Abstract
Informatics systems, particularly those that provide capabilities for data storage, specimen tracking, retrieval, and order fulfillment, are critical to the success of biorepositories and other laboratories engaged in translational medical research. A crucial item—one easily overlooked—is an efficient way to receive and process investigator-initiated requests. A successful electronic ordering system should allow request processing in a maximally efficient manner, while also allowing streamlined tracking and mining of request data such as turnaround times and numerical categorizations (user groups, funding sources, protocols, and so on). Ideally, an electronic ordering system also facilitates the initial contact between the laboratory and customers, while still allowing for downstream communications and other steps toward scientific partnerships. We describe here the recently established Web-based ordering system for the biorepository at Washington University Medical Center, along with its benefits for workflow, tracking, and customer service. Because of the system's numerous value-added impacts, we think our experience can serve as a good model for other customer-focused biorepositories, especially those currently using manual or non-Web–based request systems. Our lessons learned also apply to the informatics developers who serve such biobanks.
Introduction
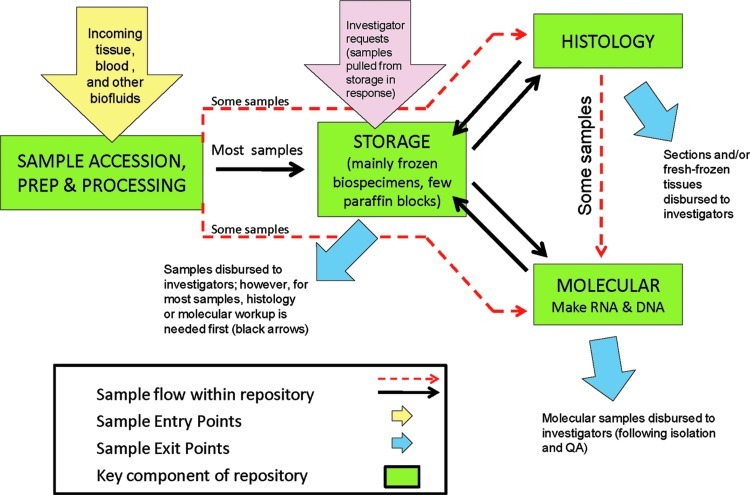
The Tissue Procurement Core (TPC) at the Washington University School of Medicine is a large and successful research biorepository that contains ∼400,000 diseased and normal specimens, employs 14 people in various scientific and administrative roles, and supports a wide variety of translational and other research programs at the School of Medicine on a request-driven basis. The traditional focus of the repository is neoplastic disease. A wide variety of study protocols and clinical trials are supported by the bank. In addition, the bank maintains a general specimen collection—not part of any specific clinical study or protocol—which is derived from surgical pathology discard tissue (ie, tissue not needed for clinical diagnosis which would otherwise be thrown away), and from which tissue is released for general research purposes in a de-identified fashion.1 The Washington University biorepository not only disburses specimens on request, but also provides a full array of laboratory services, including DNA and RNA isolation and characterization, frozen and paraffin tissue sectioning and staining, biofluid processing, pathology interpretation, and laser-capture microscopy. Consequently, the bank is organized into processing, molecular, and histology divisions, with storage as a centralized core function (Fig. 1). During the past year, the repository accessioned 34,863 specimens and disbursed 11,936 specimens for 85 different internal research programs or protocols. Most biobank functions, including accessioning, storage, specimen processing, inventory management, and distribution, are provided or managed by the caBIG™ caTissue Suite software program.2 However, this program lacks a mechanism to track the handling/processing stage of requests, or a customer request process sufficiently matched to our workflow and investigator needs.
FIG. 1.
The Washington University Medical Center biorepository workflow. This is a workflow diagram indicating the core components of the biorepository (green boxes), the entry point for biospecimens (yellow arrow), the entry point for requests (pink arrow), and the disbursal points for requested specimens and data (blue arrows). Workflow interconnections between the core laboratory areas are indicated with the black and red arrows. A color version of this figure is available in the online article at www.liebertpub.com/bio
A streamlined request process matters to customer service-oriented laboratories such as ours, since the efficiency of downstream activities depends on the request step and the capture of pertinent information. Probably, this is more relevant as the volume of work goes up. Despite its size and complexity, the Washington University biorepository, before 2011, had used mainly a nonautomated request process. Under this system, investigators typically requested specimens or services by simple e-mail communications, or by filling out a Microsoft Word-based request document (asking for contact information, project details, and so on) and attaching this form to an e-mail. Though such methods allowed the collection of needed information—and also preserved customer-focused interactions needed at the project's start—they were relatively inefficient, and prone to gaps in the communication or follow-through process. Other disadvantages are they did not allow requests to be efficiently tracked through sequential processing steps, and did not easily permit usage metrics assessments and other forms of data mining. As a result, lab personnel were unable to quickly determine the status of a request through online means, or to efficiently compile metrics needed to assess the current state of the business and the most promising areas for future growth and impact.
