Abstract
Pharmacology, in its broadest interpretation, is defined as the study of the interaction between physiological entities and drugs. In modern neuropsychopharmacology, this interaction is viewed as the drug itself on one side and signal transducer (receptor), the signal transduction cascade (effector proteins, second messengers), the cellular response (transcriptional regulation, activity modulation), the organ response (brain circuitry modulation), and, finally, the whole organism response (behavior) on the other. In other words, pharmacology has structured itself around the idea that the exogenous molecule (the drug) encodes a “signal” leading to everything on the other side including, in extreme renditions, a physiological response. The inference is that engaging a particular signal transduction pathway in a defined cell type leads inexorably to a prototypic physiological response. Thus, for instance, serotonergic activation of 5-HT2A receptors in rat aortic smooth muscle cells leads to an increase in intracellular Ca++ (via IP3 release) and smooth muscle contraction (Roth et al., 1986). Here, we suggest that the invention of synthetic ligand – GPCR pairs (aka DREADDs, RASSLS, ‘pharmacogenetics’) permits the study of pharmacology using a shifted equation: more of the signal transduction elements moved to the left and, subsequently, under experimental control. For the purposes of disambiguation and to clarify this new interpretation as a creation of pharmacological manipulation, we present the term pharmacosynthetics to describe what has heretofore been called pharmacogenetics or chemicogenetics. This review discusses this new interpretation and reviews recent applications of the technology and considerations of the approach.
Keywords: pharmacosynthetics, pharmacogenetics, DREADD, RASSL, synthetic biology
1. INTRODUCTION
The gap between receptor mediated signaling and the ultimate functional output of the brain is shrinking. The past decade has witnessed the advent of multiple technologies that allow exquisite manipulation of neurons in an intact animal, providing the opportunity to definitively determine the neuronal correlates of complex brain function. The primary technologies are optogenetic – the modulation of transgenic receptors and channels via photons-- and pharmacogenetic – the modulation of transgenic receptors via pharmacologic agents. This review will focus on pharmacogenetics.
First and foremost, the authors present an alternative name for this technology to disambiguate the topic from other uses of the word pharmacogenetic. The term “pharmacogenetic” is already an established MeSH (Medical Subject Headings) term, defined as “a branch of genetics which deals with the genetic variability in individual responses to drugs and drug metabolism.” This word has been well adopted, retrieving 3318 results from PubMed as of publishing date, has been in use for an extended period of time (Gonzalez-Vacarezza et al., 2012; La Du, 1972; Weinshilboum et al., 1999) and is the foundation of personalized medicine (Cohen, 1997; Kohane, 2012). For similar reasons, the term “chemical genetics” (or its portmanteau chemicogenetic), while not being assigned its own MeSH term, has been defined as “the study of gene-product function in a cellular or organismal context using exogenous ligands” (Stockwell, 2000). Here we present alternative terminology and reimagination of pharmacogenetics (the modulation of transgenic receptors via pharmacologic agents) as pharmacosynthetics. This term integrates the true meaning and functional mechanisms of the technology: pharmaco- meaning drug and -synthetic meaning the combination of two or more parts in an artificial manner. Pharmacosynthetics provides a clear distinction from both pharmacogenetics and chemicogenetics, and as of publishing date, retrieves no results in PubMed or Wikipedia.
We present the formal definition of pharmacosynthetics as “a branch of biology which deals with the creation of pharmacological modulation using artificial components”. While it is possible to equate conventional drugs with pharmacosynthetics (or having been developed through pharmacosynthesis), there are distinctions within the semantics that should be explored to provide clarification. A chemical is synthesized to have a particular pharmacology, and this pharmacology is based on the system that the chemical interacts with. On the other hand, a pharmacosynthetic approach creates a pharmacological response within a system using artificial components. While a pharmacological agent may be synthesized, at no point in this effort is the pharmacology of the agent created – instead, it is measured. In one way of thinking about it, a pharmacology (as defined as the study of drug action) is synthesized for an otherwise inert chemical by engineering a receptor and inserting the receptor into a living system. On the other hand, when a novel chemical is synthesized, its pharmacology in a living system is studied to determine whether or not it is a drug.
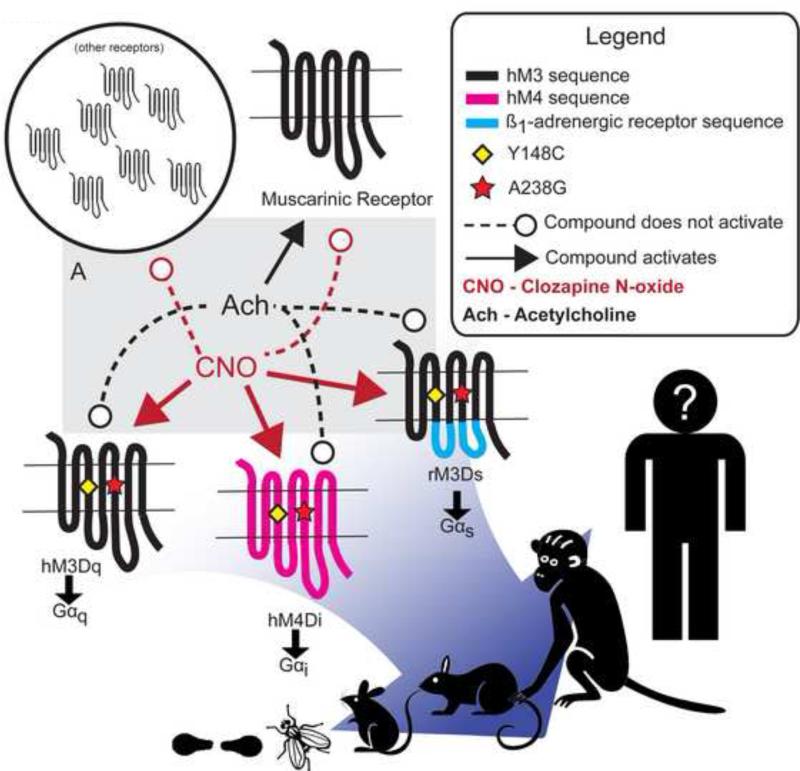
The pharmacosynthetic tools currently utilized include the Designer Receptors Exclusively Activated by Designer Drug (DREADDs), the latest iteration of a long-standing concept of creating orthologous ligand-receptor pairs to remotely control cellular GPCR signaling (Conklin et al., 2008). The original DREADDs were human muscarinic acetylcholine receptors engineered to be activated by clozapine N-oxide (CNO), an otherwise inert pharmacological agent. Additionally, DREADDs are insensitive to the endogenous ligand, acetylcholine. There are currently three DREADDs in common use – the hM3Dq that activates Gαq signaling, the hM4Di that activates Gαi signaling, and the rM3Ds that activates Gαs signaling. These three DREADDs share the same point mutations (Figure 1) that simultaneously engender CNO efficacy and acetylcholine inefficacy (Armbruster et al., 2007). The rM3Ds was engineered to couple Gαs by replacing intracellular loops 2 and 3 of the hM3Dq with those from the turkey β1-adrenergic receptor (Guettier et al., 2009). With these three DREADDs, it is possible to control the most common types of G-protein signaling found in the mammalian brain.
Figure 1. Properties and Composition of Currently Utilized DREADDs.
(A) Gray box indicates two-way selectivity component of DREADDs. Solid lines with arrows indicate that the source compound activates the receptor at the arrow's target. Dashed lines with circles indicate that the source compound does not activate the targeted receptor.
Connecting receptor mediated signaling to overt brain function is the defining challenge of neuropsychopharmacology research. The hypothesis that aberrant neuronal activity underlies neuropsychiatric disease and the knowledge that drugs modulate neuronal activity via receptors has fueled the persistence of this challenge. To date, small molecule therapeutics are the first line treatments for debilitating mental illness including schizophrenia, Parkinson's disease, and depression, to name a few. DREADDs offer a unique opportunity to study the neurophysiological correlates of therapeutic efficacy due to the nature of the DREADD technology and therapeutic mechanisms of efficacy. First and foremost, DREADDs are G-protein coupled receptors – a drug target class of which 36% of all currently approved drugs either directly or indirectly modulate (Klabunde and Hessler, 2002). Furthermore, DREADDs are modulated in a drug-like fashion since the small-molecule ligand exhibits drug-like pharmacokinetics. Finally, it has been observed that therapeutic efficacy is most often obtained through modulation of diffusely expressed albeit specific drug targets (Roth et al., 2004). These three characteristics can only be mimicked via systemic injection of drug and genomic transgene-driven dispersed expression of the DREADD. This similarity to conventional therapeutics will allow an immediate crossover of insights gleaned from research utilizing DREADDs to the physiological phenomena responsible and necessary for therapeutic efficacy.
