Abstract
Mammalian SWI/SNF complexes utilize either BRG1 or BRM as alternative catalytic subunits with DNA-dependent ATPase activity to remodel chromatin. Although the two proteins are 75% identical, broadly expressed, and have similar biochemical activities in vitro, BRG1 is essential for mouse embryonic development, while BRM is dispensable. To investigate whether BRG1 and BRM have overlapping functions during mouse embryogenesis, we performed double-heterozygous intercrosses using constitutive null mutations previously created by gene targeting. The progeny of these crosses had a distribution of genotypes that was significantly skewed relative to their combined gene dosage. This was most pronounced at the top and bottom of the gene dosage hierarchy with a 1.5-fold overrepresentation of Brg1+/+;Brm+/+ mice and a corresponding 1.6-fold underrepresentation of Brg1+/−;Brm−/− mice. To account for the underrepresentation of Brg1+/−;Brm−/− mice, timed matings and blastocyst outgrowth assays demonstrated that ~50% of these embryos failed to develop beyond the peri-implantation stage. These results challenge the idea that BRG1 is the exclusive catalytic subunit of SWI/SNF complexes in ES cells and suggest that BRM also interacts with the pluripotency transcription factors to facilitate self-renewal of the inner cell mass. In contrast to implantation, the Brm genotype did not influence an exencephaly phenotype that arises because of Brg1 haploinsufficiency during neural tube closure and that results in peri-natal lethality. Taken together, these results support the idea that BRG1 and BRM have overlapping functions for certain developmental processes but not others during embryogenesis.
Keywords: BRG1, BRM, redundancy, blastocyst, peri-implantation development
Introduction
The mammalian genome is packaged as chromatin, and the most fundamental unit is the nucleosome, which consists of a 147-bp segment of DNA wrapped around an octamer of histones (Kornberg and Lorch 1999). Because nucleosomes are separated by short (10–90 bp) DNA linkers, they are ubiquitous and number ~3 × 107 per nucleus in mammalian cells. Although nucleosomes and higher-order chromatin structures help compact the genome and maintain its organization, they present major challenges for transcription, DNA replication, and DNA repair. To counteract the effects of chromatin and facilitate these DNA-templated processes, four families of chromatin-remodeling complexes (SWI/SNF, ISWI, NuRD/Ni-2/CHD, and INO80) are recruited to numerous sites in the genome where they harness the energy of ATP hydrolysis to alter nucleosome position (Cairns 2007, Flaus and Owen-Hughes 2011, Hota and Bartholomew 2011).
Mammalian SWI/SNF complexes consist of 9–12 subunits and utilize either BRG1 (also known as SMARCA4) or BRM (also known as SMARCA2) as their catalytic subunit with DNA-dependent ATPase activity (de la Serna et al 2006, Ho and Crabtree 2010). When recruited to promoters by sequence-specific transcription factors, SWI/SNF complexes slide or evict nucleosomes away from transcriptional start sites to regulate RNA Polymerase II occupancy and transcriptional initiation (Liu et al 2011). Although BRG1 and BRM are able to slide or evict nucleosomes on their own as recombinant proteins in cell-free systems (Phelan et al 2000), their chromatin-remodeling activity is enhanced in the presence of additional SWI/SNF subunits [often referred to as BAFs (BRG1 or BRM associated factors) followed by a number corresponding to their molecular weight in kDa] (Phelan et al 1999).
Based on gene-targeting experiments in mice, the BRG1, SNF5/BAF47, BAF155, and BAF250A/ARID1A subunits are required during the peri-implantation stage (Bultman et al 2000, de la Serna et al 2006, Gao et al 2008, Guidi et al 2001, Ho and Crabtree 2010, Kim et al 2001, Klochendler-Yeivin et al 2000, Roberts et al 2000). This is compatible with the pluripotency transcription factors SOX2, OCT4, and Nanog recruiting a SWI/SNF subcomplex, called esBAF, to downstream target genes in embryonic stem (ES) cells to facilitate their pluripotency and self-renewal (Gao et al 2008, Ho et al 2009a, Ho et al 2009b, Kaeser et al 2008, Kidder et al 2009, Yan et al 2008). This complex also participates in the conversion of fibroblasts into induced pluripotent stem (iPS) cells (Singhal et al 2010). In addition, the BAF180/PBRM1 subunits and BAF60C are required for cardiovascular development and survival beyond the mid-gestation stage (Bruneau 2010, de la Serna et al 2006, Ho and Crabtree 2010, Lickert et al 2004, Wang et al 2004). In this case, it is the cardiogenic transcription factors TBX5, GATA4, and Nkx2–5 that interact with SWI/SNF to program non-cardiac mesoderm into cardiomyocytes (Bruneau 2010, Takeuchi and Bruneau 2009). SWI/SNF complexes participate in many other developmental processes based, in part, on an allelic series of Brg1 mutations generated by Cre/loxP conditional gene targeting and ENU mutagenesis (Table 1).
Table 1.
