Abstract
The chromosome 21 gene RCAN1, encoding a modulator of the calcineurin (CaN) phosphatase, is a candidate gene for contributing to cognitive disability in people with Down syndrome (DS; trisomy 21). To develop a physiologically relevant model for studying the biochemistry of RCAN1 and its contribution to DS, we generated bacterial artificial chromosome-transgenic (BAC-Tg) mouse lines containing the human RCAN1 gene with a C-terminal HA-FLAG epitope tag incorporated by recombineering. The BAC-Tg was expressed at levels only moderately higher than the native Rcan1 gene; approximately 1.5-fold in RCAN1BAC-Tg1 and 2-fold in RCAN1BAC-Tg2. Affinity purification of the RCAN1 protein complex from brains of these mice revealed a core complex of RCAN1 with calcineurin (CaN), glycogen synthase kinase 3-beta (Gsk3b), and calmodulin, with sub-stoichiometric components including LOC73419. The BAC-Tg mice are fully viable, but long-term synaptic potentiation (LTP) is impaired in proportion to BAC-Tg dosage in hippocampal brain slices from these mice. RCAN1 can act as a tumor suppressor in some systems, but we found that the RCAN1 BAC-Tg did not reduce mammary cancer growth when present at a low copy number in Tp53;WAP-Cre mice. This work establishes a useful mouse model for investigating the biochemistry and dose-dependent functions of the RCAN1 protein in vivo.
Introduction
Down syndrome (DS), with an incidence of 1 in 700 live births, is caused by trisomy of human chromosome 21 (Antonarakis et al. 2004). DS phenotypes include cognitive disability, early-onset Alzheimer's disease (AD), congenital heart defects, increased autoimmunity and mild immunodeficiency, and an increased incidence of leukemia in early childhood. In contrast to the leukemia predisposition, children and adults with DS have fewer solid tumors than the general population. The Down Syndrome Critical Region 1 (DSCR1) gene, current name Regulator of Calcineurin 1, RCAN1, maps to chromosome band 21q22.12 and encodes a regulatory protein in the calcineurin (CaN)/NFAT signaling pathway. Both positive and negative effects of RCAN1 on CaN phosphatase activity have been reported, probably reflecting a biphasic dose-response (Chan et al. 2005; Fuentes et al. 2000; Hilioti et al. 2004; Kingsbury and Cunningham 2000; Rothermel et al. 2000; Sanna et al. 2006). The RCAN1 gene consists of seven exons plus the alternative first one (Davies et al. 2007) and produces several mRNA isoforms through alternative splicing; in this report we refer to the isoform with 252 amino acids (NP_004405) as the long isoform, and the isoform (NP_981963) with 197 amino acids as the short isoform. RCAN1 is expressed in human brain, heart, mammary gland, skeletal muscle and other organs (Ermak et al. 2001; Fuentes et al. 1997). It is overexpressed in the brain of Down syndrome fetuses roughly in proportion to its 1.5-fold increased copy number (Fuentes et al. 2000) and its expression is increased with aging and in Alzheimer's disease (Cook et al. 2005).
Over-expression of RCAN1 from cDNA-based transgenes (Tg’s) in mice produced cardiac and brain phenotypes (Ma et al. 2004), while overexpression and deletion of nebula, an ortholog of human RCAN1, disrupted olfactory learning and long-term memory in Drosophila (Chang et al. 2003). Deletion of the Rcan1 and Rcan2 genes from mice also produced a behavioral phenotype, and biochemical assays in these and other studies have led to a working model for dosage-dependent function of this CaN-interacting protein, in which some expression of RCAN1 is essential for and stimulates CaN function, while higher levels can be inhibitory (Sanna et al. 2006; Vega et al. 2003). A role for RCAN1 over-expression and deficiency in establishing the “set point” for CaN-dependent cardiac hypertrophy has been extensively investigated, and several studies using cDNA expression vectors and gene knockout have implicated RCAN1 as a regulator of tumor angiogenesis and a candidate tumor suppressor gene, albeit with different effects depending on the experimental system (Hesser et al. 2004; Iizuka et al. 2004; Minami et al. 2004; Vega et al. 2003). In undertaking the current study we reasoned that to fully understand the biochemistry and physiological role of RCAN1 in normal tissues and in DS it will be essential to utilize in vivo models with the RCAN1 gene over-expressed at levels that are comparable to the moderate copy-number-dependent overexpression of this gene in human DS, and to stably express epitope-tagged versions of this protein which will allow its biochemical interactions to be studied in native tissues. Here we describe the production of BAC-Tg mice that over-express epitope-tagged human RCAN1 at physiological levels and we use tissues from these mice to study the biochemistry and function of RCAN1.
Materials and Methods
BAC recombineering and production of BAC-Tg mice
BAC DNA was prepared by double potassium acetate precipitation followed by CsCl gradient separation (Xing et al. 2007). A short tandem tag containing the sequence encoding HA and FLAG peptides (17 amino acids total) was inserted prior to the stop cordon of the last exon of RCAN1 in BAC RPCI-11-272A3 using homologous recombination. BAC recombineering was performed in EL250 (Lee et al. 2001) cells electroporated with BAC DNA; a PCR cassette was introduced into these cells, which were cultured at 42°C for 15 min to induce expression of the Flpe recombinase. The PCR cassette contains the tandem tag and kanamycin selection marker and is flanked by 60bp sequences on each side which are homologous to the sequence of the insertion site on the BAC. The following primers were used: TG-left, GACCTAAGCCAAAAATTATCCAGACCAGGAGGCCGGAGTACACGCCGATCCACCTC AGCTACCCTTATGACGTGCCCGAT; and TG-right, AGTAAAAGATTCCTCCCGTGAGTATGATTTGGAATGCGTCCTCGTCGCGTGCCAGTT CATTTGGAGGCTACCATGGAGAA. The plasmid pIGCN21 (Lee et al. 2001) was used to provide the Frt-kan-Frt cassette. Kanamycin-resistant BAC clones were induced by L-arabinose to remove the kanamycin marker so that only the tandem tag was left in the final modified BAC. The BAC DNA (1 ng/µl in circular form) was injected into 200 pronuclei of fertilized oocytes of C57BL/6×CBA mice, and the oocytes transferred to pseudopregnant Swiss Webster foster mothers. All mice used in this study were housed in an AAALAC accredited facility and all procedures relating to the care and use of animals were performed in accordance with the National Institutes of Health guidelines at Columbia University. p53flex7 (Chen et al. 2005) and Wapcre mice (Ludwig et al. 2001) were described previously.
Southern and northern blotting
Genomic DNA was prepared by lysing the tissue in SDS/proteinase K, incubating for several hours at 50°C, followed by phenol/chloroform extraction and precipitation in ethanol. The Southern blots for copy number analysis were co-hybridized with genomic probes specific to human RCAN1 and a mouse probe for the Col6a1 gene, located on mouse chromosome 16 and designed to hybridize to a conveniently sized restriction fragment in the same lanes. PCR primers for synthesizing the human RCAN1 probe for Southern blotting were: Hu-Dscr-left: 5’-CCGTCAGCTCCTCATTTCTC-3’, Hu-Dscr-right: 5’- CCCAGGAGAGCTTCCTACCT-3’. RNA was prepared after solubilizing tissues in Trizol reagent (Invitrogen, Carlsbad, CA) according to the protocol of the manufacturer. RNA was prepared from different tissues of Tg and litter mate wild type adult mice and 5ug RNA was used for northern blotting followed by hybridization of the blots with probes specific for the last exon of human RCAN1 or mouse Rcan1. The primers for generating the northern probes were mouse For, 5’- CCGTGTGGAATTGTCCTTCT -3’, mouse Rev, 5’- CAAATGTTCACCGAGTGTCG -3’, human For, 5’- AGGGCAGGTTGATCAGTGAG -3’; human Rev, 5’- TCAGGAATGAATGGGGAAAG -3’.
Western blotting
Tissues were lysed in cold RIPA buffer (50mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP40) and centrifuged to remove insoluble debris. Total cell extracts normalized for protein content were boiled for 5 minutes in a denaturing solution containing 12 mmol/L Tris (pH 6.8), 5% glycerol, 0.4% SDS, 3 mmol/L 2-mercaptomethanol, and 0.02% bromophenol blue. Of the protein lysate, 50 µg per sample was electrophoresed on PAGE gels (Invitrogen). Primary antibodies were: Anti-HA (11867423001; Roche Applied Science), anti-FLAG M2 (Sigma, F1804), anti-RCAN1 antibody (Abnova, H00001827-M0; recognizing both human and mouse RCAN1), anti-calcineurin (BD biosciences, 556350), anti- GSK3b (Chemicon, AB8687), anti-phospho-DARPP32 (RD systems, VUPO1), anti-phospho-Tau (Ser396) (Spring Biosciences, E12124), anti-phospho-CREB (Ser133) (Cell Signaling Technology, 9191S), anti-phospho-dynamin (Affinity Bioreagents, PA1-4620), anti-NFATc1(Santa Cruz Biotechnology, sc-7294), anti-NFATc2 (4G6-G5) (Santa Cruz Biotechnology, sc-7296). The signal was amplified and detected using a peroxidase-conjugated goat anti-rabbit, anti-mouse or anti-Rat immunoglobulin G and the Enhanced Chemiluminescence-Plus detection system (Amersham Pharmarcia Biotech).