As the specimen inventory, request form usage, and laboratory procedure services continued to grow in size and scope, the biorepository clearly needed a better, more automated way to receive and track requests. And so we designed our Web-based ordering system, which has provided significant benefits in the months since its early 2011 inception. To design it successfully, we needed to (i) visualize the process from the customer's perspective, especially the typical needs of our customer base, and (ii) consider the priorities imparted by our workflow, and how the request features should be designed based on those needs.
Two respective system components, the “front end” and the “back end,” were created in response. We describe here the details of our ordering mechanism and how it streamlined workflow. We think our design approach can be instructive for informatics developers and customer-focused biobanks—particularly entities that are high volume, or have services or workflow paths analogous to our own.
Our approach was consistent with the general intent of published best practice documents from the National Cancer Institute Office of Biorepositories and Biospecimen Research3 and the International Society of Biological and Environmental Repositories,4 whose respective sections “Biospecimen Collection, Processing, Storage, Retrieval, and Dissemination” (B.2) and “Inventory Systems” (H3.000) recommend the implementation of computer-based systems that facilitate tracking and data management activities in biorepositories.
Key aspects of workflow and quality control relating either to this biobank, or to biobank science in general, have been separately described.1,5,6
Materials and Methods
The system's user interface was created using open source programming languages PHP and JavaScript™, while the backbone (site management and data housing) is from Microsoft™. This union of open source and proprietary development tools allowed us to utilize the inherent access rights and security built into the Microsoft™ material, yet the freedom to deploy these tools in an existing Web presence. The chosen technologies allowed flexibility to refine or adapt the system in the future in response to changing needs or usage patterns. Before designing the system, current workflow maps (Figs. 1–3) were devised and used as a guide.
FIG. 3.
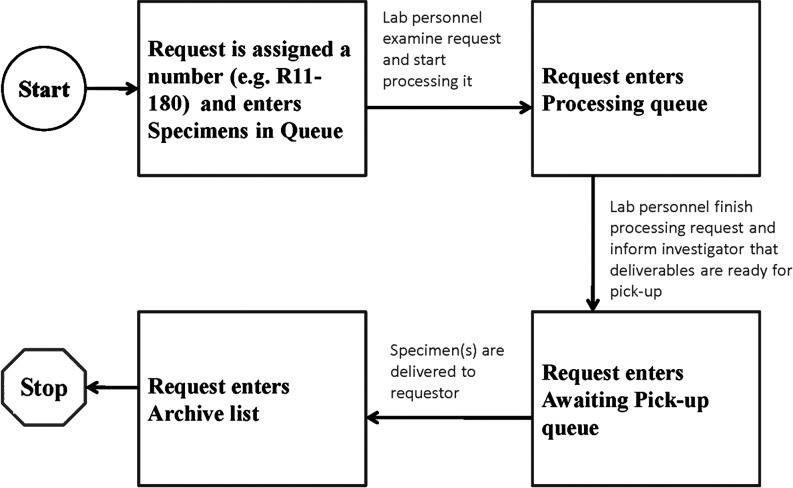
Request processing and tracking workflow for lab personnel (system's “back end”). This is a business process flow map covering the “back end” of the ordering system, the part visible only to laboratory personnel, and which is described in the Materials and Methods section.
Request form Web site (system's “front end”)
Consideration of the optimal design for a Web-based request system started with the basic workflow for the repository (Fig. 1). Requests originating from Washington University investigators, as opposed to investigators or groups external to the University, were the sole focus of the project, since all requests to the biorepository must either be for a University laboratory, or whether involving an outside collaborator, must have a clearly established University contact who is willing to enter the request and assume responsibility for it. There are 3 general categories of requests that the biobank receives:
• Submission of a user's own samples for lab procedures, such as laser-capture microscopy or nucleic acid extraction; these are represented by the large yellow arrow in Fig. 1, followed by either red arrow depending on the request.
