Abstract
Cancer is currently considered as the end point of numerous genomic and epigenomic mutations and as the result of the interaction of transformed cells within the stromal microenvironment. The present work focuses on breast cancer, one of the most common malignancies affecting the female population in industrialized countries. In this study we perform a proteomic analysis of bioptic samples from human breast cancer, namely interstitial fluids and primary cells, normal vs disease tissues, using Tandem mass Tags (TmT) quantitative mass spectrometry combined with the MudPIT technique. To the best of our knowledge this work, with over 1700 proteins identified, represents the most comprehensive characterization of the breast cancer interstitial fluid proteome to date. Network analysis was used to identify functionally active networks in the breast cancer associated samples. From the list of differentially expressed genes we have retrieved the associated functional interaction networks. Many different signaling pathways were found activated, strongly linked to invasion, metastasis development, proliferation and with a significant cross-talking rate. This pilot study presents evidence that the proposed quantitative proteomic approach can be applied to discriminate between normal and tumoral samples and for the discovery of yet unknown carcinogenesis mechanisms and therapeutic strategies.
Keywords: Interstitial fluid, breast cancer, Tandem mass Tags, MudPIT, LC-MS/MS, pathway analysis, Cytoscape
INTRODUCTION
Cancer is considered one of the fastest growing diseases in the world. In 2008, 3.2 million of new cases and 1.7 million of deaths were estimated only in Europe. Among these new cases, breast cancer is approximately 28% of all cancers, with 40% of patients with lymph nodes positive to the disease. Generally it is considered that risk of recurrence and death is related to the stage of the disease at the time of first detection and surgery. Currently, considering the many different types of screening, only mammography, with its high sensitivity and precision, is a widely accepted method for early detection of breast cancer in non-symptomatic women1. At present there is an urgent need for new biomarkers to provide a more reliable diagnosis, to improve the assessment of individuals patient’s response to therapy and to identify new drug targets2. In the light of recent findings in molecular biology and oncology, tumors are no longer considered only as mass of proliferating cells but rather as a very complex tissue system composed of different cell types that interact in many different ways to guarantee their own survival. Indeed, in the tumor mass it is possible to detect normal cells, forming the tumor associated stroma, which contributes to the development of the tumor mass itself and the progression from benign breast disease to invasive breast cancer3. In tumor tissues it is possible to distinguish two main components to analyze, namely the different cell types and the tumor microenvironment. The interstitial space is part of the microenvironment, and it is considered as the connective and supportive tissue of the body. The interstitial space can be divided into two compartments: interstitial fluid and structural parts of the extracellular matrix (ECM). Interstitial water serves as a transport medium for nutrients, waste discarding and as a storage for signaling substances produced locally or brought to the organs by circulation. Tumor interstitial fluids are characterized by high H+, CO2, lactic acid and low glucose, due to the tumor metabolism4 and proteins. These fluids bath all tumor components and may contain an enriched population of tumor specific shed and secreted proteins with respect to peripheral blood. Interstitial fluids can, therefore, be considered as a valuable source of information about the biology of tumor microenvironment5. These kinds of proteins can include growth factors, proteases, cell motility factors, cytokines, chemokines and cell surface receptors, each one having a specific role in tumor progression, invasion, metastasis and angiogenesis, regulating cell-cell and cell-ECM interaction6. Proteomic analysis of fluids may represent an attractive source of biomarkers, similarly to the study of fluids derived from ascites or pleural effusions, which, however, appear only in late stage of cancer. Gromov et al. developed a procedure to extract interstitial fluids from tissues, starting from the consideration that fluids composition may reflect the physiological and pathological state of the tissues. They used the tumor interstitial fluids as source and 2D-gel-based proteomics as discovery tool with the aim of discovering differentially expressed externalized proteins in breast cancer samples. In a first step, they analyzed samples derived from a single tumor/benign tissue pair, identifying 110 proteins up-regulated by a factor 2. In a second step, they analyzed the presence of those 110 proteins of interest in 68 tumor interstitial fluid samples by 2D-gel-based proteomics and systematic computer assisted analysis, trying to maximize the probability of finding a clinically useful biomarker7. A similar approach, based on 2D-gel/MALDI-TOF-based proteomics, was applied by Celis et al on mammary adipose tissues and corresponding interstitial fluids, with the aim to identify the role of adipocytes and the related molecular circuitry in the breast tumor microenvironment8.
Our study can be considered as a pilot study to propose an accurate and reliable method for tumor microenvironment proteomic profiling, based on the combined exploitation of mass spectrometry and bioinformatics tools. The core of this work has consisted in the identification of cancer up/down regulated proteins externalized by cells in the tumor environment. These data have been also compared with the quantitative proteomic pattern expressed in primary cells of the same biopsies9,10. We have performed mass spectrometry analysis of TmT labeled samples on Orbitrap Velos, using MudPIT technology to fractionate and decrease samples complexity. Data analysis has been performed using ProLucid/DTASelect softwares11,12, coupled to the appropriate statistical and quantitative validation methods. Specific bioinformatics tools like Gene Ontology and Cytoscape have been used to identify molecular pathway modifications. The datasets have been interfaced with the Reactome database and classified using Gene Ontology categories, extracting information about pathways activated and connected to carcinogenesis. This work yields two main novel contributions: (i) from a methodological point of view, the development of a combined experimental and computational approach to the analysis of in vivo breast cancer interstitial fluids and primary cells, based on quantitative mass spectrometry13. On the biological side, (ii) many different signaling pathways have been found activated, strongly linked to invasion, metastasis development and proliferation, like TGF-β, syndecan, glypican, integrin and hepatocyte growth factor. Hopefully, these findings can be exploited in future works as a useful source of information to guide targeted experiments, aimed at the discovery of yet unknown carcinogenesis mechanisms and therapeutic strategies.
EXPERIMENTAL PROCEDURES
Materials
The highest quality reagents were used: Peptide Calibration Standard (Thermo Scientific, San Jose, CA, USA); modified sequencing grade trypsin for mass spectrometry (Promega, Madison, WI, USA); sixteen protein mixture (Sigma Aldrich, San Louis, MO, USA); protease inhibitor cocktail EDTA-free (Roche Diagnostics, Indianapolis, IN, USA); TmT sixplex label reagent set (Thermo Scientific, San Jose, CA, USA; Bradford Assay (Promega, Madison, WI, USA); HAM (Gibco, Invitrogen, Carlsbad, CA, USA), Foetal bovine serum (Gibco, Invitrogen, Carlsbad, CA, USA), trypsin-EDTA for cell cultures (Gibco, Invitrogen, Carlsbad, CA, USA), penicillin-streptomicyn (Gibco, Invitrogen, Carlsbad, CA, USA), fungizone (Gibco, Invitrogen, Carlsbad, CA, USA), phosphate buffered saline (Gibco, Invitrogen, Carlsbad, CA, USA), collagenase III (Sigma Aldrich, San Louis, MO, USA), trypan blu (Sigma Aldrich, San Louis, MO, USA), Lys-C (Promega, Madison, WI, USA).
Patients characteristics and specimens procurement
Patients were enrolled at the “Tommaso Campanella Foundationz” - Magna Graecia University of Catanzaro. We provided all the specific information and the consensus form in accordance with the institutional guidelines.
All patients were scheduled to perform surgery to remove a previously identified breast lesion, followed by histopathological diagnosis. For each patient we obtained pathological tissues and tissues extracted in a region as far as possible from the lesion, macroscopically considered “healthy”14 by the surgeon.