Affinity purification of the RCAN1 protein complex
The tandem epitope-tag affinity purification procedure for isolating protein complexes from native tissues was slightly modified from previous reports (Gu et al. 1999; Nikolaev et al. 2003). Mouse brain tissue, 3 g, was homogenized in 30 ml HB buffer (10 mM Tris, 10 mM KCl, 1.5 mM MgCl2, PH 7.3) containing protease inhibitors (Sigma, P8340) using a tissue grinder, then kept on ice for 20 minutes followed by centrifugation at 1000xg for 12 minutes. To the collected supernatant NaCl and Triton X-100 were added to final concentrations of 133 mM and 0.2% respectively, followed by incubation on ice for one hour with occasional mixing. After centrifugation at 15,000 rpm for 30 minutes, the supernatant was passed through a 0.45 μm Millipore filter. Anti-Flag M2 affinity beads (Sigma, A2220), 200 μl of slurry, was added to the supernatant and gently rotated at 4°C overnight. The beads were spin-washed five times with 0.5 ml HB133 buffer (HB buffer, 133mM NaCl, 0.2% Triton X-100, protease inhibitors) at 1000xg, and eluted twice at 4°C with 150 μl of HB133 containing Flag peptide (Sigma, F4799) at a concentration of 200 ng/μl. The 300 μl eluate was then incubated with 30 μl slurry of anti-HA-Agarose beads (Sigma, A2095) overnight and washed as above. Proteins bound to the washed HA beads were eluted with 20 μl HA peptide (1mg/ml in HB133 buffer, Roche, 11666975001) two times at room temperature, and were resolved by SDS-PAGE in a 4%–20% gradient gel (Novex, Invitrogen). The gels were stained according to the protocols of GelCode Blue Stain Reagent (Thermo Scientific, 24590) and ProteoSilver Silver Stain Kit (Sigma, PROT-SIL1). Specific bands were cut out from the blue-stained gels and subjected to peptide sequencing by liquid chromatography-mass spectrometry (LC-MS) in the Proteomics Shared Resource of the Herbert Irving Comprehensive Cancer Center.
Size exclusion chromatography
For assessing stability of the core complex, affinity purified RCAN1 complex from BAC-Tg brain was loaded onto a Superose 12 PC 3.2/30 SMART gel-filtration column, and eluted with HB133 buffer. Each fraction, 40 μl, was collected and resolved by SDS-PAGE followed by probing of the resulting western blots with the indicated antibodies.
Measurement of CaN activity
Lysates were prepared from whole brain tissue or the hippocampal formation of BAC-Tg and wild-type control mice using the lysis buffer provided in the calcineurin assay kit (Biomol International, AK-804). Supernatant was collected after centrifugation at 20,000 × g for 15 min. This supernatant was further centrifuged at 100,000 × g for 45 min to obtain the ultra-supernatant, as well as a pellet containing organelles and membranes. The calcineurin phosphatase assay was done according to the Biomol International protocol; liberated phosphate was colorimetrically determined using a plate reader at 620 nm.
Cell culture
Jurkat cells were cultured in RPMI1640 with 10% FCS. Neuro-2a cells, 293 cells and MEF cells were cultured in DMEM with 10% FCS. To establish MEF cells, E14.5 embryos were dissected and the trunk tissues of each eviscerated embryo were separately finely cut and incubated with trypsin at 37°C for 10 minutes. The cells were plated on one 10 cm tissue culture dish per embryo and cultured for 36 hours, then split to generate multiple plates for thapsigargin (100nM) treatment.
Interaction of FLAG-RCAN1 and HA-LOC73419 in transfected cells
A FLAG-tagged expression construct for the mouse Rcan1 open reading frame (NM_001081549) was created by RT-PCR of mouse brain RNA followed by cloning in the pIRESneo vector (Clontech) with a FLAG tag at the COOH terminus, to generate the p-FLAG-Rcan1 construct. An HA-tagged expression construct for mouse LOC73419 was obtained by cloning the PCR fragment (IMAGE Mouse cDNA clone: 40131035) into pIRESneo vector with a HA tag at the COOH terminus. The final DNA constructs were sequenced. 293 cells or Neura-2a cells were transfected with expression constructs for RCAN1 and LOC73419 using Lipofectamine 2000 (Invitrogen). Transfected cells were lysed 36 hours post transfection in lysis buffer (20 mM Tris-HCl pH 7.9, 150 mM NaCl, 0.2% Triton X-100, 10% glycerol and fresh proteinase inhibitor cocktail). The whole cell extracts were immunoprecipitated with Anti-FLAG M2 affinity beads or Anti-HA agarose beads. After three washes with lysis buffer, the bound proteins were eluted using FLAG-peptide or HA-peptide. The eluted material was resolved by SDS-PAGE and detected by western blotting with antibodies as indicated.
Luciferase reporter assays
Jurkat leukemia cells (5 × 105 / 2 ml in one well) were transfected in 6-well plates with plasmid DNAs using Fugene 6 (Roche). After 24 hours of incubation, 2 ml medium was added to each transfected well, mixed and separated equally into two new wells. One set of transfected cells were stimulated with PMA (10 ng/ml) and ionomycin (1uM) for 8 hours. Cells were harvested and the luciferase activity was measured using the Dual Luciferase Reporter Assay System Kit (Promega) according the manufacturer’s protocol.
Electrophysiological studies
Electrophysiological measurements were performed essentially as previously described (Puzzo et al. 2008). Transverse hippocampal slices of a thickness of 400 micron were maintained in an interface chamber at 29°C‥ They were perfused with saline solution (124 mM NaCl, 4.4 mM KCl, 1 mM Na2HPO4, 25 mM NaHCO3, 2 mM CaCl2, 2 mM MgSO4, 10 mM glucose) continuously bubbled with 95% O2 and 5% CO2‥ Slices were permitted to recover for at least 90 minutes before recordings. Field-excitatory postsynaptic potentials (fEPSP) were monitored by placing the recording electrodes in CA1 stratum radiatum. Basal synaptic transmission was assessed by plotting the stimulus voltage (V) against slopes of fEPSP to generate input-output relations. For long-term potentiation (LTP) experiments, baseline stimulation was delivered every minute at an intensity that evoked a response of ≈ 35% of the maximum evoked response. Baseline responses were recorded for 30 minutes prior to beginning experiments in order to assure stability of the response. LTP was induced using theta-burst stimulation (4 pulses at 100 Hz, with the bursts repeated at 5 Hz and each tetanus including 3 ten-burst trains separated by 15 seconds).
Statistical analysis
Survival in mouse experiments are represented by Kaplan-Meier curves, and significance was estimated with the log-rank test using Graph-Pad Prism (version 4) software.
Results
Production of BAC-transgenic mice carrying the human RCAN1 (DSCR1) genomic locus
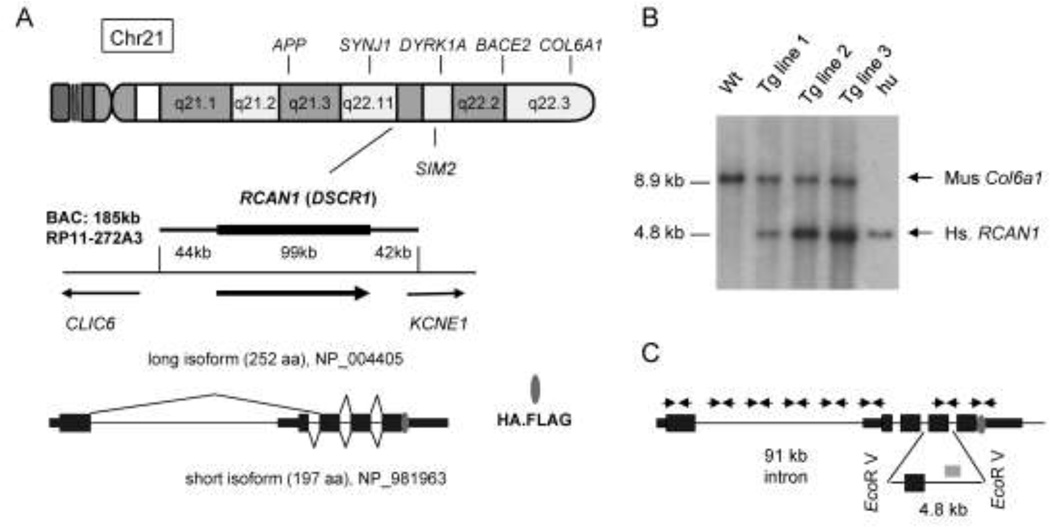
BAC RP11-272A3, from the RPCI library (Osoegawa et al. 2001) is a 185 kb clone containing the entire RCAN1 gene flanked by 44 kb of upstream DNA and 42 kb of downstream DNA (Fig. 1A). This BAC does not include any other complete genes; although the 5’ portion of the KCNE1 gene is present on the BAC these sequences encode only the first non-coding exon. We used BAC recombineering to insert a tandem HA-FLAG peptide sequence immediately preceding the stop codon in the last exon. BAC-Tg mice were created by injecting purified BAC DNA into male pronuclei of fertilized B6/CBA oocytes. To test the resulting progeny, we used eight pairs of PCR primers specific for the 5’, middle and 3’ regions of the human RCAN1 gene, as well as Southern blotting with a RCAN1 probe, which identified 3 founder mice with intact BAC DNA (Fig. 1B, C). Phosphorimaging of Southern blots simultaneously hybridized with similarly sized DNA probes specific for the human RCAN1 and the mouse Col6a1 gene allowed us to estimate that the BAC DNA copy number in the lines derived from founder #1, #2 and #3 was approximately 1, 3, and 5 (Fig. 1B). We refer to these transgenic lines as RCAN1BAC-Tg1, RCAN1BAC-Tg2 and RCAN1BAC-Tg3 respectively. The RCAN1BAC-Tg1 and RCAN1BAC-Tg2 lines transmitted the transgene in Mendelian proportions, with no evidence of embryonic or adult mortality. However, founder #3 only transmitted the BAC-Tg to males, which was presumably due to integration of the BAC on Y chromosome. Because this high-copy line over-expressed the transgene at supra-physiological levels we chose to perform most of our subsequent experiments using the lower-copy RCAN1BAC-Tg1 and RCAN1BAC-Tg2 lines. Data reported here are from these BAC-Tg mice in the C57BL/6 genetic background, after ≥5 generations of backcrossing to wild-type C57BL/6. We have not observed any deaths or morbidity over several years of work with these lines, with the mice routinely maintained up to 9 months of age.
Figure 1. The RCAN1 gene and production of BAC-Tg mice.