Mouse Brg1 allelic series
| Mutant Allele | Tissue(s) | Phenotype | Reference(s) |
|---|---|---|---|
| Null (constitutive) | All | −/− peri-implantation lethality +/− exencephaly and mammary tumors with incomplete penetrance |
Bultman et al. 2000 Bultman et al. 2008 |
| ENU1 (constitutive) Hypomorph (E1083G) | All | Null/ENU1 E12.5 lethality, anemia associated with globin and replication defects ENU1/ENU1 post-natal failure to thrive |
Bultman et al. 2005 Kim et al. 2007, 2009a, 2009b Cohen et al. 2010 |
| Zp3-Cre | Oocytes | Maternal effect: progeny exhibit 2-cell arrest associated with ZGA defect | Bultman et al. 2006 |
| Tie2-Cre | VECs | E11.5 lethality associated with yolk sac angiogenesis defects |
Griffin et al. 2008, 2011 Stankunas et al. 2008 Curtis and Griffin 2012 |
| Lck-Cre | T cells | DN-DP block in thymus, CD4 derepression, intestinal inflammation |
Gebuhr et al. 2003 Chi et al. 2003 |
| Lck-Cre Knock-in Antimorph (K785R) | T cells | T cell defect with similarities and differences compared to conditional null | Jani et al. 2008 |
| Nestin-Cre | Neuronal progenitors | Neuronal differentiation defects and early postnatal lethality |
Matsumoto et al. 2006 Lessard et al. 2007 |
| Sm22α-Cre | Cardiomyocytes | Lethality just after E11.5 associated with proliferation defect; inducible mutation prevents MHC switch from occurring in adults in response to hypertrophy | Hang et al. 2010 |
| Nkx2.5-Cre | Cardiomyocytes | Cardiac defects with lethality mostly at E10.5 but with some survivors | Takeuchi et al. 2011 |
| Mhv-Cre | Germ cells | Synaptic defects and meiotic arrest of spermatocytes | Kim et al. 2012 |
| Mx1-Cre | VEC, HSC, liver | Lethal cardiovascular defect within ~1 month of mutation induction if also Brm−/− | Willis et al. 2012 |
| Wap-Cre | Mammary, ovary, uterus | No mammary tumors but ovarian cysts and uterine tumors | Serber et al. 2012 |
| K14-Cre | Keratinocytes and epithelial cells | Peri-natal lethality due to dehydration from skin barrier permeability impairment; limb malformations due to AER defect during development | Indra et al. 2005 |
| Dhh-Cre | Schwann cells | Neuropathy due to Schwann cell differentiation and myelination defect | Welder et al. 2012 |
| Le-Cre | Eye | Microphthalmia and other eye defects | He et al. 2010 |
| smMHC-Cre | Smooth muscle | Lethality associated with smooth muscle defects in cardiovascular, pulmonary, and other sites including GI tract | Zhang et al. 2012 |
-Note: does not include RNAi experiments or expression of dominant-negative BRG1 in tissue-culture cells including ES cells, which have been important for showing a role in processes such as pluripotency and muscle cell differentiation (for example, de la Serna et al. 2001).
-All mutant alleles are conditional nulls unless indicated otherwise.
-Abbreviations: ZGA, zygotic genome activation; VEC, vascular endothelial cell; DN, double negative (CD4−CD8−); DP, double positive (CD4+CD8+); MHC, myosin; heavy chain; HSC, hematopoitic stem cells; AER, apical ectodermal ridge; consult references for gene names of promoters used to drive Cre.
From these gene-targeting experiments, the BRM subunit is a notable exception because Brm null homozygotes are viable and fertile (Reyes et al 1998). Several lines of evidence suggest that Brg1 can functionally compensate for the loss of Brm in these mice: 1, the two proteins are 75% identical and have similar biochemical activities in vitro (Chiba et al 1994, Phelan et al 2000); 2, BRG1 protein levels are elevated in Brm−/− tissues (Reyes et al 1998); 3, Brg1 is expressed at much higher levels than Brm in the early embryo (Bultman et al 2000, LeGouy et al 1998), and esBAF incorporates BRG1 as the catalytic subunit rather than BRM (Ho et al 2009b). It is currently unclear whether Brm functions during embryogenesis. On the one hand, it might not since BRG1 is expressed at higher levels in undifferentiated cells that are highly proliferative, whereas BRM is enriched in differentiated cells that are less proliferative (Reisman et al 2005). Furthermore, RNAi knockdown experiments have shown that BRG1 and BRM can have opposing functions in tissue-culture cells with BRG1 inhibiting differentiation and BRM promoting differentiation (Flowers et al 2009). These findings suggest BRM might be more important for maintaining homeostasis in adult cells as supported by a recent cardiovascular study (Willis et al 2012). On the other hand, Brm might participate in certain developmental processes during embryogenesis, but null mutants do not exhibit an obvious phenotype because of Brg1 functional compensation. To distinguish between these possibilities, we have analyzed Brg1-Brm double mutants at different stages of development. As we report here, Brm is required to prevent implantation defects but not neural tube defects in Brg1 heterozygotes. These findings suggest that Brm participates in a subset of developmental processes that are sensitive to Brg1 dosage.
Materials and methods
Mice
All mouse experiments were approved by the Institutional Animal Care and Use Committees (IACUC) review board at the University of North Carolina at Chapel Hill and were performed in accordance with federal guidelines. The Brg1 and Brm mutant lines have been characterized previously (Bultman et al 2000, Reyes et al 1998). For timed matings, noon of the day that each copulation plug was detected was considered E0.5.
Blastocyst outgrowth analyses
Double heterozygotes were intercrossed, and E3.5 blastocysts were flushed from the uterine tract. Blastocysts were placed in 10 µL of ES cell media (DMEM, 15% FCS) in Nunc dish microwells (Nalgene, Palo Alto, CA) and grown under paraffin oil in 5% CO2 at 37 °C. After 7 days, explants were scored for degree of outgrowth, photographed, and genotyped by PCR.
Genotyping
Tail biopsies, embryos, and blastocyst outgrowths were digested in 200 µg/mL Proteinase K in 50 mM Tris (pH 8.8), 1 mM EDTA (pH 8), and 0.5% Tween 20 at 55 °C. Lysates were PCR amplified with the following cycling parameters: 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min for 35 cycles. The Brg1 primers were: 5’-cagtcatagccgaatagcct-3’, 5’-gcagtttgtaagtcacaggc-3’, 5’-ttggagtggctggtgtccctgtaca-3’, 5’-tgatgaggaaaggcccgttgatgcg-3’ with the first two primers amplifying a 194-bp mutant fragment and the last two primers amplifying a 176-bp wild-type fragment. The Brm primers were: 5’-ctggactgccagctgcagag-3’, 5’-cctgagtcatttgctatagcctgtg-3’, and catcgccttctatcgccttc-3’, which amplified a 310-bp wild-type fragment and a 700-bp mutant fragment. PCR products were resolved on 2% agarose gels.
