Abstract
Caffeic acid o-methyltransferase (COMT) is one of the important enzymes controlling lignin monomer production in plant cell wall synthesis. Analysis of the genome sequence of the new grass model Brachypodium distachyon identified four COMT gene homologs, designated as BdCOMT1, BdCOMT2, BdCOMT3, and BdCOMT4. Phylogenetic analysis suggested that they belong to the COMT gene family, whereas syntenic analysis through comparisons with rice and sorghum revealed that BdCOMT4 on Chromosome 3 is the orthologous copy of the COMT genes well characterized in other grass species. The other three COMT genes are unique to Brachypodium since orthologous copies are not found in the collinear regions of rice and sorghum genomes. Expression studies indicated that all four Brachypodium COMT genes are transcribed but with distinct patterns of tissue specificity. Full-length cDNAs were cloned in frame into the pQE-T7 expression vector for the purification of recombinant Brachypodium COMT proteins. Biochemical characterization of enzyme activity and substrate specificity showed that BdCOMT4 has significant effect on a broad range of substrates with the highest preference for caffeic acid. The other three COMTs had low or no effect on these substrates, suggesting that a diversified evolution occurred on these duplicate genes that not only impacted their pattern of expression, but also altered their biochemical properties.
1. Introduction
Temperate grains like wheat and barley, along with forage grasses, contribute greatly to the human food and animal feed supply. However, the large and complex genomes in these economically important grasses present challenges for genomics studies and map-based cloning of target genes for crop improvement. Similarly, although large perennial grasses like switchgrass and Miscanthus are being developed as dedicated herbaceous energy crops, our knowledge about the biological and genetic basis of important bioenergy traits remains limited [1–4]. Brachypodium distachyon (hereafter referred as Brachypodium) is an attractive experimental system and genomics model for grass research. It has many desirable attributes (small physical stature, short generation time, easy growth requirement, etc.) and numerous freely available genomics resources (high quality genome sequence, EST collection, large-insert BAC libraries, expression/tilling microarray, T-DNA mutant population, etc.) [5]. Thus, Brachypodium can serve as a useful model system to address issues unique to grasses ranging from grain improvement to the development of superior bioenergy crops [5–7].
Plant cell walls are a complex composite structure composed of polysaccharides (cellulose and hemicellulose) that are embedded and crosslinked by other polymers and small molecules (lignin, pectin, and ferulic acid). The type of hemicellulose and other components differs significantly between the grasses and dicots [8]. Briefly, primary grass cell walls contain glucuronoarabinoxylans and mixed-linkage glucans as the hemicelluloses, high levels of ferulic acid and p-coumaric acids, and the relatively little pectin and protein. Dicot primary cell walls contain xyloglucan, mannans, and glucomannans as the hemicelluloses and high levels of pectin and structural proteins. Secondary cell walls of both grasses and dicots are composed of cellulose, glucuronoarabinoxylans, and lignin. However, the structure of the glucuronoarabinoxylans differs between dicots and grasses. Given the compositional and architectural difference between grass and dicot cell walls, we have turned to Brachypodium as our model system to study grass cell walls.
The use of cellulosic biomass crops as a feedstock for the production of transportation fuel offers significant potential environmental and economic advantages. However, due to the difficulty in converting the sugars locked in cellulose and hemicellulose into fuel, our ability to use this renewable feedstock is limited. A deeper understanding of the genes that control cell wall biosynthesis and architecture may allow us to tailor the cell wall to be more amenable to conversion into biofuel [10–14]. Lignin in particular is an impediment to the production of biofuels and forage digestibility [15–20]. Due to crosslinking, it blocks access of cell wall degrading enzymes to cellulose and hemicellulose [21–23]. This makes it necessary to employ harsh pretreatment to disrupt the cell wall structure prior to enzyme hydrolysis. This harsh pretreatment create compounds that are inhibitory to organisms used to convert the free sugars into biofuels like ethanol and butanol. However, due to its phenolic ring structure, lignin is an energy dense compound and thus a high lignin content is desirable if the biomass is used for direct combustion.