• Request for disbursal of specific specimens previously deposited by the investigator, or from another protocol that the investigator has permission to access. These are represented by the large pink arrow in Fig. 1.
• Request for general tissues or biofluids for certain organs or diseases, without a preconceived preference for specific sample numbers from specific protocols. These are also represented by the large pink arrow in Fig. 1.
The request form was placed as an easily visible link on the biorepository's Web site, which in turn was accessible from the main pathology department Web site. This assured that the ordering Web site would be sufficiently visible and accessible to its user base (though, since entry to the ordering Web site is password protected, details of a request are available to the biorepository but not to other investigators unless those details are voluntarily shared). Instructions and communication materials were prepared in advance, to help the new and existing bank users navigate the site. Also, a specific date was chosen starting from which all request-initiated activities would need to be documented by an investigator-submitted online form.
In brief, the request process functions as follows (Fig. 2), with key information in “required fields” that require the user to enter a response before being allowed to proceed.
FIG. 2.
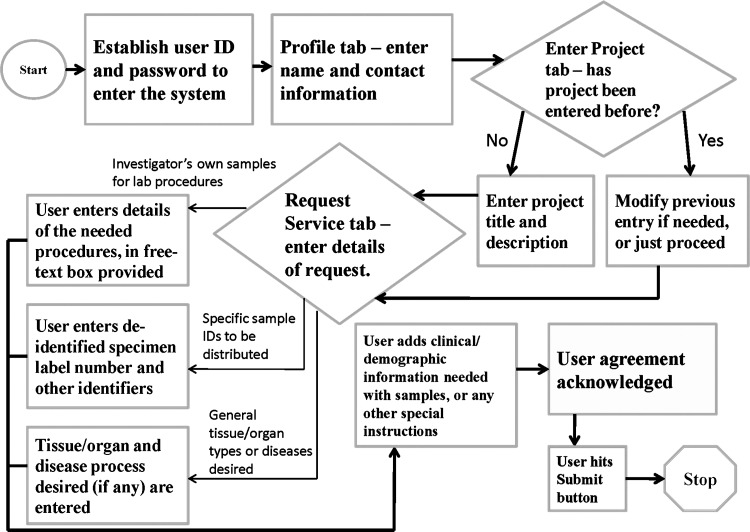
Request system process flow (system's “front end”). This is a business process flow map covering the “front end” online ordering system with which the requestor interacts (Section 3.1). Note start and stop points, and the 3-way branch in the flow process, lower left-hand corner of diagram, which depends on the type of request being initiated.
1. New users must establish a departmental user ID and password to enter the system.
2. Investigators then enter a Profile tab, where they enter (or update, as appropriate) their basic contact information.
3. Users then go to a Projects tab, where they can enter a new project title or choose a previous one. For either option, the information requested or listed is the same: project name, primary investigator, and details; billing and grant information; and Institutional Review Board (IRB) approval status. These can either be entered new or updated. The purpose of the IRB inquiry is to ascertain that the investigator has the proper approval for the project and that the proposed project scope and use for any currently banked samples is consistent with existing informed consent and/or protocol status for those same samples.
-
4. Users then go to a Request Service tab, where a pull-down menu lists all the projects previously entered by that investigator. Choosing one then initiates a new request for that project. A description and requested processing completion date (“target date”) are asked for; then the user makes a 3-way strategic choice that determines what subsequent information is requested:
• Submit investigator's own samples for lab procedures—as part of this, the user describes in a free-text box (see step 5 below) what procedures are desired.
• Request specific sample ID numbers distributed directly from the laboratory—here, the user is asked the clinical protocol name and various specimen numbers that enable the lab to retrieve the sample(s). The user would possess these numbers from an inventory search, from the lab staff, or from having used those samples in a prior study. The sample numbers being referred to here are de-identified; that is, not containing protected health information, and not traceable by the investigator to such. De-identified sample numbers are readily available to our customer base and are released with the disbursal of the samples. Protected health information is only visible to lab personnel behind the secure informatics “firewall” and is of course not accessible by users outside the firewall.
• Request general tissue/organ types or diseases—the key information entered here is the tissue or organ type desired, and the disease process desired (if any).