Cell culture, RNAi, and Western blot analyses
Normal human fibroblasts that ectopically express the catalytic subunit of telomerase (NHF1-hTERT) were cultured under standard conditions (DMEM/10%FBS at 37 °C and 5% CO2), and RNAi-mediated protein depletion was performed using validated Smartpool siRNAs (M-010431 and M-017253 from Dharmacon, Layfayette, CO) introduced by electroporation. Western blot analyses were performed following standard procedures using urea lysates and the following antibodies: J1 (kindly provided by Drs. Weidong Wang and Gerald Crabtree), BRG1 (Santa Cruz G-7), BRM (Abcam 15597), and Tubulin (Sigma T6793).
Results
Brg1+/−;Brm−/− double-mutant mice are underrepresented
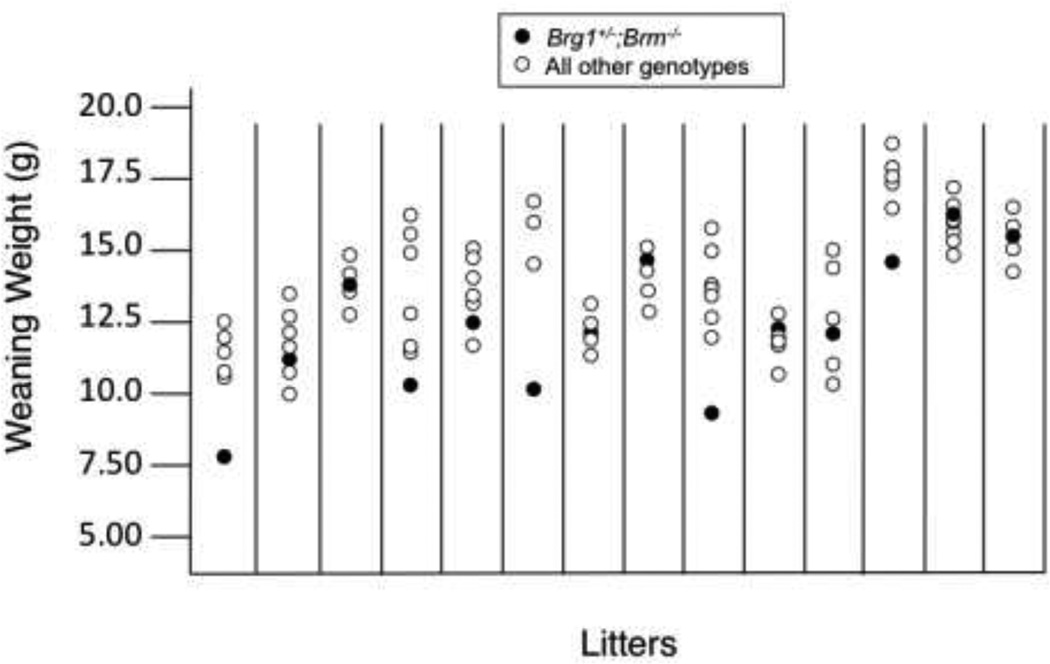
To investigate whether there is a combined gene dosage requirement for the BRG1 and BRM catalytic subunits, we performed double-heterozygous (Brg1+/−;Brm+/−) intercrosses and genotyped the progeny at weaning. Because Brg1−/− is embryonic lethal (Bultman et al 2000), 6 genotypic categories were observed instead of 9 (Table 2). The absence of Brg1−/− mice on a Brm−/− background (in addition to Brm+/+ and Brm+/− backgrounds) eliminated the possibility of a functionally antagonistic relationship. Although we did not expect the Brg1−/− embryonic-lethal phenotype to be suppressed by Brm deficiency, it had been a formal possibility, especially since BRG1 expression is often associated with undifferentiated cells that are highly proliferative, whereas BRM expression is enriched in differentiated cells that are less proliferative. Of the six genotypic categories, the one with the lowest combined gene dosage (Brg1+/−;Brm−/−) was underrepresented by 39% (Table 2). Furthermore, a significant proportion of the Brg1+/−;Brm−/− double-mutant mice that did survive to weaning were runted (Figure 1). However, the runted animals were fertile upon reaching sexual maturity (data not shown). Interestingly, the underrepresentation of the Brg1+/−;Brm−/− category did not result in overrepresentation of the other 5 categories of mice to similar extents. On the contrary, the other categories were either underrepresented or overrepresented in a manner that was correlated with their combined gene dosage (Table 2). For example, mice with the next lowest combined gene dosage (Brg1+/−;Brm+/−) were underrepresented by 17% (Table 2). In contrast, Brg1+/+;Brm+/+ mice with the highest combined gene dosage were overrepresented by 1.5-fold compared to what was expected (Table 2).
Table 2.
Genotype of live-born progeny obtained from Brg1-Brm double heterozygous intercrosses
| Genotype | Expected^ | Observed# | Observed/Expected | |
|---|---|---|---|---|
| Brg1 | Brm | |||
| +/+ | +/+ | .107 | 108 (.163) | 1.52*** |
| +/+ | +/− | .213 | 144 (.218) | 1.02 |
| +/+ | −/− | .107 | 87 (.131) | 1.22* |
| +/− | +/+ | .144 | 109 (.165) | 1.14 |
| +/− | +/− | .286 | 156 (.236) | 0.83** |
| +/− | −/− | .144 | 58 (.088) | 0.61*** |
A total of 662 mice were genotyped at weaning.
