Summary
Spatial and temporal control of actin filament barbed end elongation is crucial for force generation by actin networks. In this study, genetics, cell biology, and biochemistry were used to reveal three complementary mechanisms that regulate actin filament barbed end elongation in Arp2/3-derived networks. Aip1 inhibits elongation of aged ADP-actin filaments decorated with cofilin, and together with capping protein (CP), maintains a high level of assembly-competent actin species. We identified Abp1 and Aim3 as two additional proteins that work together to inhibit barbed end elongation. Abp1/Aim3 collaborates with CP to control elongation of newly assembled ATP-actin filaments to organize filament polarity within actin networks. Thus, three distinct mechanisms control filament elongation in different regions of Arp2/3 networks, maintaining pools of assembly-competent actin species while ensuring proper filament polarity and facilitating force production.
Introduction
Eukaryotic cells use actin filament networks as the workforce for a diverse range of cellular processes. An essential characteristic of actin networks is that they are highly dynamic. Cells continuously assemble filamentous actin in defined areas of the cytoplasm, and disassemble older parts of the network to replenish the assembly competent actin pool (Pollard and Borisy, 2003). This actin turnover cycle is controlled by many proteins and protein complexes organized in space and time depending on the cellular requirements for assembly versus disassembly (Moseley and Goode, 2006).
Identification of the minimal set of actin regulating proteins required for the maintainance of efficient actin turnover was a major breakthrough in the understanding of actin-based motility systems (Loisel et al., 1999). In the case of branched networks of actin filaments, these proteins include a nucleation promoting factor (NPF), which activates the Arp2/3 complex (Campellone and Welch, 2010). The Arp2/3 complex stimulates actin assembly by nucleating new ATP-actin filaments as branches on the sides of pre-existing filaments (Blanchoin et al., 2000a). ATP-bound subunits within the filaments hydrolyze ATP stochastically, and ADP-actin filaments become substrates of cofilin, which stimulates actin filament disassembly (Blanchoin and Pollard, 1999; Michelot et al., 2007). Another critical protein required for productive actin assembly is the heterodimeric CP complex, which binds to polymerizing actin filament barbed ends and inhibits their elongation (Wear and Cooper, 2004; Wear et al., 2003). CP depletion from in vitro systems results in dramatic defects in motility and filament organization (Loisel et al., 1999; Wiesner et al., 2003). These and other observations have led to the notion that CP plays three important functions (Pantaloni et al., 2001; Pollard et al., 2000): (1) maintainance of the polarity of actin networks by preventing filament elongation away from nucleation areas, (2) maintainance of a pool of assembly-competent actin species by funneling subunits to a limited number of free barbed ends, and (3) creating dense Arp2/3-derived networks by producing shorter filaments at a constant branching frequency.
While in vitro studies have been critical for development of the dendritic nucleation model (Pollard et al., 2000) and for identifying a role for CP in actin filament organization and force production, few studies have explored the role of CP in the context of the dendritic nucleation model in living cells. Results from some such studies seem to contradict certain aspects of the model. For example, in spreading Drosophila S2 cells, where actin filament turnover rates can be measured by tracking GFP-actin fluorescent speckles, depletion of CP reduces the size of the lamellipodium, but does not reduce the velocity of the fastest actin speckles, indicating that actin assembly occurs at the plasma membrane at a similar rate in the absence of CP (Iwasa and Mullins, 2007). This observation indicates that the polarity of the network and the pool of polymerizing actin species are not affected by the absence of CP, which contradicts a key prediction of the dendritic nucleation model. As another example, in yeast, although actin dynamics are strictly required for clathrin-actin mediated endocytosis, absence of CP does not prevent endocytic membrane internalization (Kaksonen et al., 2005).
In this manuscript, we revisit the dendritic nucleation model and demonstrate that apparent contradictions are attributable to the fact that protein complexes in addition to CP inhibit actin filament barbed end elongation. We employed live cell and extract studies of budding yeast that allowed us to detect changes in actin dynamics in living cells and to perform mechanistic tests in precisely defined in vitro assays.
Using a combination of genetics, biochemistry and cell biology, we identified three complementary mechanisms for controlling actin filament barbed end elongation in Arp2/3-derived networks, and dissected their contributions to actin filament turnover dynamics. These studies provide a broader and deeper conceptual understanding of how the dynamics of actin filament networks are controlled, and demonstrate that the repertoire of mechanisms that control actin filament dynamics in vivo is more complex than was previously appreciated.
Results
CP is not Required for the Fast Turnover of Arp2/3-Derived Networks
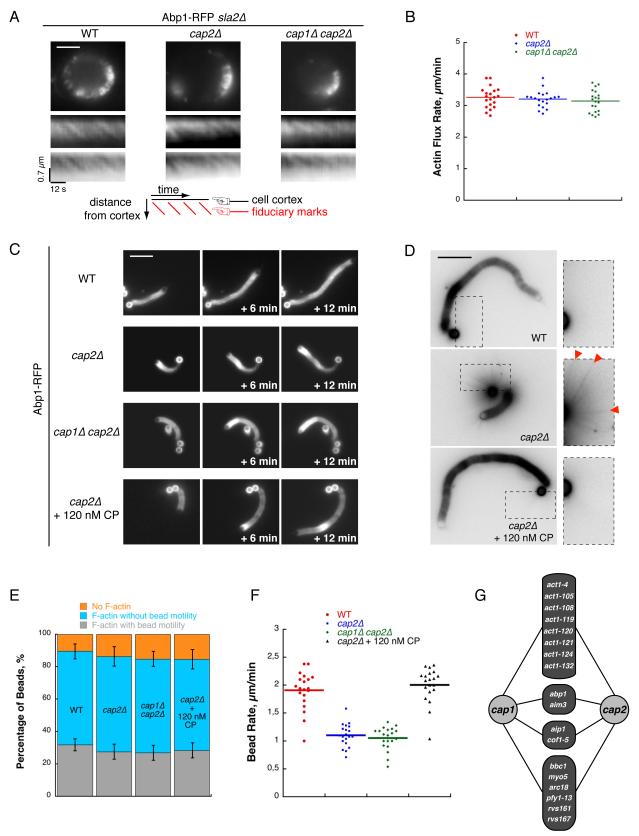
To investigate the contribution of CP function to actin assembly in vivo, we deleted the gene encoding the endocytic adapter Sla2. In the absence of this protein, actin polymerization is uncoupled from clathrin-mediated endocytosis internalization. Cells form elongated actin tails undergoing Arp2/3-dependant actin assembly at the cell cortex and cofilin-dependant disassembly more distal to the plasma membrane (Kaksonen et al., 2003; Okreglak and Drubin, 2007). When we combined the sla2Δ mutation with null mutations in one or both of the CP subunits, in cells expressing Abp1-RFP (Actin binding protein 1) as an actin marker, the spatial separation of zones of actin assembly and disassembly allowed for a quantitative determination of actin flux rates (Figure 1A and Movie S1). We imaged cells continuously to track Abp1-RFP intensity discontinuities as they progress toward the interior of the cells. Kymographs plotted along lines perpendicular to the plasma membrane show dynamics of these discontinuities, which can be used as fiduciary marks to calculate flux rates. Surprisingly, we did not measure any significant difference in flux rates for any of the CP mutants (Figure 1B; 3.2 ± 0.3 μm.min−1 for cap2Δ; 3.2 ± 0.4 μm.min−1 for cap1Δ cap2Δ), compared to the parent sla2Δ strain (3.3 ± 0.3 μm.min−1).
Figure 1. CP is not Required for the Fast Turnover of Arp2/3-Derived Networks.
(A) Top images: Single frames from time-lapse imaging of Abp1-RFP in sla2Δ,sla2Δ cap2Δ and sla2Δ cap1Δ cap2Δ cells. Scale bar: 3 μm. Bottom images: Kymographs plotted along a line perpendicular to the plasma membrane show discontinuous actin assembly as oblique dark lines, or as bright lines when intensities are inverted. Actin flux rates were derived from the slopes of these fiduciary marks (red lines in the cartoon) (see also Movie S1).
(B) Quantification of actin flux rates from (A). Each dot represents an individual measurement. Horizontal bar represents the mean value (MV).
(C) Time course of actin network assembly from incubating beads coated with 100 nM Las17 in cytoplasmic extracts prepared from Abp1-RFP, Abp1-RFP cap2Δ, Abp1-RFP cap1Δ cap2Δ yeast strains or in cytoplasmic extract prepared from an Abp1-RFP cap2Δ yeast strain and then supplemented with 120 nM CP. Images were taken at the indicated time intervals. Scale bar: 10 μm.