Surgical specimens were inspected by two different pathologists and preserved using the following method: a) one portion was fixed in buffered formalin 10% for histopathological diagnosis; b) two different portions were stored in cold phosphate buffered saline added with 1% penicillin-streptomicin and 1% amphotericin B. Stage of cancer was determined by the anatomo-pathology specialist according with the TNM stage (Tumour-Node-Metastasis, UICC-2009). Three patients, not previously treated with chemotherapy, were selected for this preliminary study, based on the type of lesion and tissues availability (Table 1, Supporting info).
Interstitial fluids
Surgical specimens, healthy and tumoral, were transported in PBS solution to the molecular biology laboratory, for further treatment, within maximum 30 min from the surgery. Tissues were washed three times with PBS solution, fragmented in small pieces (1-2 mm3) with sterile scalpels, washed again, and put in phosphate buffered saline (PBS) solution in a conical tube, in a humidified CO2 incubator at 37°C, 5% CO2 for 4-6h. After 6 hours, tubes were centrifuged at 1500 rpm 10 minutes, supernatants were isolated, put in a different tube and immediately centrifuged at 7.000 rpm 20 minutes 4°C. Proteins concentrations were measured by Bradford assay (Promega, Madison, WI, USA); aliquoted and kept at −80°C. The use of PBS was chosen in accordance to the conclusions established by Teng et colleagues5.
Primary cells
Also for primary cells culture, surgical specimens, healthy and tumoral, were transported in PBS solution from the surgical room to the molecular biology laboratory, for further treatment within maximum 30 min from the surgery. Tissues were washed three times with PBS solution, fragmented with sterile scalpels, digested in collagenase IIII (Sigma Aldrich, San Louis, MO, USA) and filtered many times by 100 m nylon cell strainers (BD Biosciences, NY, USA). This procedure was performed at room temperature. Heterogeneous population cells were obtained, and maintained in culture in HAM medium supplemented with 10% foetal bovine serum (FBS), antibiotics and 1% amphotericin B in a humidified CO2 incubator at 37°C, 5% CO2. Cells were grown in a 6-well culture dish (Corning, NY, USA). After 6 days, trypan blue exclusion test, to evaluate the growth rate for each cell stock, and phenotype cancer cell characterization by flow cytometry, using EpCAM and Fibronectin. (Abcam, Cambridge, United Kingdom) (FACS Calibur, Becton Dickinson, Franklin Lakes, NJ, USA), were performed.
Trypan blue exclusion test
After 6 days, cells were at 50-60% confluence. Cells were trypsinized with 1 ml of trypsin (Gibco, Invitrogen, Carlsbad, CA, USA), at 37°C for 2 minutes, pooled adding complete medium and centrifuged at 1500 rpm, 5 min RT. Cells were manually counted with a hemocytometer and plated again to evaluate the growth rate. This procedure was repeated several times for reproducibility, and average of the cells number was used. Cells were trypsinized at different days, stained with trypan blue and counted.
Immunofluorescence assay and phenotype characterization
Cultured primary cells were subjected to cell cycle analysis and phenotype characterization. Cell cycle analysis was performed by propidium iodide staining and phenotype characterization by EpCAM and Fibronectin antibodies (Abcam, Cambridge, United Kingdom). Flow cytometry was performed on FACS Calibur (FACS Calibur, Becton Dickinson, Franklin Lakes, NJ, USA), using Modfit (Verity Software House) for DNA distribution analysis and Cellquest (FACS Calibur, Becton Dickinson, Franklin Lakes, NJ, USA) for antigen expression. For the staining, cells were incubated with the appropriate primary antibody and with the PE conjugated goat antimouse secondary reagent (Abcam, Cambridge, United Kingdom). Statistical analysis was performed using the two-tailed two sample t-test. Correlations were assessed using Pearson correlation coefficient.
The same labeling method was performed by immunofluorescence with the following protocol. Cells were cultured on sterile cover-glasses O.N. 37°C 5% CO2. Cells were washed softly and fixed with 10% formalin 10 minutes, then washed with PBS-Tween 20 0,05% (Sigma Aldrich, San Louis, MO, USA) solution for 2 minutes; cells were stained with the appropriate antibody in solution followed by the PE conjugated goat antimouse secondary antibody (Abcam, Cambridge, United Kingdom). Cells were evaluated on confocal microscopy (Leica Camera AG, Solms, Germany).
Validation of proteomics data- Western Blotting
Primary cells were trypsinized, washed and pooled according to the described protocol. Lysis Buffer (100 μl; TEAB 200mM, SDS 0,5%, NaCl 100 mM) (Sigma Aldrich, San Louis, MO, USA) was added to each sample and incubated in ice, vortexing, for 30 minutes with three freeze/thaw steps −20°C. Samples were centrifuged at 13000 rpm for 30 min, 4°C and the supernatant was extracted in a different tube. Protein concentration of each sample was measured using the Bradford assay15, with three different measurements and extracting the related averages. Interstitial fluids were extracted from the surgical specimens as described in the previous paragraph, and the proteins concentrations were measured using the Bradford assay with three different measurements.
Total extracts (30 μg of proteins both for primary cells lysates and interstitial fluids) were used, added with 4× LDS and 10× Reducing Agent (Invitrogen, Carlsbad, CA, USA), heated at 70°C for 10 minutes and loaded on 4-12% polyacrylamide gels (Invitrogen, Carlsbad, CA, USA), according to the manufacturer instruction. Silver staining was performed according to the manufacturer protocol of SilverExpress kit (Invitrogen, Carlsbad, CA, USA).
Proteins were transferred on nitrocellulose filter membrane (Amersham Biosciences), for 2 hours at 100V, in 192 mM glycine/25 mM Tris-HCl (pH 7.5) and 10% methanol. Loading normalization on the membranes was checked by ponceau red (Sigma Aldrich, San Louis, MO, USA). Membranes were blocked with 5% non fat dried milk and incubated with primary antibodies O.N. 4°C. After three washes in PBS-Tween 20, 0,05%, membranes were incubated with the appropriate secondary HRP antibodies 1h RT. Proteins were visualized by enhanced chemiluminescence (Pierce, Idaho, USA). Primary antibodies were: anti 14-3-3 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) rabbit 1:2000; GDI-1 home made mouse 1:500; GAPDH mouse 1:1000 (Millipore, Billerica, MA, USA); AUF-1 rabbit 1:1000 (Upstate, Charlottesville, VA, USA); PCNA mouse 1:5000 (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Secondary antibodies were goat antimouse and antirabbit 1:5000 (Amersham Biosciences, GE Healthcare, Uppsala, Sweden).
Trypsin digestion and TmT labeling
As previously indicated, we analyzed two different samples for each patient (n=3), healthy and tumoral tissues. Tandem mass tags (TmT) kit (Pierce, Idaho, ID, USA) was used. 100 μg of each sample were precipitated in cold acetone (1:6), −20°C O.N. Six samples were dissolved in 45 μl of TEAB 200mM pH 8.0; 2.5 μl SDS 2% and 10 μl TCEP 200mM were added. Reaction was performed for 60 min at 55°C. According to the manufacturer instruction, 5 μl/sample of iodoacetamide (IAA) 375mM was added and the mixtures were reacted for 30 min RT in the dark. We performed a double digestion in order to improve protein identification and characterization16. Lys-C (Promega, Madison, WI, USA) was added for each sample in 40:1 relation, and digestion was performed at 37°C, O.N. As second enzymatic digestion, an amount of 2.5 μl (1 μg/μl) of tripsyn was added per 100 μg protein. The digestion was performed O.N. at 37°C.
TmT reagents were reconstituted according to the manifacturer instructions. Tubes containing the different isobaric chemical tags (0,8 mg each) were added with 41 μl anhydrous acetonitrile at room temperature. Reagents were dissolved by vortexing 5 min and the solutions were gathered by centrifugation. Protein samples were labeled by adding 41 μl of TmT isobaric tag, followed by an incubation step for 1 hour at room temperature. To quench the reaction, 5% hydroxylamine (8 μl per sample) was added followed by a 15 min incubation RT. Samples were finally pooled, divided into six aliquots and stored at −80°C. At the time of the analysis, the aliquotes were thawed at room temperature and acidified with Buffer A (see below) 1:6, before MudPIT analysis.