A, The RP11-272A3 BAC contains the complete RCAN1 gene flanked by 44 kb upstream and 42 kb downstream sequence (nucleotides 34,953,503-34,768,332 of the 2006 NCBI36/hg18 human genome assembly). There are three transcripts of RCAN1 according to Entrez Gene; the two major mRNA isoforms are diagrammed here. The HA.Flag epitope tag consisting of 17 amino acids was inserted before the stop codon of the last exon of the RCAN1 gene using BAC recombineering prior to creation of the Tg mice. B, Copy number analysis of the BAC-Tg in the three mouse lines by Southern blotting with a human RCAN1 probe and a control probe for the mouse Col6a1 gene. Mouse tail DNA and human genomic DNA was digested with EcoRV and used for Southern blotting. The human probe (841 bp long; 44% GC) recognizes a 4.8 kb fragment, while the mouse probe (550 bp long; 58% GC) recognizes a 8.9kb fragment. Wt: wild type mouse DNA; Hu: human DNA. The BAC copy number is estimated as 1, 3 and 5 for the RCAN1BAC-Tg1, RCAN1BAC-Tg2 and RCAN1BAC-Tg3 lines respectively. C, Eight PCR primer pairs were used for verifying integrity of the BAC Tg in each mouse line. The position of the human RCAN1 probe used for Southern blotting is also indicated on this map (grey rectangle).
Copy number-dependent RCAN1 protein expression in RCAN1 BAC transgenic mice
To ask whether the major regulatory elements for RCAN1 expression are present on the RP11-272A3 BAC, we surveyed mRNA expression in multiple adult tissues of the three transgenic lines. We hybridized northern blots sequentially with cDNA probes matching the human and murine RCAN1 orthologues. Since we designed the probes to span regions of these genes that have divergent nucleotide sequences, there was no cross-hybridization on these blots. For each of the three Tg lines the mRNA expression pattern of the human RCAN1 gene in the BAC-Tg mice was found to be similar, though not identical, to that of the endogenous murine Rcan1 gene (Suppl. Fig. S1). A transcript of the expected size was seen in all organs except the liver, which showed a shorter transcript (but normal protein size, see below).
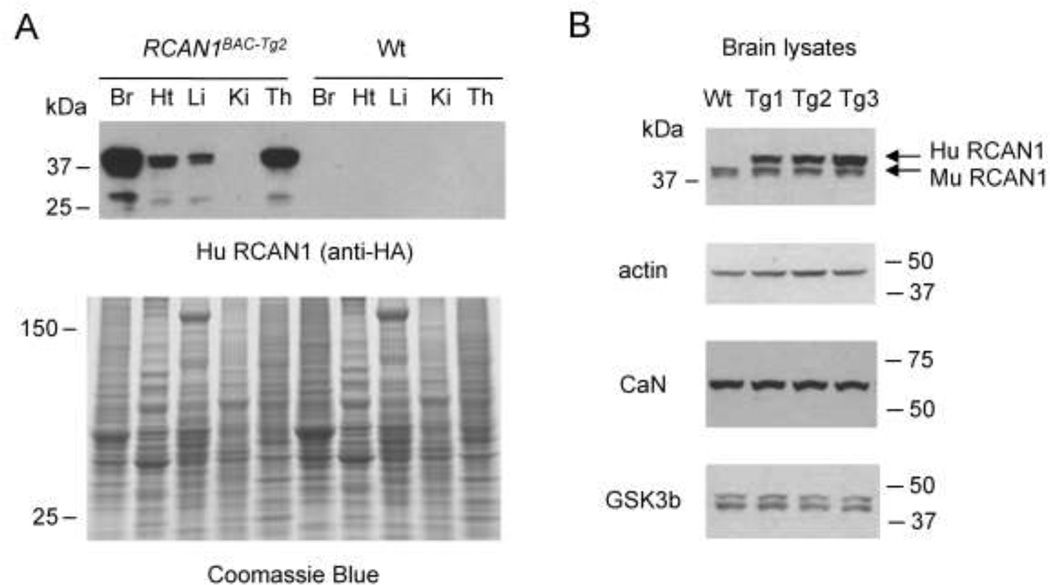
To verify that the observed expression of human RCAN1 mRNA in the transgenic mice in fact led to overproduction of RCAN1 protein, we performed western blots using total protein lysates from adult tissues of the BAC-Tg mice and their wild type non-transgenic littermates. We first probed these blots with anti-HA antibody recognizing the tagged human RCAN1. The highest expression of the tagged RCAN1 protein was found in the brain, and we also found significant expression in other relevant tissues, namely the thymus and heart (Fig. 2A). Although the liver mRNA is a shorter size, it gives rise to the correct size protein. In addition, the BAC Tg gives rise to both the long 38 kDa and short 27 kDa isoforms of RCAN1 protein. Western blotting of brain lysates in successive generations of mice has shown that expression of the epitope-tagged RCAN1 protein has been stably maintained in these mouse lines over more than 14 generations.
Figure 2. Western blot analysis confirming epitope-tagged RCAN1 expression in brain, thymus, and heart, with expression proportional to copy number of the BACTg.
A, Total proteins were prepared from BAC-Tg and wild type mouse tissues and blotted with anti-HA antibody to recognize the epitope-tagged human RCAN1 protein. The HA antibody detects the expression of two isoforms of RCAN1 in brain, thymus and heart. The Coomasie blue stained gel is shown as a loading control. B, Western blot probed with a RCAN1-specific antibody that recognizes RCAN1 proteins of both human and mouse. Comparison of the three BAC-Tg mouse lines shows copy-number dependent RCAN1 protein expression.
In addition to tissue-specificity, another criterion for testing the inclusion of key regulatory elements in a BAC-Tg construct is copy number-dependent expression of the gene under consideration. If the observed expression increases with increased copy number, this suggests that the transgene is at least partially insulated from the cis-effects of chromatin at its sites of integration. By western blotting of total protein from adult brains, we in fact observed copy number-dependent expression, with the RCAN1 protein levels paralleling the BAC DNA copy numbers (1X, 3X and 5X) in the three BAC-Tg lines (Fig. 2B and Suppl. Fig. S2). Probing these blots with an antibody recognizing RCAN1 protein of both human and mouse origin, we could establish the ratio of RCAN1 expressed from the BAC Tg to that of the endogenous mouse protein, leading to estimates of 1.5X, 2X and 5X in the RCAN1BAC-Tg1, RCAN1BAC-Tg2 and RCAN1BAC-Tg3 lines.
Purification of the RCAN1 protein complex
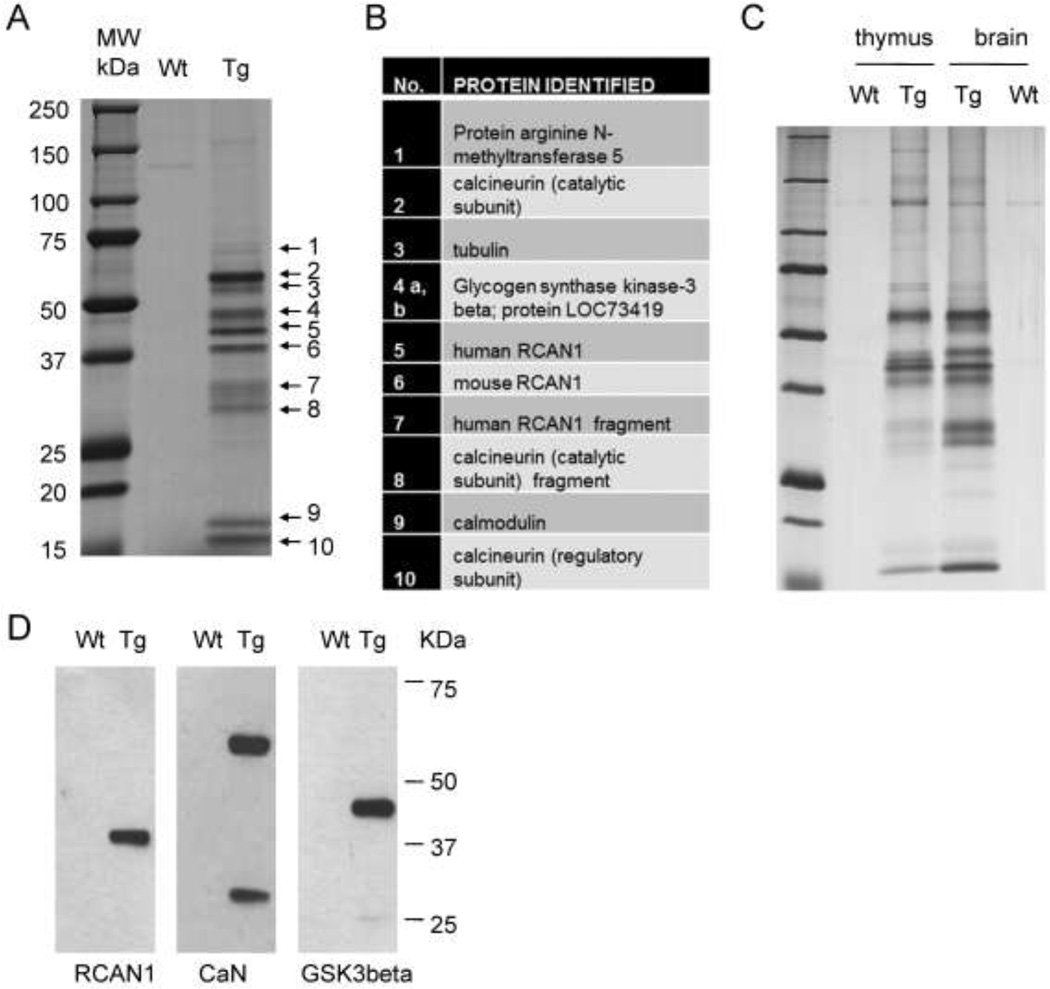
Using BAC recombineering we had tagged the transgenic RCAN1 with an HA-FLAG peptide epitope, thus making it possible to affinity purify RCAN1 protein complexes from native tissues of the BAC-Tg mice. To reduce the possibility of unspecific binding due to overexpression of the bait in these experiments we used brain lysates from the low copy number RCAN1BAC-Tg1 mice. After sequential FLAG and HA affinity pull-downs and elutions, the final protein preparations were separated on SDS-PAGE gels (Fig. 3A) and the individual bands were cut out from the gel and subjected to LC-MS analysis.
Figure 3. Purification of the RCAN1 complex from low-copy BAC-Tg mouse brain and identification of its major protein components.
A, Wild type and RCAN1BAC-Tg1 whole mouse brain lysates in hypotonic buffer were subjected to tandem affinity purification using anti-Flag- and anti-HA-beads. The final protein preparation was resolved by SDS-PAGE and individual bands were excised and analyzed by LC-MS. B, List of proteins identified in each band by LC-MS. C, Silver-stained gel of tandem affinity purified material from thymus and brain lysates, showing that the RCAN1 complex is similar in these two tissues. D, Western blots of the purified complex, independently confirming that the bait RCAN1-HA-Flag and two major interacting proteins, CaN and GSK3b, were correctly identified by LC-MS.