(D) Actin network assembled on beads coated with 200 nM Las17 and incubated in cytoplasmic extracts prepared from Abp1-RFP, Abp1-RFP cap2Δ or in cytoplasmic extract prepared from Abp1-RFP cap2Δ yeast strain and then supplemented with 120 nM CP. Images are taken after 1 h incubation of the beads in the extracts. Intensities are inverted to highlight radial array of actin filaments, examples of which are indicated by red triangles. Scale bar: 10 μm.
(E) Percentage of beads in (C) after a 40 min incubation that are either motile with an actin tail (grey), or non motile with an actin shell (blue), or non motile with no polymerized actin network (orange). Error bars indicate standard deviation (SD).
(F) Bead rates for (C). Each dot represents an individual measurement. Horizontal bar represents the MV.
(G) Genome-wide synthetic genetic array (SGA) screen with a cap1Δ or cap2Δ query allele, scored on the basis of a defined confidence threshold. Negative genetic interactions are defined by the intermediate cutoff in (Costanzo et al., 2010). Figure shows actin patch proteins that genetically interact with both cap1Δ and cap2Δ. Protein hits are grouped as actin alleles, aip1 and cof1-5 (for rationale, see Figures 2 and 3), aim3 and abp1 (for rationale, see Figures 4 and 5), or other hits (see also Table S1).
To further test the role of CP in actin dynamics, we used a previously described assay, which uses microbeads functionalized with the yeast WASP homologue Las17, to mimmick actin patch assembly (Michelot et al., 2010). Importantly, we demonstrated previously that the protein composition of the reconstituted actin patch networks is very similar to the composition of actin patches in vivo. When we used extracts generated from cap2Δ or cap1Δ cap2Δ yeast strains for this assay, we observed that beads are still able to assemble dense actin networks, and that some beads break network symmetry and become motile (Figure 1C). We observed that in CP mutant extracts, beads that have initiated actin filament assembly have slight defects in network polarity, because a sub-population of actin filaments elongates away from the beads or from the actin tails (indicated by red triangles in Figure 1D). This phenomenon is similar to what was observed for Arp2/3-based bead motility with purified proteins and low CP concentrations (Wiesner et al., 2003), and reflects unproductive elongation of actin filaments away from the beads due to a lack of actin filament barbed end capping. For this study, we quantified defects in bead movements by measuring the following parameters: (1) the percentage of beads that have nucleated actin filaments and broken network symmetry after a 40 minute incubation in the extract (Figure 1E - grey histogram), (2) the percentage of beads that have nucleated a symmetrical cloud of actin filaments but have not broken network symmetry (Figure 1E - blue histogram), (3) the percentage of beads that have not nucleated any actin filaments (Figure 1E - orange histogram); and (4) the rate of the bead motility (Figure 1F). We observed that beads assemble actin networks and break symmetry at a similar frequency in all CP mutant extracts (Figure 1E), but that the bead motility rate is slower in all of the CP mutant extracts (Figure 1F; 1.9 ± 0.4 μm.min−1 in wild type extracts; 1.1 ± 0.3 μm.min−1 in cap2Δ extracts; 1.1 ± 0.3 μm.min−1 in cap1Δ cap2Δ extracts), suggesting that actin assembly and/or force production is defective in the CP mutant extracts, even though productive assembly is not abolished. We tested whether the motility defects in the CP mutant extracts are due to the absence of CP by adding purified CP to the mutant extracts. A concentration of 120 nM of CP, lower than the concentration of CP in wild type extracts (Figure S1; 465 ± 50 nM), was sufficient to restore bead motility to the same level as in wild-type extracts (Figure 1C, 1D and 1F; 2.0 ± 0.4 μm.min−1).
In total, these results indicate that CP is not required for the fast turnover of actin networks in vivo and in vitro, but is required for normal force production in vitro.
Searching for Proteins that Might Compensate for Loss of CP Function
The observation that actin filament turnover dynamics are unperturbed in sla2Δ cells lacking CP is surprising given that CP is the only yeast protein known to be able to bind to ATP, ADP-Pi and ADP actin filament barbed ends in order to sterically prevent monomer addition (Blanchoin et al., 2000b; Wear and Cooper, 2004). We therefore hypothesized that additional proteins or protein complexes might inhibit actin filament elongation to enable force generation by Arp2/3-based actin assembly.
We used a genetic approach to search for proteins that have an overlapping function with CP. A genome-wide synthetic genetic array (SGA) screen was performed using either a cap1 or a cap2 deletion allele as a query mutant to uncover CP genetic interactions (Costanzo et al., 2010). Quantitative assessment of double mutant fitness phenotypes enabled us to identify mutants showing pronounced growth defects specifically when they were combined with cap1 and cap2 deletion alleles (Table S1). Since CP functions as a heterodimer with both Cap1 and Cap2 as essential subunits, we considered that relevant hits should have a negative genetic interaction with both cap1 and cap2. Eighteen of forty such mutants identified correspond to actin patch proteins (Figure 1G). While 8 of the identified mutants correspond to various actin alleles, 10 mutants correspond to other patch proteins.
In the following sections, we describe how this list enabled us to identify proteins that collaborate with CP to organize Arp2/3-based actin networks at the plasma membrane and to maintain a high concentration of assembly-competent actin species in the cytoplasm through inhibition of filament elongation. We exploited the fact that a yeast mutant extract is a sensitive diagnostic system for detection and analysis of actin assembly defects in vitro, and we focused on the mutants that when combined with CP mutants, induced more pronounced bead motility defects in extracts.
CP Collaborates with Aip1 to Maintain a High Level of Low Molecular Weight Actin Species
Our genetic screen identified an aip1 null mutant and a cofilin point mutant as having negative genetic interactions with cap1 and cap2 null mutants (Figure 1G). In vitro, Aip1 has a dual effect on actin filaments in the presence of cofilin. Aip1 enhances the severing activity of cofilin (Okada et al., 2002; Rodal et al., 1999), and together, these proteins inhibit the barbed end elongation of actin filaments and prevent the reannealing of severed fragments (Balcer et al., 2003; Okada et al., 2002). Although these observations predict that Aip1 only inhibits the elongation of cofilin-bound ADP actin filaments, we considered Aip1 a candidate to inhibit actin filament elongation in cells lacking CP.
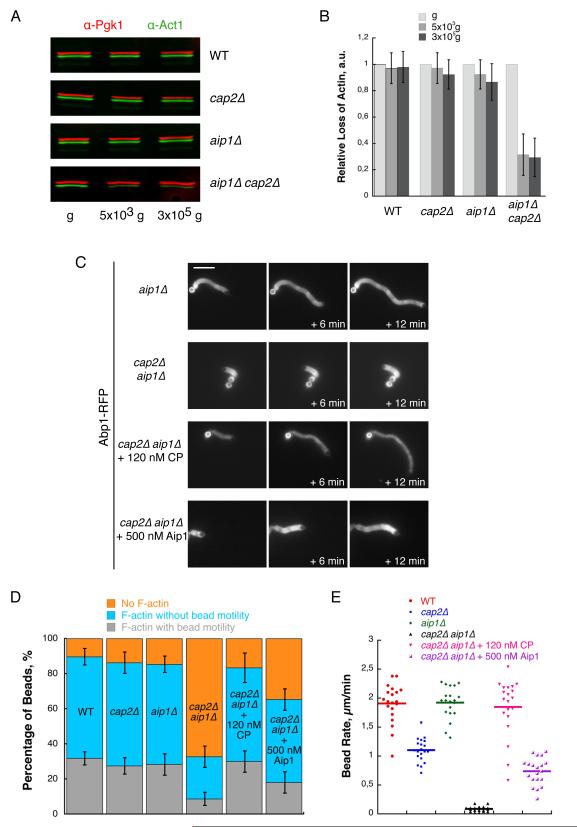
We observed that during the preparation of yeast cytoplasmic extracts from a cap2Δ aip1Δ strain, a large fraction of actin is lost (Figure 2A and 2B). We determined that while loss of actin in wild-type, cap2Δ or aip1Δ cell extracts is comparable during a low or high speed centrifugation of the cell lysates, about 70% of the actin in cap2Δ aip1Δ extracts pellets (Figure 2A and 2B). Loss of actin at this step indicates that most of the actin in cap2Δ aip1Δ extracts is composed of high molecular weight actin filaments or assemblies of actin filaments, while wild-type, cap2Δ or aip1Δ extracts presumably are principally composed of actin monomers or low molecular weight actin species such as short multimers. In a previous study, we demonstrated that Aip1 has a role in maintaining the actin monomer pool (Okreglak and Drubin, 2010), but the analysis in Figure 2A and 2B reveals a previously undiscovered collaboration between CP and Aip1 to maintain a high level of low molecular weight actin species.