DREADDs are capable of providing non-invasive temporal control of neuronal signaling for three important reasons that clearly distinguish DREADDs from previous pharmacosynthetic technology. The first is the two-way selectivity of the receptor-ligand pair, in that CNO does not modulate other known effectors in a biological system and that the engineered receptor is not activated by effectors present in the biological system. Secondly, DREADDs do not exhibit constitutive activity – i.e., in the absence of CNO, the DREADDs do not modulate neuronal signaling. The variant rM3Ds has been shown to exhibit constitutive activity in pancreatic beta cells (Guettier et al., 2009), though constitutive activity was not observed in striatal neurons (Farrell, In revision). Finally, the drug used to activate the DREADD is bio-available and drug-like, meaning that a simple administration method (injection, drinking water, food, etc.) can be used to modulate DREADD activity. These advancements are perhaps the most important in terms of the ultimate goal of neuropsychopharmacology, as it permits the investigation of specific signaling states on changes in overt animal behavior with minimal invasiveness.
For a review of the primary research and development of pharmacosynthetics, the reader is directed to Rogan & Roth (2011) in which the hM3Dq and hM4Di neuronal validation is reviewed in addition to the original technology development. The reader is also directed to other published reviews on DREADDs and RASSLs (Conklin et al., 2008; Dong et al., 2010; Pei et al., 2008). This review will focus on the uses of DREADDs in the intervening years and will also provide a novel perspective on this technology.
2. Gs-DREADD (rM3Ds) neuronal validation
The most recent of the DREADDs to be validated is the Gs-DREADD (rM3Ds). The rM3Ds was originally validated in pancreatic beta islet cells by Guettier et al., 2009. To determine whether the rM3Ds could modulate neuronal Gs-type signaling, Farrell et al, (In revision) created a transgenic mouse expressing the rM3Ds in the striatum. The striatum is a nucleus of the basal ganglia, an area of the brain responsible for volitional movements and reward processes (DeLong and Wichmann, 2009). The nucleus is comprised of two distinct populations of medium spiny neurons (MSNs) that project to unique downstream basal ganglia nuclei. Additionally, these two populations have unique cellular expression profiles. The striatonigral MSNs project to the internal globus pallidus and substantia nigra pars reticulata and express dopamine D1 receptors, substance P and dynorphin peptides, whereas the striatopallidal MSNs project to the external globus pallidus and express dopamine D2 receptors and enkephalin peptides (Gerfen et al., 1990; Svenningsson et al., 1998). Activity of the striatopallidal MSNs inhibits behavior (DeLong, 1990; Kravitz et al., 2010) and Gs-type signaling in MSNs is thought to promote neuronal excitability (Centonze et al., 2001; Surmeier et al., 2007). The rM3Ds was expressed in the striatopallidal MSNs using the adenosine A2A bacterial artificial chromosome to create the adora2A-rM3Ds mice. The transgene contains a mCherry fluorescent reporter downstream of an internal ribosomal entry site (IRES), and expression targeting of the transgene was confirmed by crossing the adora2A-rM3Ds mice with the GENSAT Drd2-GFP and Drd1a-GFP reporter mice. Biochemical studies confirmed that the rM3Ds signals through canonical Gs-signaling cascades in these mice by quantifying the phosphorylation of DARPP-32 at Thr34, a well-studied downstream target of the Gs / cAMP / PKA signaling cascade in MSNs (Ouimet et al., 1984; Svenningsson et al., 2004). Administration of CNO caused a significant inhibition of novelty-induced locomotor activity and a dose-dependent decrease in spontaneous locomotor activity during the dark phase. These studies confirm that the rM3Ds signals via canonical Gs-type cascades in a neuronal environment.
The validation study was extended by testing the effects of rM3Ds activation in an animal model of addiction. Drugs of abuse modulate the biochemical processes of the basal ganglia, with dopaminergic signaling being of unique significance for psychostimulants. This class of drugs, which include the drugs methamphetamine, cocaine, and recently popular mephedrone, directly increase the amount of dopamine present in the basal ganglia (McKenzie and Szerb, 1968; Sulzer, 2011). Psychostimulants cause a hyperlocomotor response in rodents, and repeated administration of a constant dose causes an enhanced locomotor response. This increased locomotor activity in response to a constant dose of psychostimulant is a manifestation of behavioral sensitization, which has become a well-studied animal model of addiction (Steketee and Kalivas, 2011). While the exact neuronal correlates of sensitization are unknown, the role of dopaminergic signaling in the MSNs has been heavily implicated (Narendran and Martinez, 2008) . Overactivity of the D2 receptor in the striatopallidal MSNs could, for example, induce neuronal modifications that cause these neurons to become less sensitive to neuronal input, thus causing an increased behavioral response to the same dose of drug. The dopamine D2 receptor inhibits cAMP production via Gi modulation, so it was hypothesized that increasing cAMP signaling via rM3Ds-Gs activity would counteract the effects of repeated amphetamine treatment. When CNO was co-administered with amphetamine for 5 consecutive days, the adora2A-rM3Ds mice did not develop locomotor sensitization. After 10 days of no drug treatment, mice were injected on day 15 with amphetamine alone and their locomotor activity recorded. The adora2A-rM3Ds mice that had been treated with amphetamine and CNO did not show sensitization expression. Furthermore, measurements of striatopallidal MSN excitability confirmed that CNO co-administration reversed changes in membrane conductance caused by amphetamine treatment. The behavioral data recapitulate endogenous Gs signaling phenomena, as it has previously been shown that pharmacological modulation of Gs signaling in these neurons via the adenosine A2A receptor can inhibit amphetamine sensitization (Shimazoe et al., 2000). Thus, these data provide further validation that the rM3Ds signals through canonical Gs-signaling pathways in a neuronal context and demonstrate the utility of the DREADD technology in the study of disease etiology.
3. Application of pharmacosynthetics
3.1. Selective mimicry of endogenous receptors
One application of pharmacosynthetics is to use them as very selective pharmacological agents. In this manner, DREADDs can be expressed in a neuronal population that matches a pharmacologically intractable endogenous receptor. The Gs-DREADD validation study described above can be viewed as an example of this type of application, in which the DREADD enabled selective modulation of a distinct population of neurons residing in a nucleus of heterogenous neuronal composition. In this study, the Gs-DREADD was driven by the adora2A gene in a bacterial artificial chromosome. In the mouse genome, the native adora2A gene drives the expression of the adenosine A2A receptor, which itself is a Gs-coupled receptor. To date, the availability of selective adenosine A2A agonists is limited, and CGS 21680 is the most commonly used. Thus, by mimicking the expression of the adenosine A2A receptor with the Gs-DREADD, we essentially created a highly selective adenosine A2A receptor ligand-pair. Utilizing the DREADD as a selective mimicry is limited, though, in that the DREADD must be of the same coupling-type as the endogenous receptor for which it is mimicking. In the instance above, the Gs-DREADD and the adenosine A2A receptor both couple to Gs signaling pathways. Indeed, the advent of functional selectivity (a.k.a agonist-directed trafficking, biased agonism) introduces complications (or potential benefits) that will be discussed in section 5.1. One can imagine applying pharmacosynthetics in this manner to study the effects of orphan receptor modulation, receptors with unique neuronal expression patterns with no known modulators.
3.2. Creating drug-like modulation where none exists
Additionally, the DREADD system can be conceptualized of as a way to pharmacologically modulate spatially defined neuronal populations for which no pharmacological modulatory agents exist. Krashes et al. (2011) used DREADDs in this fashion to study the arcuate nucleus (ARC) of the hypothalamus. This nucleus has been implicated in regulating energy homeostasis and has therefore been a focus of study for the understanding and treatment of obesity, with particular focus on agouti-related protein (AgRP) neurons expressed in this nucleus. To date, investigative efforts into the function of these neurons have been limited to conventional genetic, invasive, and ablative approaches: overexpression of AgRP in transgenic mice, central administration of peptides, and ablation of AgRP neurons. The first approach removes the temporal specificity required for definitive experimentation, whereas the latter approaches introduce confounds associated with non-reversible and invasive administration techniques. Krashes et al. (2011) utilized DREADD technology to study the acute effects of AgRP neuronal activity. The hM3Dq DREADD was targeted to AgRP neurons using a Cre-recombinase dependent adeno-associated virus (AAV) injected into AgRP-Ires-cre mice. Following i.p. administration of CNO (0.3 mg/kg), hM3Dq-expressing mice began feeding and ate almost four times as much food than control mice in the first half hour. Additionally, this group infected the same neurons with the hM4Di to induce neuronal silencing and observed a decrease in food intake. This study demonstrates that DREADDs can be used to introduce pharmacological modulation to nuclei for which drug-like compounds do not exist.