Because Brg1−/− is lethal, no mice were observed for this genotype regardless of Brm genotype (+/+, +/−, or −/−), and the absence of these mice were taken into account when determining the expected genotypic frequencies. In addition, Brg1 heterozygotes are underrepresented due to perinatal lethality associated with exencephaly. This incompletely penetrant phenotype was also taken into account when determining the expected genotypic frequencies.
p < 0.05,
p < 0.005,
p < 0.001 based on chi-squared goodness-of-fit statistical tests.
Fig. 1.
Runting of Brg1+/−;Brm−/− mice. Weight of weaning-age mice from 14 litters of double-heterozygous intercrosses. The weights of Brg1+/−;Brm−/− mice are compared to the other 5 genotypes.
Brm compensates for Brg1 haploinsufficiency during development in a tissue-specific manner
To determine at which stage of development Brg1+/−;Brm−/− double mutants die, we performed additional double-heterozygous intercrosses but as timed matings and genotyped embryos. Dissections at embryonic day (E) E15.5–18.5 of gestation yielded results similar to the genotype data at weaning because those embryos with the lowest combined gene dosage were significantly underrepresented while embryos with the highest combined gene dosage were significantly overrepresented (Table 3). A subset of Brg1+/− embryos are known to exhibit exencephaly (Bultman et al 2000), but the incidence of this incompletely penetrant phenotype was neither increased nor decreased in Brg1+/−;Brm−/− double-mutants compared to Brg1+/− embryos on a Brm+/+ or Brm+/− background (Table 3). These findings indicate that Brg1+/−;Brm−/− double mutants do not have an increased incidence of either exencephaly or peri-natal lethality.
Table 3.
Genotype of E15.5–18.5 embryos obtained from Brg1-Brm double heterozygous intercrosses
| Genotype | Expected^ | Observed# | Exencephalic | Observed/Expected | |
|---|---|---|---|---|---|
| Brg1 | Brm | ||||
| +/+ | +/+ | 1/12 (.083) | 23 (.144) | 0/23 | 1.7** |
| +/+ | +/− | 2/12 (.167) | 22 (.138) | 0/22 | 0.8 |
| +/+ | −/− | 1/12 (.083) | 15 (.094) | 0/15 | 1.1 |
| +/− | +/+ | 2/12 (.167) | 36 (.225) | 3/36 | 1.3 |
| +/− | +/− | 4/12 (.333) | 47 (.294) | 3/47 | 0.88 |
| +/− | −/− | 2/12 (.167) | 17 (.106) | 1/17 | 0.63* |
A total of 160 embryos were genotyped.
Because Brg1−/− is lethal, no embryos were observed for this genotype regardless of Brm genotype (+/+, +/−, or −/−), and the absence of these mice were taken into account when determining the expected genotypic frequencies.
p < 0.05 and
p < 0.005 based on chi-squared goodness-of-fit statistical tests
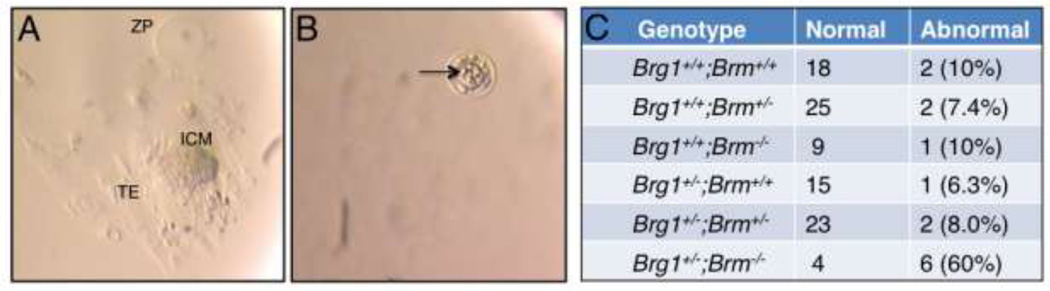
Dissections at E5.5–8.5 demonstrated that Brg1+/−;Brm−/− double mutants are already significantly underrepresented at this early stage of embryogenesis (Table 3). In addition to the 140 embryos that were genotyped at this stage, 11 empty deciduae were observed that could not be genotyped because of maternal contamination. If these empty deciduae were from Brg1+/−;Brm−/− embryos that failed to implant properly, then this genotypic class would have been represented at the expected frequency (25/151 = .166 versus .167 expected). To explore this possibility, we performed blastocyst outgrowths. Following double-heterozygous intercrosses, blastocysts were isolated, cultured for 7 days ex vivo, photographed under a dissection microscope, and then genotyped. Two types of outgrowths were observed in these experiments. Normal outgrowths hatched from the zona pellucida, trophoblast giant cells from the trophectoderm attached to and spread out across the tissue culture wells, and inner cell masses (ICM) underwent extensive cell proliferation (Figure 2A). These events recapitulate certain aspects of peri-implantation development. In contrast, abnormal outgrowths did not hatch from the zona pellucida and neither the trophectoderm nor the ICM showed any signs of growth (Figure 2B). Unlike the other genotypic classes, in which >90% of the blastocyst outgrowths were normal, only 40% of the Brg1+/−;Brm−/− double mutant blastocysts gave rise to normal outgrowths, whereas 60% were abnormal (Figure 2C). These results support the idea that Brg1+/− blastocysts on a Brm-deficient background are susceptible to implantation defects. This situation is similar to the implantation defect of Brg1−/− blastocysts except the penetrance was ~50% instead of 100%.
Fig. 2.