Lignins are complex heteropolymers derived from three monolignols, p-coumaryl, coniferyl, and sinapyl alcohol. These monolignols produce p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units, respectively, when incorporated into the lignin polymer. Although composed of only three building blocks, the composition and structure of lignin varies considerably within and among plants because the ratio of monomers varies and there is no repeating linkage structure. Rather, the lignin monomers undergo what appears to be a random cross-linking in the cell wall via several linkages. The S content and the S to G ratio are critical parameters that measure for characterizing lignin composition in the cell wall of angiosperm plants [15, 20]. Enzymes involved in the lignin biosynthesis pathway have been characterized in different plant species [24]. o-methylation modulates the chemical and physical properties of the lignin polymer. o-methylation mediated by o-methyltransferases (OMTs) transfers a methyl group from S-adenosyl-L-methionine (SAM) to the hydroxyl group of a methyl acceptor molecule. Plants contain a large family of OMT enzymes [24]. One of the essential OMTs in lignin biosynthesis is caffeoyl CoA 3-o-methyltransferase (CCoAOMT; EC 2.11.104) which is primarily responsible for the initial o-methylation of the 3-hydroxyl group and specifically methylates the 3,4-dihydroxy substrate as a CoA-linked thioester. A second important OMT termed caffeic acid o-methyltransferase (COMT; EC 2.1.1.68) generally catalyzes the o-methylation of the 5-hydroxyl group of 3-methoxy-4,5-dihydroxy precursors. The function of both OMTs has been inferred from the effects on lignin composition through the downregulation of these enzymes in transgenic plants as well as in mutant lines affecting the expression of these genes. For instance, the COMT gene was first cloned in maize by differential screening of a root cDNA library [25]. Characterization of the maize COMT promoter indicated that it directed GUS expression to the xylem and other tissues undergoing lignifications [26]. Transgenic plants with a downregulation of the maize COMT showed a strong decrease of Klason lignin content, a decrease in syringyl units, lower p-coumaric acid contents, and the occurrence of an unusual 5-OH guaiacyl unit [27]. These features are similar to those observed in the maize bm3 (brown-midrib3) mutant lacking the functional COMT gene [27, 28]. Furthermore, it appeared that a decrease in COMT activity led to improved forage digestibility, suggesting that the downregulation of COMT might alters the overall cell wall organization in a way that walls become more accessible to bacterial enzymes [27]. The effects of caffeic acid o-methyltransferase (COMT) gene modification on reduction of S monomer level, which is linked directly to the lignin reduction, have been shown in other modified plant species [29–36].
OMT genes have been cloned and characterized in a number of plant species and provide valuable information for the study of the evolution, expression, and function of OMTs in plants [29, 32, 33, 36–40]. However, the characterization of OMT genes in Brachypodium has not been reported. In this study, we identified, isolated, and characterized four Brachypodium COMT genes. Comparative and phylogenetic analyses revealed that while related grass genomes (rice and sorghum) contain only one copy of COMT, Brachypodium possesses four copies located in three different chromosomes. Functional characterization indicated that only the orthologous copy has the biochemical properties similar to other COMTs in grass species although all the Brachypodium COMT genes are expressed during plant development. The isolation and characterization of Brachypodium COMTs will permit us to develop an efficient system to analyze these enzymes in the lignin biosynthesis pathway through forward and reverse genetics approaches in this tractable model species.
2. Results
2.1. Genomic Organization of COMT Genes in Brachypodium
To identify COMT genes in Brachypodium, the rice COMT gene (XP480185) and Arabidopsis COMT gene (AAB96879) were used in a BLASTP search against the Brachypodium genome database v1.0 annotation (http://www.brachypodium.org/). The top four hits matched annotated Brachypodium genes: Bradi1g14870, Bradi2g02380, Bradi2g02390, and Bradi3g16530, with e-values lower than 1e − 100 (data not shown). These four genes were renamed BdCOMT1, BdCOMT2, BdCOMT3, and BdCOMT4, respectively, in this study. From the fifth hit, the e-values dropped considerably to be higher than e −50 and are, therefore, considered distantly related to COMT. BdCOMT1 and BdCOMT4 are located on Brachypodium chromosomes 1 and 3, respectively, while both BdCOMT2 and BdCOMT3 are located on chromosome 2. Interestingly, the BdCOMT2 and BdCOMT3 genes are next to each other with inverted orientation (Figure 1). When the sequences of the four BdCOMT genes were blasted against both the rice and sorghum genome databases, they all retrieved a single strong match to the corresponding COMT gene, suggesting that only Brachypodium contains multiple copies of COMT gene.