The twelve samples (six interstitial fluids and six primary cells), were labeled by TmT as described in Table 2 (Supporting Information).
Multidimensional protein identification technology analysis (MudPIT)
Each pool of labeled samples was split in six aliquots of 100 μg each, analyzing four of them. Each sample was subjected to MudPIT analysis according to the literature17,18. Analysis of TmT samples was performed on LTQ Orbitrap Velos (Thermo Scientific, San Jose, CA, USA) interfaced at the front end with a quaternary HP 1100 series HPLC pump (Agilent Technology, Santa Clara, CA, USA). The analytical column consisted of a 100 m diameter fused-silica capillary (J/W Scientific, Agilent Technology, Santa Clara, CA, USA), pulled with a P-2000 laser (Sutter Instrument Co., Novato, CA, USA) and packed with 12 cm of 5 m C18 resin (Aqua, Phenomenex, Torrence, CA, USA). The biphasic microcapillary trapping column (5 cm of 250 m diameter), consisted of a fritted capillary with Kasil 1624, packed with 2.5 cm reversed-phase C18 (Aqua, Phenomenex, Torrence, CA, USA) and 2.5 cm of strong cation exchange (5 m Partisphere, Whatman, Maidstone, Kent, UK) packing material. The biphasic column, loaded offline with sample by a pressure pump, was connected to a peek microcross as previously described19 in order to split the gradient pump flow to 0.15-0.20 μl/min and supply a spray voltage of 1.8kV. The LTQ Orbitrap Velos was operated via Instrument Method files in the Sequence Setup window of Excalibur. The heated desolvation capillary was set to 180°C.
A fully automated 11-cycle chromatographic run was performed on each sample using a three mobile phases system consisting of Buffer A (5% acetonitrile (ACN); 0,1% acid formic (FA)) (Sigma Aldrich, San Louis, MO, USA), Buffer B (80% ACN, 0,1% FA), Buffer C (500mM ammonium acetate, 5% ACN, 0,1% FA). The first cycle consisted of a 120 min linear gradient from 0 to 100 % buffer B. Steps 2-9 show the following profile: 2 min 100% buffer A; 4 min (100-X)% buffer A, X% Buffer C; 45 min from 100% buffer A to 50% buffer A and 50% buffer B, 10 min from 50% to 100% buffer B, 1 minute from 100% buffer B to 100% A, 10 min at 100% buffer A. For buffer C, X % was, respectively, 10%, 20%, 30%, 40%, 50%, 60%, 70%, 100%. Steps 10 and 11 show the following profile: 2 min 100% buffer A; 4 min 10% buffer B, 90% Buffer C; 45 min from 100% buffer A to 50% buffer A and 50% buffer B, 10 min from 50% to 100% buffer B, 1 minute from 100% buffer B to 100% A, 10 min at 100% buffer A. A schematic representation of the gradient profile is reported in Table 3 Supporting Information.
The application of the distal voltage of 1.8kV electrospayed the eluted peptides directly into LTQ Orbitrap Velos (Thermo Scientific, San Jose, CA, USA). A cycle of one full scan was applied (300-1600 m/z, resolution 60.000) followed by 10 data dependent CID-HCD dual MS/MS scans acquired for TmT reporter ions, and repeated continuosly through each MudPIT step. Collisional-induced dissociation (CID) scans were acquired in LTQ; full scans and higher energy collisionally activated dissociation (HCD) scans in Orbitrap respectively at resolution 60.000 and 7.500. Normalized collision energy of 35% in CID and 45% in HCD with 20 ms activation time was used to acquire MS/MS spectra. The following parameters for dynamic exclusion were applied: 1 repeat count, 30 msec repeat duration, 500 exclusion list size, 120 sec exclusion duration, exclusion width low energy 0.51; exclusion width high energy 1.51.
Database Searching for Protein Identification
The resulting files were analyzed by ProLuCID algorithm v. 1.3.1 to interpret MS/MS, and protein identification as previously described11, using tandem mass spectrometry and protein sequence databases (EBI-IPI_Human 3.71 of March 24-2010 reversed fasta). Variable amino acid modifications were: carbamidomethylation of cysteines (57.021464 C) and TmT-labeled peptides in primary amino groups (229.1629 K, N-ter). Trypsin was selected as enzyme. The precursor peptide mass tolerance was 50 ppm with 5 number of isotopic peaks and 600 ppm of fragment mass tolerance. The database search was filtered using DTASelect software v. 2.0.47 with a false positive rate less than 0.05, precursor delta mass cut off 10 ppm, minimum Sp score 412.
All accepted results had a Cn of 0.1 or greater and cross-correlation score (Xcorr) had to be greater than 1.9. To perform the quantitative analysis we used Census20, a software tool for quantitave proteomic analysis. Starting from the raw data, Census was able to calculate the relative intensities of reporter ions from a specific identified tandem mass spectrum (a threshold intensity rate of 10000 for the sum of the reporter ion intensities of HCD spectra was applied). Protein intensities resulted from the average of the single TmT reporter ion intensities, obtained for each peptide associated to a specific protein.
Biostatistics
A first statistical evaluation of these large datasets was performed by computing, for each experiment, the distribution of m/z measurements, to check that the Healthy vs. Pathologic log2 fold change follows a Gaussian distribution. For each patient, the ratios healthy/pathological were transformed as log2 terms and the statistical distribution and box plot were computed. Values assume a gaussian distribution with some asymmetry; median and standard deviation were calculated and ratio values corrected for the median, to account for variability among different experiments. Subsequently, data derived from mass spectrometry analysis were examined using Anova Test and Benjamini Hochberg correction for false discovery rate (FDR), assuming as significative threshold FDR = 0,0521.
Bioinformatic Analysis- Pathway Analysis
Pathway analysis was performed using Cytoscape 2.822,23, associated to two different plugins. The Reactome FI plugin version 1.124,25 was used to interface experimental datasets to the Reactome database, extracting data regarding pathways enrichment, GO cellular components and GO molecular functions. CentiScaPe version 1.2 was used to compute nodes centralities26, in order to gain further insight and identify the hubs and the most relevant proteins in the interaction networks, which can be considered as the most promising candidates for further biological validation experiments.
RESULTS AND DISCUSSION
Here we present the quantitative proteomic profiles of three breast cancer patients: two invasive ductal carcinomas and one phylloides tumor. Ductal carcinomas have a high incidence in the female population and commonly occur as a result of neoplastic proliferation arising from the luminal epithelial cells, disrupting the basement membrane and the myoepithelial cells3. Phylloides tumors, also called cistosarcoma phylloides, are very uncommon types of neoplasia (1% of total breast cancers) with a very high rate of proliferation. They occur predominantly in connective tissue (stroma) rather than in epithelial tissue (ducts and lobes). We chose to analyze these two types of cancer due to their impact on the population and to the different origin of the cancer, epithelial and stromal respectively, which could identify differences in the active molecular networks. However, our principal aim was the creation of a reliable and accurate method for biomarker discovery, by the combination of mass spectrometry and bioinformatic tools.
For each patient, primary cancer cells and interstitial fluids, both from tumoral and healthy counterpart, were extracted. Primary cells were tested for EpCAM and Fibronectin expression by flow cytometry and immunofluorescence and for growth rate (Figure 1). For simplicity, in Figure 1, we show only the staining panel relative to a single patient (Pt1). To quantitate protein expression changes we applied Tandem mass Tags together with tandem mass spectrometry to determine differences between the normal and disease samples. Samples were analyzed in quadruplicate on an LTQ-Orbitrap Velos.