A similar approach was used to modulate orexin neurons in the lateral hypothalamic area. These neurons have been implicated in sleep and wakefulness and produce neuropeptides orexin A and orexin B. As mentioned above, the pharmacological modulation of neuropeptide systems is historically difficult due to issues of invasiveness (local microinjection of purified peptide), lack of temporal control (genetic modulation) or off-target effects (physical ablation). Using virally mediated gene transfer, Sasaki et al. (2011) was able to express the hM3Dq and hM4Di in the orexin neurons of the lateral hypothalamic area. Following intraperitoneal administration of CNO (5.0 mg/kg) during the light phase (when mice typically sleep), the percent of wakefulness during the following hour was significantly greater and the NREM time was significantly shorter. Similarly, administration of CNO during the dark phase (when mice are typically awake), caused significant increase in wakefulness. Conversely, administration of CNO to mice expressing the hM4Di (the inhibitory DREADD) in the orexin neurons decreased wakefulness during the dark phase and the light phase.
Introducing pharmacological modulation to non-druggable neuronal populations was also demonstrated by Ray et al. (2011). Here, the hM4Di (referred to as Di in the report) was expressed in serotonergic neurons of the brainstem using conditional intersectional genetics. In normal mice, an increase in the amount of carbon dioxide inhaled causes an increase in breathing. Upon administration of CNO (10 mg/kg), the hM4Di transgenic mice had a blunted response to increased CO2 concentrations. These data indicate that the serotonergic neurons of the brainstem are involved in the modulation of respiratory activity. Additionally, they found that CNO injection decreased oxygen consumption in room air independent of ventilation modulation, indicating that these neurons can modulate metabolic rate independent of respiration. Furthermore, this group found that modulation of these neurons caused long-term changes in body temperature (up to 10 hours in duration), and that this effect became desensitized following repeated CNO treatment.
3.3. Encoding and modulating diffuse neuronal ensembles
A very elaborate application of pharmacosynthetics was performed by Garner et al. (2012) in which the neuronal ensemble encoding a memory trace was captured and recreated by DREADDs. To achieve this engram capturing ability, a transgenic mouse was designed to express the hM3Dq in active neurons during a conditioning task. This was achieved using a c-fos promoter-driven tTa transgenic mouse crossed with the TRE-hM3Dq mouse. The protein c-fos is a transcription factor that is upregulated in response to increases in neuronal activity. Thus, by using the c-fos promoter sequence, the active neurons of transgenic mice are labeled with hM3Dq receptor. These labeled neurons can then be activated via CNO administration to re-create the neuronal activity pattern responsible for the initial neuronal labeling. Garner et al. used these mice to determine whether a synthetic memory trace could be formed and re-created using pharmacosynthetics. To do this, they used a fear conditioning protocol in which a mouse is electrically shocked in a novel environment. In this paradigm, a mouse learns very quickly that particular environments are associated with an electric shock, and when later placed in the shock-paired environment will “freeze” – a well characterized behavior associated with learned fear. During these sessions, the neurons involved in the formation of these memories will become more active and thus express the hM3Dq. Interestingly, they found that the synthetic memory trace encoded by the DREADD-tagging could not produce a fear response on its own. The DREADD-encoded memory trace, however, was powerful enough to interfere with a naturally encoded memory trace. This study exemplifies the ability of pharmacosynthetics to modulate a diffuse but specific population of neurons.
3.4. Long-lasting specific neuronal modulation
A useful aspect of pharmacological modulation is the duration of activity inherent in the pharmacological approach. In addition to the pharmacokinetics of CNO activation, the hM3Dq, hM4Di, and rM3Ds have been shown to produce long-lasting neuronal modulation exceeding what can be explained by pharmacokinetics alone (see table 1 for summary). This longevity of action can be utilized in studies requiring chronic modulation of neuronal activity. For example, to determine the influence of neuronal circuit activity on excitatory synaptogenesis in medium spiny neurons, Kozorovitskiy et. al (2012) administered a chronic regimen of CNO (1.0 mg/kg, twice-daily, from post-natal day 8-15) to mice pups that had been infected at post-natal day 0-1 with an AAV carrying a cre-dependent hM4D. In this study, D1 and D2-Cre mice were used to target expression of the hM4Di in the two primary neuron populations of the striatum. The chronic regimen of CNO administration, theorized to produce long-lasting neuronal activity modulation, dampened synaptogenesis of direct or indirect pathway MSNs as measured by mEPSC frequency and spine density on day 15 in mice expressing hM4Di throughout the striatum. In mice expressing the hM4Di unilaterally in a more dispersed manner (theorized to not effect circuitry activity), there was no effect of CNO on synaptogenesis. To determine whether the corticostriatal neurons (those that project to the striatum from the cortex) could influence synaptogenesis, the hM4D AAV was injected to the Rbp4-cre mouse, a mouse expressing Cre in corticostriatal neurons. These mice were treated with the same chronic regimen of CNO. On day 15, decreased mEPSC frequency and spine density was observed, indicating a decrease in synaptogenesis. Similar results were also observed when measured at postnatal day 25-28, indicating these changes persist into adulthood. This study demonstrates the ability of the pharmacosynthetic approach to provide chronic neuronal modulation.
Table 1. Summary of neuronal DREADD applications to date.
This table provides a quick reference for all neuroscience research performed to date that utilize DREADD pharmacosynthetic technology.
| Publication | DREADD used | Expression System | Vector Tissue Targeting Mechanism | Expression Localization | Dose used (mg/kg) | Behavior effect at highest dose | Physiological effect reported at highest dose (in vivo) | Longest Measured Duration of Activity |
|---|---|---|---|---|---|---|---|---|
| Alexander 2009 | hM3Dq | genomic transgene | CamKII-alpha promoter | Cerebral neurons | 0.03 - 5.0 | Hyperlocomot or activity (0.3) | Status epilepticus (> 0.5) | 9 hours |
| Ferguson 2011 | hM4Di | Viral | Enkephalin promoter in virus | Striatopallidal MSNs | 1.0 | Enhanced amphetamine sensitization development and expression | Decreased number of evoked action potentials | |
| Dynorphin promoter in virus | Striatonigral MSNs | 1.0 | No effect on development, but mice did not express sensitization on challenge day | |||||
| Localized injection | VTA | 3.0 | Dopamine transmission inhibition | |||||
| Geuttier 2009 | hM3Dq rM3Ds |
Genomic transgene | 650 bp fragment of Rat insulin promoter | pancreatic Beta cells pancreatic Beta cells |
0.0001 - 10.0 | None reported | Decreased blood glucose, increased plasma insulin levels | 8 hours |
| Krashes 2011 | hM3Dq | Viral transgene hybrid | Cre-dependent virus in Agrp-ires-Cre transgenic mice | Agouti-related peptide expressing neurons in arcuate nucleus | 0.3 | Increased feeding, increased breakpoint for food reward, increased food-seeking behavior | Increased c-fos | 4 hours |
| hM4Di | Decreased feeding | 4 hours | ||||||
| Sasaki 2011 | hM3Dq | Viral transgene hybrid | Cre-dependent virus in orexin-cre transgenic mice | Orexin expressing neurons of lateral hypothalamis area | 5.0 | Increased wakefulness | Increased c-fos, decreased NREM and REM sleep time. | 1-2 hours |
| hM4Di | 5.0 | Decreased wakefulness | Decreased c-fos, increased NREM sleep time | 1-2 hours | ||||
| Ferguson 2012 | hM4Di | Viral | Enkephalin promoter on virus | Striatopallidal MSNs | 1.0 | Impaired action-outcome learning | None reported | 1 hour |
| Garner 2012 | hM3Dq | Genomic transgene | c-fos driver | Diffuse c-fos expressing neurons | 5.0 | Memory interference | Increased spiking activity | 1 hour |
| Farrell 2012 | rM3Ds | Genomic transgene | adora2A-Bac | Striatopallidal MSNs | 0.01 - 5.0 | Decreased locomotor activity, inhibited amphetamine sensitization development | Increased DARPP-32 phosphorylation | 2 hours |
| Kozorovitskiy 2012 | hM4Di | Viral transgene hybrid | Dl-Cre, D2-Cre, adora2A-Cre mice, Rbp4-cre | Striatonigral, striatopallidal, corticostriatal neurons | 1.0 | None reported | Decreased synaptogenesis | |
| Ray 2011 | hM4Di | Genomic transgene | Slc6a4-Cre | Serotonergic neurons | 10.0 | None reported | Reduced ventilation response, hypothermia | 10 hours |
4. Considerations and Implications of the above studies
Pharmacosynthesis requires consideration of multiple factors to be utilized effectively. The key elements to be considered are the expression of DREADD and the dose of CNO required for experimental manipulation. Here we provide a primer on the consideration of these elements.