Brg1+/−;Brm−/− blastocysts give rise to an increased percentage of abnormal outgrowths. (A, B) Brightfield photographs of normal (A) and abnomal (B) blastocyst outgrowths after 7 days in culture. (A) Normal blastocysts outgrowths hatched from the zona pellucida (ZP), the trophectoderm (TE) attached to and spread out across the bottom of the tissue culture wells, and the inner cell masses (ICM) underwent extensive cell proliferation. (B) Abnormal blastocyst outgrowths failed to hatch from the zona pellucida and died. Pyknotic blastomeres are evident (arrow). (C) Normal and abnormal blastocyst outgrowths for each of the 6 genotypic categories. The percentage value in the abnormal column refers to the percentage of abnormal outgrowths.
Another difference between Brg1+/−;Brm−/− and Brg1−/− embryos is the specific timing of their peri-implantation phenotype. When we performed double-heterozygous intercrosses and flushed embryos from the uterine tract after the onset of implantation at E5.0, Brg1+/−;Brm−/− embryos were not underrepresented (7 observed compared to 8 expected). This suggests that Brg1+/−;Brm−/− embryos do not exhibit delayed implantation or a failure to implant. In contrast, previous experiments with Brg1 intercrosses indicated that Brg1−/− embryos were overrepresented under the same conditions (Bultman et al 2000), suggesting that they do not implant normally.
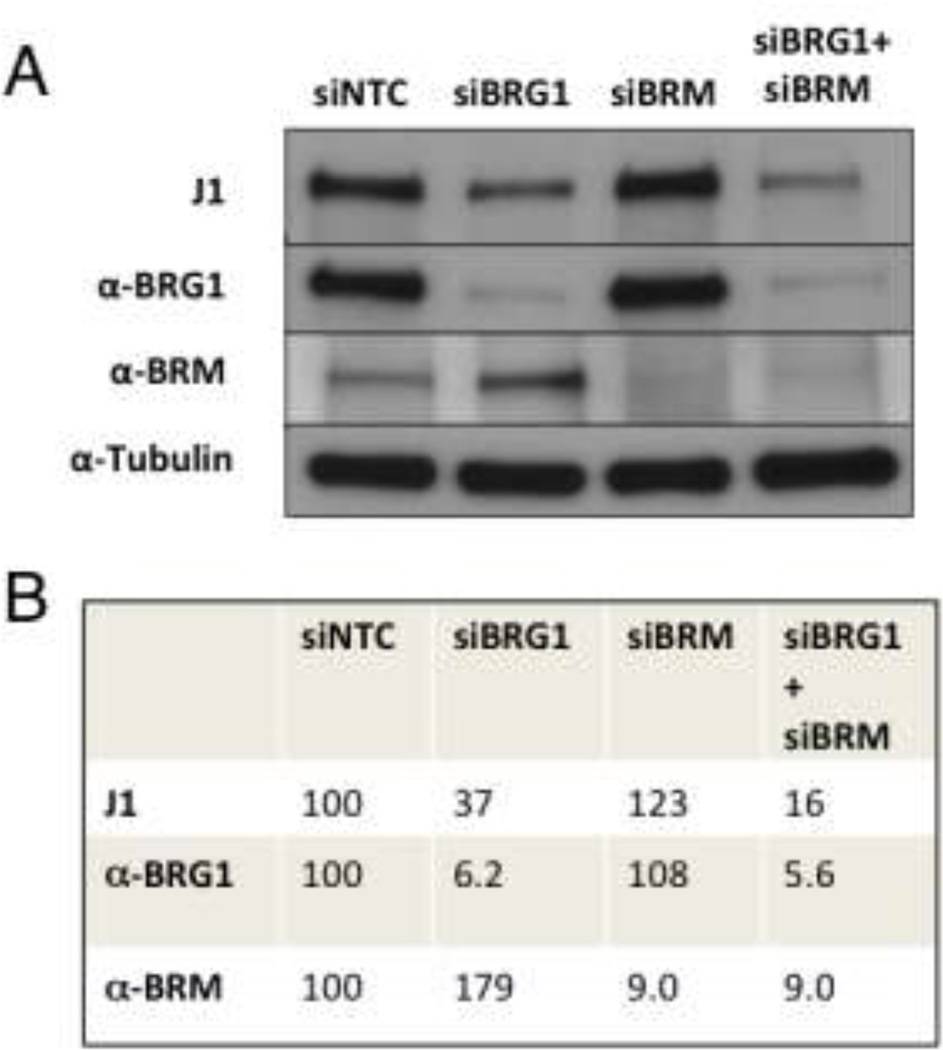
The J1 antibody used to biochemically purify esBAF binds more strongly to BRG1 than BRM Our blastocyst outgrowth experiments suggest that the combined gene dosage requirement for Brg1 and Brm applies to both the ICM and the trophectoderm. The ICM result was initially surprising because SWI/SNF complexes in ES cells, referred to as esBAF, incorporate only BRG1 as the catalytic subunit rather than either BRG1 or BRM (Ho et al 2009a, Ho et al 2009b). To reconcile this apparent discrepancy, we hypothesized that a subset of esBAF complexes might be catalyzed by BRM but were not detected in the biochemical purification experiments for biological and technical reasons. Biologically, Brm is expressed at much lower levels than Brg1 in ES cells and in the ICM from which they are derived (Bultman et al 2000, LeGouy et al 1998). Technically, the biochemical purification of esBAF involved an affinity purification step using a BRG1 antibody (J1), and its capacity to cross-react with BRM has not been characterized to our knowledge. Therefore, we performed western blots on normal human fibroblasts (NHF1-hTERT) following the RNAi-mediated knockdown of BRG1, BRM, or both BRG1 and BRM simultaneously (herein referred to as double knockdowns). A validated BRG1-specific antibody (Santa Cruz G-7) demonstrated that siBRG1 cells had only 6% of BRG1 levels compared to non-targeted control (siNTC) cells after normalizing with tubulin (Figure 3, second row). This low signal represented residual BRG1 protein rather than BRM cross-reactivity because the signal was not further diminished in double-knockdown cells. In contrast, J1 yielded higher signal in siBRG1 cells (37% relative to siNTC compared to 6% for α-BRG1 G-7), and some of this signal was due to cross-reactivity to BRM because the signal dropped to 16% in double-knockdown cells (Figure 3, top row). However, J1 clearly binds to BRG1 with higher affinity than BRM, and quantification of these results suggest that J1 binds to BRM only ~15% as strongly as to BRG1. The limited cross-reactivity of J1 to BRM is reinforced by the strong signal observed in siBRM cells. These cells had only 9% of BRM compared to siNTC using a validated BRM-specific antibody (Abcam) after normalizing with tubulin (Figure 3, third row), yet J1 signal was 107% in siBRM compared to siNTC. These results strongly suggest that J1 would have preferentially isolated BRG1-catalyzed complexes in ES cells even if BRG1 and BRM were of equal abundance (which they are not). Taken together, these results suggest that BRM catalyzes a minority fraction of esBAF complexes that are functional if BRG1 dosage is diminished.