Figure 1.
(A) Breast tumoral biopsy from a ductal carcinoma in situ. (B) Expression of EpCAM assayed by flow cytometry in control (HS) and pathological (PS) sample of primary cells. (C) Relative growth rate of cancer primary cells for the three patients, obtained by trypan blue exclusion test, at days 0,1,5, 16. (D) Fluorescent image showing the cancer primary cells in bright field and the corresponding staining, in the green channel, indicating respectively EpCAM and Fibronectin.
Protein Identification by MudPIT
Protein mixtures derived from interstitial fluids are characterized by a very high complexity, which is further complicated by the presence of blood contamination. Blood is a notoriously complex proteomic sample due to the overwhelming presence of albumin and immunoglobulins. Both proteins can, in principle, be removed by an immunodepletion step. Nevertheless, we decided not to proceed with a depletion step, principally to avoid introducing additional experimental variability. For both interstitial fluids and primary cells 100 μg of lysates for each sample (3 healthy, 3 diseased) were labeled with TmT, pooled, and divided into aliquots. Four aliquots were analyzed on the Orbitrap Velos by MudPIT. First, we evaluated analysis performance in terms of: (i) MW and pI range of detected proteins; (ii) MudPIT peptide fractionation efficiency, both for interstitial fluids and primary cells (Figures 1s, 2s). As shown, we were able to identify proteins having a wide range of MW, which spanned mainly from 10.000 to 500.000 Da. Most proteins had a pI between 5 and 7, both for interstitial fluids and primary cells. As shown, more than 50% of the peptides were identified in the last three fractions for both sample typologies. In order to improve analytical precision, in the case a peptide was identified in multiple fractions (redundant peptide) all quantitative data associated to that particular peptide were used for deriving protein quantitation tables. A total of more then 104.000 spectra were matched to peptide sequences and used for protein quantitation in interstitial fluid quadruplicate analysis, excluding spectra deriving from contaminants and reversed protein sequences. An average of 1324 non-redundant proteins were identified and quantified per sample. The quantification efficiency varied from 79% to 86%, with some fluctuation in the replicates essentially due to mass spectrometer performance limitation.
In primary cells, we obtained an average of more than 31.000 spectra for each patient, corresponding to an average of 1886 protein identified and quantified per sample. Quantification efficiency fluctuated from 93% to 95%. The number of identified and quantified proteins was higher in primary cells than in interstitial fluid, presumably because the latter partially resembles serum/plasma in terms of protein composition. Indeed, it is well-known that the number of identified proteins is usually higher for cellular proteomes than for undepleted biofluid proteomes, when the same analytical method is used and the same sample amount is analysed27.
Identification of Proteins exhibiting Significant Modulation
Here we propose a discussion of the results concerning each patient separately, according to the nomenclature introduced in Table 1 (Supporting Information).
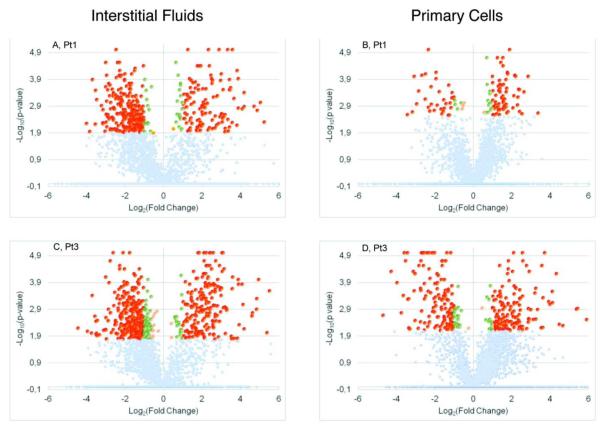
Considering a fold-change ≥ 1.5 and FDR = 0.05, we singled out 399 proteins showing a up/down modulation in the interstitial fluids of Patient 1 (Pt1), as shown in the Volcano plots reported in Figure 2A. The same thresholds have been applied to Pt1 primary cell data, obtaining 116 modulated proteins (Figure 2B). In both interstitial fluids and primary cells, we identified a number of singleton proteins, i.e. proteins identified with at least three quantifiable peptides only in the healthy samples or only in the diseased sample per patient.
Figure 2.
Volcano plots reporting the −log10 (p-value) on y-axes and log2 (fold change) on the x-axes. Colors denote different modulation and significance values: Benjamini-Hochberg adjusted p value (BH-p)<0,05 & abs(fold change)>2 red; green BH-p<0,05 & 1,5<abs(fold change) >2 in green; BH-p<0,05 & abs(fold change)<1,5 in yellow; BH-p>0,05 in gray.
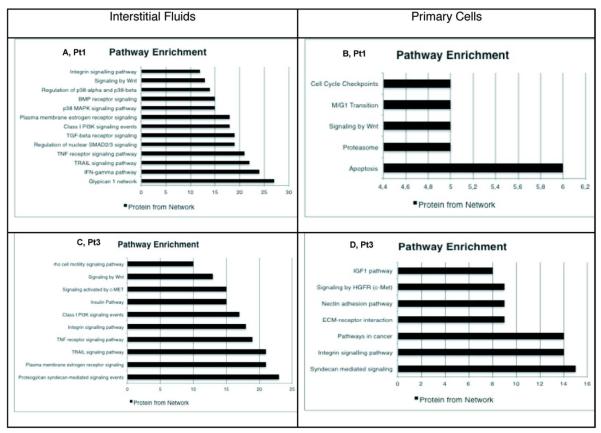
We performed a comparison between interstitial fluids and cell lysate datasets showing that only 11 proteins were in common, with PSBM, NME3, PTX328 and VIM (vimentin) showing a different modulation between fluids and cell lysates. In particular, vimentin was found highly down-regulated in healthy cells suggesting a lack of differentiation29, Tenascin C was also found up-regulated in cancer cells, supporting the hypothesis of a epithelial-mesenchymal process during tumor progression30. We imported into Cytoscape the list of modulated proteins, generating separate graphs for interstitial fluids and primary cells, integrated with the list of singleton proteins. The Reactome FI plugin was exploited to derive the functional interaction networks associated to our protein lists, along with the functional interaction annotations, according to the information contained in the Reactome database (formerly known as Genome Knowledgebase31). By visual inspection of the resulting graphs, shown in Figure 3, one can rapidly perform a first evaluation of the differential expression results in healthy and pathological samples, focusing on the most relevant proteins and their interactions. In order to better investigate the properties of the network, and to evaluate the relevance of the single proteins, we have conducted further analysis by using other functions of the Reactome Fl and the CentiScaPe plugin. In particular, we have applied the Cluster FI network algorithm to divide the network into several modules, chosen on the basis of topological properties of the distinct modules. After this step the pathway enrichment function has been applied both to the entire network and to the single modules. The module-wise analysis reveals a strong correlation between the topological and biological properties of the network. In particular, nodes belonging to the same module often happen to belong to the same pathway (Figure 3s). Using the Cytoscape Network Analysis tool, we have analyzed the molecular function/localization of the protein datasets according to Gene Ontology (GO) functional annotations and categories. GO analysis showed that most of the modulated proteins in Pt1 interstitial fluids have cytoplasmic origin, followed by extracellular region, splicesome and proteasome complexes; FDR=0.05 has been chosen for the classification in the GO categories (data not shown). The main molecular functions associated to the modulated proteins are: nucleotide binding (specifically ATP binding), RNA binding and peptidase inhibitor. Finally, as found by means of pathway enrichment, the most active pathways are those of glypican, interferon gamma, SMAD 2/3, TGF-β, Wnt. Statistical significance was, also in this case, set to FDR=0.05. Most of the above mentioned pathways are strictly connected to growth factors response, invasion, motility, cell survival and adhesion32,33,34 (Figure 4A).