Figure 2. CP Collaborates with Aip1 to Maintain a High Level of Low Molecular Weight Actin Species.
(A) Equal volumes of yeast cell extracts collected before centrifugation (g), in the supernatant after a low speed centrifugation (5 × 103 g for 20 min) or in the supernatant after a high speed centrifugation (3 × 105 g for 20 min) were analyzed by western-blot. Actin and Pgk1 (used as a loading control) levels were assessed using goat anti-yeast actin antibodies and mouse anti-Pgk1 antibodies. Immunoblot signals are revealed using anti-goat (green signal) and anti-mouse (red signal) secondary antibodies.
(B) Quantification of (A). Histograms represent actin intensity signals normalized for each lane with the corresponding Pgk1 intensity signal. Error bars indicate SD from three independant experiments.
(C) Time course of actin network assembly on beads coated with 100 nM Las17- and incubated in cytoplasmic extracts prepared from Abp1-RFP aip1Δ or Abp1-RFP cap2Δ aip1Δ yeast strains, or in cytoplasmic extract prepared from an Abp1-RFP cap2Δ aip1Δ yeast strain and then supplemented with 120 nM CP or 500 nM Aip1. Images were taken at the indicated time intervals. Scale bar: 10 μm.
(D) Percentage of beads in (C) after a 40 min incubation that are either motile with an actin tail (grey), or non motile with an actin shell (blue), or non motile with no polymerized actin network (orange). Error bars indicate SD.
(E) Bead rates in (C). Each dot represents an individual measurement. Horizontal bar represents the AV.
Consistant with the observed depletion of actin from the cap2Δ aip1Δ extract, we observed that the percentage of Las17-functionalized microbeads nucleating an actin network is lower in these extracts (Figure 2D; 90 ± 4 % in wild-type extracts; 87 ± 5 % in cap2Δ extracts; 85 ± 5 % in cap1Δ cap2Δ extracts; 33 ± 6 % in cap2Δ aip1Δ extracts), and that beads move at a significantly slower rate (Figure 2C and 2E; 1.9 ± 0.3 μm.min−1 for aip1Δ; 0.1 ± 0.1 μm.min−1 for cap2Δ aip1Δ). These results suggest that actin filament nucleation and elongation are reduced in this extract as a consequence of a lower abundance of assembly-competent actin species. However, addition of purified CP or Aip1 restored bead motility to rates equivalent to those observed for single mutant extracts (Figure 2C, 2E and S1; 1.8 ± 0.5 μm.min−1 for cap2Δ aip1Δ extracts + 120 nM CP; 0.8 ± 0.3 μm.min−1 for cap2Δ aip1Δ extracts + 500 nM Aip1), indicating that the slow actin assembly in cap2Δ aip1Δ extracts is due to the absence of Aip1 and CP, and can not be accounted for by the lower total amount of actin (or by the absence of any protein that might have pelleted with the actin filaments). Collectively, these data indicate that Aip1 and CP function together to maintain high levels of assembly-competent actin species.
Combined Loss of CP and Aip1 Impacts Actin Assembly In Vivo
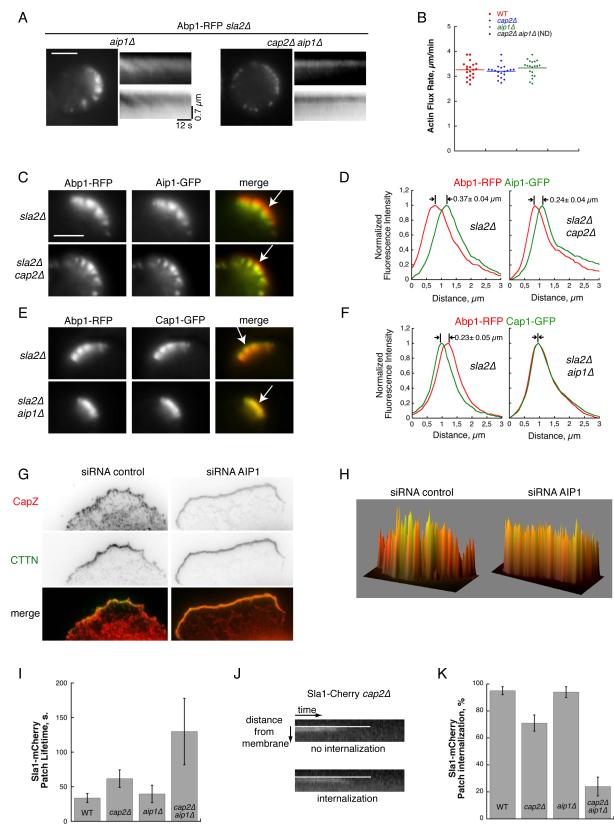
We further interrogated the collaborative roles of Aip1 and CP in mediating fast actin filament turnover. Indeed, the two known biochemical effects of Aip1 on cofilin-decorated actin filaments could account for the increase of low molecular weight actin species. As an actin filament severing complex, Aip1/cofilin enhances the rate of actin filament disassembly, and as a barbed end capping complex, it blocks unproductive elongation of ADP-actin filaments, and helps to maintain a large pool of polymerizing actin species. We attempted to determine the nature of the defect that accounted for the decreased level of low molecular weight actin species observed in the absence of CP and Aip1 by looking at actin tails in sla2Δ cap2Δ aip1Δ mutants. Since the length of the tails is controlled by the balance between assembly rates at the membrane and disassembly rates inside the cells, if loss of Aip1 and CP activities was mostly causing defects in actin disassembly, we would expect to observe elongated and non-dynamic actin tails as we previously observed in sla2Δ cof1-22 cells (Okreglak and Drubin, 2007). On the contrary, we observed a different phenotype, wherein sla2Δ cap2Δ aip1Δ cells form very short actin tails (Figure 3A, 3B and Movie S2). We could not determine actin flux rates in this strain because the tails were too short. Nevertheless, this result is a first indication that in the absence of CP and Aip1, assembly defects are more severe than disassembly defects.
Figure 3. CP and Aip1 Regulate Distinct Populations of Actin Filaments, Maintain Fast Actin Assembly and Efficient Endocytosis.
(A) Left images: Single frames from time-lapse imaging of Abp1-RFP in sla2Δ aip1Δ and sla2Δ cap2Δ aip1Δ cells. Scale bar: 3 μm. Right images: Kymographs plotted along a line perpendicular to the membrane show discontinuous actin assembly as oblique dark lines, or as bright lines when intensities are inverted (see also Movie S2).
(B) Quantification of actin flux rates from (A). Each dot represents an individual measurement. Horizontal bar represents the AV.
(C) Two-color imaging showing the localization of Abp1-RFP and Aip1-GFP in sla2Δ and sla2Δ cap2Δ yeast strains. Tips of white arrows indicate the position of the plasma membrane. Scale bar: 3 μm.
(D) Quantification of normalized fluorescence intensities along a line perpendicular to the membrane. Distance between the peak intensities in (C) is indicated by the MV ± SD (n > 10).
(E) Two-color imaging showing the localization of Abp1-RFP and Cap1-GFP in sla2Δ and sla2Δ aip1Δ yeast strains (see also Figure S2). Tips of white arrows indicate the position of the plasma membrane.
(F) Quantification of normalized fluorescence intensities along a line perpendicular to the membrane. Distance between the peak intensities in (E) is indicated by the MV ± SD (n > 10).
(G) Two-color imaging showing the immuno-localization of endogenous CP (CapZ), in red and cortactin (CTTN), in green in PtK1 cells treated with control siRNA or AIP1 siRNA.
(H) Three-dimensional dual-color representation of CP (in red) and cortactin (in green) fluorescence intensity signals in the lamellipodial area of control siRNA or AIP1 siRNA treated cells (coefficients of Pearson characterizing the correlation between the red and green signal intensities are r = 0.68 ± 0.03 (n = 7) for control siRNA treated cells, and r = 0.80 ± 0.05 (n = 5) for AIP1 siRNA-treated cells).
(I) Histogram of coat protein Sla1-Cherry lifetimes in wild-type, cap2Δ, aip1Δ, and cap2Δ aip1Δ strains. Mean lifetimes are calculated for n > 25 patches for all strains. Error bars indicate SD.
(J) Examples of two distinct actin patch behaviors (one does not internalize and the other does internalize) from a single Sla1-Cherry cap2Δ yeast cell. White line represents the approximate position of the plasma membrane.
(K) Percentage of internalizing patches in Sla1-Cherry, Sla1-Cherry cap2Δ, Sla1-Cherry aip1Δ and Sla1-Cherry cap2Δ aip1Δ cells. Measurements were derived by tracking n > 40 patches for each strain. Error bars indicate SD.