Figure 1.
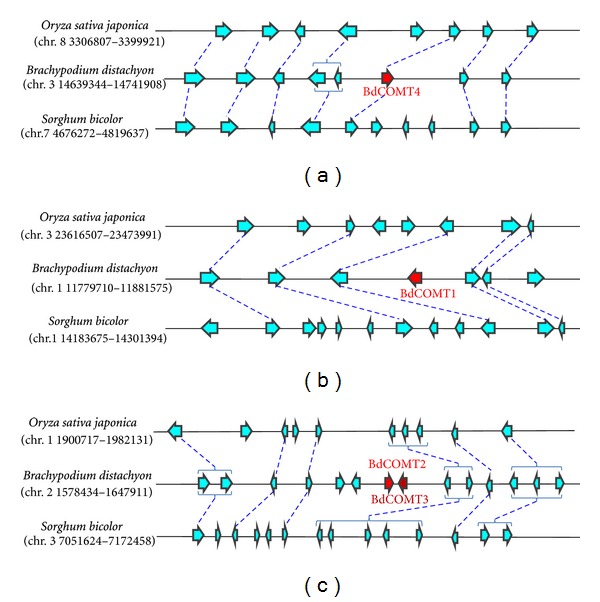
Collinearity analysis of Brachypodium COMT gene-containing regions with the orthologous regions of rice and sorghum genomes. Genomic regions harboring BdCOMT4 (a), BdCOMT1 (b), BdCOMT2 (c), and BdCOMT3 (c) were extracted and the annotated genes in these regions were used to identify orthologous regions from rice and sorghum genomes for collinearity analyses. Annotated genes are represented by arrows with the direction of arrow heads indicating the orientation of transcription. Brachypodium COMT genes are labeled in red. Orthologous genes that are shared in two or more genomes are connected with dashed lines. Duplicate genes in a genome are bracketed.
To analyze the genomic organization of these Brachypodium genes, genomic regions surrounding the four BdCOMT genes were used to search for orthologous regions in the rice and sorghum genomes. The collinearity of these regions was compared to the orthologous regions in the rice and sorghum genomes (Figure 1). The rice and sorghum regions orthologous to the BdCOMT4 region contained their single COMT genes (Figure 1(a)), indicating that BdCOMT4 is the orthologous counterpart of the rice and sorghum COMT genes. The rice and sorghum regions orthologous to the other Brachypodium COMT gene regions did not contain orthologous COMT genes although the surrounding genes showed general collinearity (Figures 1(b) and 1(c)). This collinearity analysis supports the sequence blast search results indicating that multiple COMT genes exist only in Brachypodium.
2.2. Phylogenetic Analysis of COMTs in Brachypodium and Other Species
To further confirm the evolutionary relatedness of the Brachypodium COMT genes, previously characterized COMTs from both dicot and monocot species were used to perform phylogenic analysis (Figure 2). Dicot COMTs formed one clade and monocot COMTs were grouped into another clade. BdCOMT4 is more closely related to the other grass COMTs than the other three Brachypodium COMTs, which formed a distinct clade separated from the orthologous COMT of different grass species. Nevertheless, the observation that BdCOMT1, BdCOMT2, and BdCOMT3 were grouped into monocot COMT clade suggests that duplicated Brachypodium COMT genes were generated after the divergence of dicot and monocot.
Figure 2.

Phylogenetic analysis of COMTs from different plant species. COMT sequences represented by their GenBank or gene IDs from different plant species were extracted from NCBI database and used to construct a phylogenetic tree with phylogenic tree using neighbor-joining method.