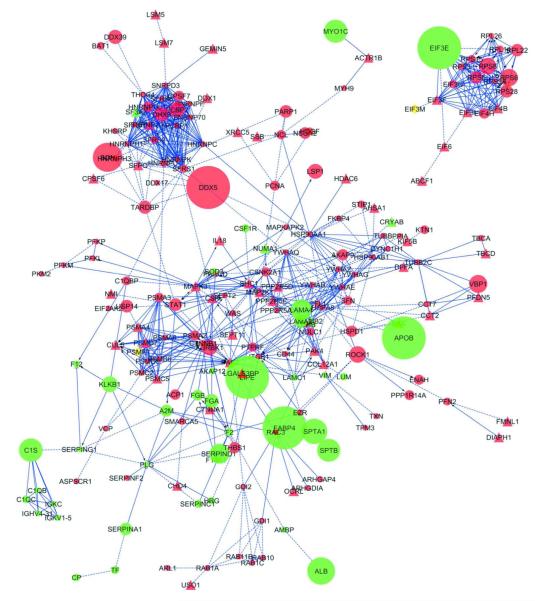
Figure 3.
Cytoscape diagram of the functional interaction network representing proteins modulation in Pt1 interstitial fluid. The network was edited in order to get an effective visualization of the fold-change of each protein/node. In particular, we have used different colors for up- and down-regulated proteins, displayed in red and green, respectively, whereas the node size was made proportional to the fold-change of the corresponding modulated protein. Red circles represent up-regulated proteins in the pathological samples; green circles, up-regulated proteins in the healthy samples; red triangles, singleton proteins identified in the pathological samples; green triangles, singleton proteins identified in the healthy samples.
Figure 4.
Cytoscape pathway enrichment tool, applied to the modulated proteins in the functional interaction networks. Classification as the number of proteins enriched per network.
To get a quantitative evaluation of the relative importance of the modulated proteins with respect to their functions in the network, as opposed to the sole fold-change value, we computed the nodes centralities by using the CentiScaPe plugin. Focusing on the node degree, we have identified as “hubs” the following proteins SFRS1, HNRNPH1, PAK2, MAPK3, HNRNPU, HNRNPK, SFRS2, SFRS7, connected to spliceosome complex and tumor cell invasion35. Koh et al. demonstrated that the endothelial cell collagen matrix interactions result in integrin-dependent signaling, leading to activation of Pak2 and Pak4, regulated by Src and Yes signaling during lumen formation in angiogenesis. Pak2 and Pak4 act as signals integration systems for a multicomponent kinase signaling pathway downstream of integrin-matrix interaction, involving PKCε, Src, Yes, Pak2, Pak,4, B-Raf, C-Raf and Erk 1/2 (also called MAPK3/1) to control endothelial cells lumen in angiogenesis36.
The same approach was used to analyze the primary cells protein dataset: the functional interaction network between the modulated proteins (117 proteins) was computed using Cytoscape (Figure 4s). The network shows that the main modulated signaling pathways are those connected to apoptosis, proteasome constituents and Wnt (Figure 4B). These pathways are involved in cell degradation, apoptosis and cell cycle transitions according to GO categories32.
In Pt2, invasive ductal carcinoma G2, score 7, as shown in the Volcano plots, only one protein was found significantly modulated in the interstitial fluids and none in the primary cells. We surmise that there was no difference between the healthy and pathological samples from the beginning (Figure 5s). More specifically, the most likely explanation for this absence of statistical difference between the samples is that the tumoral invasion process from the neoplastic lesion to the surrounding tissue had already reached an advanced phase, though not macroscopically visible at the time of the surgical intervention. This hypothesis is also supported by the difference in the growth rate behavior of Pt2 healthy cells, compared to the healthy cells of Pt1 and Pt3 (Figure 1), sustained also by previous studies reporting that one of the major issues is the possibility of obtaining right quantity of well-defined normal tissues2.
In Pt3 samples, phylloides breast tumor, we detected 564 up/down-modulated proteins in the interstitial fluids considering again a fold-change≥1.5 and a FDR=0.05 (Figure 2C, 6s). The resulting list of modulated proteins has been analyzed, performing clustering, Pathway Enrichment, GO Analysis and network topological analysis, as described for Pt1 (Figure 4C). The network has been edited and the resulting graph is shown in Figure 5s. The GO analysis evidences cellular components similar to those found in the first patient with most of the proteins derived from the cytoplasm. The GO molecular functions are fundamentally connected to protein and nucleotide binding (data not shown). Concerning the pathway analysis, the data are reported as the histogram with FDR=0.05 statistical significance. The most enriched signaling pathways in the interstitial fluids are: syndecan, integrin, Wnt and insulin. These pathways are strictly connected to different functions, e.g. survival/apoptosis, proliferation, migration37,38,39. In the primary cells, 336 proteins were found modulated (Figure 2D) and the resulting graph was computed (Figure 7s). GO analysis shows that most of the proteins identified and quantified have extracellular and mitochondrial origin, with main molecular functions being calcium ion binding and oxidoreductase activity. From the pathway enrichment analysis (figure 4D), it is possible to observe that the main modulated pathways are connected to proteoglycan syndecan-mediated signaling, integrin, ECM-receptor, c-MET. All these pathways are connected to proliferation, survival and motility40,41.
A comparison between fluids and cells in Pt3 protein datasets was performed, identifying 49 common proteins, some of them with a different modulation pattern, like GAPDH (down-regulated in healthy sample, as demonstrated by western blot) and SOD2 (data not shown). In our data we can highlight the contemporary downregulation of caveolin-1 both in fluids and in primary cells, associated with SOD2, which is up-regulated in primary cells (but down-regulated in fluids) according to the “autophagic tumor stroma model of cancer metabolism”. Indeed, Trimmer et al. proposed that tumor cells can induce autophagy in the adjacent cancer-associated fibroblasts via the loss of caveolin-1, which is sufficient to promote oxidative stress in stromal fibroblasts42. This stress activates the autophagic program in the tumor microenvironment, which results in lysosomal degradation of caveolin-1 and the production of recycled nutrients to feed cancer cells43. In a subsequent work Trimmer et al. restored in MDA-MB-231 cells the expression of SOD2 (mitochondrial enzyme that de-activate superoxide effects), inhibiting the tumor growth effect of caveolin loss44. This process is associated to ROS production for mitochondrial disfunction43. This may be considered as a possible input for further studies about the implication of mitochondrial oxidative stress for tumor growth in cancer biopsies
A comparison of modulated protein datasets between Pt1 and Pt3 interstitial fluids was performed, highlighting that 213 proteins were in common. Pathway enrichment was separately applied to the common and non-common protein sets. The analysis has shown that the common enriched molecular pathways were prevalently connected to BMP receptor signaling, FoxO signaling, p-38 MAPK, and several metabolic pathways. As for the non common enriched pathways, instead, we have found that they were prevalently involved in maturation of mRNA and apoptosis in Pt1; and in focal adhesion, integrin signaling and cell cycle transition in Pt3. Proteins which have shown a different modulation between the two interstitial fluids pools were TSTD1, PP1R14A, ANXA3, IDI1, DDHD2, RBP1, CSF1R, EIF3E (data not shown). Concerning primary cells analysis, we compared our results with other reports recently appearing in the literature. In particular, we focused our attention on a very interesting approach for normal and cancerous breast tissues analysis described by Sutton and colleagues. The study was based on a dual lysis buffer method and iTRAQ labeling followed by preparative IEF and RPnanoHPLC, Maldi MS/MS and database search. Comparing our list of modulated proteins with the one identified by Sutton and colleagues14, we have found that most of the proteins in common belong to the histone family, heat shock proteins and hepatoma-derived growth factors.