4.1. Expression systems
The primary challenge in pharmacosynthetics is inserting the DREADD receptor into the desired tissue of the model organism. To date, this has been achieved using virally mediated gene transfer and genomic insertion of a transgene. Although a full review of each approach is beyond the scope of this manuscript, the benefits and complications of each approach will be briefly discussed.
4.1.1. Virally Mediated Gene Transfer
Virally mediated gene transfer utilizes viral mechanisms to insert the DREADD DNA into a neuronal population. There are many different types of viruses available, each having their own benefits and drawbacks. The reader is directed to reviews and applications of viral vectors and their utility (de Silva and Bowers, 2011; Foust et al., 2009; Teschemacher et al., 2005; Weinberg et al., 2012; Wu et al., 2006). The primary benefit of the viral approach is the relative turnaround between project conception and functional animal model expressing the DREADD. In addition to the expediency provided by this approach, the local microinjection of viral particles enhances the spatial specificity of DREADD expression. Finally, the viral approach permits utilization of the DREADD in model organisms for which transgenic approaches are not available or widespread (e.g., rats, monkeys). The drawbacks of the viral approach arise from the nature of local microinjections and the size limitations of viral packaging. This vector delivery method is invasive, potentially inducing an immune response and causing damage to tissue, including cell populations either directly or indirectly involved in the scrutinized output. Additionally, the spatial resolution provided can also be a limitation, in that DREADD expression is limited to the number of microinjection sites and the spread of viral particles. This latter point, however, can be a benefit depending on the goals of the study. Furthermore, the expression pattern of DREADDs between animal subjects will not be precisely identical due to differential stereotactic coordinate alignment, inconsistent viral diffusion, and experimental variation. Finally, viruses are only capable of carrying a certain quantity of DNA, potentially limiting the addition of desirable vector traits including targeting information (promoter sequences) and cell-type markers (fluorescent proteins).
4.1.2. Genomic Insertion of Transgene
Genomic insertion of a DREADD-encoding transgene involves the creation of a new line of genetically modified mice or the selective breeding of extant transgenic lines. The benefits of the transgenic approach are inherent to genome-level insertion of a transgene. First and foremost, the transgenic line of mice carries the transgene and expresses the DREADD in accordance to the genetic information in the transgene. Thus, no invasive interventions are necessary to obtain DREADD expression, providing a truly non-invasive means of cell-type specific signaling. Secondly, DREADDs can be expressed in a more dispersed manner using a transgenic approach. Whereas virally mediated gene transfer can only provide expression of the DREADD in the range of viral diffusion, the transgene inserted into the genome is present in all cells. Expression of the DREADD is dependent upon the information contained in the transgene, and while the genetic sequences that confer cell-type specificity of expression are still a matter of research, certain promoter sequences have been determined. Thus, while the noninvasive component is definitively beneficial, the utility of dispersed expression patterns is dependent upon the research goals.
One type of transgenic approach is to create a new transgenic line to suit the needs of the intended research. When creating a new line of DREADD transgenic mice, the DREADD-encoding construct is inserted downstream of a promoter that is useful for the research goal. For instance, the Gs-DREADD mice described above utilized the adenosine A2A receptor BAC to drive DREADD expression in striatopallidal medium spiny neurons, an area where the adenosine A2A receptor is naturally enriched. This permitted for the selective activation of Gs signaling in those neurons in mice carrying a single transgene. One benefit of the single transgene approach is breeding efficiency: hemizygous breeding strategies can produce 50% transmission with the end result being 50% of the litter can be used for study. Additionally, because the cell-type specificity is transmitted in one transgene, these mice can be easily crossbred with other types of engineered mice. The downside of this approach is that these single-transgene mice can only be used to study the cell population originally intended. An additional downside is that efforts to identify the genetic information responsible for tissue targeting have been, for the most part, unsuccessful, though a select few promoters have been commonly used in neuroscience research. One way to overcome this lack of knowledge is to knock-in the transgene, such that the endogenous promoter for a given neuronal subtype carries the DREADD sequence in addition to the endogenous gene product. To avoid interfering with a particular genomic locus, an alternative transgenic strategy is to create a transgene containing the entirety of the genetic information associated with a particular cell-type specific protein. This approach can be achieved by using bacterial artificial chromosomes (BACs), which are capable of carrying 200-300 kb of genetic information, a drastically larger amount than other transgenic approaches utilize (Heintz, 2001). For instance, the camKII-alpha promoter sequence is 8.5 kb (Tsien et al., 1996), whereas the adora2A BAC is 175 kb. Finally, creating a transgenic mouse is both resource intensive and the effort has no guaranteed yield, depending on the strategy.
Alternatively, targeting DREADD expression to a cell type of interest can be achieved using conditional intersectional genetics. This approach involves the selective breeding of extant transgenic lines to produce mice carrying multiple transgenes; i.e., polytransgenic mice. In these mice, the expression specificity of the DREADD depends on the design of the transgenes, an approach called “intersectional genetics”. A handful of technologies are available for this approach and the number of transgenic mice carrying these technologies is ever expanding. Prominently in use are the Cre, Flp, and Tet technologies, all of which are based on proteins and DNA sequences exogenous to the targeted model organism (in this case, the mouse) (Mallo, 2006). The Cre and Flp systems are based on the recombinases and their associated DNA targeting sequences. In these systems, one mouse carries a transgene for the Cre or FLP protein in which the protein sequence is downstream of a promoter that drives expression in a particular cell population (cell population A). A second mouse has a separate transgene, in which the DREADD sequence is inserted downstream of a ubiquitous promoter followed by a stop cassette that is flanked by either the Cre excision sites (LoxP) or the Flp excision site (Flpe). Thus, in the cell population that expresses Cre or Flp, the stop cassette is removed from the genetic sequence. The nuclear expression machinery can then translate the DREADD sequence into protein in that cell population. In the cells that do not express Cre or Flp, the DREADD transgene remains silent due to the presence of the stop cassette. The Cre / Flp systems can also take advantage of a second phenomenon of the recombinatorial proteins in that they can reverse the direction of the sequence between the excision sequences. Dependent upon the orientation of the excision sequences, the Cre and Flp can either excise the bookended DNA or flip the direction. The “reversal” approach is less leaky than the excision method; i.e., the intended specificity of expression is more likely to occur.
In the Tet system, one mouse is designed with a transgene containing the tet trans-activator (Tta) sequence downstream of a cell-type specific promoter. A second mouse is designed with a transgene containing the DREADD sequence downstream of the tet-response element (TRE) sequence. When the two mice are interbred to create double-transgenic offspring, the tet trans activator binds to the tet response element and permits DREADD expression. This system also permits for temporal control by the administration of doxycycline to the organism. Doxycycline binds to the trans activator and blocks its interaction with the tet response element, ultimately inhibiting expression of the DREADD. Alternatively, the reverse trans activator (rTta) can be used, in which doxycycline administration is necessary for the interaction of the trans activator and the response element. In this version of the system, doxycycline can be administered to induce the expression of the DREADD. The tet system has been enhanced recently with the tet-on 3G system (Clontech), but the underlying principles remain.
4.1.3. Combinatorial approaches
Already the world of neuroscience research is seeing the full implementation of these technologies and the benefits of combining them. For example, the Krashes et al. (2011) and Sasaki et al. (2011) studies combined the specificity of expression provided by the genomic transgene approach with the spatial resolution and quick turnaround of the viral approach to achieve cell-type specific neuronal modulation. The Ray et al. (2011) study used intersectional genetics to increase the specificity of DREADD expression with minimal invasiveness. At this point, the ability to target DREADD expression to specific tissue populations depends on the transgenic state of the art.
4.2. CNO Doses and Routes of Administration
In the articles mentioned above, a range of CNO doses is used to obtain effects at the whole-organism level, and these doses are summarized in Table 1. The dose of CNO required varies depending on the expression system, localization of expression and type of DREADD. For example, the hM3Dq DREADD is very effective at depolarizing neurons, and thus, relatively low doses can be used to elicit an effective neuronal response (Alexander et al., 2009) . On the other hand, the hM4Di is reportedly less effective at inhibiting neuronal firing, so higher doses of CNO are occasionally used (Ray et al., 2011; Sasaki et al., 2011). Beyond the inherent differences between Gq and Gi modulation of neuronal excitability, the effects of the CNO-DREADD mediated manipulation depend on the tissue distribution of the DREADD. For example, the hM3Dq mice originally characterized express the Gq DREADD in all neurons of the cerebrum using the calmodulin II kinase alpha promoter, creating a condition in which low doses of CNO could influence neuronal activity on a large scale (Alexander et al., 2009). Conversely, the transgenic mice used in Garner (2012) used the c-fos promoter to drive expression of the hM3Dq, so the DREADD was expressed at lower levels and higher doses of CNO were necessary. Furthermore, Krashes (2011) used a viral approach to express the hM3Dq in a small nucleus and administered 5.0 mg/kg CNO to elicit a response in these mice. From the body of work performed with DREADDs to date, it can be seen that the dose of CNO is variable and dependent on the type of DREADD and the expression system used.