We also tested the possibility that BdCOMT1, BdCOMT2, and BdCOMT3 belong to other related o-methyltransferases, caffeoyl-CoA o-methyltransferase (CCoAOMT) groups, by including a number of characterized CCoAOMTs in our phylogenic analysis. These Brachypodium COMTs did not group with CCoAOMTs (see Supplemental Figure S1 available at http://dx.doi.org/10.1155/2013/423189), providing further supporting evidence that these Brachypodium COMT genes are likely to be paralogs of BdCOMT4 that resulted from gene duplication.
2.3. Sequence Analyses of BdCOMTs
ClustalW analysis was used to align the Brachypodium COMT genes at both nucleotide and protein sequence levels. The results showed that they shared over 70% nucleotide identity between any two sequences (data not shown). At the amino acid level, these COMTs shared at least 60% sequence similarity between any two sequences. As expected, BdCOMT2 and BdCOMT3 are the most closely related of the four BdCOMTs. The physical proximity indicated that they are derived from a tandem duplication event (Figure 1(c)). Sequence alignment with COMTs from other species indicated that BdCOMT4 protein shares 58%, 59%, 78%, and 83% sequence similarity with Arabidopsis thaliana, Medicago, Zea mays, and Oryza sativa COMT proteins, respectively.
From examination of crystal structure of the alfalfa COMT protein, amino acids responsible for catalytic sites and substrate binding sites have been inferred [41]. There are three catalytic sites predicted to be conserved within COMT genes. Two catalytic sites, E297 and E329, in MsCOMT are conserved among all four BdCOMT proteins (Figure 3). The other catalytic site H269 in MsCOMT is conserved in BdCOMT4 (H266) and BdCOMT1 (H270). However, in BdCOMT2 and BdCOMT3, it was replaced with N at this site. Thirteen substrate binding sites have been previously predicted based on MsCOMT crystal structure; twelve of these sites are conserved in BdCOMT4. The only difference in sites is a change at I316 in MsCOMT to V313 in BdCOMT4. It seems this mutation occurred in MsCOMT since OsCOMT, ZmCOMT, and AtCOMT all encode V at this position. However, this site was changed to D in BdCOMT1, and it was mutated to encode I in BdCOMT2. In comparison with BdCOMT4, more changes in these substrate binding sites were observed in BdCOMT2 (8 sites), BdCOMT3 (8 sites), and BdCOMT1 (7 sites). Thus, although BdCOMTs have an overall higher amino acid similarity among themselves when each is compared to MsCOMT, BdCOMT4 and MsCOMT showed higher conservation in the substrate binding sites. The methyl donor (SAM) binding sites were also analyzed. Among all five donor binding regions, there is only one site (W271 in MsCOMT) where BdCOMT2 and BdCOMT3 are similar to each other but different from other COMTs. Otherwise, all sites are generally conserved among BdCOMTs and COMT genes from other species (Figure 3).