Fig. 3.
Characterization of J1 antibody binding to BRG1 and BRM. (A)Representative western blot of protein lysates from normal human fibroblasts (NHF1-hTERT) following depletion of BRG1 and BRM, alone and in combination, by RNAi using specific siRNAs as indicated at the top. The first lane is a non-targeted control (NTC) siRNA. Protein lysates were probed with the J1 antibody (top panel), antibodies specific for BRG1 (second panel) or BRM (third panel), and Tubulin (bottom panel) as a loading control. (B) Quantification of BRG1 and BRM levels from western blot above. Protein levels detected by J1, α-BRG1, and α-BRM in siNTC, siBRG1, siBRM, and siBRG1 + siBRM cells are shown. All values were normalized with α-Tubulin and are relative to the siNTC control (set at 100).
Discussion
The results presented here demonstrate that the combined gene dosage of Brg1 and Brm is crucial for mammalian development. When the combined gene dosage is diminished from four copies in wild-type (Brg1+/+;Brm+/+) embryos to one copy in Brg1+/−;Brm−/− double-mutant embryos, it falls below a threshold required for implantation to occur successfully on a consistent basis. This finding is compatible with the idea that SWI/SNF complexes play a particularly important role in peri-implantation development based on the phenotype of Brg1, Snf5/Baf47, Baf155, and Baf250a/Arid1a null homozygotes. The Brg1+/−;Brm−/− embryos that fail to develop appear to undergo the decidualization reaction and die shortly after implantation. This phenotype is similar to Snf5/Baf47, Baf155, and Baf250a/Arid1a null homozygotes but a little later than Brg1 null homozygotes that do not implant.
The combined gene dosage requirement for Brg1 and Brm applies to both the ICM and the trophectoderm based on blastocyst outgrowth assays. The ICM result was seemingly at odds with biochemical data that BRG1 is the exclusive subunit of esBAF complexes. However, our experiments demonstrating that the J1 antibody (which was used to biochemically purify esBAF complexes) binds to BRG1 more strongly than BRM provides a plausible explanation for this discrepancy. We propose that a minority fraction of esBAF complexes are catalyzed by BRM. To keep this catalytic subunit heterogeneity in perspective, however, BRG1 is clearly more important than BRM for esBAF function. Not only is BRG1 more abundant than BRM in ES cells and in the early embryo, but 100% of Brg1−/− blastocysts fail to implant compared to 0% for Brm−/−. It is only when Brg1 is heterozygous that Brm is a factor with ~50% of Brg1+/−;Brm−/− blastocysts failing to develop beyond the implantation stage.
The results presented here support a previous study demonstrating that Brg1−/− and Brm−/− mouse embryo fibroblasts (MEFs) can survive, whereas Brg1−/−;Brm−/− MEFs are not viable (Wang et al 2009). Therefore, in addition to Brg1 being able to compensate for Brm, Brm can compensate for Brg1. However, the combined gene dosage of Brg1 and Brm are functionally important in certain cell types but not in others. For example, Brg1 heterozygotes are susceptible to exencephaly (Bultman et al 2000), which results in peri-natal lethality, but Brm heterozygosity or homozygosity did not increase the penetrance or expressivity of this phenotype. It is not clear why this is the case. Baf155 heterozygotes are also predisposed to exencephaly, and this phenotype was associated with decreased cell proliferation during neural tube closure (Kim et al 2001). One possibility is that BRG1-catalyzed complexes are primarily utilized in highly proliferative cells during embryonic development, while BRG1- and BRM-catalyzed complexes are utilized to a similar extent in more differentiated cell types in adult tissues. This idea is consistent with the relative expression levels of BRG1 and BRM since the former is enriched in highly proliferative cell types that self renew, while the later is enriched in more differentiated cell types that do not proliferate to the same extent (Reisman et al 2005). This idea is further supported by functional studies of Brg1 and Brm in vascular endothelial cells (VEC) during their development, when they are relatively highly proliferative, as well as in adults when they are less proliferative. Brg1Tie2-Cre conditional mutants are embryonic lethal (Griffin et al 2008, Stankunas et al 2008), and the phenotype of the developing VECs was not more severe on a Brm-deficient background (Griffin et al 2008). In contrast, an inducible mutation of Brg1 in adult VECs did not confer an observable phenotype on a wild-type background but resulted in rapid cell death and organismal death on a Brm-deficient background (Willis et al 2012).