To date, a majority of studies performed have used the intraperitoneal route of administration, though other routes of administration are possible. Our lab has demonstrated that CNO can be administered through the drinking water to create chronic administration conditions (10 mg/kg/day, unpublished observations).
5. Untapped Utility of pharmacosynthetic Cell-type specific modulation
Pharmacosynthetics has untapped potential. The utilities not yet applied are inherent in the nature of GPCR signaling in general and that of the pharmacosynthetic approach itself. With the advent of more specific cell-type expression and measurement systems, DREADD technology can be utilized to probe the mechanisms of pharmacotherapeutic efficacy and the nature of GPCR-induced neuronal modulation. Here we will discuss currently underutilized aspects of pharmacosynthetics.
5.1 Non-interfering modulation
An overlooked aspect of the pharmacosynthetic approach is the lack of interference with endogenous signaling. Whereas the noninvasive aspect relates to the physiological benign approaches to DREADD expression (transgenic mice), this lack of interference relates to the nature of the experimental manipulation.
It has been posited elsewhere that ultimate function of the brain arises from the collection, transmission, and integration of information (deCharms and Zador, 2000; Rolls and Treves, 2011). The brain encodes this information in the biochemicoelectric phenomena of neurons, with the transmission and integration occurring through the function of action potentials, neurotransmitters, and receptors. Various nuclei in the brain have been implicated in the etiology of disease and the mechanism of action of therapeutics. Due to the limitations of conventional approaches, it is difficult to separate the role of a nuclei's transmission, integration, or generation of information in the etiology of the associated diseases. For example, in a standard pharmacological approach, a small molecule ligand would be used to modulate a particular receptor. In addition to inherent off-target confounds this approach, receptor theory posits that any small molecule will compete with the endogenous ligand for that receptor, ultimately functioning as an antagonist of the endogenous tone. This confound applies to allosteric modulation as well. Whereas measurements from such a study would implicate the role of receptor-mediated changes in the postsynaptic neuron, the phenomena observed may be due, in part, to interference with the endogenous tone of ligand-receptor signaling. Thus, the interpretation of such a study would not be able to resolve whether the experimental manipulation modulated the integrator and transmitter of information (the post-synaptic cell receiving input) or the information itself (the endogenous tone).
Pharmacosynthetics circumvents this confound by utilizing an exogenous receptor, leaving the endogenous tone intact. This aspect of the DREADDs has been studied in vitro, where the hM3Dq (with or without CNO) had minimal or no effect (dependent on measurement system used) on the quaternary organization of wild-type and DREADD variants (hM3 and hM3Dq) of the human muscarinic receptor (Alvarez-Curto et al., 2010). Furthermore, the inert clozapine N-oxide does not interfere with endogenous receptor signaling. In this manner, the experimenter can specifically modulate the neuronal nuclei in question, independent of the information transfer. This non-interference of experimental manipulation has yet to be explicitly utilized or considered in experimental design using the DREADDs.
5.2 Cell-type specific GPCR signaling vs. “activation” and “silencing”
A majority of the studies to date have been designed and the data interpreted in the context of DREADD-induced activation or silencing of neuronal activity. Whereas a result of Gαq-coupled GPCR activation is depolarization and a result of Gαi-coupled GPCR activation is hyperpolarization, these electrophysiological endpoints are only one result of GPCR signaling pathways (Beaulieu and Gainetdinov, 2011; Deneris and Wyler, 2012). In the pharmacosynthetic field, other physiological endpoints have heretofore been, for the most part, overlooked. G-protein pathways are involved in a myriad of neuronal functions, including gene regulation (West et al., 2002). Indeed, whereas the straightforward interpretation and design of Gαq-mediated depolarization and Gαi-mediated hyperpolarization is pragmatic for studies to date, the inherently metabotropic nature of GPCR signaling needs to both be utilized and taken into account when considering pharmacosynthetics for experimental manipulation.
An application of pharmacosynthetics that would utilize this facet of DREADDs is determining the particular G-protein signaling necessary for therapeutic efficacy. The advent of functional selectivity has caused a sea change in our understanding of GPCR function, in that the simple distinction of ligands as agonists, antagonists, and inverse agonists no longer exists (Allen and Roth, 2011; Urban et al., 2007). Instead, it is now appreciated that a particular small molecule can impart intracellular signaling entirely dependent on the signaling machinery present in a given cell type. Thus, a given “agonist” to a receptor in cell population A can induce receptor-mediated signaling, whereas the same “agonist” at the same receptor in cell population B can have no effect or a different effect entirely. Furthermore, the observed phenomena to date suggest that the signaling induced upon ligand binding is dependent on the small molecule - a particular “agonist” X to a receptor can induce a particular GPCR-mediated signaling phenomenon (such as cAMP accumulation), whereas another “agonist” Y at the same receptor can cause entirely different GPCR-mediated signaling phenomenon (such as beta-arrestin signaling). This “functional selectivity” of small molecule ligands opens a new chapter in small molecule drug discovery for G-protein coupled receptors, an already well-validated drug targeted.
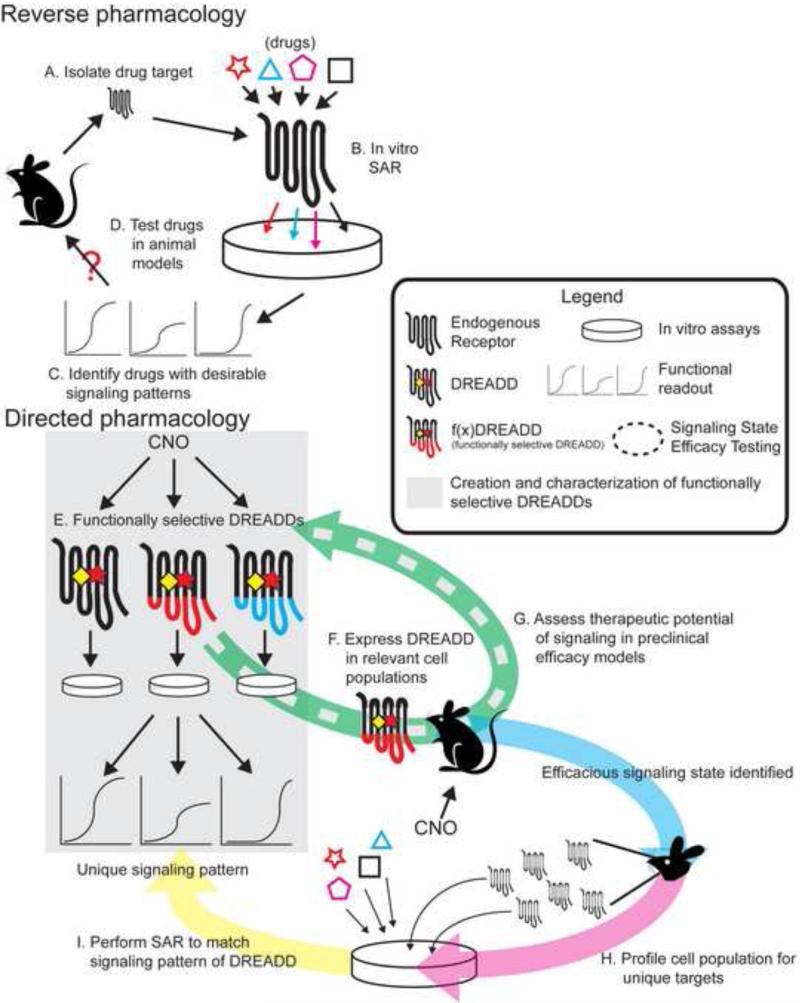
This new effort is hindered by an unfortunate fusion of the phenomena to be exploited and the nature of modern drug discovery efforts. To date, a majority of drug discovery efforts use cultured cell populations as a model system to study GPCR signal transduction. In what is termed “reverse pharmacology”, a receptor is isolated from the organism and its signal transduction properties are studied in the cultured cell populations (Figure 2, top). Due to the implications of functional selectivity, these cultured cell populations, in combination with the type of GPCR being studied and the chemical space of the drug, synergize to produce the signaling phenomena observed. Thus, the observed phenomena are dependent upon the model cell system used. Any subsequent translation of a drug's function to the whole organism is due to fortuitous similarity of cellular phenotype between the model system and the whole organism. While reverse pharmacology has created a wealth of information regarding the relationship between the structure of a small molecule ligand and the response of the receptor, the translation of these findings to the whole organism, in terms of therapeutic efficacy, has been less fruitful. A well-known contributor to inverse pharmacology's lack of success is that these chemicals can have off-target effects when reintroduced to the whole organism. While this confound is measurable and perhaps rectified with further compound development, a second unmeasurable confound is the differential receptor function in the native cellular environment compared to the cultured cell. Whereas a chemical may induce a unique signaling state when it is bound to a receptor in the model cell culture system, the native neuronal environment of the receptor may not have cellular factors capable of recognizing the signal being transduced by the chemical-receptor complex (for review see Allen and Roth 2011).