Figure 3.
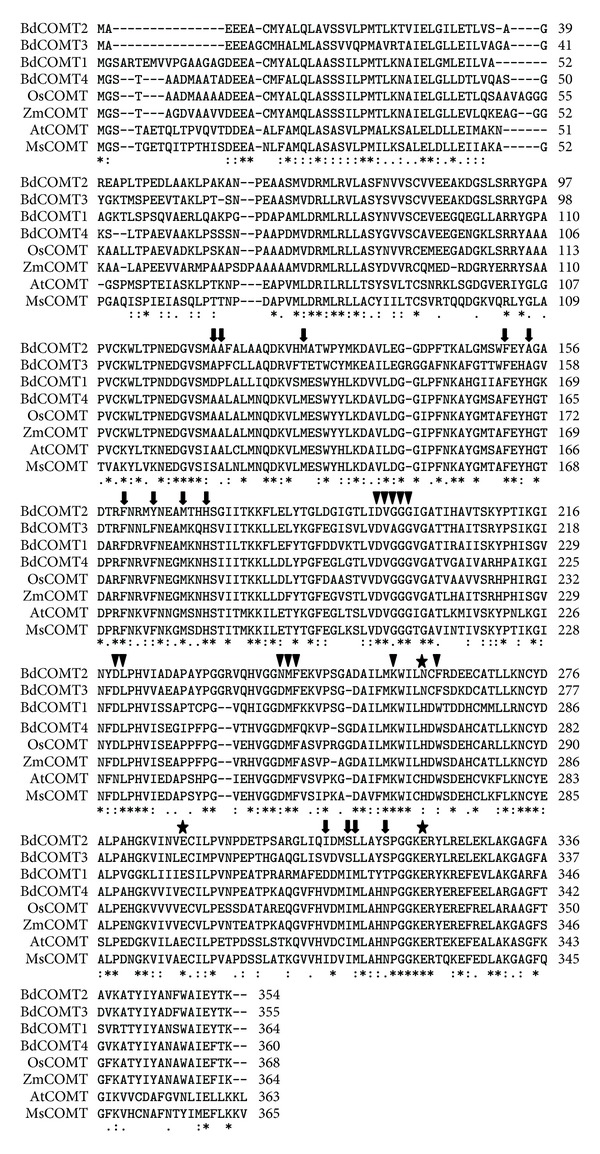
Comparison of amino acid sequences of four BdCOMTs (BdCOMT1, BdCOMT2, BdCOMT3, and BdCOMT4) with COMTs from Oryza sativa (OsCOMT), Zea mays (ZmCOMT), Arabidopsis thaliana (AtCOMT), and Medicago sativa (MsCOMT). Inferred from MsCOMT crystallographic structure, substrate binding residues are indicated by arrows, methyl donor binding sites by triangles, and catalytic residues by stars. An asterisk “∗” indicates identical residues in all sequences. A semicolon “:” indicates strongly conserved residues (score >0.5), and a dot “.” indicates weaker conserved residues (score <0.5) [9].
2.4. Tissue-Specific Expression of BdCOMT Genes
PCR primers specific for each Brachypodium COMT gene were designed to characterize the expression of these genes in various tissues including root, stem node, stem internode, leaf sheath, leaf blade and callus (Figure 4). In this semiquantitative RT-PCR analysis experiment, BdCOMT4 was found to be expressed uniformly and constitutively in all the tissues examined as compared to the ubiquitous Brachypodium actin gene expression. In contrast, the other three COMT genes showed very different expression patterns. For example, BdCOMT2 was only expressed in the internode, leaf sheath and leaf blade, but absent in the other tissues, whereas BdCOMT3 appeared to be expressed in all the tissues with the highest expression level detected in root and leaf blade, suggesting that these tandemly duplicated genes are now regulated differently at the transcriptional level. BdCOMT1 gene was strongly expressed in the stem node and internode but barely detected in the other tissues (Figure 4).
Figure 4.
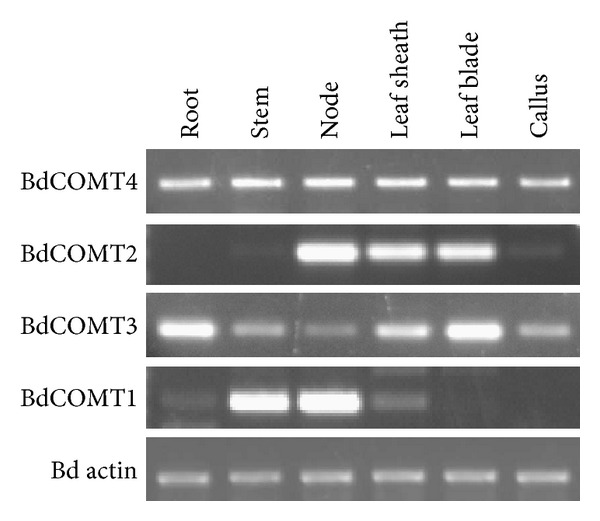
Tissue-specific expression of BdCOMT genes. Total RNAs were extracted from different Brachypodium tissues as indicated and used for synthesis of cDNA for RT-PCR. Gene-specific primers for each Brachypodium COMT genes were designed and used to amply corresponding transcripts. Primers specific for actin expression are used as the internal control to indicate that equal amount of cDNAs was used in each RT-PCR assay.