We have also performed a comparison with respect to the modulated protein reported in Gromov7 and Celis8 in their works. Interestingly, most of the common proteins belong to the families of 14-3-3 proteins, heat shock proteins, PCNA, vimentin, GDI-2, annexins.
In view of the important biological role of those proteins commonly identified in breast cancer outcome, we may hypothesize that this coincidence is linked to the advanced cancer phenotype. It is remarkable the common identification of vimentin, a well-known epithelial-mesenchymal transition marker with an essential role in cell signaling. In many studies it has been shown that vimentin is able to interact with phosphorylated Erk1 (MAPK3) to protect it from inhibiting phosphatases, maintaining the active status. A second important interaction is the one with 14-3-3 proteins: this binding prevents the association between Raf and 14-3-3, suggesting a link to the regulation of many pathways connected to 14-3-3 family members. Another relevant family of proteins, identified in common with Sutton et al., is represented by the heat shock proteins, often found overexpressed in cancer tissues blocking apoptosis and replicative senescence pathways. Of course, further analyses by means of low-throughput molecular biology techniques are required to confirm the significance of these data and to exclude possible technical artifacts.
Western blot validation
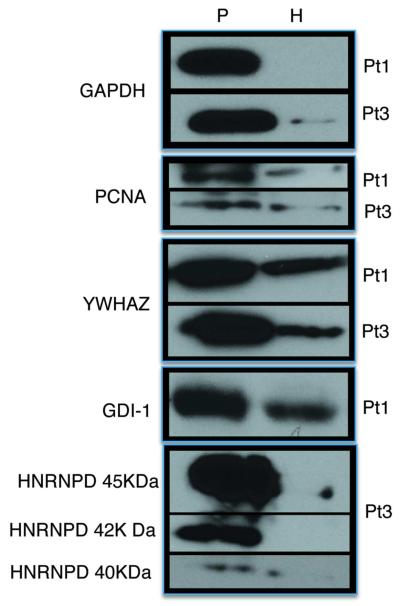
The final phase of our work consisted in the experimental validation of mass spectrometry data by using a complementary low-throughput experimental technique. To this aim, we performed on the same lysates, western blot analysis of five proteins identified via mass spectrometry from the interstitial fluids dataset, choosing to analyze proteins with different functions as component of spliceosome, cell-cycle checkpoint etc. The validation was performed on the interstitial fluids of Pt1 and Pt3. Proteins confirmed by western blot were: GAPDH, YWHAZ, GDI-1 (which is commonly known to be involved in mental retardation45), HNRNPD (a component of the spliceosome, identified as a “hub” by topological analysis), PCNA (Figure 5). All of the proteins analyzed by western blot substantially confirm the quantitative data obtained by mass spectrometry. In Supporting information section (Figures 8s, 9s, 10s, 11s,12s respectively), we show the MS/MS spectrum, the relative sequence and the zoomed view for the corresponding TmT reporter ions
Figure 5.
Validation of the differential expression of GAPDH, PCNA, YWHAZ, GDI-1 and HNRNPD in Pt1, Pt3, pathological (P) vs healthy (H) samples, by western blotting.
CONCLUSIONS
One of the biggest challenges in the management of cancer remains the lack of prognostic and predictive biomarkers that can help in the design of therapeutic strategies as well as in monitoring the tumor response. A new approach toward biomarker discovery is emerging, where pathways instead of individual proteins are monitored and targeted. There are two important challenges for the success of this approach. First, biomarker discovery should be oriented toward secreted proteins, which have a better chance of entering the bloodstream. Second, the methodology used to discover such biomarkers should be high-throughput to allow the analysis of a sufficient number of samples during the discovery phase46. Mass spectrometry techniques are currently considered as one of the election methods for high-performance analysis, in association with bioinformatic and systems biology tools to integrate experimental data in a cellular signaling context47. In the present work we illustrate our strategy, based on combined high-resolution quantitative proteomic experiments and computational tools to identify new sources for biomarker candidates in interstitial fluids and primary cancer cells. It is worth remarking that, to the best of our knowledge, the set of over 1700 proteins identified by the shotgun proteomics approach represents the most comprehensive analysis of the interstitial fluids proteome to date. Here, we demonstrated that interstitial fluids could be considered more informative than primary cells regarding pathway analysis. In Pt1, it is possible to underline an important cross-talk signaling, based on TGF-β induction of cytokines mediated by non canonical Smad and p38 MAPK pathways, as described by Gupta and colleagues48. TGF-β is also known to collaborate with BM (bone morphogenetic protein) for tumor invasion and metastasis49 and to transmit biological signals to regulate the quantitative output of the pathway for crosstalk with other signal transduction pathways, governing the complex life of the cells50. In the same way, in Pt3, pathways identified in interstitial fluids were principally connected to cell-cycle progression, adhesion and migration (syndecans, cytokines, Wnt), and extracellular signaling (cMET), focusing our attention on syndecans, a family of proteins regulating tumorigenesis and tumor progression interacting with cytokines and chemokines51. They can also interact with specific types of integrin, enhancing invasion phenotype and tumor growth, regulating also ErbB2 activation52,53.
The results presented in this work show that the interstitial fluids proteome is characterized by a high cellular content. It is our opinion that part of this cellular protein content is due to the invasive experimental procedure, more specifically to the fragmentation step with the scalpel. However, in a recent work, Park and colleagues have shown that, in A431 cells, hypoxic or reoxigenated cells can produce a cellular protein secretion phenomenon that modulates the tumor microenvironment composition to accelerate angiogenesis and metastatic sprouting. This hypothesis could be considered as an interesting object of future works54.
The goal of the pathway analysis was to set up a hypothesis generating approach to reveal new protein connections between the tissue-surrounding fluids and intracellular signaling pathways. Functional testing will be essential for the validation of individual protein interactions deduced by the proposed approach. In the light of the obtained results, we can conclude that the proposed approach represents an accurate and reliable method to discriminate and narrow down not only the list of proteins but also the signaling pathways that are deregulated in cancer. It is also possible to consider interstitial fluids as a very useful and rich type of sample to study cancer microenvironment, also in the light of the heterogeneity of the signaling pathways identified with respect to primary cells.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH P41 RR011823, and R01 MH067880.