Figure 2. An application of pharmacosynthetics in drug discovery.
(A-D) Conventional drug discovery efforts – reverse pharmacology. In this approach, a drug target is selected (usually based on previous success at that particular target) and isolated from the organism. A particular type of signaling is hypothesized to be efficacious, and chemicals are created that cause the receptor to produce that signaling state in cultured cells. The drug is then reintroduced to the model organism and tested for therapeutic efficacy. (E-I) A theoretical workflow of drug discovery efforts using pharmacosynthetics, here termed direct pharmacology. In this approach, functionally selective DREADDs ( f(x)DREADD, where x=signaling type) are created by modifying DREADDs to modulate distinct signaling phenomena in cultured cells while maintaining their DREADD properties. These f(x)DREADDs are then expressed in therapeutically relevant neuronal populations. These mice are then tested in animal models of therapeutic efficacy. If a particular f(x)DREADD in a particular neuronal population is efficacious, then that cell population is profiled to find druggable targets. These druggable targets are then isolated (expressed in cultured cells) and chemicals are created that modulate these targets to reproduce the signaling state created by the f(x)DREADD. Chemicals that successfully recapitulate the f(x)DREADD-induced signaling are then tested in animal models of efficacy.
Pharmacosynthesis creates a means to overcome this confound. The opposite approach to reverse pharmacology can be termed “directed” pharmacology, in which the signaling of a particular receptor is designed and characterized in the cultured cell system and then introduced into the whole organism (Figure 2, bottom). An aspect of the DREADDs not fully appreciated is the fact that mutations used to engender CNO modulation and rectify endogenous neurotransmitter modulation are functionally benign. That is, the receptor behaves for all intents and purposes identical to the wild-type receptor (Alvarez-Curto et al., 2011). This facet of the DREADDs permits manipulation of intracellular receptor components that can affect the coupling of the receptor to downstream effectors. Guettier et. al. (2009) recognized this and swapped the intracellular loops of the hM3Dq with the loops of the turkey β1-adrenergic receptor to confer Gs-coupling of the DREADD. Although the exact mechanism of ligand induced GPCR activation is unknown, is has been hypothesized to be a shared mechanism across GPCRs. For instance, many groups have reported creating chimeras of the transmembrane and extracellular portions of GPCR type A with the intracellular portions of GPCR type B to confer type B receptor signaling following application of ligands for receptor type A (Kim et al., 2005; Marion et al., 2006; Oh et al., 2010). If this holds true across receptor types, one can hypothesize that the intracellular components of a DREADD can be replaced with those of a different receptor to confer CNO modulation of that receptor's signaling. DREADDs could be designed to modulate unique and specific signal transduction pathways in cell culture systems, and then these DREADD variants could be inserted in vivo to determine whether the designed functional selectivity translates into usefully different neuronal signaling and function (Figure 2E-G). Because the specificity of CNO for the DREADD is pharmacologically unprecedented, this approach would allow us to introduce designer signaling into specific cell types and study the subsequent physiological response to this signaling. Indeed, recent efforts have culminated in the creation of optically activated β-arrestin functionally-selective DREADDs (Lee and Roth, in preparation) which allow for the precise spatio-temporal control of arrestin signaling.
In one sense, this potential application can invert our standard means of molecular pharmacology. Instead of isolating the signal transduction device (the GPCR) and studying its effects on non-native signal transduction cascades (Figure 2A,B), we can now study the ligand-receptor complex as a single entity. The results of these studies will be correlative information between the signaling transduction induced by CNO/DREADDs in model cell systems and the physiological response of the CNO/DREADDs in the whole organism (Figure 2E,F). These data could then be used to design small-molecule ligands (or collections of small molecule ligands) that mimic the CNO/DREADD signaling phenomena observed in the cultured cell system (Figure 2I). This approach would essentially invert the process of drug discovery: instead of designing a chemical to elicit an efficacious physiological response, we could directly design an efficacious signaling state and then create a chemical (or chemicals) that can recapitulate the designed state. To our knowledge, only the DREADD technology provides this utility, capitalizing on the drug-like modulation of selective but dispersed neuronal populations.
6 Future Developments Necessary for Increased Utility
6.1 A non CNO-based DREADD
First and foremost, the development of a second, non CNO-based DREADD would be the most advantageous development to further our neuropharmacological understanding of the brain. The availability of an additional DREADD could permit the mapping of functional neuronal circuits, for example, by placing an excitatory non CNO-based DREADD upstream of a nuclei modulated by the hM4Di. In this fashion, one could determine the functional involvement of a series of nuclei posited to be integral for a given neuronal circuit. Indeed, we have recently created a new Gi-biased DREADD to this end (Vardy et al, in preparation).
6.2 Enhanced genetic expression strategies
The cell-type specificity afforded by the pharmacosynthetic approach is dependent upon the available genetic targeting approaches. As mentioned above, the virally mediated gene transfer provides for the most effective targeting of small nuclei, whereas the genomic modification approach is the most truly non-invasive means to obtain cell-type specific DREADD expression, though the expression patterns obtained can be off-target (due to the nature of the gene promoter used). Furthermore, existing technologies have been designed to be versatile as opposed to specific, creating scenarios in which multiple transgenes must be present in a mouse to confer DREADD expression or generations of germline recombination must be undertaken to obtain a useable mouse. Ideally, a DREADD could be expressed in the intended neuronal population using as few transgenes as possible. This both simplifies mouse breeding requirements and would “future proof” a given mouse line. I.e., if a single transgene mouse was created that expressed a DREADD in a specific population, a second transgenic mouse carrying a non CNO-based DREADD could be crossed with this mouse. Thus, one could have two different DREADDs expressed using only two transgenes.
6.3 Complete experimental control of signal transduction
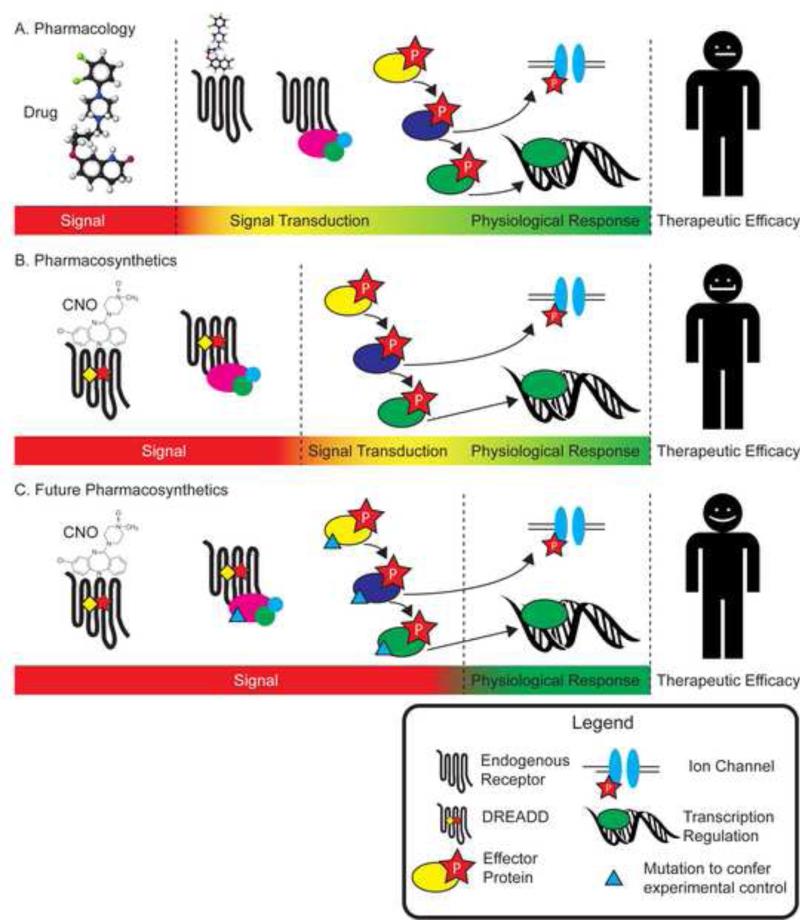
The supposition that signal transduction can be separated from the physiological response, in addition to the implications of functional selectivity, necessitate a further rightward shift in our control and understanding of the pharmacological equation. Whereas the pharmacosynthetic state of the art currently provides for control of the ligand-receptor pair, the physiological response is still dependent on the effectors present in a given cell type. In the future, it may be possible to control the ligand-receptor-effector triplet (or the ligand-receptor-effector-effector quartet, etc), providing unprecedented depth of pharmacological manipulation as was recently described by Yagi et al, (2011). For instance, one can imagine creating polycistronic transgenes that contain custom effectors designed to solely interact with designer receptors (Fig 3c). In this manner, one could truly synthesize signaling states in specific cell populations to create an end-goal for pharmacotherapeutic development. These custom signaling cascades will provide an unprecedented level of signaling control and definitively determine the type of signaling required for a particular physiological response.