CP and Aip1 Function in Defined Areas of Actin Networks, but Have Overlapping Function
The previous results suggest that an important part of the Aip1 and CP collaboration to maintain a high level of assembly-competent actin is achieved through inhibition of unproductive actin filament barbed end elongation. To test this possibility, we sought direct evidence that Aip1 interacts with actin filament barbed ends in vivo.
Aip1 only interacts with ADP-actin filaments found in an area of the actin tails distal from the plasma membrane in sla2Δ cells (Figure 3C and 3D) (Okreglak and Drubin, 2010). In sla2Δ cap2Δ cells where actin assembles robustly at the plasma membrane (Figure 1A and 1B), Aip1 still localizes in a distal region of the actin tails, because of the inability of cofilin to bind to ATP or ADP-Pi actin filaments (Figure 3C and 3D). In contrast to Aip1, we found that CP localizes to region of the actin tails that is proximal to the plasma membrane (Figure 3E, 3F, S2A and S2B), similar to what was observed for CP in the lamellipodium of some motile cell types (Iwasa and Mullins, 2007; Lai et al., 2008). However, in sla2Δ aip1Δ cells, we found that CP relocalizes along the entire length of the actin tails and colocalizes with Abp1 (Figure 3E and 3F). Likewise, in cof1-19 mutant cells, where Aip1 does not localize to cortical actin networks (Rodal et al., 1999), CP colocalizes with Abp1 (Figure S2C and S2D). These results suggest that actin filament barbed ends in the ADP-actin rich region of actin tails are left uncapped in the absence of Aip1 and become available for binding by CP. These results demonstrate that Aip1 and CP compete for binding to barbed ends of aged, ADP-actin filaments. In wild-type cells, Aip1 prevents CP from binding in these regions and therefore segregates CP to ATP-actin filament regions.
To investigate whether these results are specific to yeast cells, we extended our study to mammalian cells. In PtK1 cells, CP is enriched near the cell periphery (Schafer et al., 1998), and the CP localization pattern is distinct from the pattern of our actin marker, cortactin (Figure 3G and 3H). In agreement with the results obtained in yeast, when we depleted Aip1 in these cells by siRNA, we observed a more complete colocalization of the CP and cortactin signals (Figure 3G and 3H). These results suggest that competitive interaction between CP and Aip1 is a conserved feature of Arp2/3-derived actin filament networks.
Simultaneous Loss of CP and Aip1 Functions Induces Strong Endocytic Defects
Since Aip1 and CP work together to maintain optimal conditions for actin polymerization in Arp2/3 derived networks, we next wanted to test the importance of these proteins for the efficiency of cellular processes in which these types of networks function. In yeast, one of the main roles of Arp2/3-derived actin networks is to provide a force essential for membrane invagination during clathrin-actin mediated endocytosis (Aghamohammadzadeh and Ayscough, 2009; Kaksonen et al., 2003). Individual endocytic events can be studied by following the behavior of the coat protein Sla1 at the plasma membrane. The first defect we observed is that while the average lifetimes of Sla1 patches in cap2Δ and aip1Δ cells are longer than in wild-type cells, the average lifetime of Sla1 patches in cap2Δ aip1Δ cells is significantly increased (Figure 3I; 34 ± 7 s for wild-type; 62 ± 13 s cap2Δ; 40 ± 13 s for aip1Δ; 130 ± 48 s for cap2Δ aip1Δ). In addition, when we tracked movements of Sla1 patches, we observed a clear accentuated defect in the ability of the cap2Δ aip1Δ cells to internalize coats (Figure 3J and 3K; 95 ± 3 % internalization for wild-type; 71 ± 6 % cap2Δ; 94 ± 4 % for aip1Δ; 24 ± 7 % for cap2Δ aip1Δ), indicating that the actin cytoskeleton is defective in driving efficient endocytic internalization when CP and Aip1 are simultaneously absent.
Aim3 and Abp1 Collaborate to Inhibit Elongation of ATP-Bound Actin Filaments and to Maintain the Polarity of the Actin Network
Although combined loss of CP and Aip1 has dramatic effects in vitro and in vivo, Las17-functionalized microbeads are still able to assemble dense and polarized actin filament networks in the absence of these two proteins in vitro, leading to the formation of short actin tails and bead motility (Figure 2C and 2D). In light of our current understanding of Arp2/3-derived motile processes, this observation suggests either that additional factors in yeast inhibit actin filament elongation, or that yeast have another mechanism that enables cells to overcome effects of a loss of capping activity. Since cofilin only has a high affinity for ADP-actin filaments (Blanchoin and Pollard, 1999; Michelot et al., 2007), and since both cofilin and Aip1 localize in the aged part of the actin networks in vivo (Okreglak and Drubin, 2010), we expected that such hypothetical additional factors should impact actin polymerization in both ATP and ADP-actin regions.
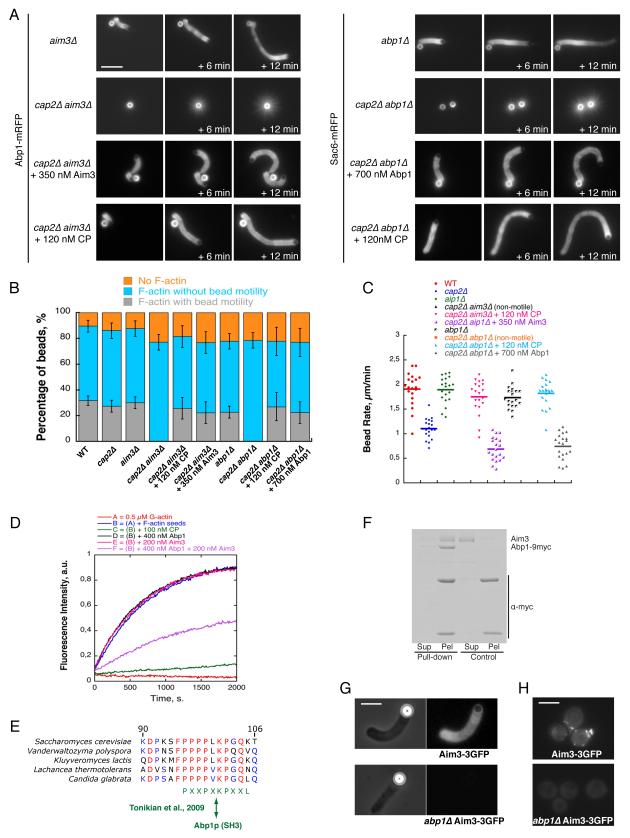
We therefore searched for additional proteins whose absence from a cap2Δ extract would cause an enhanced bead motility defect. Among the mutants identified in our genetic screen (Figure 1G), those in genes encoding Aim3 and Abp1 lead to similar defects in actin assembly in the bead assay when CP was absent. In both cap2Δ aim3Δ and cap2Δ abp1Δ extracts, levels of low molecular weight actin species were nearly equivalent as in single mutant extracts (Figure S3A and S3B) and beads assembled actin filament networks, but failed to become motile (Figure 4A, 4B and 4C). Instead, we observed a dramatic mispolarization of the actin filament networks around the beads, leading to the formation of star-shaped networks (Figure 4A). We confirmed that the respective absences of Aim3 and Abp1 are responsible for this phenomenon, since addition of purified Aim3 or Abp1 in these extracts to levels equivalent to those in wild-type extracts restored the formation of actin tails (Figure 4A, 4B, 4C and S1; 0.7 ± 0.3 μm.min−1 for cap2Δ aim3Δ extracts + 350 nM Aim3; 0.8 ± 0.3 μm.min−1 for cap2Δ abp1Δ extracts + 700 nM Abp1). Also, CP was sufficient to restore bead motility to rates observed in wild-type extracts (Figure 4A, 4B and 4C; 1.8 ± 0.4 μm.min−1 for cap2Δ aim3Δ extracts + 120 nM CP; 1.8 ± 0.3 μm.min−1 for cap2Δ abp1Δ extracts + 120 nM CP).
Figure 4. Aim3 and Abp1 Collaborate to Inhibit the Elongation of Actin Filaments and to Maintain the Polarity of the Actin Network.
(A) Time course of actin network assembly on beads coated with 100 nM Las17 and incubated in cytoplasmic extracts prepared from Abp1-RFP aim3Δ, Abp1-RFP cap2Δ aim3Δ, Sac6-RFP abp1Δ, Sac6-RFP cap2Δ abp1Δ yeast strains, or in cytoplasmic extracts prepared from Abp1-RFP cap2Δ aim3Δ or Sac6-RFP cap2Δ abp1Δ yeast strains, and then supplemented with 350 nM Aim3, 700 nM Abp1 or 120 nM CP. Images were taken at the indicated time intervals. Scale bar: 10 μm.