ABBREVIATIONS
- TmT
tandem mass tag
- MudPIT
Multidimensional protein identification technology
- ECM
extracellular matrix
- MALDI-TOF
matrix assisted laser desorption/ionization time-of-flight mass spectrometry
- RP nano-LC
reverse phase nano-liquid chromatography
- FBS
foetal bovine serum
- IEF
capillary isoelectric focusing
- PE
phycoerithrin
- TEAB
triethylammonium bicarbonate buffer
- TNM
tumor-node-metastasis grade
- PBS
phosphate buffered saline solution
- MW
molecular weight
- pI
isoelectric point
- CID
collision induced dissociation
- HCD
higher energy collisionally activated dissociation
- EpCAM
epithelial cell ashesion molecule
- AUF
heterogeneous nuclear ribonucleoprotein
- PCNA
proliferating cell nuclear antigen
- NME3
nucleoside diphosphate kinase 3
- PTX3
pentraxin related protein
- VIM
vimentin
- SMAD
mothers against decapentaplegic homolog
- TGFβ
transforming growth factor beta
- HNRNP
heterogeneous nuclear ribonucleoprotein
- MAPK
mitogen-activated protein kinases
- PAK2
serin/threonin-protein kinase
- c-MET
hepatocyte growth factor receptor
- RPP1
ribosomal protein
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- SOD2
superoxide dismutase2
- BMP
bone morphogenic protein
- TSTD1
thiosulphate sulfurtransferase/rhodanese-like domain-containing protein
- ANXA3
annexin A3
- PDI1
protein disulfide isomerase 1
- DDHD2
phospolipase DDHD2
- CSF1R
macropahge colony-stimulating factor 1 receptor
- EIF3E
eukaryotic translation initiation factor 3
REFERENCES
- 1.Lee CH, Dershaw DD, Kopans D, Evans P, Monsees B, Monticciolo D, Brenner RJ, Bassett L, Berg W, Feig S, Hendrick E, Mendelson E, D’Orsi C, Sickles E, Burhenne LW. Breast cancer screening with imaging: recommendations from the Society of Breast Imaging and the ACR on the use of mammography, breast MRI, breast ultrasound, and other technologies for the detection of clinically occult breast cancer. J. Am. Coll. Radiol. 2010;7(1):18–27. doi: 10.1016/j.jacr.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 2.Teng PN, Bateman NW, Hood BL, Conrads TP. Advances in proximal fluid proteomics for disease biomarker discovery. J. Proteom. Res. 2010;9(12):6091–100. doi: 10.1021/pr100904q. [DOI] [PubMed] [Google Scholar]
- 3.Cichon MA, Degnim AC, Visscher DW, Radisky DC. Microenvironmental influences that drive progression from benign breast disease to invasive breast cancer. J. Mammary Gland. Biol. Neoplasia. 2010;15(4):389–397. doi: 10.1007/s10911-010-9195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiig H, Tenstad O, Iveren PO, Kalluri R, Bijerkvig R. Interstitial fluid: overlooked component of the tumor microenvironment? Fibrogenesis & Tissue Repair. 2010;3:12. doi: 10.1186/1755-1536-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teng P, Rungruang BJ, Hoods BL, Sun M, Flint MS, Bateman NW, Dhir R, Bhargava R, Richard SD, Edwards RP, Conrads TP. Assessment of buffer systems for harvesting proteins from tissue interstitial fluid for proteomic analysis. J. Proteom. Res. 2010;9(8):4161–9. doi: 10.1021/pr100382v. [DOI] [PubMed] [Google Scholar]
- 6.Wu CC, Hsu CW, Chen CD, Yu CJ, Chang KP, Tai DI, Liu WH, Chang YS, Yu JS. Candidate serological biomarker for cancer identified from the secretome of 23 cancer cell lines and the human protein atlas. Mol. Cell. Proteom. 2010;9(6):1100–1117. doi: 10.1074/mcp.M900398-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gromov P, Gromov I, Bunkenborg J, Cabezon T, Moreira JMA, Timmermans-Wielenga V, Roepstorff P, Fritzank F, Celis JE. Up-regulated proteins in the fluid bathing the tumour cell microenvironment as potential serological markers for early detection of cancer of the breast. Molecular Oncology. 2010;4(1):65–89. doi: 10.1016/j.molonc.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celis JE, Moreira JMA, Cabezon MT, Gromov P, Friis E, Rank F, Gromova I. Identification of extracellular and intracellular signaling components of the mammary adipose tissue and its interstitial fluid in high risk breast cancer patients. Mol. Cell. Proteomics. 2005 Apr.(4):570–81. doi: 10.1074/mcp.M500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Mirza SP, Olivier M. Methods and approaches for the comprehensive characterization and quantification of cellular proteomes using mass spectrometry. Physiol. Genom. 2008;33(1):3–11. doi: 10.1152/physiolgenomics.00292.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu C, McCoss M. Shotgun proteomics: tools for the analysis of the complex biological systems. Curr. Opin. Mol. Ther. 2002;4(3):242–250. [PubMed] [Google Scholar]
- 11.Xu T, Venable JD, Park SK, Cociorva D, Lu B, et al. ProLuCID, a fast and sensitive tandem mass spectra-base protein identification program. Mol. Cell. Proteom. 2006;5:S174. [Google Scholar]
- 12.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tool for assembling and comparing protein identifications form shotgun proteomics. J. Proteome Res. 2002;1(1):21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutton CW, Rustogi N, Gurkan C, Scally A, Loizidou MA, Hadjisavvas A, Kyriacou k. Quantitative proteomic profiling of matched normal and tumor breast tissues. J. Proteome Res. 2010;9(8):3891–3902. doi: 10.1021/pr100113a. [DOI] [PubMed] [Google Scholar]
- 15.Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 16.Fonslow BR, Yates JR., 3rd . Proteolytic digestion approach for shotgun proteomics. In: Pawliszyn Janusz., editor. Comprehensive Sampling and Sample Preparation: Analytical Techniques for Scientist. Vol (x) Elsevier Inc; USA: 2012. pp. (a)–(b). [Google Scholar]
- 17.Washburn MP, Ulaszek R, Ceciu C, Schieltz DM, Yates JR., 3rd Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal. Chem. 2002;74(7):1650–1657. doi: 10.1021/ac015704l. [DOI] [PubMed] [Google Scholar]
- 18.Motoyama A, Vaneble JD, Ruse CI, Yates JR., 3rd Automated ultra-high-pressure multidimensional protein identification technology (UHP-MudPIT) for improved peptide identification of proteomic samples. Anal. Chem. 2006;78(14):5109–5118. doi: 10.1021/ac060354u. [DOI] [PubMed] [Google Scholar]
- 19.Gatlin CL, Kleemann GR, Hays LG, Link AJ, Yates JR., 3rd Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal. Biochem. 1998;263(1):93–101. doi: 10.1006/abio.1998.2809. [DOI] [PubMed] [Google Scholar]
- 20.Park SK, Venable JD, Xu T, Yates JR., 3rd A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods. 2008;5(4):319–22. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glantz, Stanton A. Primer of Biostatistics. 6th Ed McGraw-Hill; 2007. [Google Scholar]
- 22.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smoot M, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croft D, O’Kellu G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jasal B, Jupe S, Kalatskaya I, Mahajan S, May B, Ndegwa N, Schmidt E, Shamovsky V, Yung C, Birney E, Hermjakob H, D’Eustachio P, Stein L. Reactome: a database of reaction, pathways and biological processes. Nucleic Acids Research. 2011;39 doi: 10.1093/nar/gkq1018. Database issue: D691-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu G, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biology. 2010;11(5):R53. doi: 10.1186/gb-2010-11-5-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scardoni G, Petterlini M, Laudanna C. Analyzing biological network parameters with CentiScaPe. BioInformatics. 2009;25(21):2857–2859. doi: 10.1093/bioinformatics/btp517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu C, Rudnick PA, Martinez MY, Cheek KL, Stein SE, Slebos RJC, Liebler DC. Depletion of abundant plasma proteins and limitations of plasma proteomics. J. Proteom. Res. 2010;9(10):4982–4991. doi: 10.1021/pr100646w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tafani M, Russo A, Di Vito M, Sale P, Pellegrini L, Schito L, Gentileschi S, Bracaglia R, Marandino F, Garagi E, Russo MA. Up-regulation of pro-inflammatory genes as adaptation to hypoxia in MCF7 cells and in human mammary invasive carcinoma microenvironment. Cancer Sci. 2010;101(4):1014–23. doi: 10.1111/j.1349-7006.2010.01493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bell CD, Tischler EM, Laroye GJ. The relationship of cytoplasmic intermediate filaments and membrane antigens with hormone receptors, nuclear staining density, and mode of stromal invasion in human breast cancer. Breast Cancer Res Treat. 1995;33(2):147–62. doi: 10.1007/BF00682722. [DOI] [PubMed] [Google Scholar]