Figure 3. Shifting the pharmacological equation.
(A) The conventional understanding of pharmacology posits that drugs interact with physiological systems to produce a physiological response. This interaction takes place through the transduction of a signal (either an endogenous chemical or an exogenous drug) via signal transduction mechanisms. The current depth of pharmacology defines the signal as the ligand itself, freeing the signal transducer and the signal transduction cascade to blend with the physiological response. (B) Pharmacosynthetic depth of pharmacology. Using pharmacosynthetics, both the ligand and the signal transducer (receptor) shift to the left side of the equation, creating a more defined signal with less signal transduction noise. (C) Future Pharmacosynthetics. With further development, a deeper level of pharmacosynthetic manipulation can be obtained. With the entirety of the signal transduction cascade under experimental control, we could potentially understand the type of signal needed to evoke a particular physiological response.
7 Conclusions
7.1 Key Difference Between Optogenetics and Pharmacosynthetics
It is important to note the differences between the pharmacosynthetic approach and the optogenetic approach. Using optogenetics, one can gain precise spatio-temporal control of neuronal firing using a combination of light and transgenic expression of engineered receptors. Among the optogenetic tools available, one difference is the level of invasiveness required for experimental manipulation when compared to pharmacosynthetics. Using optogenetics, one must deliver light to neurons- a process that currently requires implantation of fiber optics into the brain. Notwithstanding the proper implementation of control conditions, experiments can be imagined for which survival surgeries (and the subsequent hardware attachment) present insurmountable confounds. In its most noninvasive form - genomic transgene – pharmacosynthetic manipulation can be achieved with minimal invasiveness to the organism using peripheral administration of CNO (or even via food or drinking water). However, the prevalent use of virally mediated gene transfer in pharmacosynthetics (Table 1) would indicate that the level of initial invasiveness between approaches is similar. A second difference is the type of neuronal modulation afforded. Currently, the most widespread variation of optogenetics utilizes the channelrhodopsins, a family of light activated ion channels that directly modulate the ion conductance of neuronal membranes and either hyperpolarize or depolarize neurons. In contrast, DREADDs modulate G-protein mediated signaling – signaling cascades for which neuronal hyperpolarization or depolarization are only one outcome. However, there are optogenetic tools available – the OptoXRs - that modulate G-protein mediated signaling using light, though these have not gained widespread use.
Whereas the type of signaling afforded by these two approaches is not considerably different, nor the level of invasiveness considering the use of viral-mediated gene transfer, it can be argued that the nature of the neuronal modulation provided by these two technologies is what clearly separates them as distinct experimental tools with considerably different utility. The nature of the neuronal modulation created using pharmacosynthetics more closely resembles the hormonal signaling mechanisms found in the brain, in that the modulatory agent (CNO) “lingers” in the extraneuronal space and activates signal transduction mechanisms (GPCRs) dependent on diffusion, clearance, and receptor regulatory mechanisms (internalization, desensitization). This nature of activity more closely resembles the nature of metabotropic neurotransmitter or drug activity than any other process. On the other hand, optogenetics more closely resembles the electrochemical functionality of neurons. That is, the manipulation of membrane conductance more closely resembles the end result of factor-induced neuronal changes. In other words, pharmacosynthetics permits for the study of neuronal modulation itself, whereas optogenetics permits for the study of what a modulated neuron actually does in the brain. While these differences should fundamentally affect the type of experimentation performed with the respective technologies, pharmacosynthetics has yet to be fully implemented in this fashion.
7.2 Concluding remarks
Pharmacosynthetics provides an effective means to study the physiological effects of artificial, drug-like modulation of distinct neuronal populations. The specificity and exclusivity of GPCR modulation afforded by the technology can be utilized to explore mechanisms of therapeutic efficacy to further the development of small-molecule therapeutics. Further development of pharmacosynthetics will create levels of experimental manipulation that will challenge our conventional understanding of signal transduction and pharmacology. With this reimagination of this technology, it can be seen that the pharmacosynthetic approach provides unprecedented ability to truly answer the defining question of neuropharmacology – how do drugs modulate brain activity?
Acknowledgements
This work was funded by NIH Grant # 1F31MH091921 to MSF and RO1MH61887, U19MH82441, the NIMH Psychoactive Drug Screening Program and the Michael Hooker Chair in Pharmacology to BLR
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander GM, et al. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron. 2009;63:27–39. doi: 10.1016/j.neuron.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JA, Roth BL. Strategies to discover unexpected targets for drugs active at G protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2011;51:117–44. doi: 10.1146/annurev-pharmtox-010510-100553. [DOI] [PubMed] [Google Scholar]
- Alvarez-Curto E, et al. Ligand regulation of the quaternary organization of cell surface M3 muscarinic acetylcholine receptors analyzed by fluorescence resonance energy transfer (FRET) imaging and homogeneous time-resolved FRET. J Biol Chem. 2010;285:23318–30. doi: 10.1074/jbc.M110.122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Curto E, et al. Developing chemical genetic approaches to explore G protein-coupled receptor function: validation of the use of a receptor activated solely by synthetic ligand (RASSL). Mol Pharmacol. 2011;80:1033–46. doi: 10.1124/mol.111.074674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster BN, et al. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci U S A. 2007;104:5163–8. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, Gainetdinov RR. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol Rev. 2011;63:182–217. doi: 10.1124/pr.110.002642. [DOI] [PubMed] [Google Scholar]
- Centonze D, et al. Dopaminergic control of synaptic plasticity in the dorsal striatum. Eur J Neurosci. 2001;13:1071–7. doi: 10.1046/j.0953-816x.2001.01485.x. [DOI] [PubMed] [Google Scholar]
- Cohen J. Developing prescriptions with a personal touch. Science. 1997;275:776. doi: 10.1126/science.275.5301.776. [DOI] [PubMed] [Google Scholar]
- Conklin BR, et al. Engineering GPCR signaling pathways with RASSLs. Nat Methods. 2008;5:673–8. doi: 10.1038/nmeth.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva S, Bowers WJ. Targeting the central nervous system with herpes simplex virus / Sleeping Beauty hybrid amplicon vectors. Curr Gene Ther. 2011;11:332–40. doi: 10.2174/156652311797415845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deCharms RC, Zador A. Neural representation and the cortical code. Annu Rev Neurosci. 2000;23:613–47. doi: 10.1146/annurev.neuro.23.1.613. [DOI] [PubMed] [Google Scholar]
- DeLong M, Wichmann T. Update on models of basal ganglia function and dysfunction. Parkinsonism Relat Disord. 15 Suppl. 2009;3:S237–40. doi: 10.1016/S1353-8020(09)70822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong MR. Primate models of movement disorders of basal ganglia origin. Trends Neurosci. 1990;13:281–5. doi: 10.1016/0166-2236(90)90110-v. [DOI] [PubMed] [Google Scholar]
- Deneris ES, Wyler SC. Serotonergic transcriptional networks and potential importance to mental health. Nat Neurosci. 2012;15:519–27. doi: 10.1038/nn.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Rogan SC, Roth BL. Directed molecular evolution of DREADDs: a generic approach to creating next-generation RASSLs. Nat Protoc. 2010;5:561–73. doi: 10.1038/nprot.2009.239. [DOI] [PubMed] [Google Scholar]