The major proteins identified in this core RCAN1 complex are listed in Figure 3B. Both the catalytic and regulatory subunits of the CaN phosphatase are major components of the complex, a finding that is consistent with extensive previous evidence that RCAN1 is a regulator of CaN (Fuentes et al. 2000; Rothermel et al. 2000). Consistent with CaN being a Ca++/calmodulin activated enzyme, we found that calmodulin is also present in the core RCAN1 complex. Glycogen synthase kinase 3 beta (GSK3b) is also present in substantial amounts in the complex. A previous report showed that GSK3b can phosphorylate RCNs, which are homologues of human RCAN1, in yeast (Hilioti et al. 2004), and in mammalian cells it has also been shown that RCAN1 (Calcipressin-1) is a phosphoprotein (Genesca et al. 2003).
These data show a highly stable physical interaction between GSK3b and RCAN1. The bait human RCAN1 appears as one of the major bands on the gels of this complex as expected and, importantly, the endogenous mouse RCAN1 protein co-exists in the complex, thus indicating that the epitope-tagged human RCAN1 protein has properties very similar to the endogenous mouse RCAN1. The LC-MS data further revealed that >90% of the GSK3b in the complex is phosphorylated at Tyr-216, a known activating mark for this enzyme, and that approximately 60–70% of human and mouse RCAN1 in the complex is phosphorylated at Ser-112 (QFLISPPASPPVGWK). Lastly, to ask whether the binding partners of RCAN1 are similar in different tissues, we used the same protocol to purify the core RCAN1 complex from thymus. The results (Fig. 3C) showed that the pattern of binding partners is very similar in these two different tissues. Lastly, separate experiments using direct IP-western blotting from brain lysates independently confirmed the physical associations of epitope-tagged RCAN1 with CaN and GSK3b in mouse brain (Fig. 3D).
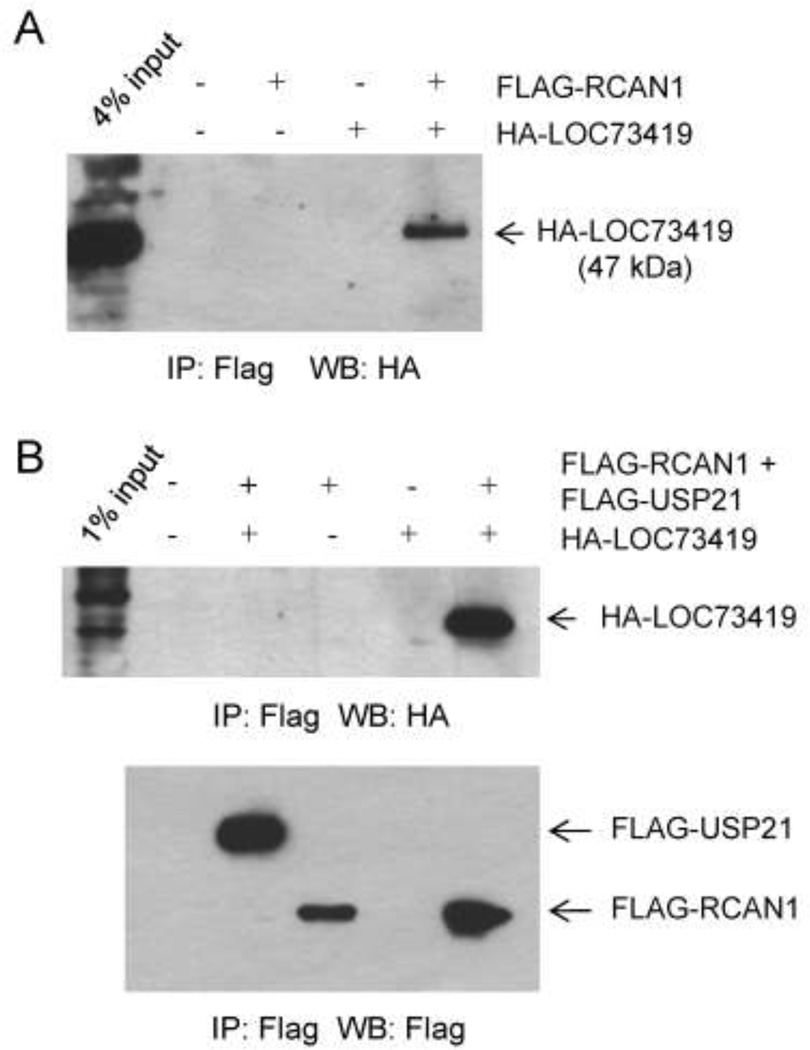
The LOC73419 protein interacts sub-stoichiometrically with RCAN1
An evolutionarily conserved but as yet poorly characterized protein, LOC73419 (1700052N19Rik; human homologue C6orf211), with molecular weight 47 kDa was detected sub-stoichiometrically in the RCAN1 complex (Fig 4A). To verify whether the interaction was specific we expressed fusion proteins FLAG-RCAN1 and HA-LOC73419 in 293 transformed kidney cells. HA-LOC73419 co-immunoprecipitated with FLAG-RCAN1 in protein lysates from these cells (Fig 4A). Considering that the affinity purification of the RCAN1 complex was done from brain lysates, we also expressed these two tagged proteins by transfection into Neuro-2a neuroblastoma cells, which revealed a more efficient co-IP than in the non-neuronal 293 cells (Fig 4B). As a negative control in these transfections we included a plasmid encoding an unrelated protein, FLAG-Usp21, which did not bind to HA-LOC73419 at all (Fig. 4B). These results support a bona fide albeit sub-stoichiometric interaction between these two proteins; however, the biochemical and physiological importance of this interaction remain to be determined. Relevant to this question, when we carried out promoter-reporter assays in Neuro-2a cells co-transfected with pIL2-luciferase and increasing amounts of the HA-LOC73419 plasmid we found a small but statistically significant increase in reporter activity, indicating a modest net positive effect of LOC73419 on the activity of this CaN-sensitive promoter (Suppl. Fig. S3).
Figure 4. The protein encoded by the LOC73419 gene interacts substoichiometrically with RCAN1.
A, Plasmids encoding HA-LOC73419 and Flag-RCAN1 were co-expressed in 293 cells. After IP of Flag-RCAN1, HA- LOC73419 is detected. The input lysate for the IP (4% of total) has been loaded in the first lane; comparison with the IP lane shows that the interaction of these two proteins in 293 cells is sub-stoichiometric. B, HA-LOC73419 and Flag-RCAN1 plasmids were co-transfected into Neuro-2a cells, with an unrelated control expression plasmid, producing Flag-USP21, serving as a negative control. IP was performed as in (A). Ha-LOC73419 is more efficiently pulled down by IP of Flag-RCAN1. The negative control Flag-USP21 shows no binding to Ha-LOC73419. The first lane shows 1% input; comparison with the results in (A) suggests that HA-LOC73419 interacts more efficiently with Flag-RCAN1 in Neuro-2a cells than in 293 cells.
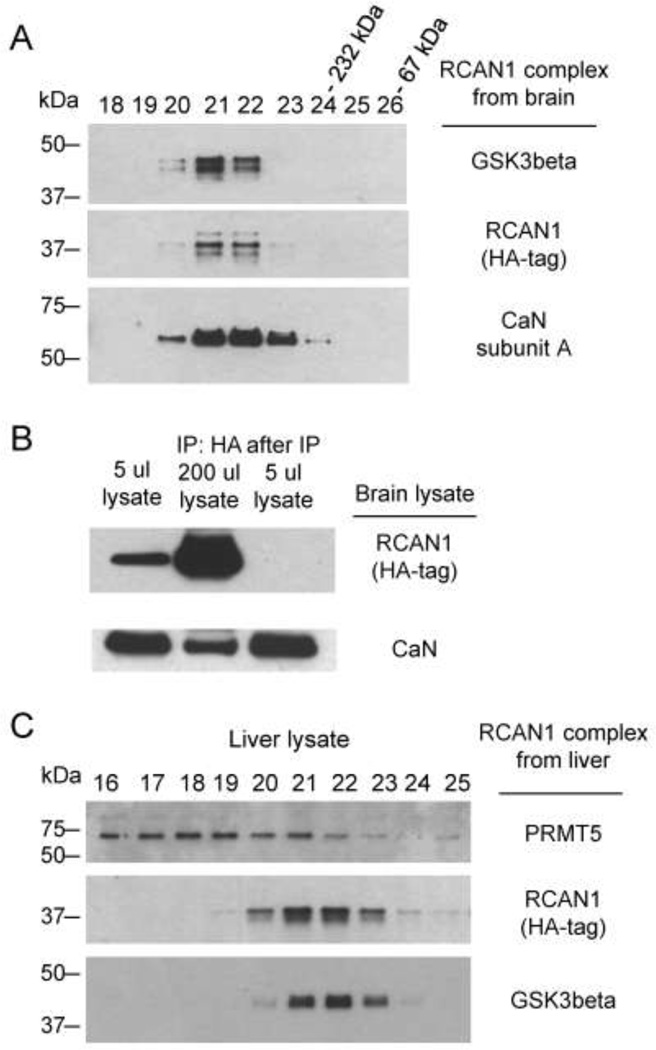
Stability of the RCAN1 core complex with CaN and GSK3b
When we subjected the purified RCAN1 protein complex to gel-filtration chromatography on Superose 6, we found that RCAN1, CaN and GSK3b co-eluted in the same fractions (Fig 5A), indicating that the three proteins exist in a stable complex that remains intact through several affinity and gel filtration steps carried out over several days in buffers with physiological salt concentrations. There is apparently a higher order stoichiometry in the complex, as the predicted molecular weight corresponding to the peak fractions is substantially greater than the sum of these three proteins. Conversely, since GSK3b has diverse signaling functions in the cell, involving a large number of protein substrates other than RCAN1 and CaN, we next asked what percentage of total cellular GSK3b is bound in the RCAN1 complex. Transgenic brain lysates were prepared using lysis buffer with physiological NaCl concentration (HB133; see Methods) and separated on the same gel-filtration column. This procedure revealed that only a small fraction of total cellular GSK3b, crudely estimated from the immunoblots as about 1%, is in the RCAN1 complex (data not shown). Similarly, although CaN is a major component of the RCAN1 complex, by comparing the amount of RCAN1-bound CaN in the IP material to the total input we estimate that only about 1% of total cellular CaN is stably associated with RCAN1 (Fig 5B). Lastly, the protein PRMT5, which constituted a minor but reproducible band (#1) in the affinity purified material, is a known false-positive interactor in tandem epitope tag affinity purification procedures, and is probably not a specific binding partner of RCAN1 since in gel filtration it does not elute in the same fractions as RCAN1 and GSK3b (Fig 5C).