(B) Percentage of beads in (A) that after a 40 min incubation are either motile with an actin tail (grey), or non motile with an actin shell (blue), or non motile with no polymerized actin network (orange). Error bars indicate SD.
(C) Bead rates in (A). Each dot represents an individual measurment. Horizontal bar represents the MV.
(D) Kinetics of yeast actin filament barbed end elongation in the absence or in the presence of CP, Aim3 and/or Abp1 (see also Figure S3C, S3D and S3E). Preformed yeast actin filaments (0.5 μM) were incubated with indicated concentrations of Aim3 and/or Abp1 or CP, 0.45 μM of yeast actin monomers and 0.05 μM of pyrene-labeled rabbit skeletal muscle actin monomers.
(E) Sequence alignment of conserved region (90-106) from budding yeast Aim3 with homologues from other yeasts. Red letters indicate conserved amino acids; Blue letters indicate less conserved amino acids. The conserved PXXPXKPXXK motif is most similar to PXXPXKPXXL motif predicted to bind to Abp1’s SH3 domain (Tonikian et al., 2009).
(F) Abp1 interacts with Aim3 in vitro. Pull-down experiment of 200 nM Aim3 by control beads (2 right lanes) or beads activated with Abp1-9myc (2 left lanes). SDS-Page gel shows protein composition of supernatants (Sup) and pelleted beads (Pel).
(G) Aim3-3GFP localization on motile Las17-functionalized microbeads in extracts prepared from Aim3-3GFP or Aim3-3GFP abp1Δ yeast strains. Scale bar: 5 μm.
(H) Cellular localization of Aim3-3GFP in wild-type or abp1Δ yeast cells. Scale bar: 5 μm.
Abp1 is known to interact with yeast actin filaments with a high affinity in vitro (Drubin et al., 1988), and with the Arp2/3 complex as a Nucleation Promoting Factor (NPF) or as an inhibitor of Las17 NPF activity (Goode et al., 2001; Moseley and Goode, 2006). However, inhibition of actin filament elongation by this protein has not been reported. Aim3 was identified as a patch protein (Michelot et al., 2010), but its biological function and biochemical effect on actin have not been investigated. We used pyrene fluorescence to measure barbed end elongation from yeast actin filament seeds, and determined that neither Aim3 nor Abp1 alone is able to inhibit elongation (Figure 4D). Interestingly, we found that one of the conserved regions of Aim3 corresponds to a poly-proline sequence (Figure 4E). Because such sequences in other proteins interact with SH3 domains, we searched in our published yeast SH3 domain interactome (Tonikian et al., 2009) to determine whether Aim3’s conserved PXXPXKPXXL sequence is predicted to bind to any SH3 domains. We found that two yeast SH3 domains have a high probability to interact with PXXPXKPXXL: the SH3 domain of Hse1, predicted to bind PXXPXK, and the SH3 domain of Abp1, predicted to bind PXXPXKP (Tonikian et al., 2009) (Figure 4E). Because Hse1 is not known to be a patch protein, and because the consensus for Abp1 is longer, we considered Abp1 to be the most probable candidate among yeast SH3 proteins to interact with Aim3. We tested for binding between these two proteins in vitro and found that Abp1 can pull down Aim3 (Figure 4F). Next we examined the localization of each protein in the absence of the other. We previously observed that in the absence of Aim3, Abp1 still localizes at actin patches (Michelot et al., 2010). In contrast, Aim3-3GFP was not detectable in vitro along actin tails in a bead assay performed using an abp1Δ extract (Figure 4G). In agreement with this result, Aim3-3GFP did not localize at actin patches in abp1Δ cells (Figure 4H), demonstrating that Abp1 is responsible for the recruitement of Aim3 to actin patches in vivo. Most importantly, in our pyrene assay, we observed that elongation of actin filament barbed ends was inhibited only by the simultaneous presence of Aim3 and Abp1 (Figure 4D).
While concentrations of CP, Abp1 and Aim3 are of the same orders of magnitude in extracts (Figure S1), absence of CP affects bead movements, while neither Abp1 nor Aim3 absence does (Figure 1C, 1F, 4A and 4C). To explain this observation, we compared the efficiency of both CP and Abp1/Aim3 in inhibiting barbed end assembly in vitro. Since pyrene actin assembly assays are the most sensitive way to quantify such effects, we titrated CP and of Abp1/Aim3 in a seeded actin elongation assay (Figure S3C, S3D and S3E). We observed that at a 2:1 Abp1:Aim3 ratio (a ratio similar to the ratio of these proteins measured in the extracts), the apparent affinity of the Abp1/Aim3 complex is approximately an order of magnitude lower than the affinity of CP for actin filament barbed ends (Kd,CP = 10 ± 2 nM; Kd,app,Aim3 = 125 ± 15 nM). In this context, it is expected that actin network elongation is more sensitive to loss of CP than to loss of Abp1/Aim3 function.
Simultaneous Loss of CP and Aim3 Function Disorganizes Actin Networks In Vivo and Induces Pronounced Endocytic Defects
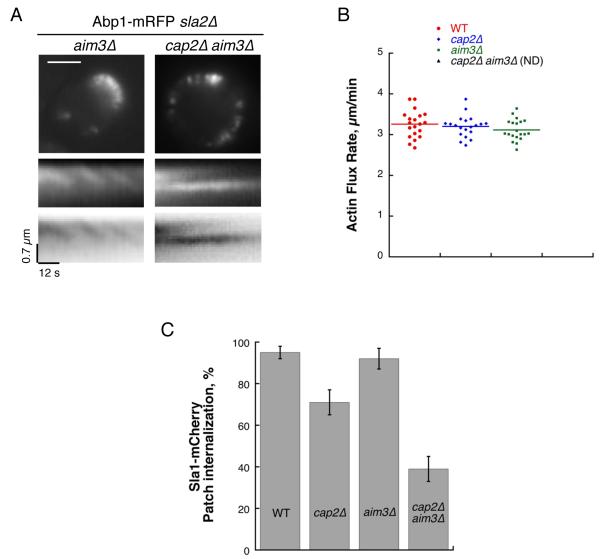
Results using our bead assembly assay predict an in vivo requirement for CP and Abp1 or Aim3 to maintain polarity of Arp2/3-derived actin networks. We tested this prediction in vivo by creating a sla2Δ cap2Δ aim3Δ yeast strain. We observed that these cells are still able to generate elongated actin structures at the membrane, but found that actin assembly in these cells is highly irregular and unpolarized (Figure 5A, 5B and Movie S3).
Figure 5. CP and Aim3/Abp1 Collaborate to Maintain Fast Actin Turnover and Efficient Endocytosis.
(A) Top images: Single frames from time-lapse imaging of Abp1-RFP in sla2Δ aim3Δ and sla2Δ cap2Δ aim3Δ cells. Scale bar: 3 μm. Bottom images: Kymographs plotted along a line perpendicular to the plasma membrane show discontinuous actin assembly as oblique dark lines, or as bright lines when intensities are inverted (see also Movie S3).
(B) Quantification of actin flux rates from (A). Each dot represents an individual measurment. Horizontal bar represents the MV.
(C) Percentage of internalizing patches in Sla1-Cherry, Sla1-Cherry cap2Δ, Sla1-cherry aim3Δ and Sla1-Cherry cap2Δ aim3Δ cells. Measurements were derived by tracking n > 40 patches for all strains. Error bars indicate SD.
Finally, we determined the importance of these proteins for driving efficient clathrin-actin mediated endocytosis. When we tracked the movement of Sla1 patches in these cells, we observed a clear accentuated defect in the ability of the cap2Δ aim3Δ cells to internalize coats, indicating that in the simultaneous absence of CP and Aim3 the actin cytoskeleton is unable to mediate efficient endocytic internalization (Figure 5C; 92 ± 5 % internalization for aim3Δ; 39 ± 6 % for cap2Δ aim3Δ).
Discussion
Actin Filament Elongation in Arp2/3-Derived Actin Networks: Evidence for 3 Inhibitory Mechanisms
In this study, we screened for proteins displaying overlapping function with CP in Arp2/3-based actin assembly. By using a combination of genetic, biochemical and cell biology techniques, we identified three distinct factors in yeast that control actin filament barbed end elongation. We found that in addition to CP, two sets of proteins, Aip1 and Abp1/Aim3, are able to inhibit actin filament elongation in vitro and in vivo.