- Farrell MS, Pei Y, Wan Y, Yadav P, Daigle T, Sciaky N, Simmons A, Nonneman R, Huang XP, Hufeisen S, Guettier J, Moy SS, Wess J, Caron MG, Calakos N, Roth BL. A Gαs DREADD mouse for selective modulation of cAMP production in striatopallidal neurons. Neuropsychopharmacology. doi: 10.1038/npp.2012.251. In revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, et al. Transient neuronal inhibition reveals opposing roles of indirect and direct pathways in sensitization. Nat Neurosci. 2011;14:22–4. doi: 10.1038/nn.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, Neumaier JF. Grateful DREADDs: engineered receptors reveal how neural circuits regulate behavior. Neuropsychopharmacology. 2012;37:296–7. doi: 10.1038/npp.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust KD, et al. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner AR, et al. Generation of a synthetic memory trace. Science. 2012;335:1513–6. doi: 10.1126/science.1214985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, et al. D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science. 1990;250:1429–32. doi: 10.1126/science.2147780. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Vacarezza N, et al. Use of pharmacogenetics in bioequivalence studies to reduce sample size: an example with mirtazapine and CYP2D6. Pharmacogenomics J. 2012 doi: 10.1038/tpj.2012.29. [DOI] [PubMed] [Google Scholar]
- Guettier JM, et al. A chemical-genetic approach to study G protein regulation of beta cell function in vivo. Proc Natl Acad Sci U S A. 2009;106:19197–202. doi: 10.1073/pnas.0906593106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. BAC to the future: the use of bac transgenic mice for neuroscience research. Nat Rev Neurosci. 2001;2:861–70. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- Kim JM, et al. Light-driven activation of beta 2-adrenergic receptor signaling by a chimeric rhodopsin containing the beta 2-adrenergic receptor cytoplasmic loops. Biochemistry. 2005;44:2284–92. doi: 10.1021/bi048328i. [DOI] [PubMed] [Google Scholar]
- Klabunde T, Hessler G. Drug design strategies for targeting G-protein-coupled receptors. Chembiochem. 2002;3:928–44. doi: 10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Kohane IS. (Mis)treating the pharmacogenetic incidentalome. Nat Rev Drug Discov. 2012;11:89–90. doi: 10.1038/nrd3659. [DOI] [PubMed] [Google Scholar]
- Kozorovitskiy Y, et al. Recurrent network activity drives striatal synaptogenesis. Nature. 2012;485:646–50. doi: 10.1038/nature11052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes MJ, et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Invest. 2011;121:1424–8. doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravitz AV, et al. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature. 2010;466:622–6. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Du BN. Pharmacogenetics: defective enzymes in relation to reactions to drugs. Annu Rev Med. 1972;23:453–68. doi: 10.1146/annurev.me.23.020172.002321. [DOI] [PubMed] [Google Scholar]
- Mallo M. Controlled gene activation and inactivation in the mouse. Front Biosci. 2006;11:313–27. doi: 10.2741/1799. [DOI] [PubMed] [Google Scholar]
- Marion S, et al. A beta-arrestin binding determinant common to the second intracellular loops of rhodopsin family G protein-coupled receptors. J Biol Chem. 2006;281:2932–8. doi: 10.1074/jbc.M508074200. [DOI] [PubMed] [Google Scholar]
- McKenzie GM, Szerb JC. The effect of dihydroxyphenylalanine, pheniprazine and dextroamphetamine on the in vivo release of dopamine from the caudate nucleus. J Pharmacol Exp Ther. 1968;162:302–8. [PubMed] [Google Scholar]
- Narendran R, Martinez D. Cocaine abuse and sensitization of striatal dopamine transmission: a critical review of the preclinical and clinical imaging literature. Synapse. 2008;62:851–69. doi: 10.1002/syn.20566. [DOI] [PubMed] [Google Scholar]
- Oh E, et al. Substitution of 5-HT1A receptor signaling by a light-activated G protein-coupled receptor. J Biol Chem. 2010;285:30825–36. doi: 10.1074/jbc.M110.147298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouimet CC, et al. DARPP-32, a dopamine- and adenosine 3':5' monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. III. Immunocytochemical localization. J Neurosci. 1984;4:111–24. doi: 10.1523/JNEUROSCI.04-01-00111.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y, et al. Engineered GPCRs as tools to modulate signal transduction. Physiology (Bethesda) 2008;23:313–21. doi: 10.1152/physiol.00025.2008. [DOI] [PubMed] [Google Scholar]
- Ray RS, et al. Impaired respiratory and body temperature control upon acute serotonergic neuron inhibition. Science. 2011;333:637–42. doi: 10.1126/science.1205295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogan SC, Roth BL. Remote control of neuronal signaling. Pharmacol Rev. 2011;63:291–315. doi: 10.1124/pr.110.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls ET, Treves A. The neuronal encoding of information in the brain. Prog Neurobiol. 2011;95:448–90. doi: 10.1016/j.pneurobio.2011.08.002. [DOI] [PubMed] [Google Scholar]
- Roth BL, et al. 5-Hydroxytryptamine2 receptors coupled to phospholipase C in rat aorta: modulation of phosphoinositide turnover by phorbol ester. J Pharmacol Exp Ther. 1986;238:480–5. [PubMed] [Google Scholar]
- Roth BL, Sheffler DJ, Kroeze WK. Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nat Rev Drug Discov. 2004;3:353–9. doi: 10.1038/nrd1346. [DOI] [PubMed] [Google Scholar]
- Sasaki K, et al. Pharmacogenetic modulation of orexin neurons alters sleep/wakefulness states in mice. PLoS One. 2011;6:e20360. doi: 10.1371/journal.pone.0020360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazoe T, et al. Roles of adenosine A(1) and A(2A) receptors in the expression and development of methamphetamine-induced sensitization. Eur J Pharmacol. 2000;388:249–54. doi: 10.1016/s0014-2999(99)00899-7. [DOI] [PubMed] [Google Scholar]
- Steketee JD, Kalivas PW. Drug wanting: behavioral sensitization and relapse to drug-seeking behavior. Pharmacol Rev. 2011;63:348–65. doi: 10.1124/pr.109.001933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell BR. Chemical genetics: ligand-based discovery of gene function. Nat Rev Genet. 2000;1:116–25. doi: 10.1038/35038557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulzer D. How addictive drugs disrupt presynaptic dopamine neurotransmission. Neuron. 2011;69:628–49. doi: 10.1016/j.neuron.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmeier DJ, et al. D1 and D2 dopamine-receptor modulation of striatal glutamatergic signaling in striatal medium spiny neurons. Trends Neurosci. 2007;30:228–35. doi: 10.1016/j.tins.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Svenningsson P, et al. Activation of adenosine A2A and dopamine D1 receptors stimulates cyclic AMP-dependent phosphorylation of DARPP-32 in distinct populations of striatal projection neurons. Neuroscience. 1998;84:223–8. doi: 10.1016/s0306-4522(97)00510-1. [DOI] [PubMed] [Google Scholar]
- Svenningsson P, et al. DARPP-32: an integrator of neurotransmission. Annu Rev Pharmacol Toxicol. 2004;44:269–96. doi: 10.1146/annurev.pharmtox.44.101802.121415. [DOI] [PubMed] [Google Scholar]
- Teschemacher AG, et al. Targeting specific neuronal populations using adeno- and lentiviral vectors: applications for imaging and studies of cell function. Exp Physiol. 2005;90:61–9. doi: 10.1113/expphysiol.2004.028191. [DOI] [PubMed] [Google Scholar]
- Tsien JZ, et al. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–26. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- Urban JD, et al. Functional selectivity and classical concepts of quantitative pharmacology. J Pharmacol Exp Ther. 2007;320:1–13. doi: 10.1124/jpet.106.104463. [DOI] [PubMed] [Google Scholar]
- Weinberg MS, Samulski RJ, McCown TJ. Adeno-associated virus (AAV) gene therapy for neurological disease. Neuropharmacology. 2012 doi: 10.1016/j.neuropharm.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinshilboum RM, Otterness DM, Szumlanski CL. Methylation pharmacogenetics: catechol O-methyltransferase, thiopurine methyltransferase, and histamine N-methyltransferase. Annu Rev Pharmacol Toxicol. 1999;39:19–52. doi: 10.1146/annurev.pharmtox.39.1.19. [DOI] [PubMed] [Google Scholar]
- West AE, Griffith EC, Greenberg ME. Regulation of transcription factors by neuronal activity. Nat Rev Neurosci. 2002;3:921–31. doi: 10.1038/nrn987. [DOI] [PubMed] [Google Scholar]
- Wu Z, Asokan A, Samulski RJ. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol Ther. 2006;14:316–27. doi: 10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Yagi H, et al. A synthetic biology approach reveals a CXCR4-G13-Rho signaling axis driving transendothelial migration of metastatic breast cancer cells. Sci Signal. 2011;4:ra60. doi: 10.1126/scisignal.2002221. [DOI] [PMC free article] [PubMed] [Google Scholar]