Figure 5. Stability of the interaction of RCAN1 with CaN and GSK3b.
A, CaN and GSK3b remain associated with RCAN1 through Superose chromatography of the purified complex. Based on molecular weight markers co-eluting in later fractions, the molecular weight corresponding to Superose fractions 21 and 22 is more than the sum of all these three proteins in the core complex, indicating higher order stoichiometry. B, Less than one percent of CaN associates with RCAN1 protein in whole brain lysate from RCAN1BAC-Tg1 mice. The amount of CaN in RCAN1 complex from 200 ul of lysate is compared with the CaN in 5 μl of the original lysate (2.5% input of the IP amount). C, PRMT5 is not a stable binding partner in the core RCAN1 complex. A preparation of the RCAN1 complex from RCAN1BAC-Tg1 liver lysate was loaded on the same Superose column as in (A). PRMT5, a known non-specific binder to affinity resin in HA-Flag tandem purification experiments, is not stably bound to the RCAN1 complex, as indicated by lack of co-elution.
RCAN1 enhances CaN activity in the RCAN1 protein complex
RCAN1 has been extensively studied as a regulator of CaN – a protein phosphatase that is crucial for the nuclear factor of activated T-cells (NFAT) signaling pathway. However, there have been ostensibly contradictory findings in terms of its stimulatory versus inhibitory effects on CaN activity (Fuentes et al. 2000; Sanna et al. 2006). Initially, we used an enzymatic assay to measure total CaN activity in extracts from whole brain and hippocampus. This assay measures the activity of all cellular CaN, including both RCAN1-bound CaN and the large amount of intracellular non-RCAN1-associated CaN. At the level of sensitivity of this colorimetric phosphatase assay, we did not detect significant differences between wild type and Tg mice (data not shown). We therefore used the same enzymatic assay to measure the activity of total cellular CaN by IP with anti-CaN antibody, and comparing the results with assay of the activity of transgenic RCAN1-bound CaN, immunoprecipated from Tg mouse tissues with anti-HA antibody. To control for possible effects of the anti-CaN antibody on CaN activity, the whole cell lysates in the latter procedure were pre-incubated with anti-CaN antibody prior to the IP with anti-HA antibody. These experiments showed that the activity of RCAN1-bound CaN is dramatically higher than that of total cellular CaN (Suppl. Fig. S4). This result is consistent with previous conclusions from Rcan1/Rcan2 double-KO mice, in which CaN activity is abrogated (Sanna et al. 2006), and it suggests that one fraction of total CaN in the mouse brain exists in a low activity state, not bound to RCAN1, while another fraction exists in a high-activity RCAN1-bound state.
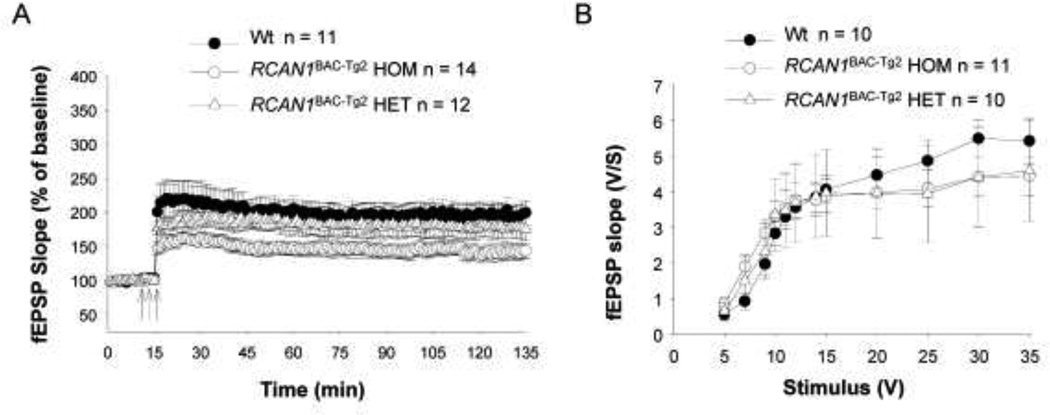
Deficient hippocampal LTP in RCAN1 BAC-Tg mice
RCAN1 is a candidate gene for contributing to cognitive disability in people with DS via its dose-dependent over-expression and consequent effects on CaN activity in neurons. In particular, the CaN phosphatase, which is regulated in part by RCAN1, is well known to be essential for synaptic plasticity (Winder and Sweatt 2001; Zeng et al. 2001). As our RCAN1 BAC-Tg lines showed appropriate expression of the BAC-Tg in whole brain and in isolated hippocampus, we undertook experiments to test long term potentiation (LTP), an electrophysiological surrogate for memory, in hippocampal slices from the RCAN1BAC-Tg2 mice. As shown in Figure 6A, this experiment revealed a clear gene dosage dependent impairment of LTP, with the greatest impairment in brains from mice that had inherited the BAC-Tg from both parents (estimated Tg copy number 6) and a more modest impairment, though not achieving statistical significance, in mice with the BAC-Tg inherited from only one parent (estimated Tg copy number 3). Basal synaptic transmission was not affected (Fig. 6B), indicating that the over-expression of RCAN1 specifically affects synaptic plasticity. We interpret these results as showing perturbation of synaptic plasticity by relatively modest over-expression of the RCAN1 gene and protein, at levels that are slightly increased relative to the situation in human DS brains, where the RCAN1 gene is triplicated. These findings can be reconciled to the lack of increased crude CaN activity in total brain lysates in vitro, and with our observation that a large fraction of total cellular CaN is not RCAN1-bound, by postulating a role for RCAN1 in regulating a specific functionally important sub-cellular fraction of CaN in neuronal synapses. Importantly, the phenotype of deficient LTP in these BAC-Tg mice proves that the epitope-tagged RCAN1 construct is biologically active, which in turn supports the relevance of our biochemical findings with this tagged construct.
Figure 6. Deficient LTP in RCAN1BAC-Tg2 mouse hippocampal brain slices.
A, CA1-LTP was reduced in RCAN1BAC-Tg2 mice. The reduction was significant for mice that had inherited the BAC-Tg from both parents [F(1,23)=8.218 P= 0.0087] and more modest for mice with the BAC-Tg inherited from only one parent [F(1,21)=0.6022 P= 0.4461]. The arrows indicate time and pattern of the tetani. B, Basal synaptic transmission at the Schaffer collateral/CA1 connection of hippocampal slices was not affected in RCAN1BAC-Tg2 mice (two-way ANOVA F(1,19) = 0.01114, p > 0.05 for mice that had inherited the BAC-Tg from both parents vs WT littermates; and F(1,18) = 0.00001, p > 0.05 for mice that had inherited the BACTg from only one parent vs. WT littermates ).
RCAN1over-expression alters the expression of NFATc1
We next investigated the effect of expression of RCAN1 on the transcription factor NFAT. NFAT is a family including NFATc1, NFATc2, NFATc3 and others, which are substrates of CaN (Hogan et al. 2003; Macian 2005). We prepared mouse embryonic fibroblast (MEF) cells from both Tg and litter-mate wild-type embryos and used thapsigargin to alter Ca++ levels in these cells. Thapsigargin activates the calcium/CaN signaling pathway (Stewart et al. 1998), acting by inhibiting the ATPase pumps on Ca++ storage organelles, which maintain homeostasis by pumping Ca++ from the cytosol into these organelles, including the endoplasmic reticulum. Inhibition of pump action causes emptying of intracellular calcium stores and a resultant Ca++ influx across the plasma membrane via capacitative entry. After exposure to thapsigargin, RCAN1-Tg MEFs reproducibly showed increased expression of NFATc1 (Suppl. Fig. S5), an effect that was consistent across all three BAC-Tg lines. This increased expression of NFATc1 in turn strongly induced the expression of the short isoform of Tg RCAN1 (Suppl. Fig. S5), a finding that is consistent with previous reports that only the expression of short form RCAN1 is NFAT-dependent (Lange et al. 2004; Yang et al. 2000).
These data further demonstrate that the epitope-tagged RCAN1 protein produced from the BAC-Tg is active in the cellular context. However, when we carried out western blotting of whole brain lysates from the BAC-Tg mice (Tg1 and Tg2 lines) to test for effects of RCAN1 overexpression in vivo on the phosphorylation levels of several known substrates of the CaN phosphatase, namely DARPP32, Tau, CREB, and dynamin, we found no change in steady state levels of the phosphorylated forms of these proteins (data not shown). These negative results using whole brain lysates suggest that the effects of changes in RCAN1 levels, within the physiological range relevant to DS, on the phosphorylation states of these CaN target proteins are either subtle or, more likely, highly localized to specific anatomical regions and/or subcellular structures (e.g. synapses, neurites, etc.); possibilities that can be examined in future experiments with the BAC-Tg mice.
RCAN1 over-expression slightly accelerates or has no effect on Myc-driven mammary tumorigenesis
The RCAN1 protein has been implicated in inhibition of tumor growth via an inhibitory role in endothelial cell signaling and tumor angiogenesis – findings that have supported the involvement of RCAN1 gene dosage in the well-known DS phenotype of resistance to solid tumors (for example, (Baek et al. 2009; Minami et al. 2004; Qin et al. 2006)). However, other experimental approaches have given results that seem to argue against this hypothesis (Ryeom et al. 2008) and other chromosome 21-linked genes, notably ETS2, have also been proposed as tumor suppressors (Reynolds et al. 2010; Sussan et al. 2008; Wolvetang et al. 2003).