CP is well-characterized as a heterodimer able to bind to actin filament barbed ends, sterically preventing actin monomer addition (Wear and Cooper, 2004; Wear et al., 2003). In Arp2/3-nucleated actin tails present in sla2Δ mutants, CP localizes in areas proximal to the plasma membrane, corresponding to areas where actin assembles into ATP filaments. This localization is similar to what was reported in other eukaryotic cell types (Iwasa and Mullins, 2007; Lai et al., 2008). The fact that CP is found associated with a subset of actin filaments was striking, given the fact that CP affinity for actin filament barbed ends is not dependant on the nucleotide state (Blanchoin et al., 2000b). Several mechanisms could explain such specificity. Previously it was proposed that CP localization proximal to the plasma membrane is due to the fact that more actin filament barbed ends are present at the leading edge. Indeed, actin networks seem to be composed of short, densely branched actin filaments in the lamellipodium, while more distal areas such as the lamella seem to be composed of longer, linear actin filament arrays (Svitkina et al., 1997). However this explanation presents a chicken and egg problem, because filaments are believed to be short near the plasma membrane partly because of the presence of CP. Also, this explanation does not account for the absence of CP from regions distal to the plasma membrane, where cofilin is believed to generate numerous new barbed ends by severing. Another study suggested that loss of CP from the distal leading edge occurs via filament severing, to effectively “uncap” the distal filament barbed ends (Miyoshi et al., 2006). Our work supports an alternative model in which CP is excluded from certain areas of actin networks by competition with other barbed end binding factors. Several factors, including for example formins, gelsolin and VASP, are known to compete in vitro with CP for binding to actin filament barbed ends. In our study, we found that absence of Aip1 causes CP to localize along the entire length of actin tails in sla2Δ yeast cells, and had a similar effect in PtK1 cells, implying a conserved mechanism. For these reasons, we favor a competition model wherein Aip1 and CP compete for binding to ADP-actin filament barbed ends.
Aip1 was shown ten years ago to be an inhibitor of elongation of cofilin-decorated actin filaments (Balcer et al., 2003; Okada et al., 2002). However, whether Aip1 has a similar activity in vivo has never been definitively tested. Here, we provide evidence that Aip1 affects actin filament barbed ends in vivo; it prevents CP from gaining access to ADP-actin filament rich areas of the cytoskeleton. This observation raises the question of why actin filament barbed ends in ADP-rich regions preferentially bind to Aip1 rather than to CP. CP in various species binds to actin filament barbed ends with affinities in the 0.08 to 24 nM range (Huang et al., 2003; Wear et al., 2003), while Aip1 has an apparent affinity for actin filament barbed ends saturated with cofilin in the 25 nM range (Okada et al., 2002). The fact that CP generally shows a higher affinity for barbed ends than Aip1 indicates that a simple competition effect between Aip1 and CP is not likely to account for the localization of Aip1. Also CP has a low dissociation constant (around 0.0005 s−1, equivalent to one dissociation event every 2000 s) (Wear et al., 2003). Given the fact that the average actin tail length is about 1 μm long (Figure 1A), and that flux rates are 3.3 ± 0.5 μm.min−1 (Figure 1B), actin subunits spend approximately 18 s in the network during each turnover cycle. This fact indicates that a model in which CP binds to actin filament barbed ends first and is then displaced by Aip1, would not work because CP is not be released fast enough for Aip1 to bind. This calculation demonstrates that the association constant for Aip1 with ADP-actin filaments in cellular networks must be much higher than the association constant for CP. In other words, Aip1 must bind to ADP-actin filament barbed ends before CP. This conclusion is in agreement with a model in which Aip1 binds to actin filaments decorated with cofilin before or during severing events, before CP can access them. Such a mechanism has the advantage of preventing newly generated barbed ends from undergoing unproductive elongation, which would otherwise happen for a short time if diffusion of CP was necessary to cap the fragments after severing.
Our study identified a third cellular mechanism for inhibiting actin filament elongation, which depends on the simultaneous presence of Abp1 and Aim3. Twenty years ago, before the interaction of Abp1 with the Arp2/3 complex was discovered, Abp1 was identified as a protein that binds to yeast actin filaments (Drubin et al., 1988). The function of this interaction had not been determined, but our study demonstrates a function in control of actin filament elongation, positively regulated through an interaction of Aim3’s proline rich domain with the SH3 domain of Abp1. It is of note that Abp1 localizes along the entire length of Arp2/3-derived networks in sla2Δ cells, suggesting that the function of Abp1 is not dependant on the filament nucleotide state. Abp1 is widely conserved among eukaryotes, suggesting that this activity might be conserved in other organisms. We tried to identify homologues of Aim3 in other organisms, but this protein seems to have diverged rapidly during evolution. However, given the fact that Aim3 appears to interact with Abp1 through a short poly-proline sequence, and that the SH3 domain of Abp1 is conserved across diverse species, the existence of proteins with poly-proline sequences that play the same role as Aim3 in other organisms is plausible. In support of this possibility, our recent unpublished data shows conservation in other eukaryotes of the poly-proline motif that binds to Abp1’s SH3 domain.
Three Ways to Inhibit Actin Filament Barbed End Elongation: Distinct Properties, but Overlapping Functions
Importantly, although the three protein complexes investigated here share a common activity of inhibiting actin filament elongation, they have distinctive features giving them specific functions in vivo. This notion is supported by the observation that loss of function of CP is enhanced differently when combined with a loss of function of the other two inhibitors of actin filament barbed end elongation.
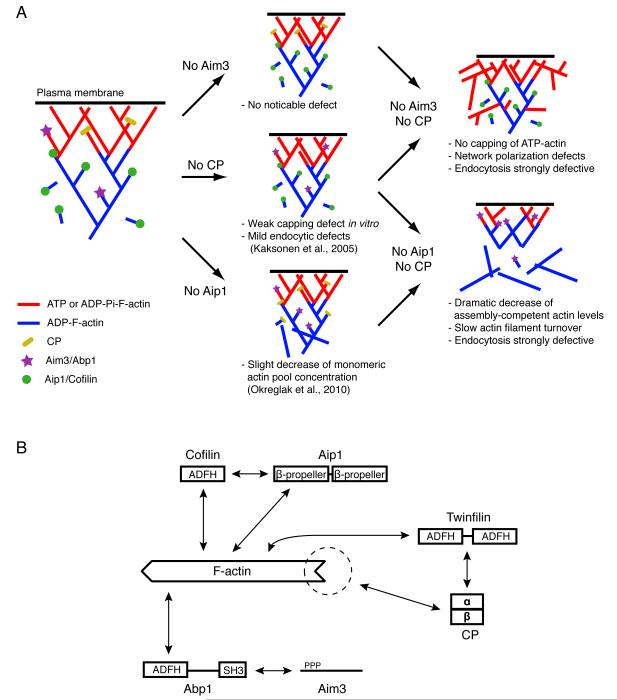
We attempted to synthesize the results of our study in a model (Figure 6A). Elimination of any one of the three mechanisms for inhibiting actin filament barbed end elongation leads to only minor defects in actin network regulation and endocytosis. Our analyses showed that the subtlety of the effects is because the loss of function of any single protein complex is compensated by another complex. Hence, the effects observed in double mutants are much more dramatic. In a previous study, we showed that in aip1Δ cells, levels of low molecular weight assembly-competent actin species are significantly reduced (Okreglak and Drubin, 2010). Our current study shows that this effect is enhanced if CP is simultaneously not expressed. In cap2Δ aip1Δ double mutants, Abp1/Aim3 does not seem to inhibit unproductive filament elongation sufficiently to maintain a high pool of assembly-competent species, because its affinity for actin filament barbed ends is low. This leads to severe defects in actin assembly observed in vitro and in vivo and defective endocytosis. In cap2Δ aim3Δ double mutants, the situation is different. In this case, because Aip1 only binds to ADP-actin filament barbed ends, cells do not have a mechanism to inhibit elongation of actin filaments at the plasma membrane where newly formed filaments have ATP or ADP-Pi subunits at their barbed ends. This lack of capping leads to a loss of normal actin network polarization and severe endocytic defects because elongation of actin filaments is not controlled.
Figure 6. A Model for how CP, Aip1 and Aim3/Abp1 Work in Specific Regions of the Actin Network and Collaborate to Maintain the Polarity and Fast Turnover of Arp2/3-Derived Networks.
(A) See Discussion for description.
(B) Schematic representation of demonstrated physical interactions between the proteins referenced in this article and their interactions with F-actin. Dotted circle indicates known interaction with actin filament barbed ends ((Moseley and Goode, 2006) and this article).
Molecular Mechanisms: Similarities and Differences
Evidence that three protein complexes in yeast can inhibit actin filament elongation raises the fundamental question of their molecular mechanisms.
