p53 gene mutation and protein overexpression in colorectal carcinomas. British Journal of Cancer. 1994;70:585–590. doi: 10.1038/bjc.1994.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nature Reviews Genetics. 2009;10:233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellqvist M, Mahlapuu M, Samuelsson L, Enerback S, Carlsson P. Differential activation of lung-specific genes by two forkhead proteins, FREAC-1 and FREAC-2. Journal of Biological Chemistry. 1996;271:4482–4490. doi: 10.1074/jbc.271.8.4482. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA: a cancer journal for clinicians. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Kaiser S, Park YK, Franklin JL, Halberg RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG, Kong S, Sakthivel B, Xu H, Reichling T, Azhar M, Boivin GP, Roberts RB, Bissahoyo AC, Gonzales F, Bloom GC, Eschrich S, Carter SL, Aronow JE, Kleimeyer J, Kleimeyer M, Ramaswamy V, Settle SH, Boone B, Levy S, Graff JM, Doetschman T, Groden J, Dove WF, Threadgill DW, Yeatman TJ, Coffey RJ, Jr, Aronow BJ. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome biology. 2007;8:R131. doi: 10.1186/gb-2007-8-7-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinichenko VV, Bhattacharyya D, Zhou Y, Gusarova GA, Kim W, Shin B, Costa RH. Foxf1 +/− mice exhibit defective stellate cell activation and abnormal liver regeneration following CCl4 injury. Hepatology. 2003;37:107–117. doi: 10.1053/jhep.2003.50005. [DOI] [PubMed] [Google Scholar]
- Kalinichenko VV, Zhou Y, Bhattacharyya D, Kim W, Shin B, Bambal K, Costa RH. Haploinsufficiency of the mouse Forkhead Box f1 gene causes defects in gall bladder development. Journal of Biological Chemistry. 2002;277:12369–12374. doi: 10.1074/jbc.M112162200. [DOI] [PubMed] [Google Scholar]
- Katoh M, Katoh M. Human FOX gene family (Review) International Journal of Oncology. 2004;25:1495–1500. [PubMed] [Google Scholar]
- Lai E, Clark KL, Burley SK, Darnell JE., Jr Hepatocyte nuclear factor 3/fork head or “winged helix” proteins: a family of transcription factors of diverse biologic function. Proceedings of the National Academy of Sciences U S A. 1993;90:10421–10423. doi: 10.1073/pnas.90.22.10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo PK, Lee JS, Liang X, Han L, Mori T, Fackler MJ, Sadik H, Argani P, Pandita TK, Sukumar S. Epigenetic inactivation of the potential tumor suppressor gene FOXF1 in breast cancer. Cancer Research. 2010;70:6047–6058. doi: 10.1158/0008-5472.CAN-10-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo PK, Lee JS, Sukumar S. The p53-p21WAF1 checkpoint pathway plays a protective role in preventing DNA rereplication induced by abrogation of FOXF1 function. Cellular signalling. 2012;24:316–324. doi: 10.1016/j.cellsig.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlapuu M, Enerback S, Carlsson P. Haploinsufficiency of the forkhead gene Foxf1, a target for sonic hedgehog signaling, causes lung and foregut malformations. Development. 2001a;128:2397–2406. doi: 10.1242/dev.128.12.2397. [DOI] [PubMed] [Google Scholar]
- Mahlapuu M, Ormestad M, Enerback S, Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development. 2001b;128:155–166. doi: 10.1242/dev.128.2.155. [DOI] [PubMed] [Google Scholar]
- Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nature Reviews Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- Nilsson J, Helou K, Kovacs A, Bendahl PO, Bjursell G, Ferno M, Carlsson P, Kannius-Janson M. Nuclear Janus-activated kinase 2/nuclear factor 1-C2 suppresses tumorigenesis and epithelial-to-mesenchymal transition by repressing Forkhead box F1. Cancer Research. 2010;70:2020–2029. doi: 10.1158/0008-5472.CAN-09-1677. [DOI] [PubMed] [Google Scholar]
- Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, Miura N, Carlsson P. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development. 2006;133:833–843. doi: 10.1242/dev.02252. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito RA, Micke P, Paulsson J, Augsten M, Pena C, Jonsson P, Botling J, Edlund K, Johansson L, Carlsson P, Jirstrom K, Miyazono K, Ostman A. Forkhead box F1 regulates tumor-promoting properties of cancer-associated fibroblasts in lung cancer. Cancer Research. 2010;70:2644–2654. doi: 10.1158/0008-5472.CAN-09-3644. [DOI] [PubMed] [Google Scholar]
- Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D, Pachlewski J, Oledzki J, Ostrowski J. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010;5:e13091. doi: 10.1371/journal.pone.0013091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Welsh JB, Sapinoso LM, Kern SG, Dimitrov P, Lapp H, Schultz PG, Powell SM, Moskaluk CA, Frierson HF, Jr, Hampton GM. Molecular classification of human carcinomas by use of gene expression signatures. Cancer Research. 2001;61:7388–7393. [PubMed] [Google Scholar]
- van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nature Reviews Molecular Cell Biology. 2007;8:440–450. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- Watson JE, Doggett NA, Albertson DG, Andaya A, Chinnaiyan A, van Dekken H, Ginzinger D, Haqq C, James K, Kamkar S, Kowbel D, Pinkel D, Schmitt L, Simko JP, Volik S, Weinberg VK, Paris PL, Collins C. Integration of high-resolution array comparative genomic hybridization analysis of chromosome 16q with expression array data refines common regions of loss at 16q23-qter and identifies underlying candidate tumor suppressor genes in prostate cancer. Oncogene. 2004;23:3487–3494. doi: 10.1038/sj.onc.1207474. [DOI] [PubMed] [Google Scholar]