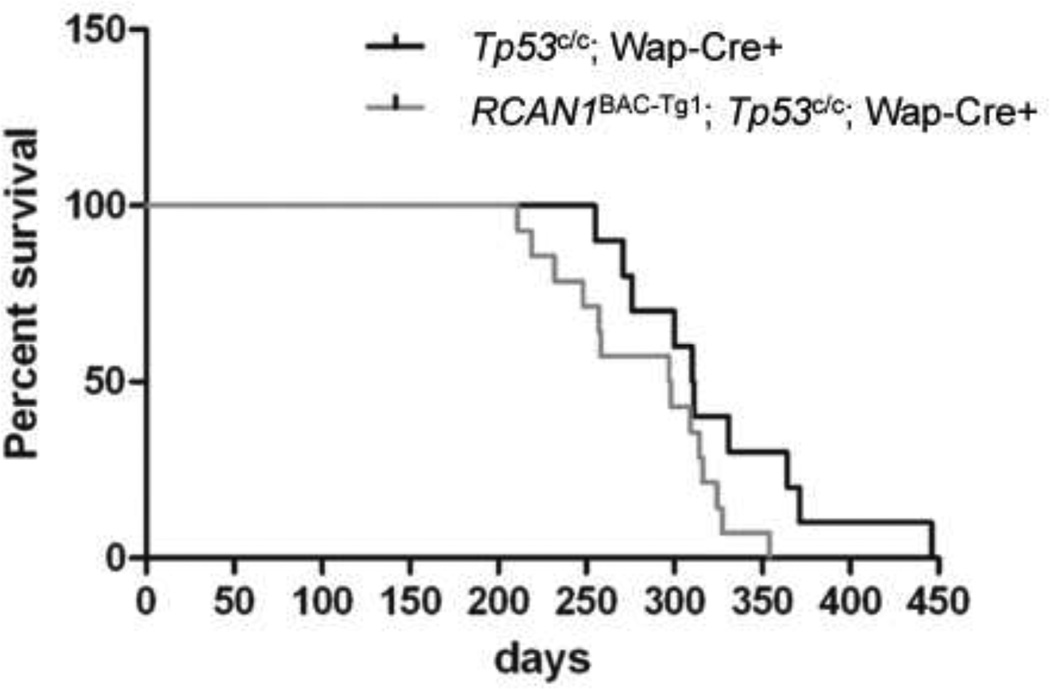
To test the effects of RCAN1 gene dosage on tumorigenesis in our BAC-Tg mice we first crossed these mice with Tp53 conditional knockout (CKO) mice, p53flex7 (Chen et al. 2005). The resulting RCAN1BAC-Tg1; p53flex7/+ mice were then crossed with p53flex7/flex7; WAPcre/cre mice to obtain two types of female mice: Tg-RCAN1; p53flex7/flex7; WAPcre/+, and p53flex7/flex7; WAPcre/+. These females were mated with wild type males to induce pregnancy and cre expression, leading to inactivation of the Tp53 tumor suppressor gene specifically in epithelial cells of the mammary glands. Thus the only variable in the resulting cohorts of tumor-predisposed mice was RCAN1 gene dosage (either wt or BAC-Tg). We followed a cohort of 14 female BAC-Tg mice and 10 wild-type female for the kinetics (latency) of mammary tumor formation, and obtained tumor tissue from each mice at necropsy. As shown in Figure 7, tumorigenesis in the p53-deficient background was either unaltered or slightly accelerated in the BAC-Tg mice (RCAN1BAC-Tg1; p53flex7/flex7; WAPcre/+ (T50=283 days); p53flex7/flex7; WAPcre/+ (T50=325 days); p=0.07, log-rank test).
Figure 7. Mammary gland tumor formation is unaltered or slightly accelerated by the RCAN1BAC-Tg1 allele crossed into p53flex7/flex7; Wapcre/+.
A total of 14 female experimental mice (RCAN1BAC-Tg1; p53flex7/flex7; Wapcre/+) and 10 matched control female mice (p53flex7/flex7; Wapcre/+) were followed for tumor formation. The BAC-Tg mice showed a shorter average survival (RCAN1BAC-Tg1; p53flex7/flex7; Wapcre/+ (T50=283 days) versus p53flex7/flex7; Wapcre/+ (T50=325 days)) but the difference was not statistically significant (p=0.07, log-rank test). Time is depicted in days after birth of the first litter.
The adenocarcinomas that form in this mouse model are clonal proliferations of p53-null mammary epithelial cells, which are surrounded by variable amounts of (p53-wild-type) reactive stromal cells. To verify the Tp53 exon 7 deletion in the tumors from our mouse cohort we used PCR across the floxed exon to assess its deletion in genomic DNA from each tumor. This procedure revealed, as expected, a strong recombined band in all tumors, regardless of the presence or absence of the RCAN1 Tg (Suppl. Fig. S6). However, there was a modest but reproducible relative increase in the minor non-recombined p53flex7 PCR product from the p53flex7:RCAN1Tg tumors. This result suggests that the p53flex7:RCAN1Tg tumors may have modestly greater numbers of stromal cells (non-recombined Tp53 CKO allele) compared to the tumors which form in the absence of the RCAN1 Tg – a possibility that we plan to test in the future using quantitative histological analysis. However, as all of the mice in our experiment died from mammary cancers, the survival data clearly show that RCAN1 is not a tumor suppressor when over-expressed at low levels in this in vivo model, without additional potentially cooperating chromosome 21-derived genes.
Discussion
The RCAN1 (DSCR1) gene encodes a key regulator of the cytoplasmic phosphatase CaN, and it is one of several genes on chromosome 21 that are thought to make important contributions to cognitive disability, heart defects, and possibly other phenotypes such as resistance to solid tumors, including breast cancer and neuroblastoma, in human DS. DS is caused by an extra copy of multiple genes on human chromosome 21, and BAC transgenic mice (Table 1) are excellent models to investigate the contributions of individual chromosome 21-linked genes to the constellation of phenotypic features in this disorder. Here we have reported the generation of three BAC-Tg mouse lines expressing epitope-tagged human RCAN1 in relevant tissues, including brain, heart and thymus, in a copy number-dependent manner. Our initial set of phenotypic studies described here highlight the full viability of these mice, without major anatomical defects, which is consistent with prior data showing that restoration of the Rcan1 gene to disomy has no impact on developmental anomalies, including heart defects, in trisomy 16 mice, which are a model for DS (Lange et al. 2005). Nonetheless, we have shown here that the extra dosage of RCAN1 from the BAC-Tg produces quantitative abnormalities including impairment of hippocampal LTP in brain slices and increased calcium-dependent induction of NFATc1 expression in MEFs. Deficient LTP in these mice adds to multiple other lines of evidence that cumulatively support a role for RCAN1 triplication in the cognitive disability phenotype of human DS.
Table 1. BAC- and YAC-Tg mice for testing chromosome 21 gene function.
Moderate over-expression refers to 1.5- to 3-fold greater than in wild type littermates. Learning deficits were scored by Morris water maze and fear conditioning protocols. Anxiety/exploratory behavior were assessed in the open field and male intruder test. LTP, hippocampal long-term potentiation; LTD, hippocampal long-term depression; WB, western blotting. Relevant studies utilizing cDNA-transgenic mice, Ts65Dn and its engineered derivatives including Ms1Rhr/Ts65Dn and Ts1Rhr, and the Tc1 trans-chromosomal mouse line are not listed here but are discussed and referenced in the main text.
| Tg | Gene(s) | Expression | Phenotypes | Additional Results | Refs. |
|---|---|---|---|---|---|
| BAC | Human COL6A1 (21q22.3) |
Tissue-specificity similar to native Col6a1 gene; BAC copy number-dependent moderate over-expression by WB. |
Viable; normal histology of heart and skin. |
(Xing et al. 2007) | |
| YAC | Human DYRK1A/MNB; (21q22.13); additional genes |
Widely expressed; similar to endogenous Dyrk1a gene. Moderate over-expression in YAC-Tg mice by RT-PCR. |
Locomotor hypo-activity. Learning impairment. Normal LTP. |
Increased neuronal cell numbers in cerebral cortex. |
(Chabert et al. 2004; Smith and Rubin 1997; Smith et al. 1997) |
| YAC | Human DYRK1A/MNB; (21q22.13); additional genes |
Expressed in brain; moderate over- expression by WB. |
Learning impairment and locomotor hyper-activity. |
Increased brain weight and neuronal size. Increased CREB and CREM phosphorylation. |
(Branchi et al. 2004) |
| BAC | Human DYRK1A/MNB; (21q22.13) |
Expressed in brain; high in hippocampus. Moderate over-expression by WB. |
Learning deficits and deficiencies in both LTP and LTD. |
(Ahn et al. 2006) | |
| BAC | Human SIM2 (21q22.13) |
Tissue-specificity similar to endogenous Sim2 gene; moderate over-expression by RT-PCR. |
Increased anxiety/decreased exploratory behavior; normal learning. |
No histopathology in brain or several other organs examined. |
(Chrast et al. 2000) |
| BAC | Human SYNJ1 (21q22.11) |
Expressed in brain; other organs not tested. Moderate over-expression by WB. |
Learning impairment. | PtdIns(4,5)P2 metabolism is altered in the brain. |
(Voronov et al. 2008) |
| YAC | Human APP (21q21.3) |
Expressed in brain (neurons); lower in other organs. Moderate over-expression by RT-PCR and WB. |
Expression levels of the YAC-Tg vary greatly across different mouse strains. |
(Kulnane and Lamb 2001; Lamb et al. 1993; Lehman et al. 2003) |
|
| BAC | Human RCAN1/DSCR1 (21q22.11) |
Highest expression in brain; similar to endogenous Rcan1 gene. Moderate over- expression by WB. |
Impaired LTP; Tg dosage- dependent. |
BAC-Tg produces an epitope- tagged protein facilitating isolation of RCAN1 protein complexes. |
Current study. |
The above phenotypic measurements also indicate that the epitope-tagged RCAN1 BAC-Tg construct is biologically active, and our biochemical studies by tandem affinity purification show that RCAN1 forms a stable complex with CaN and Gsk3b, plus several other proteins. We have further shown that the CaN in this complex is highly active. Prior discordant findings in the literature of both stimulatory and inhibitory effects of RCAN1 on CaN may be due to differences in the experimental systems utilized, to tissue-specific effects, and to a dose-dependence in which RCAN1 becomes inhibitory only at a high RCAN1/CaN ratio. We also found a novel candidate interacting protein, encoded by the LOC73419 gene, which is a component of the RCAN1 complex in vivo. Based on our transfection experiments, this protein-protein association is stronger in Neuro-2a neural lineage cells than in 293T kidney cells, which suggests that cellular context (other proteins and/or post-translational modifications) is important for the interaction. The LOC73419 protein contains a DUF89 domain, for which the function remains to be determined. However, a recent study showed evidence for a growth-promoting role of its human orthologue, C6orf211, in MCF7 breast cancer cells (Dunbier et al. 2011).