For all the 3 categories just detailed, the user selects the specimen type (fluid, molecular, tissue, or cells) through a pull-down menu, along with other options that enable the laboratory to carry out its function. For example, for molecular requests, DNA, RNA, total nucleic acid, protein, and others maybe selected. For disbursals of banked specimens, the user is asked milligram quantities, tissue section thicknesses, or other relevant parameters. Also, for all categories, an option is provided to iteratively ask for more samples or services without having to start a new electronic form. Multiple requests over time can be added for a given project.
5. Using a free-text box, users then can request clinical, demographic, or other database information that they want, or other special or pertinent instructions.
-
6. The investigator must then electronically acknowledge a user agreement, in which they assure the following details:
• The samples and data will only be used according to the relevant IRB project approval.
• Careful safeguards will be maintained to prevent inappropriate information or data release.
• Samples will not be distributed to unauthorized individuals.
• Responsibility for processing charges will be assumed by the requestor or primary investigator, and any changes in funding status will be communicated to the repository.
7. User officially submits the request, which is then automatically assigned a request number, for example, R11-134. Users are provided with contact information for any subsequent questions.
8. When the user reenters the system, a “past requests” tab shows all previous requests by that user. The status of these is automatically updated by the “back end” component so as to communicate the current handling status of a request.
Request processing and tracking within laboratory (system's “back end”)
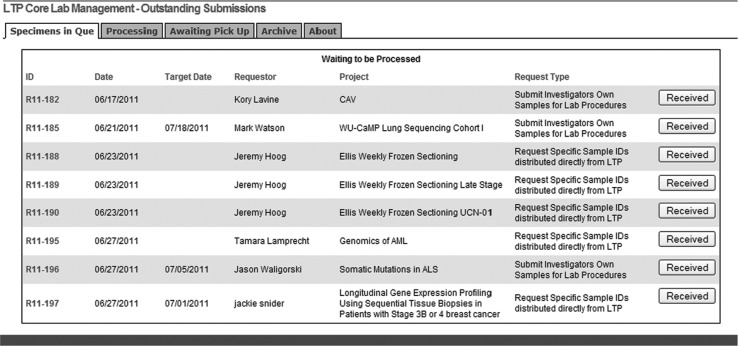
This component can be accessed only by lab personnel and informatics support specialists, and enables tracking and monitoring of requests within the laboratory. Its fundamental characteristic is a sequential listing of all requests by assigned number (see step 7 in the previous section), arranged according to 4 tabs that reflect various stages of the handling process: specimens in queue, processing, awaiting pick up, and archiving. As personnel act on requests, they move them from one stage tab to the next by clicking a button, thus providing the capability to track them by processing stage (Fig. 3, flow diagram; Fig. 4, computer display). Along with requests listed by their assigned number, all display tabs show basic request information (Fig. 4): date of request, target date (the “need by” date), and the requestor, project, and request type.
FIG. 4.
Request processing and tracking display for lab personnel (system's “back end”). This is the actual monitor display seen when laboratory personnel access the “back end” component of the ordering system; this is further described in the Materials and Methods section.
All requests sit within one and only one of the 4 tabs, meaning that there is no ambiguity about where they stand. Throughout the “back end” system, the request number (eg, R11–134) is itself a link that, when selected, takes the user to an electronic version of the form previously submitted by the investigator, with all of the supplied information. This form can be saved or printed if desired. The tabs reflective of request stage are sequentially arranged as follows (see Fig. 4):
• Specimens in queue—This first tab lists all requests that have been submitted but have not yet been acted on by laboratory personnel. This display is the one shown as an example in Fig. 4, with the other tabs having the same horizontal headings.
• Processing—This next tab contains a list of requests that are currently being processed in the laboratory according to the instructions provided by the requestor.
• Awaiting pick up—This next tab contains all requests that have completed processing, and are ready for investigators to pick up from the laboratory. Typically at this stage, laboratory personnel call or e-mail investigators to tell them their processed specimens are available.
• Archive—This final tab contains a list of all completed requests, as a useful reference.
Results and Discussion
As of this writing, ∼270 requests during 2011 have been submitted through the new system, providing a basis by which to judge its early successes, and its potential areas for enhancement. The new system has benefited us in numerous ways compared with the previous method (Table 1), and these benefits serve as “lessons learned” for informatics developers who might be thinking of modeling their approach after ours. The key ingredients for success were as follows:
Table 1.