2.5. Expression and Biochemical Characterization of Recombinant BdCOMT Proteins
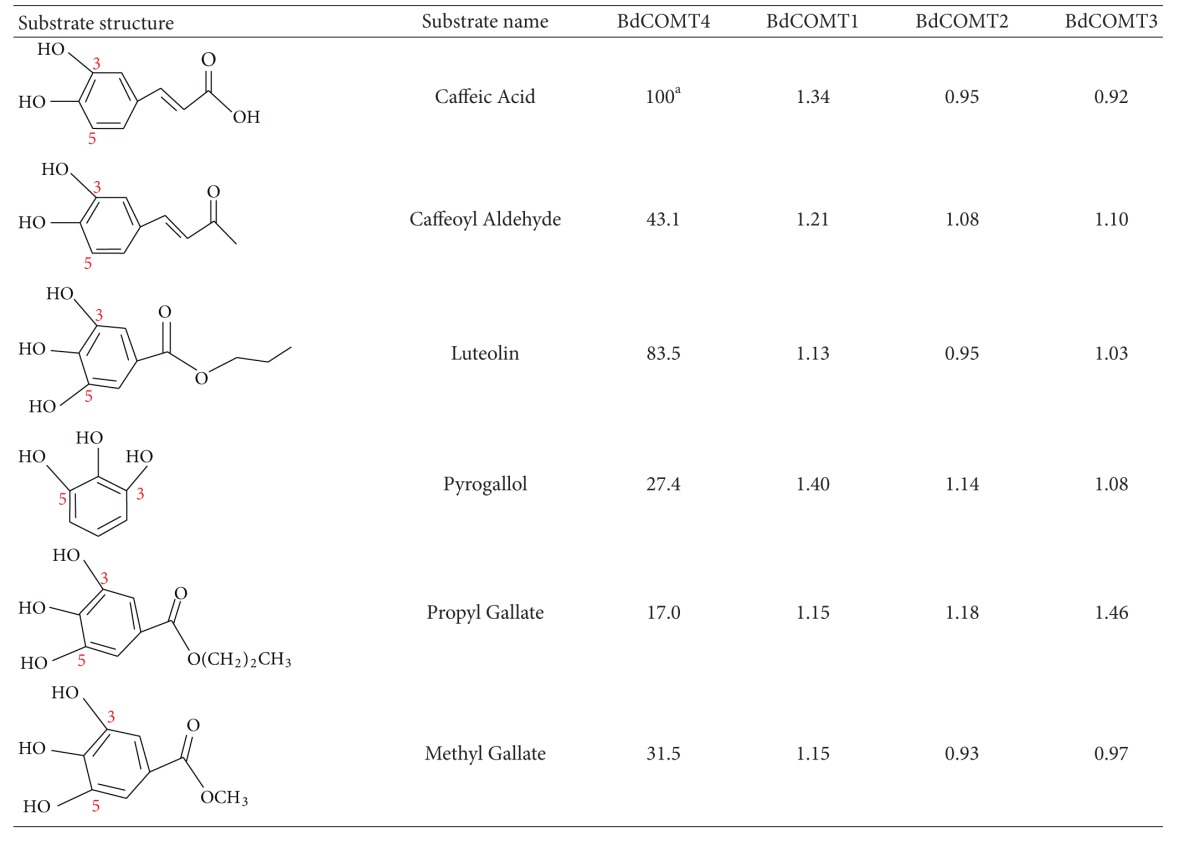
The gene expression analysis indicated that all the Brachypodium COMT genes are active genes. To biochemically characterize these Brachypodium COMTs, full-length cDNA for each COMT gene was isolated and cloned in frame into His-tagged pQE-T7 expression vector. The recombinant proteins were expressed in E. coli and purified with nickel-conjugated agarose beads (Figure 5(a)). Minor nonspecific protein contamination was detected in purified recombinant proteins despite imidazole concentrations in the wash series up to 50 mM (Figure 5(b)). Nevertheless, we estimated that the purified proteins reached at least 85% purity. The purified recombinant proteins were used for enzymatic activity assays with S-[methyl- 14C]-adenosyl-L-methionine (SAM) as the methyl donor and six substrates including caffeic acid, caffeoyl aldehyde, and four flavonoid compounds (Table 1). Among these substrates, BdCOMT4 showed the highest activity on caffeic acid, and this activity was set as 100% for comparison with other substrates. BdCOMT4 activity on caffeoyl aldehyde was less than half of that on caffeic acid (41.1%). BdCOMT4 also showed high catalytic activity on luteolin (85.0%). This flavonoid compound was a preferred substrate for rice COMT [38]. In addition, BdCOMT4 displayed activity on three other flavonoid substrates at different levels, ranging from 17.0 to 31.1% (Table 1). These results suggest that BdCOMT4 prefers the most likely in vivo substrate, caffeic acid, but is also active on a range of substrates. Kinetic parameters of purified recombinant BdCOMT4 were determined with caffeic acid as the substrate. The apparent K m and V max values calculated based on the nonlinear Michaelis-Menten plot were 187 μM and 44 nkat/mg protein, respectively. Compared to the kinetic parameter of rice COMT on caffeic acid [38], BdCOMT4 has a higher K m (rice COMT; K m 69 μM) and a greater V max (rice COMT; V max 5.5 nkat/mg protein). In contrast to BdCOMT4, the three other purified Brachypodium COMTs failed to show significant enzymatic activity on the substrates examined (Table 1). Further studies will be required to determine if these proteins possess enzymatic activity with different substrates. However, based on sequence similarity and phylogenetic analysis result, currently, we still classified them as COMT genes in this study.
Figure 5.