Insights into this question may come from the observation that each of the three complexes is composed of one protein having conserved ADF-homology (ADFH) domains (Figure 6B) (Lappalainen et al., 1998). Cofilin consists of one ADFH domain, Abp1 has an ADFH domain at its N-terminus, and it has been shown that CP interacts with twinfilin (Palmgren et al., 2001), a protein composed of two tandem ADFH domains. These proteins and ADFH domains are conserved among eukaryotes. In yeast, cofilin, Abp1 and twinfilin are in fact the only proteins with identified ADFH domains (Moseley and Goode, 2006), suggesting a potential conserved function for these domains in binding actin filaments for the inhibition of barbed end elongation. Each of these proteins containing ADFH domains is able to bind to the sides of actin filaments with high affinity, and this interaction is conserved among species. Cofilin has long been known to interact with ADP-actin filaments (Blanchoin and Pollard, 1999; Michelot et al., 2007). Abp1 was discovered because of its strong binding to actin, altough the role of this interaction was not determined (Drubin et al., 1988). Twinfilin has been shown to bind to actin filaments in vitro. It has also been demonstrated that mammalian twinfilin alone can inhibit actin filament elongation (Helfer et al., 2006; Paavilainen et al., 2007). Whether this property is conserved with yeast twinfilin needs to be tested using yeast actin. In this study, we did not consider twinfilin as a possible factor to inhibit actin filament elongation in the absence of CP, because CP is necessary to recruit twinfilin to actin patches in vivo (Palmgren et al., 2001), because twinfilin does not show a negative genetic interaction with CP (Figure 1G), and because extracts from cap2Δ twf1Δ yeast strains do not show more severe bead motility defects than cap2Δ mutants (data not shown). Abp1 and cofilin each requires a cofactor (Aim3 and Aip1, respectively) for inhibiting actin filament barbed end elongation.
It is well established that the CP heterodimer, composed of Cap1 and Cap2 in yeast, binds to actin filament barbed ends and sterically blocks their elongation (Wear and Cooper, 2004; Wear et al., 2003). Structural studies and mutants of CP have given rise to a clear picture of this interaction and the mechanism for blocking elongation. However, no previous study to our knowledge provided evidence that such a capping mechanism could account for the biological activities of Aip1 and Abp1/Aim3. For this reason, in our study we used the term [mgn] inhibition of elongation [grh] instead of capping, because the term capping implies sterical blocking of actin monomer addition to actin filament barbed ends. Aip1 has been shown by yeast two-hybrid and pelleting assays to interact with actin (Rodal et al., 1999), but it does not by itself inhibit barbed end elongation (Okada et al., 2002). On one hand, binding of cofilin and Abp1 to the sides of actin filaments might make barbed ends accessible to Aip1 and Aim3, respectively. On the other hand, ADFH domains are known to bind to the sides of actin filaments, and a large body of literature describes effects of ADFH domains in altering actin filament conformation (Michelot and Drubin, 2011), raising the possibility that binding of these domains could also induce a change in conformation of actin filament barbed ends by allosteric effects. Whether addition of actin monomers can be influenced by the conformation of an actin filament barbed end is not known, but should be tested in the future.
Experimental Procedures
Yeast Lysate and Protein Extract Preparation
Yeast strains were cultured in rich media (YPD) at 30°C to an OD 600 of 1-1.5. Cells were harvested by centrifugation, resuspended in cold water, then centrifuged again. Cell pellets were frozen in liquid N2 and ground to powder in a Waring Blender. To each gram of yeast powder, 100 μl of 10x HK buffer (1x contains 10 mM Hepes, pH 7.8, 50 mM KCl) and protease inhibitors (Cocktail Set IV, Calbiochem, Merck4Biosciences) were added, and yeast lysates were obtained by thawing and homogenizing the mixture on ice. Yeast protein extracts used in the bead motility assays were obtained by collecting the clear supernatant after a 20 minutes centrifugation at 300,000 x g. Extracts were kept on ice and used within 3 h. For western-blot analysis, equal volumes of yeast cell extracts were loaded on 12 % SDS-page gels and transferred to a nitrocellulose membrane. Proteins were immuno-blotted with primary antibodies and then with secondary antibodies conjugated with a fluorescent moiety. Immunoblots were imaged using a Typhoon 9400 Imaging System (GE Healthcare Life Sciences) and analyzed with ImageJ 1.39t (http://rsb.info.nih.gov/ij/) and Kaleida Graph 4.1.3.
Microbead Functionalization and Motility Assays
Microbeads were functionalized with Las17 as described in (Michelot et al., 2010) and kept on ice for up to 12 h. For motility assays, 1 μl of bead stock solution was mixed with 19 μl of extract. 2 μl of the mixture was then placed between a slide and a 20 mm × 20 mm coverslip, and the coverslips were sealed with VALAP (1:3 vaseline, 1:3 lanolin, 1:3 paraffin). Actin networks were observed by following the fluorescence signal of Abp1-RFP or Sac6-RFP using an Olympus IX71 microscope equipped with a 60x PlanApo objective and a CCD camera (Orca II; Hamamatsu Photonics). Images were processed using Metamorph 7.1.7.0 or ImageJ. Bead rates were calculated by measuring bead displacements over time. Mean values and standard deviations were calculated with Kaleida Graph.
Deletion Mutants Genome-Scale Genetic Interactions Analysis
This analysis was performed and analyzed as described in (Costanzo et al., 2010). The genetic interaction scores were derived from a genome-wide dataset scored in May 2010.
Mammalian Cell Culture and siRNA Transfection
PtK1 cells were grown in DMEM/F-12 supplemented 10% FCS. Cells at 25 % of confluency were transfected using lipofectaminRNAiMax (Invitrogen) and 50 nM of siRNA. Non-targeting siRNA control (ON-TARGETplus SMARTpool siRNA #4) or siRNA against AIP1 (also referenced as WDR1) (ON-TARGETplus SMARTpool siRNA) were purchased by Dharmacon-Thermo Fisher. Sequences used against AIP1 were: GGAAAGUGCGUCAUCCUAA, GGUGGGAUUUACGCAAUUA, GCGGCAAGUCCUACAUUUA and UUGUCAACUGUGUGCGAUU.
Cell Imaging
Yeast Cell Observations
Cells were cultured in imaging media (1.7 g.l−1 yeast nitrogen base, 5 g.l−1 ammonium sulfate, 5 g.l−1 casamino acids, 0.1 g.l−1 uracil, 0.1 g.l−1 adenine, 2% dextrose) at an OD = 0.1 1-2 h prior to imaging. Cells were immobilized on coverslips coated with 0.1 g.l−1 of concanavalin A (Sigma), and imaged using an Olympus IX81 microscope equipped with a 100x PlanApo objective, a two-channel simultaneous imaging system (DV2, Photometrics) and a CCD camera (Orca R2; Hamamatsu Photonics). Images were processed and analyzed using Metamorph 7.1.7.0 or ImageJ. Mean values and standard deviations of all measurments were calculated with Kaleida Graph.
Mammalian Cell Observations
Three days post transfection, cells were plated on coverslips, fixed in 4 % paraformaldehyde and permeabilized in PBS supplemented with 0.5% BSA and 0.05% saponin. Cells were stained with rabbit anti-CapZB (Millipore, dilution 1:500) and mouse anti-cortactin (Upstate, 1:500) primary antibodies, Alexa488 anti-mouse and Alexa568 anti-rabbit (Molecular Probes, 1:1000) secondary antibodies. Images were acquired using an Olympus IX81 microscope equipped with a 60x PlanApo objective and a CCD camera. Images were processed using Metamorph or ImageJ. For red and green intensity signal correlation analysis, we calculated Pearson coefficients r for multiple cropped lamellipodia using the plugin JACoP, available on ImageJ (http://rsbweb.nih.gov/ij/plugins/track/jacop.html). Values and standard deviations derive from the average of the measurments.
Protein Sequence Analysis
Homologues of Aim3 were identified using Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to screen the protein database. Sequence Alignments were plotted with MultAlin (http://multalin.toulouse.inra.fr/multalin/).
Protein Production and Labeling
Purification of Yeast Actin
Yeast actin was purified as described in (Goode, 2002) and stored in G-buffer (5 mM Tris-HCl, pH 8, 0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM DTT, 1 mM NaN3).
Purification of Rabbit Skeletal Actin and Pyrene Labeling
Rabbit skeletal muscle actin was isolated from acetone powder (Spudich and Watt, 1971). Monomeric Ca-ATP-actin was purified by gel-filtration chromatography on Sephacryl S-300 at 4°C in G-buffer. Actin was label ed on Cys-374 with pyrene iodoacetamide as described in (Kouyama and Mihashi, 1981).