Benefits of Electronic Request Form System Compared with Previous Method
| Issue | Electronic request form system's benefit | Situation using previous manual request process |
|---|---|---|
| Accessibility to research community | Link to departmental Web site, and proactive outreach process, insure that order form is highly visible and accessible. Projects in this system are also visible in corresponding order systems for other technology-driven cores (eg, histology, immunology, and sequencing) | Availability of request form depended on interpersonal communications and e-mail, which was less ideal |
| Capture of required information | Needed information is automatically captured in electronic form through required fields, saving lab personnel time | Needed information could inadvertently be omitted, costing lab personnel time and effort to track it down |
| Documentation | Request-driven lab activities are all backed up by an electronic form, allowing accountability, and facilitating the billing process | Documentation of what was requested, and who requested it, could be inadvertently misplaced when the process depended on e-mails or phone calls |
| Tracking of stage within laboratory | “Back end” of request system allows online assessment of the general laboratory stage where a request currently is (awaiting processing, in processing, awaiting pick up, or archive) | Before the current system, there was no way to track project/request stage in the laboratory other than verbal and e-mail communications |
| Metrics | System allows the assessment of useful metrics related to request process, including request and user group categorizations, turnaround times, and personnel productivity | Metrics assessments for requests were more cumbersome without the automatic time and user stamping of status updates in the current system |
| Additional levels of monitoring and review | Current system allows for the possibility of additional needed tiers of review (eg, a utilization review committee) by adding new tabs to the “back end” display layout | Required new levels of monitoring/review would be more difficult without a formalized system listing all requests and their process flow stages in one place |
| Status communications to requestors | Users can look up the status of their requests by finding their project in the Past Requests tab in the “front end” component | Under the previous system, users had to call or e-mail the laboratory to find out the status of their requests |
• Establishing accurate business process and/or workflow diagrams beforehand;
• Thoroughly understanding the needs and objectives of the customer base;
• Designing two broad parts—one for the users, and one for the lab personnel—with separate but interrelated objectives and automatic updating of one from the other;
• Designing all details—including the choice of “required-entry” fields—according to the information the laboratory most needs to streamline its workflow;
• Creating tracking methods within the system for key metrics that drive the business or are useful for accounting purposes;
• Implementing strategies that help ensure acceptance and broad usage of the system.
Through our efforts, the research community at Washington University now has an easily accessible, standardized way to make requests to the biobank, and thus research needs can be identified and filled more quickly. This, of course, requires the user community to be aware of the site, and to know how to use it. Therefore, at the system's inception, proactive communication was needed. Outreach measures are (and will be) continually needed on an ongoing basis, as current investigators acquire interest, and as periodic turnover yields users who are new to the system. We have found it beneficial to include the lab Web site and ordering mechanism as part of a general outreach program promoting the biorepository as a valuable resource for the medical community.
The standardization inherent in the online approach also ensures that required information, such as IRB approval status, grant funding mechanisms, and delivery address, are automatically captured, since these are required fields in the order form. This saves time relative to the previous, less standardized system, where needed information was sometimes inadvertently omitted, thus consuming valuable time on the part of lab personnel to track it down.
Documentation is also another key benefit. With an electronic request now required before any investigator-driven procedure is started in the biobank, performed work and the resulting billing charges can much more clearly be attributed to a specific request and user, with minimal ambiguity regarding what was ordered, or who ordered it (Note: investigators are not charged for the biosamples themselves, but rather for the lab storage and processing charges they incur when they make requests. Patients are not charged for tissue bank submissions.). Also, because project details are requested as part of the electronic form, the biorepository learns the science and rationale behind requests, and can proceed with that understanding in mind.
The system's “back end” benefits laboratory personnel in key ways. All requests are accounted for in 1 of the 4 sequential stage tabs; thus, they cannot inadvertently be lost or drop out of the queue. Requests having prolonged times in one of the queues can immediately be identified and tracked to the accountable lab personnel, or to the investigator if located in the “awaiting pick up” queue, and proper notification to the investigator has previously been made. As status updates (through the stage tabs) are made by lab personnel, the “front end” is updated automatically—as previously noted—therefore giving the requestor useful information.
The system allows various metrics to be tracked and assembled in a way that was either not possible or was less efficient under the old request scheme. Besides cumulative request numbers, the “back end” component can track request date, investigator, project, request type, and other data provided with the electronic form, so that requests can be numerically categorized by such parameters. This provides a valuable snapshot of current usage patterns, and opportunities for future growth and outreach.