There is agreement in multiple studies that children and adults with DS have a reduced incidence of solid tumors including neuroblastoma and breast cancer (Hasle 2001), but the role of RCAN1 as an anti-angiogenic tumor suppressor gene in DS needs more investigation (Hesser et al. 2004; Iizuka et al. 2004; Minami et al. 2004; Ryeom et al. 2008). Our finding that RCAN1 over-expression has no effect or slightly accelerates tumorigenesis in p53-null mouse mammary glands argues against the RCAN1 gene being a breast cancer suppressor, but these data do not rule out such a role. For example, the suppressor activity may only be evident in tumor types that have a stronger vascular dependence or, more likely, it may require additive or synergistic interaction with other chromosome 21 genes, such as ETS2, SIM2 and others (Kwak et al. 2007; Sussan et al. 2008; Yang and Reeves 2011), that are also present at an increased dosage in DS. This consideration of gene-gene interactions is crucial, not only for explaining the decreased solid tumor incidence in DS but also for gaining a better molecular understanding of the other important aspects of the syndrome, including heart defects, cognitive disability and early-onset Alzheimer’s disease. The stable RCAN1 BAC-Tg lines that we have reported here will be useful for crossing to other transgenic, trans-chromosomal and knockout mouse lines (Herault et al. 2012; Liu et al. 2011a; Liu et al. 2011b; Wiseman et al. 2009) to identify the critical combinations of genes that account for these phenotypes.
Supplementary Material
Acknowledgments
This work was supported by grants from the N.I.H. (P01-HD035897 to B.T. and W.S., and R01- AG034248 to O.A.).
References
- Ahn KJ, Jeong HK, Choi HS, Ryoo SR, Kim YJ, Goo JS, Choi SY, Han JS, Ha I, Song WJ. DYRK1A BAC transgenic mice show altered synaptic plasticity with learning and memory defects. Neurobiology of disease. 2006;22:463–472. doi: 10.1016/j.nbd.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S. Chromosome 21 and down syndrome: from genomics to pathophysiology. Nature reviews. 2004;5:725–738. doi: 10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]
- Baek KH, Zaslavsky A, Lynch RC, Britt C, Okada Y, Siarey RJ, Lensch MW, Park IH, Yoon SS, Minami T, Korenberg JR, Folkman J, Daley GQ, Aird WC, Galdzicki Z, Ryeom S. Down's syndrome suppression of tumour growth and the role of the calcineurin inhibitor DSCR1. Nature. 2009;459:1126–1130. doi: 10.1038/nature08062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branchi I, Bichler Z, Minghetti L, Delabar JM, Malchiodi-Albedi F, Gonzalez MC, Chettouh Z, Nicolini A, Chabert C, Smith DJ, Rubin EM, Migliore-Samour D, Alleva E. Transgenic mouse in vivo library of human Down syndrome critical region 1: association between DYRK1A overexpression, brain development abnormalities, and cell cycle protein alteration. J Neuropathol Exp Neurol. 2004;63:429–440. doi: 10.1093/jnen/63.5.429. [DOI] [PubMed] [Google Scholar]
- Chabert C, Jamon M, Cherfouh A, Duquenne V, Smith DJ, Rubin E, Roubertoux PL. Functional analysis of genes implicated in Down syndrome: 1. Cognitive abilities in mice transpolygenic for Down Syndrome Chromosomal Region-1 (DCR-1) Behav Genet. 2004;34:559–569. doi: 10.1007/s10519-004-5584-3. [DOI] [PubMed] [Google Scholar]
- Chan B, Greenan G, McKeon F, Ellenberger T. Identification of a peptide fragment of DSCR1 that competitively inhibits calcineurin activity in vitro and in vivo. Proc Natl Acad Sci U S A. 2005;102:13075–13080. doi: 10.1073/pnas.0503846102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KT, Shi YJ, Min KT. The Drosophila homolog of Down's syndrome critical region 1 gene regulates learning: implications for mental retardation. Proc Natl Acad Sci U S A. 2003;100:15794–15799. doi: 10.1073/pnas.2536696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrast R, Scott HS, Madani R, Huber L, Wolfer DP, Prinz M, Aguzzi A, Lipp HP, Antonarakis SE. Mice trisomic for a bacterial artificial chromosome with the single-minded 2 gene (Sim2) show phenotypes similar to some of those present in the partial trisomy 16 mouse models of Down syndrome. Hum Mol Genet. 2000;9:1853–1864. doi: 10.1093/hmg/9.12.1853. [DOI] [PubMed] [Google Scholar]
- Cook CN, Hejna MJ, Magnuson DJ, Lee JM. Expression of calcipressin1, an inhibitor of the phosphatase calcineurin, is altered with aging and Alzheimer's disease. J Alzheimers Dis. 2005;8:63–73. doi: 10.3233/jad-2005-8108. [DOI] [PubMed] [Google Scholar]
- Davies KJ, Ermak G, Rothermel BA, Pritchard M, Heitman J, Ahnn J, Henrique-Silva F, Crawford D, Canaider S, Strippoli P, Carinci P, Min KT, Fox DS, Cunningham KW, Bassel-Duby R, Olson EN, Zhang Z, Williams RS, Gerber HP, Perez-Riba M, Seo H, Cao X, Klee CB, Redondo JM, Maltais LJ, Bruford EA, Povey S, Molkentin JD, McKeon FD, Duh EJ, Crabtree GR, Cyert MS, de la Luna S, Estivill X. Renaming the DSCR1/Adapt78 gene family as RCAN: regulators of calcineurin. Faseb J. 2007;21:3023–3028. doi: 10.1096/fj.06-7246com. [DOI] [PubMed] [Google Scholar]
- Dunbier AK, Anderson H, Ghazoui Z, Lopez-Knowles E, Pancholi S, Ribas R, Drury S, Sidhu K, Leary A, Martin LA, Dowsett M. ESR1 is co-expressed with closely adjacent uncharacterised genes spanning a breast cancer susceptibility locus at 6q25.1. PLoS Genet. 2011;7:e1001382. doi: 10.1371/journal.pgen.1001382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermak G, Morgan TE, Davies KJ. Chronic overexpression of the calcineurin inhibitory gene DSCR1 (Adapt78) is associated with Alzheimer's disease. The Journal of biological chemistry. 2001;276:38787–38794. doi: 10.1074/jbc.M102829200. [DOI] [PubMed] [Google Scholar]
- Fuentes JJ, Genesca L, Kingsbury TJ, Cunningham KW, Perez-Riba M, Estivill X, de la Luna S. DSCR1, overexpressed in Down syndrome, is an inhibitor of calcineurin-mediated signaling pathways. Human molecular genetics. 2000;9:1681–1690. doi: 10.1093/hmg/9.11.1681. [DOI] [PubMed] [Google Scholar]
- Fuentes JJ, Pritchard MA, Estivill X. Genomic organization, alternative splicing, and expression patterns of the DSCR1 (Down syndrome candidate region 1) gene. Genomics. 1997;44:358–361. doi: 10.1006/geno.1997.4866. [DOI] [PubMed] [Google Scholar]
- Genesca L, Aubareda A, Fuentes JJ, Estivill X, De La Luna S, Perez-Riba M. Phosphorylation of calcipressin 1 increases its ability to inhibit calcineurin and decreases calcipressin half-life. Biochem J. 2003;374:567–575. doi: 10.1042/BJ20030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Molecular cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- Hasle H. Pattern of malignant disorders in individuals with Down's syndrome. Lancet Oncol. 2001;2:429–436. doi: 10.1016/S1470-2045(00)00435-6. [DOI] [PubMed] [Google Scholar]
- Herault Y, Duchon A, Velot E, Marechal D, Brault V. The in vivo Down syndrome genomic library in mouse. Prog Brain Res. 2012;197:169–197. doi: 10.1016/B978-0-444-54299-1.00009-1. [DOI] [PubMed] [Google Scholar]
- Hesser BA, Liang XH, Camenisch G, Yang S, Lewin DA, Scheller R, Ferrara N, Gerber HP. Down syndrome critical region protein 1 (DSCR1), a novel VEGF target gene that regulates expression of inflammatory markers on activated endothelial cells. Blood. 2004;104:149–158. doi: 10.1182/blood-2004-01-0273. [DOI] [PubMed] [Google Scholar]
- Hilioti Z, Gallagher DA, Low-Nam ST, Ramaswamy P, Gajer P, Kingsbury TJ, Birchwood CJ, Levchenko A, Cunningham KW. GSK-3 kinases enhance calcineurin signaling by phosphorylation of RCNs. Genes & development. 2004;18:35–47. doi: 10.1101/gad.1159204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes & development. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- Iizuka M, Abe M, Shiiba K, Sasaki I, Sato Y. Down syndrome candidate region 1,a downstream target of VEGF, participates in endothelial cell migration and angiogenesis. Journal of vascular research. 2004;41:334–344. doi: 10.1159/000079832. [DOI] [PubMed] [Google Scholar]
- Kingsbury TJ, Cunningham KW. A conserved family of calcineurin regulators. Genes & development. 2000;14:1595–1604. [PMC free article] [PubMed] [Google Scholar]
- Kulnane LS, Lamb BT. Neuropathological characterization of mutant amyloid precursor protein yeast artificial chromosome transgenic mice. Neurobiology of disease. 2001;8:982–992. doi: 10.1006/nbdi.2001.0446. [DOI] [PubMed] [Google Scholar]
- Kwak HI, Gustafson T, Metz RP, Laffin B, Schedin P, Porter WW. Inhibition of breast cancer growth and invasion by single-minded 2s. Carcinogenesis. 2007;28:259–266. doi: 10.1093/carcin/bgl122. [DOI] [PubMed] [Google Scholar]