Purification of Las17, CP, Aip1, Abp1 and Aim3
DNA fragments corresponding to gene coding sequences were obtained by PCR amplification (Phusion High-Fidelity DNA Polymerase, Finnzymes) of yeast genomic DNA. Fragments were cloned, proteins overexpressed in yeast and purified as described in (Rodal et al., 2003) or in (St-Pierre et al., 2009).
For purification of GFP, see supplementary experimental procedures
Pyrene-Actin Elongation Assays
Mg-ATP-actin monomers were prepared by a 5 minute incubation on ice of Ca-ATP-actin with 0.2 mM EGTA and an 11-fold excess of MgCl2 over actin. Actin was polymerized by addition of one-tenth volume of 10x KMEI (1x contains 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM Imidazole, pH 7). Polymerization was followed by measuring changes in pyrene fluorescence using a Safas Monaco Xenius fluorimeter.
Protein Pull-Down Assay
ProteinG-Sepharose 4 Fast Flow beads (GE Healthcare) were activated overnight at 4°C in the presence of 9E10 anti-myc antibodies wit h TBST buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.1 % Triton X-100), and unbound antibodies were washed from beads using HK buffer (20 mM Hepes, pH 7.5, 50 mM KCl). Abp1-9myc was bound to the beads and purified from a yeast extract as described in (Rodal et al., 2003). Beads were incubated with 200 nM Aim3 in HK buffer for 30 min at 25°C under stirring, then sedimented. Supernatants were collected, pelleted beads were resuspended in an equivalent volume of HK buffer, and protein compositions were analyzed on a 12 % SDS-Page gel.
Supplementary Material
Highlights.
Capping protein is dispensable for the fast turnover of Arp2/3-actin networks
Capping protein and Aip1 control levels of assembly-competent actin species
Capping protein and Aip1 compete for ADP-actin filament barbed end binding
Capping protein and Abp1/Aim3 control Arp2/3-actin network polarity
Acknowledgments
AM was supported by a Human Frontier Science Program fellowship LT00565/2008-L. DGD was supported by NIH grant R01 GM42759. We thank Damien d’Amours for sharing new DNA plasmid constructs useful for protein purification.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aghamohammadzadeh S, Ayscough KR. Differential requirements for actin during yeast and mammalian endocytosis. Nat Cell Biol. 2009;11:1039–1042. doi: 10.1038/ncb1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcer HI, Goodman AL, Rodal AA, Smith E, Kugler J, Heuser JE, Goode BL. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr Biol. 2003;13:2159–2169. doi: 10.1016/j.cub.2003.11.051. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000a;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J Biol Chem. 1999;274:15538–15546. doi: 10.1074/jbc.274.22.15538. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD, Mullins RD. Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr Biol. 2000b;10:1273–1282. doi: 10.1016/s0960-9822(00)00749-1. [DOI] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Miller KG, Botstein D. Yeast actin-binding proteins: evidence for a role in morphogenesis. J Cell Biol. 1988;107:2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL. Purification of yeast actin and actin-associated proteins. Methods Enzymol. 2002;351:433–441. doi: 10.1016/s0076-6879(02)51862-0. [DOI] [PubMed] [Google Scholar]
- Goode BL, Rodal AA, Barnes G, Drubin DG. Activation of the Arp2/3 complex by the actin filament binding protein Abp1p. J Cell Biol. 2001;153:627–634. doi: 10.1083/jcb.153.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer E, Nevalainen EM, Naumanen P, Romero S, Didry D, Pantaloni D, Lappalainen P, Carlier MF. Mammalian twinfilin sequesters ADP-G-actin and caps filament barbed ends: implications in motility. EMBO J. 2006;25:1184–1195. doi: 10.1038/sj.emboj.7601019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Blanchoin L, Kovar DR, Staiger CJ. Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J Biol Chem. 2003;278:44832–44842. doi: 10.1074/jbc.M306670200. [DOI] [PubMed] [Google Scholar]
- Iwasa JH, Mullins RD. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr Biol. 2007;17:395–406. doi: 10.1016/j.cub.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- Lai FP, Szczodrak M, Block J, Faix J, Breitsprecher D, Mannherz HG, Stradal TE, Dunn GA, Small JV, Rottner K. Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 2008;27:982–992. doi: 10.1038/emboj.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P, Kessels MM, Cope MJ, Drubin DG. The ADF homology (ADF-H) domain: a highly exploited actin-binding module. Mol Biol Cell. 1998;9:1951–1959. doi: 10.1091/mbc.9.8.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Michelot A, Berro J, Guerin C, Boujemaa-Paterski R, Staiger CJ, Martiel JL, Blanchoin L. Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr Biol. 2007;17:825–833. doi: 10.1016/j.cub.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Michelot A, Costanzo M, Sarkeshik A, Boone C, Yates JR, 3rd, Drubin DG. Reconstitution and protein composition analysis of endocytic actin patches. Curr Biol. 2010;20:1890–1899. doi: 10.1016/j.cub.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelot A, Drubin DG. Building distinct actin filament networks in a common cytoplasm. Curr Biol. 2011;21:R560–569. doi: 10.1016/j.cub.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Tsuji T, Higashida C, Hertzog M, Fujita A, Narumiya S, Scita G, Watanabe N. Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. J Cell Biol. 2006;175:947–955. doi: 10.1083/jcb.200604176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Goode BL. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70:605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Blanchoin L, Abe H, Chen H, Pollard TD, Bamburg JR. Xenopus actin-interacting protein 1 (XAip1) enhances cofilin fragmentation of filaments by capping filament ends. J Biol Chem. 2002;277:43011–43016. doi: 10.1074/jbc.M203111200. [DOI] [PubMed] [Google Scholar]
- Okreglak V, Drubin DG. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J Cell Biol. 2007;178:1251–1264. doi: 10.1083/jcb.200703092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V, Drubin DG. Loss of Aip1 reveals a role in maintaining the actin monomer pool and an in vivo oligomer assembly pathway. J Cell Biol. 2010;188:769–777. doi: 10.1083/jcb.200909176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavilainen VO, Hellman M, Helfer E, Bovellan M, Annila A, Carlier MF, Permi P, Lappalainen P. Structural basis and evolutionary origin of actin filament capping by twinfilin. Proc Natl Acad Sci U S A. 2007;104:3113–3118. doi: 10.1073/pnas.0608725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren S, Ojala PJ, Wear MA, Cooper JA, Lappalainen P. Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J Cell Biol. 2001;155:251–260. doi: 10.1083/jcb.200106157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni D, Le Clainche C, Carlier MF. Mechanism of actin-based motility. Science. 2001;292:1502–1506. doi: 10.1126/science.1059975. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Rodal AA, Manning AL, Goode BL, Drubin DG. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr Biol. 2003;13:1000–1008. doi: 10.1016/s0960-9822(03)00383-x. [DOI] [PubMed] [Google Scholar]
- Rodal AA, Tetreault JW, Lappalainen P, Drubin DG, Amberg DC. Aip1p interacts with cofilin to disassemble actin filaments. J Cell Biol. 1999;145:1251–1264. doi: 10.1083/jcb.145.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Welch MD, Machesky LM, Bridgman PC, Meyer SM, Cooper JA. Visualization and molecular analysis of actin assembly in living cells. J Cell Biol. 1998;143:1919–1930. doi: 10.1083/jcb.143.7.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- St-Pierre J, Douziech M, Bazile F, Pascariu M, Bonneil E, Sauve V, Ratsima H, D’Amours D. Polo kinase regulates mitotic chromosome condensation by hyperactivation of condensin DNA supercoiling activity. Mol Cell. 2009;34:416–426. doi: 10.1016/j.molcel.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonikian R, Xin X, Toret CP, Gfeller D, Landgraf C, Panni S, Paoluzi S, Castagnoli L, Currell B, Seshagiri S, et al. Bayesian modeling of the yeast SH3 domain interactome predicts spatiotemporal dynamics of endocytosis proteins. PLoS Biol. 2009;7:e1000218. doi: 10.1371/journal.pbio.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wear MA, Cooper JA. Capping protein: new insights into mechanism and regulation. Trends Biochem Sci. 2004;29:418–428. doi: 10.1016/j.tibs.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Wear MA, Yamashita A, Kim K, Maeda Y, Cooper JA. How capping protein binds the barbed end of the actin filament. Curr Biol. 2003;13:1531–1537. doi: 10.1016/s0960-9822(03)00559-1. [DOI] [PubMed] [Google Scholar]
- Wiesner S, Helfer E, Didry D, Ducouret G, Lafuma F, Carlier MF, Pantaloni D. A biomimetic motility assay provides insight into the mechanism of actin-based motility. Journal of Cell Biology. 2003;160:387–398. doi: 10.1083/jcb.200207148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.