SDS-PAGE gel analysis of purification of recombinant BdCOMTs in E. coli. (a) Expression and purification of BdCOMT4 in E. coli. Lane M, protein size standards. Lane 1, total lysate of BL21(DE2) pLysS E.coli. containing pQE-BdCOMT1 before IPTG induction. Lane 2, total lysate of BL21(DE3)pLysS E. coli. containing pQE-BdCOMT1 with 1 mM IPTG induction for 4 hrs. Lane 3, flow-through lysate. Lanes 4–8, BdCOMT1 protein samples collected in different 100 μL fractions after adding the elution buffer. (b) SDS-PAGE gel analysis of purified BdCOMT proteins. Lane M, Protein size standards. Lane 1, 3 μg BdCOMT4; Lane 2, 3 μg BdCOMT2; Lane 3, 3 μg BdCOMT3; Lane 4, 3 μg BdCOMT1.
Table 1.
Relative activity (%) of BdCOMT1 recombinant protein purified with six substrates. (a34 nkat mg−1).
|
3. Discussion
To characterize COMT in the model grass Brachypodium, four COMT genes were identified at different locations in three chromosomes. Phylogenetic analysis suggests that they can be grouped into the COMT family. Comparative analysis indicated that BdCOMT4 is the orthologous copy of COMT genes in the rice and sorghum, while the other three Brachypodium COMT genes do not have orthologous copies in rice and sorghum genomes, suggesting that they are unique to Brachypodium (Figure 1). Phylogenetic analysis also indicated that they do not belong to CCoAOMTs, a different type of OMT closely related to COMT [24]. Therefore, BdCOMT4 is the ancestral COMT gene since its counterpart is present in rice and sorghum. The other Brachypodium COMT genes are likely to be paralogs of BdCOMT4, resulting from gene duplications and translocations into different locations. It has been shown that gene duplication followed by translocation often results in the presence of noncollinear genes in syntenic regions of two closely related genomes [42]. The fact that both rice and sorghum do not have these paralogs suggests that it is most likely that the gene duplications occurred after Brachypodium lineage diverged from the rice and sorghum lineages. Evolutionarily, Brachypodium is more closely related to wheat and barley since they both belong to the subfamily Pooideae. Rice belongs to the subfamily Ehrhartoideae and sorghum to the subfamily Panicoideae [5]. Collinearity analyses of Brachypodium with wheat and barley are very limited because their genome sequences are not available yet. Since wheat is polyploid species, we searched diploid barley EST collection for expressed COMT transcripts. Phylogenetic tree analysis indicated the coding regions of four barley EST contigs aligned with four Brachypodium COMT genes (Supplemental Figure S2), suggesting both Brachypodium and Triticeae developed multiple COMT genes. However, a direct orthologous relationship among the four BdCOMTs and the four barley genes cannot be not determined here.