However, arguing against this possibility is the data presented here on peri-implantation embryos and blastocyst outgrowths since the epiblast is highly proliferative and sensitive to the dosage of both Brg1 and Brm. Additional evidence for the importance of BRM dosage in development comes from whole exome sequencing projects that recently identified recurrent loss-of-function mutations in BRM and BAF250B/ARID1B that cause human syndromes with similar phenotypic spectrums that include intellectual disability, altered craniofacial features, and distal limb anomalies (Santen et al 2012, Van Houdt et al 2012). All of these mutations occurred de novo and were therefore heterozygous, which implies that the corresponding gene products are dosage sensitive. These findings support the idea that the dosage of SWI/SNF subunits is crucial for human development. This may explain why germline homozygous null mutations of human SWI/SNF genes have not been reported; they are likely so deleterious that they would be lethal in utero as they are in mice.
Table 4.
Genotype of E5.5–8.5 embryos obtained from Brg1-Brm double heterozygous intercrosses
| Genotype | Expected^ | Observed# | Observed/Expected | |
|---|---|---|---|---|
| Brg1 | Brm | |||
| +/+ | +/+ | 1/12 (.083) | 21 (.150) | 1.8** |
| +/+ | +/− | 2/12 (.167) | 31 (.221) | 1.3 |
| +/+ | −/− | 1/12 (.083) | 14 (.100) | 1.2 |
| +/− | +/+ | 2/12 (.167) | 21 (.150) | 0.90 |
| +/− | +/− | 4/12 (.333) | 39 (.279) | 0.84 |
| +/− | −/− | 2/12 (.167) | 14 (.100) | 0.60* |
A total of 140 embryos were genotyped. 11 empty deciduae were observed that could not be genotyped due to maternal contamination.
Because Brg1−/− is lethal prior to implantation, no embryos were observed for this genotype regardless of Brm genotype (+/+, +/−, or −/−), and the absence of these mice were taken into account when determining the expected genotypic frequencies.
p < 0.05 and
p < 0.005 based on chi-squared goodness-of-fit statistical tests.
Acknowledgements
We thank Drs. Weidong Wang and Gerald Crabtree for kindly providing the J1 antibody. We also thank Dr. William K. Kaufman for use of the NHF1-hTERT line. This work was supported by the NIH (CA125237 to S.J.B.).
Footnotes
The authors declare no conflicts of interest.
References
- Bruneau BG. Chromatin remodeling in heart development. Current Opinion in Genetics & Development. 2010;20:505–511. doi: 10.1016/j.gde.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Molecular Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Magnuson T. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in beta-globin expression and erythroid development. Genes & Development. 2005;19:2849–2861. doi: 10.1101/gad.1364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes & Development. 2006;20:1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman SJ, Herschkowitz JI, Godfrey V, Gebuhr TC, Yaniv M, Perou CM, et al. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2008;27:460–468. doi: 10.1038/sj.onc.1210664. [DOI] [PubMed] [Google Scholar]
- Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nature Structural & Molecular Biology. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi TH, Wan M, Lee PP, Akashi K, Metzger D, Chambon P, et al. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 2003;19:169–182. doi: 10.1016/s1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Research. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM, Chastain PD, 2nd, Rosson GB, Groh BS, Weissman BE, Kaufman DG, et al. BRG1 co-localizes with DNA replication factors and is required for efficient replication fork progression. Nucleic Acids Research. 2010;38:6906–6919. doi: 10.1093/nar/gkq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis CD, Griffin CT. The chromatin-remodeling enzymes BRG1 and CHD4 antagonistically regulate vascular Wnt signaling. Molecular and Cellular Biology. 2012;32:1312–1320. doi: 10.1128/MCB.06222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nature Genetics. 2001;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nature Reviews Genetics. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling: the means to the end. The FEBS Journal. 2011;278:3579–3595. doi: 10.1111/j.1742-4658.2011.08281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers S, Nagl NG, Jr, Beck GR, Jr, Moran E. Antagonistic roles for BRM and BRG1 SWI/SNF complexes in differentiation. The Journal of Biological Chemistry. 2009;284:10067–10075. doi: 10.1074/jbc.M808782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebuhr TC, Kovalev GI, Bultman S, Godfrey V, Su L, Magnuson T. The role of Brg1, a catalytic subunit of mammalian chromatin-remodeling complexes, in T cell development. The Journal of Experimental Medicine. 2003;198:1937–1949. doi: 10.1084/jem.20030714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin CT, Brennan J, Magnuson T. The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development. 2008;135:493–500. doi: 10.1242/dev.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin CT, Curtis CD, Davis RB, Muthukumar V, Magnuson T. The chromatin-remodeling enzyme BRG1 modulates vascular Wnt signaling at two levels. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2282–2287. doi: 10.1073/pnas.1013751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi CJ, Sands AT, Zambrowicz BP, Turner TK, Demers DA, Webster W, et al. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Molecular and Cellular Biology. 2001;21:3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Pirity MK, Wang WL, Wolf L, Chauhan BK, Cveklova K, et al. Chromatin remodeling enzyme Brg1 is required for mouse lens fiber cell terminal differentiation and its denucleation. Epigenetics & Chromatin. 2010;3:21. doi: 10.1186/1756-8935-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proceedings of the National Academy of Sciences of the United States of America. 2009a;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proceedings of the National Academy of Sciences of the United States of America. 2009b;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hota SK, Bartholomew B. Diversity of operation in ATP-dependent chromatin remodelers. Biochimica et Biophysica Acta. 2011;1809:476–487. doi: 10.1016/j.bbagrm.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra AK, Dupe V, Bornert JM, Messaddeq N, Yaniv M, Mark M, Chambon P, Metzger D. Temporally controlled targeted somatic mutagenesis in embryonic surface ectoderm and fetal epidemal keratinocytes unveils two distinct developmental functions of BRG1 in limb morphogenesis and skin barrier formation. Development. 2005;132:4533–4544. doi: 10.1242/dev.02019. [DOI] [PubMed] [Google Scholar]