- Lamb BT, Sisodia SS, Lawler AM, Slunt HH, Kitt CA, Kearns WG, Pearson PL, Price DL, Gearhart JD. Introduction and expression of the 400 kilobase amyloid precursor protein gene in transgenic mice [corrected] Nat Genet. 1993;5:22–30. doi: 10.1038/ng0993-22. [DOI] [PubMed] [Google Scholar]
- Lange AW, Molkentin JD, Yutzey KE. DSCR1 gene expression is dependent on NFATc1 during cardiac valve formation and colocalizes with anomalous organ development in trisomy 16 mice. Developmental biology. 2004;266:346–360. doi: 10.1016/j.ydbio.2003.10.036. [DOI] [PubMed] [Google Scholar]
- Lange AW, Rothermel BA, Yutzey KE. Restoration of DSCR1 to disomy in the trisomy 16 mouse model of Down syndrome does not correct cardiac or craniofacial development anomalies. Dev Dyn. 2005;233:954–963. doi: 10.1002/dvdy.20433. [DOI] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Lehman EJ, Kulnane LS, Gao Y, Petriello MC, Pimpis KM, Younkin L, Dolios G, Wang R, Younkin SG, Lamb BT. Genetic background regulates beta-amyloid precursor protein processing and beta-amyloid deposition in the mouse. Hum Mol Genet. 2003;12:2949–2956. doi: 10.1093/hmg/ddg322. [DOI] [PubMed] [Google Scholar]
- Liu C, Belichenko PV, Zhang L, Fu D, Kleschevnikov AM, Baldini A, Antonarakis SE, Mobley WC, Yu YE. Mouse models for Down syndrome-associated developmental cognitive disabilities. Dev Neurosci. 2011a;33:404–413. doi: 10.1159/000329422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Morishima M, Yu T, Matsui S, Zhang L, Fu D, Pao A, Costa AC, Gardiner KJ, Cowell JK, Nowak NJ, Parmacek MS, Liang P, Baldini A, Yu YE. Genetic analysis of Down syndrome-associated heart defects in mice. Hum Genet. 2011b;130:623–632. doi: 10.1007/s00439-011-0980-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig T, Fisher P, Murty V, Efstratiadis A. Development of mammary adenocarcinomas by tissue-specific knockout of Brca2 in mice. Oncogene. 2001;20:3937–3948. doi: 10.1038/sj.onc.1204512. [DOI] [PubMed] [Google Scholar]
- Ma H, Xiong H, Liu T, Zhang L, Godzik A, Zhang Z. Aggregate formation and synaptic abnormality induced by DSCR1. Journal of neurochemistry. 2004;88:1485–1496. doi: 10.1046/j.1471-4159.2003.02294.x. [DOI] [PubMed] [Google Scholar]
- Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- Minami T, Horiuchi K, Miura M, Abid MR, Takabe W, Noguchi N, Kohro T, Ge X, Aburatani H, Hamakubo T, Kodama T, Aird WC. Vascular endothelial growth factor- and thrombin-induced termination factor, Down syndrome critical region-1, attenuates endothelial cell proliferation and angiogenesis. J Biol Chem. 2004;279:50537–50554. doi: 10.1074/jbc.M406454200. [DOI] [PubMed] [Google Scholar]
- Nikolaev AY, Li M, Puskas N, Qin J, Gu W. Parc: a cytoplasmic anchor for p53. Cell. 2003;112:29–40. doi: 10.1016/s0092-8674(02)01255-2. [DOI] [PubMed] [Google Scholar]
- Osoegawa K, Mammoser AG, Wu C, Frengen E, Zeng C, Catanese JJ, de Jong PJ. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 2001;11:483–496. doi: 10.1101/gr.169601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzzo D, Privitera L, Leznik E, Fa M, Staniszewski A, Palmeri A, Arancio O. Picomolar amyloid-beta positively modulates synaptic plasticity and memory in hippocampus. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:14537–14545. doi: 10.1523/JNEUROSCI.2692-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Zhao D, Liu X, Nagy JA, Hoang MV, Brown LF, Dvorak HF, Zeng H. Down syndrome candidate region 1 isoform 1 mediates angiogenesis through the calcineurin-NFAT pathway. Mol Cancer Res. 2006;4:811–820. doi: 10.1158/1541-7786.MCR-06-0126. [DOI] [PubMed] [Google Scholar]
- Reynolds LE, Watson AR, Baker M, Jones TA, D'Amico G, Robinson SD, Joffre C, Garrido-Urbani S, Rodriguez-Manzaneque JC, Martino-Echarri E, Aurrand-Lions M, Sheer D, Dagna-Bricarelli F, Nizetic D, McCabe CJ, Turnell AS, Kermorgant S, Imhof BA, Adams R, Fisher EM, Tybulewicz VL, Hart IR, Hodivala-Dilke KM. Tumour angiogenesis is reduced in the Tc1 mouse model of Down's syndrome. Nature. 2010;465:813–817. doi: 10.1038/nature09106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothermel B, Vega RB, Yang J, Wu H, Bassel-Duby R, Williams RS. A protein encoded within the Down syndrome critical region is enriched in striated muscles and inhibits calcineurin signaling. The Journal of biological chemistry. 2000;275:8719–8725. doi: 10.1074/jbc.275.12.8719. [DOI] [PubMed] [Google Scholar]
- Ryeom S, Baek KH, Rioth MJ, Lynch RC, Zaslavsky A, Birsner A, Yoon SS, McKeon F. Targeted deletion of the calcineurin inhibitor DSCR1 suppresses tumor growth. Cancer cell. 2008;13:420–431. doi: 10.1016/j.ccr.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Sanna B, Brandt EB, Kaiser RA, Pfluger P, Witt SA, Kimball TR, van Rooij E, De Windt LJ, Rothenberg ME, Tschop MH, Benoit SC, Molkentin JD. Modulatory calcineurininteracting proteins 1 and 2 function as calcineurin facilitators in vivo. Proc Natl Acad Sci U S A. 2006;103:7327–7332. doi: 10.1073/pnas.0509340103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Rubin EM. Functional screening and complex traits: human 21q22.2 sequences affecting learning in mice. Hum Mol Genet. 1997;6:1729–1733. doi: 10.1093/hmg/6.10.1729. [DOI] [PubMed] [Google Scholar]
- Smith DJ, Stevens ME, Sudanagunta SP, Bronson RT, Makhinson M, Watabe AM, O'Dell TJ, Fung J, Weier HU, Cheng JF, Rubin EM. Functional screening of 2 Mb of human chromosome 21q22.2 in transgenic mice implicates minibrain in learning defects associated with Down syndrome. Nat Genet. 1997;16:28–36. doi: 10.1038/ng0597-28. [DOI] [PubMed] [Google Scholar]
- Stewart MP, McDowall A, Hogg N. LFA-1-mediated adhesion is regulated by cytoskeletal restraint and by a Ca2+-dependent protease, calpain. The Journal of cell biology. 1998;140:699–707. doi: 10.1083/jcb.140.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussan TE, Yang A, Li F, Ostrowski MC, Reeves RH. Trisomy represses Apc(Min)- mediated tumours in mouse models of Down's syndrome. Nature. 2008;451:73–75. doi: 10.1038/nature06446. [DOI] [PubMed] [Google Scholar]
- Vega RB, Rothermel BA, Weinheimer CJ, Kovacs A, Naseem RH, Bassel-Duby R, Williams RS, Olson EN. Dual roles of modulatory calcineurin-interacting protein 1 in cardiac hypertrophy. Proc Natl Acad Sci U S A. 2003;100:669–674. doi: 10.1073/pnas.0237225100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronov SV, Frere SG, Giovedi S, Pollina EA, Borel C, Zhang H, Schmidt C, Akeson EC, Wenk MR, Cimasoni L, Arancio O, Davisson MT, Antonarakis SE, Gardiner K, De Camilli P, Di Paolo G. Synaptojanin 1-linked phosphoinositide dyshomeostasis and cognitive deficits in mouse models of Down's syndrome. Proc Natl Acad Sci U S A. 2008;105:9415–9420. doi: 10.1073/pnas.0803756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder DG, Sweatt JD. Roles of serine/threonine phosphatases in hippocampal synaptic plasticity. Nat Rev Neurosci. 2001;2:461–474. doi: 10.1038/35081514. [DOI] [PubMed] [Google Scholar]
- Wiseman FK, Alford KA, Tybulewicz VL, Fisher EM. Down syndrome--recent progress and future prospects. Human molecular genetics. 2009;18:R75–R83. doi: 10.1093/hmg/ddp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolvetang EJ, Wilson TJ, Sanij E, Busciglio J, Hatzistavrou T, Seth A, Hertzog PJ, Kola I. ETS2 overexpression in transgenic models and in Down syndrome predisposes to apoptosis via the p53 pathway. Human molecular genetics. 2003;12:247–255. doi: 10.1093/hmg/ddg015. [DOI] [PubMed] [Google Scholar]
- Xing L, Salas M, Lin CS, Zigman W, Silverman W, Subramaniyam S, Murty VV, Tycko B. Faithful tissue-specific expression of the human chromosome 21-linked COL6A1 gene in BAC-transgenic mice. Mamm Genome. 2007;18:113–122. doi: 10.1007/s00335-006-0082-y. [DOI] [PubMed] [Google Scholar]
- Yang A, Reeves RH. Increased survival following tumorigenesis in Ts65Dn mice that model Down syndrome. Cancer Res. 2011;71:3573–3581. doi: 10.1158/0008-5472.CAN-10-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Rothermel B, Vega RB, Frey N, McKinsey TA, Olson EN, Bassel-Duby R, Williams RS. Independent signals control expression of the calcineurin inhibitory proteins MCIP1 and MCIP2 in striated muscles. Circulation research. 2000;87:E61–E68. doi: 10.1161/01.res.87.12.e61. [DOI] [PubMed] [Google Scholar]
- Zeng H, Chattarji S, Barbarosie M, Rondi-Reig L, Philpot BD, Miyakawa T, Bear MF, Tonegawa S. Forebrain-specific calcineurin knockout selectively impairs bidirectional synaptic plasticity and working/episodic-like memory. Cell. 2001;107:617–629. doi: 10.1016/s0092-8674(01)00585-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.