- 30.Dandachi N, Hauser-Kronberger C, Morè E, Wiesener B, Hacker GW, Dietze O, Wirl G. Co-expression of tenascin-C and vimentin in human breast cancer cells indicates phenotypic transdifferentiation during tumour progression: correlation with histopathological parameters, hormone receptors, and oncoproteins. J. Pathol. 2001;193(2):181–9. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH752>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 31.Joshi-Tope G, Astrik I, Gopinath GR, Matthews L, Schmidt E, Gillespie M, D’Eustachio P, Jassal B, Lewis S, Wu G, Birney E, Stein L. The Genome Knowledgebase: a resource for biologists and bioinformaticists. Cold Spring Harb Symp Quant Biol. 2003;68:237–43. doi: 10.1101/sqb.2003.68.237. [DOI] [PubMed] [Google Scholar]
- 32.Mohinta S, Wu H, Chaurasia P, Watabe K. Wnt pathway and breast cancer. Front Biosci. 2007;12:4020–33. doi: 10.2741/2368. [DOI] [PubMed] [Google Scholar]
- 33.Moses H, Barcellos-Hoff MH. TGF-β biology in mammary development and breast cancer. Cold Spring Harb Perspect Biol. 2011;3(1):a003277. doi: 10.1101/cshperspect.a003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stigliano I, Puricelli L, Filmus J, Sogayar MC, Bal de Kier Joffè E, Peters MG. Glypican-3 regulates migration, adhesion and actin cytoskeleton organization in mammary tumor cells through Wnt signaling modulation. Breast Cancer Res. Treat. 2009;114(2):251–62. doi: 10.1007/s10549-008-0009-2. [DOI] [PubMed] [Google Scholar]
- 35.Coniglio SJ, Zavarella S, Symons MH. Pak1 and Pak2 mediate tumor cell invasion through distinct signaling mechanism. Moll. Cell. Biol. 2008;28(12):416–72. doi: 10.1128/MCB.01532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koh W, Sachidanandam K, Stratman AN, Sacharidou A, Mayo AM, Murphy EA, Cheresh DA, Davis GE. Formation of endothelial lumens requires a coordinated PKCepsilon-, Src-, Pak- and Raf kinase-dependent signaling cascade downstream of cdcd42 activation. J. Cell Sci. 2009;122(Pt 11):1812–22. doi: 10.1242/jcs.045799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turashvill G, Bouchal J, Burkeze G, Kolar Z. Wnt signaling pathway in mammary gland development and carcinogenesis. Pathobiology. 2006;73(5):213–23. doi: 10.1159/000098207. [DOI] [PubMed] [Google Scholar]
- 38.Taherian A, Li X, Liu Y, Haas TA. Differences in integrin expression and signaling within human breast cancer cells. BMC Cancer. 2011 Jul 13;11:293. doi: 10.1186/1471-2407-11-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Couchman JR, Chen L, Woods A. Syndecans and cell adhesion. Int. Rev. Cytol. 2001;207:113–150. doi: 10.1016/s0074-7696(01)07004-8. [DOI] [PubMed] [Google Scholar]
- 40.Elliot BE, Hung WL, Boag AH, Tuck AB. The role of hepatocyte growth factor (scatter factor) in epithelial-mesenchymal transition and breast cancer. Can. J. Physiol. Pharmacol. 2002;80(2):91–102. doi: 10.1139/y02-010. [DOI] [PubMed] [Google Scholar]
- 41.Yang N, Mosher R, Seo S, Beebe D, Friedi A. Syndecan-1 in breast cancer stromal fibroblasts regulates extracellular matrix fiber organization and carcinoma cell motility. Am. J. Pathol. 2011;178(1):325–35. doi: 10.1016/j.ajpath.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trimmer C, Whitaker-Menezes D, Bonuccelli G, Milliman JN, Daumer KM, Aplin AE, Pestell RG, Sotgia F, Lisanti MP, Capozza F. CAV1 inhibits metastatic potential in melanomas through suppression of the integrin/Src/ FAK signaling pathway. Cancer Res. 2010;70(19):7489–99. doi: 10.1158/0008-5472.CAN-10-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Outschoorn UE, Pavlides S, Howell A, Pestell RG, Tanowitz HB, Sotgia F, Lisanti MP. Stromal-epithelial metabolic coupling in cancer: integrating autophagy and metabolism in the tumor microenvironment. Int. J. Biochem. Cell. Biol. 2011;43(7):1045–51. doi: 10.1016/j.biocel.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trimmer C, Sotgia F, Whitaker-Menezes D, Balliet RM, Eaton G, Martinez-Outschoorn UE, Pavlides S, Howell A, Iozzo RV, Pestell RG, Scherer PE, Capozza F, Lisanti MP. Caveolin-1 and mitochondrial SOD2 (MnSOD2) function as tumor suppressor in the stromal microenvironment: a new genetically tractable model for human cancer associated fibroblasts. Cancer Biol. Ther. 2011;11(4):383–94. doi: 10.4161/cbt.11.4.14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D’Adamo P, Menegon A, Lo Nigro C, Grasso M, Gulisano M, Tamanini F, Bienvenu T, Gedeon AK, Oostra B, Wu SK, Tandon A, Valtorta F, Balch WW, Chelly J, Toniolo D. Mutations in GDI1 are responsible for X-linked non-specific mental retardation. Nat.Genet. 1998;19(2):134–139. doi: 10.1038/487. [DOI] [PubMed] [Google Scholar]
- 46.Lawlor K, Nazarian A, Lacomis L, Tempst P, Villanueva J. Pathway-based biomarker search by high-throughput proteomics profiling of secretomes. J. Proteom. Res. 2009;8(3):1489–1503. doi: 10.1021/pr8008572. [DOI] [PubMed] [Google Scholar]
- 47.Cosentino C, Bates DG. Feedback control in systems biology. CRC Press; Boca Raton, FL: 2011. [Google Scholar]
- 48.Gupta J, Robbins J, Jilling T, Seth P. TGF-β -dependent induction of interleukin-11 and interleukin 8 involved Smad and p38 MAPK pathways in breast tumor models with varied bone metastases potential. Cancer Biol. Ther. 2011;11(3):311–6. doi: 10.4161/cbt.11.3.14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katsuno Y, Hanyu A, Kanda H, Ishikawa Y, Aklyama F, Iwase T, Ogata E, Ehata S, Miyazono K, Imamura T. Bone morphogenetic protein signaling enhances invasion and bone metastasis of breast cancer cells through Smad pathway. Oncogene. 2008;27(49):6322–33. doi: 10.1038/onc.2008.232. [DOI] [PubMed] [Google Scholar]
- 50.Joshi A, Cao D. TGF-β signaling tumor microenvironment and tumor progression: the butterfly effect. Frontiers in Biosciences. 2010;15:180–194. doi: 10.2741/3614. [DOI] [PubMed] [Google Scholar]
- 51.Tkachenko E, Rhodes JM, Simons M. Syndecans: new kids on the signaling block. Circ Res. 2005;96(5):488–500. doi: 10.1161/01.RES.0000159708.71142.c8. [DOI] [PubMed] [Google Scholar]
- 52.Wang H, Leavitt L, Ramaswamy R, Rapraeger A. Interaction of syndecan and α6β4 integrin cytoplasmic domains. Regulation of ErbB2-mediated integrin activation. J. Biol. Chem. 2010;285(18):13569–13579. doi: 10.1074/jbc.M110.102137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iozzo R, Sanderson RD. Proteoglycans in cancer biology, tumor microenvironment and angiogenesis. J. Cell. Mol. Med. 2011;15(5):1013–31. doi: 10.1111/j.1582-4934.2010.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park JE, Tan HS, Datta A, Lai RC, Zhang H, Meng W, Lim SK, Sze SK. Hypoxic cell modulates its microenvironment to enhance angiogenesis and metastatic potential by secretion of protein and exosomes. Mol Cell Proteomics. 2010;9:1085–1099. doi: 10.1074/mcp.M900381-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.