- Jani A, Wan M, Zhang J, Cui K, Wu J, Preston-Hurlburt P, et al. A novel genetic strategy reveals unexpected roles of the Swi-Snf-like chromatin-remodeling BAF complex in thymocyte development. The Journal of Experimental Medicine. 2008;205:2813–2825. doi: 10.1084/jem.20080938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser MD, Aslanian A, Dong MQ, Yates JR, 3rd, Emerson BM. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. The Journal of Biological Chemistry. 2008;283:32254–32263. doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- Kim JK, Huh SO, Choi H, Lee KS, Shin D, Lee C, et al. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Molecular and Cellular Biology. 2001;21:7787–7795. doi: 10.1128/MCB.21.22.7787-7795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SI, Bultman SJ, Jing H, Blobel GA, Bresnick EH. Dissecting molecular steps in chromatin domain activation during hematopoietic differentiation. Molecular and Cellular Biology. 2007;27:4551–4565. doi: 10.1128/MCB.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SI, Bresnick EH, Bultman SJ. BRG1 directly regulates nucleosome structure and chromatin looping of the alpha globin locus to activate transcription. Nucleic Acids Research. 2009a;37:6019–6027. doi: 10.1093/nar/gkp677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SI, Bultman SJ, Kiefer CM, Dean A, Bresnick EH. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proceedings of the National Academy of Sciences of the United States of America. 2009b;106:2259–2264. doi: 10.1073/pnas.0806420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Fedoriw AM, Magnuson T. An essential role for a mammalian SWI/SNF chromatin-remodeling complex during male meiosis. Development. 2012;139:1133–1140. doi: 10.1242/dev.073478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Reports. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- LeGouy E, Thompson EM, Muchardt C, Renard JP. Differential preimplantation regulation of two mouse homologues of the yeast SWI2 protein. Developmental Dynamics. 1998;212:38–48. doi: 10.1002/(SICI)1097-0177(199805)212:1<38::AID-AJA4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Liu N, Balliano A, Hayes JJ. Mechanism(s) of SWI/SNF-induced nucleosome mobilization. Chembiochem. 2011;12:196–204. doi: 10.1002/cbic.201000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, et al. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev Biol. 2006;289:372–383. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Molecular Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- Phelan ML, Schnitzler GR, Kingston RE. Octamer transfer and creation of stably remodeled nucleosomes by human SWI-SNF and its isolated ATPases. Molecular and Cellular Biology. 2000;20:6380–6389. doi: 10.1128/mcb.20.17.6380-6389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman DN, Sciarrotta J, Bouldin TW, Weissman BE, Funkhouser WK. The expression of the SWI/SNF ATPase subunits BRG1 and BRM in normal human tissues. Applied Immunohistochemistry & Molecular Morphology. 2005;13:66–74. doi: 10.1097/00129039-200503000-00011. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) The EMBO Journal. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13796–13800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santen GW, Aten E, Sun Y, Almomani R, Gilissen C, Nielsen M, et al. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nature Genetics. 2012;44:379–380. doi: 10.1038/ng.2217. [DOI] [PubMed] [Google Scholar]
- Serber DW, Rogala A, Makarem M, Rosson GB, Simin K, Godfrey V, et al. The BRG1 Chromatin Remodeler Protects Against Ovarian Cysts, Uterine Tumors, and Mammary Tumors in a Lineage-Specific Manner. PloS One. 2012;7:e31346. doi: 10.1371/journal.pone.0031346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal N, Graumann J, Wu G, Arauzo-Bravo MJ, Han DW, Greber B, et al. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI, et al. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Developmental Cell. 2008;14:298–311. doi: 10.1016/j.devcel.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguin P, Holloway AK, et al. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nature Communications. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt JK, Nowakowska BA, Sousa SB, van Schaik BD, Seuntjens E, Avonce N, et al. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nature Genetics. 2012;44:445–449. doi: 10.1038/ng.1105. [DOI] [PubMed] [Google Scholar]
- Wang X, Sansam CG, Thom CS, Metzger D, Evans JA, Nguyen PT, et al. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer Research. 2009;69:8094–8101. doi: 10.1158/0008-5472.CAN-09-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhai W, Richardson JA, Olson EN, Meneses JJ, Firpo MT, et al. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes & Development. 2004;18:3106–3116. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weider M, Kuspert M, Bischof M, Vogl MR, Hornig J, Loy K, et al. Chromatin - remodeling factor BRG1 is required for schwann cell differentiation and myelination. Developmental Cell. 2012;23:193–201. doi: 10.1016/j.devcel.2012.05.017. [DOI] [PubMed] [Google Scholar]
- Willis MS, Homeister JW, Rosson GB, Annayev Y, Holley D, Holly SP, et al. Functional Redundancy of SWI/SNF Catalytic Subunits in Maintaining Vascular Endothelial Cells in the Adult Heart. Circulation Research. 2012;111:e111–e122. doi: 10.1161/CIRCRESAHA.112.265587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Wang Z, Sharova L, Sharov AA, Ling C, Piao Y, et al. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–1165. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Chen M, Kim JR, Zhou J, Jones RE, Tune JD, Kassab GS, Metzger D, Ahlfeld S, Conway SJ, Herring PR. SWI/SNF complexes containing Brahma or Brahma-Related Gene 1 play distinct roles in smooth muscle development. Molecular and Cellular Biology. 2011;31:2618–2631. doi: 10.1128/MCB.01338-10. [DOI] [PMC free article] [PubMed] [Google Scholar]