These capabilities have been recently used to determine the number of 2011 requests that fell into various categories as part of internal strategic discussions. Another potential benefit is as follows: currently, the formation of a utilization review committee is under consideration that would help assess the scientific merits of individual requests and sample submissions against the increasingly limited tissue bank infrastructure resources at our institution. The layout and function of the new system greatly facilitates discussion and planning, since the highly visible stages and tracking of the system's “back end” would allow easy viewing by a committee—possibly with the addition of a new electronic stage early in the processing sequence, requiring the committee's approval before the request can move forward.
All status updates on the “back end”—between the specimens-in-queue, processing, pick-up, and archive stages—are time stamped and user stamped automatically. This will allow not only turnaround time reporting, but can also provide insight into personnel productivity. In practice, analysis of turnaround times from the online system is possible only by manually analyzing captured data, and so the design of an automated turnaround time calculator/reporter is a future opportunity for the system's growth and refinement. The system also can quickly identify requests whose processing time is approaching the requestor's “need by” date when such has been provided.
One current limitation pertains to the submission of the investigator's samples for lab procedures, or the withdrawal of specific sample IDs from the biorepository archives. Though as previously described, the order form provides an iterative mechanism to add successive sample IDs to the same general order form, this is obviously impractical if dozens or hundreds of samples need to be described or referenced. This scenario is currently addressed by users creating their own Excel files for large specimen groups, and submitting to the lab separately from the online system (perhaps through an e-mail communication). However, a “bulk upload” feature that allows the user to populate embedded tables within the request form, with those data then being automatically uploaded into the caTissue Suite inventory on receipt, would be more ideal, and is in fact undergoing development currently.
Finally, the TPC is one of several technology-driven core groups which further translational medicine at the Washington University Medical Center. The system is designed such that projects entered in the biorepository request system are then visible in similar request sites for the other cores, for example, histology, immunology, and sequencing. This has obvious benefits for efficiency, and the synergistic union of multiple approaches for a project.
Conclusions
The implementation of an electronic ordering and tracking mechanism, based on both the typical needs of our customer base, and the layout and priorities of our laboratory workflow, has been a highly beneficial approach for the Washington University Medical Center biorepository. Our rationale, design, and approach could be useful as a model for biobanks and potentially any customer-focused laboratories who are considering streamlining their request process. The specific capabilities, work volume, and process flows of other biorepositories will, of course, influence how adaptable the features described here will be.
Acknowledgments
The authors wish to acknowledge the Siteman Cancer Center (Grant #P30 CA91842) and the Institute for Clinical and Translational Science (Grant #UL1RR024992) at the Washington University Medical Center, which has partially funded the biorepository at the Washington University School of Medicine.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.McDonald SA. Chernock RD. Leach TA. Kahn AA. Yip JH. Rossi J. Pfeifer JD. Procurement of human tissues for research banking in the surgical pathology laboratory: prioritization practices at Washington University Medical Center. Biopreserv Biobank. 2011;9:245–251. doi: 10.1089/bio.2011.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDonald SA. Watson MA. Nagarajan R. Mulvihill DA. Brink A. caTissue Suite: an open-access, feature-rich tool for biospecimen annotation and data sharing. FASEB J. 2011;25:lb338. [Google Scholar]
- 3.Office of Biorepositories and Biospecimen Research. 2010: [Aug 15;2011 ]. National Institutes of Health, U.S. Department of Health and Human Services. National Cancer Institute (NCI) Best Practices for Biospecimen Repositories. Revised Version. [Google Scholar]
- 4.International Society for Biological and Environmental Repositories (ISBER) 2008 Best Practices for Biorepositories: Collection, Storage, Retrieval, and Distribution of Biological Materials for Research. www.isber.org/bp/BestPractices2008.pdf. [Aug 15;2011 ];Cell Preserv Technol. 2008 6:3–58. [Google Scholar]
- 5.McDonald SA. Principles of research tissue banking and specimen evaluation from the pathologist's perspective. Biopreserv Biobank. 2010;8:197–201. doi: 10.1089/bio.2010.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDonald SA. Velasco E. Ilasi NT. Business process flow diagrams in tissue bank informatics system design, and identification and communication of best practices: the pharmaceutical industry experience. Biopreserv Biobank. 2010;8:203–209. doi: 10.1089/bio.2010.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]