On the other hand, we also cannot exclude the possibility that COMT gene duplications occurred prior to the divergence of the Brachypodium, rice, and sorghum genomes, and duplicate paralogs were then deleted in the rice and sorghum lineages, resulting in their different evolutionary fates. The different fates of duplicated genes in different species can be explained by the birth-death evolution model in which new genes are created by gene duplication and some of these duplicate genes stay in the genome over time, whereas others are inactivated or deleted from the genome [43].
The birth-death model provides a plausible explanation for the evolution of the OMT gene family. Plant OMTs constitute a large family of enzymes that methylate the oxygen atom of a variety of secondary metabolites including phenylpropanoids, flavonoids, and alkaloids [24]. These enzymes play a key role in lignin biosynthesis, stress tolerance, and disease resistance in plants. Duplication of OMT genes provides raw materials for diversified evolution by mutations to create new OMT genes with different structures and functions. The four BdCOMT genes and those barley COMTs could represent a process of diversified evolution of duplicate genes in Brachypodium. These duplicate Brachypodium genes now display different expression patterns and tissue specificities in development compared to the ancestral BdCOMT4 (Figure 4). In addition, sequence analysis indicated that the amino acids responsible for enzyme catalytic activity and donor binding are highly conserved in these proteins. Less conservation was observed in substrate binding sites (Figure 3), suggesting that they have evolved or are still evolving to have altered substrate binding properties. This notion is in agreement with the low or lack of activity of these proteins on the common COMT substrates (Table 1). Genes with high sequence similarity, but distinct substrate specificity, have been observed in the OMT gene family [33, 44]. Due to the changes in substrate specificity, they are often reclassified into different OMT classes [33]. Although the preferred substrates for these three Brachypodium COMTs were not identified in this study, the expression and conservation of amino acids responsible for enzyme activity suggest that they are likely to be active genes. The differential expression pattern in different tissues might suggest that these genes might have different roles in plant development. Further characterization of BdCOMT1, BdCOMT2, and BdCOMT3 by examining more substrates or gene silencing through transgenic approaches is needed to understand their functions in Brachypodium. It is also interesting to note that Brachypodium has the most compact genome of all the species examined and it tends to have fewer members within gene family [6]. These facts suggest that the BdCOMT1-3 have been retained because they might have adopted some other functions.
Enzymatic assays of the Brachypodium COMTs revealed that BdCOMT4 had strong COMT activity on caffeic acid and caffeoyl aldehyde substrates with > twofold preference for caffeic acid over caffeoyl aldehyde (Table 1). In addition, BdCOMT4 showed high activities on several flavonoid compounds. These results indicate that BdCOMT1's substrate specificity is quite similar to that of rice COMT [38], but differs from the specificity of the COMT (MsCOMT) from alfalfa, a dicot species. MsCOMT exhibited the highest activity on 5-hydroxyconiferaldehyde and caffeoyl aldehyde and the lowest activity on caffeic acid [45]. Low activity on caffeic acid substrates was also observed for COMTs from other dicots such as sweetgum and Vanilla planifolia [45–48]. In Vanilla planifolia, Van COMT showed higher preference for caffeoyl aldehyde to caffeic acid as a substrate. In addition, Van COMT has low activity on 1,2,3-trihydroxybenzene derivatives such as propyl gallate and methyl gallate, but these two substrates were preferred by Van OMT-2 and Van OMT-3, which are closely related to Van COMT at the sequence level [33]. Interestingly, BdCOMT4 also showed considerable activity on flavonoid substrates such as propyl gallate and methyl gallate substrates, although the most preferred substrate is caffeic acid (Table 1). A similar preference for these different types of substrates was also observed in rice COMT [38]. Whether rice COMT and BdCOMT4 have roles in pathways involved in flavonoid compound biosynthesis is not known. It appears that the substrate preference for COMTs might even not be shared by the monocot species. It has been shown that several monocot COMTs have substrate preference for aldehydes over alcohols and acids such as ryegrass and sorghum [49, 50]. It is also interesting to note that the expression pattern of COMT is different among monocot species. For example, while BdCOMT4 and wheat COMT have a similar expression level in different tissues [51], COMT from maize is highly expressed only in roots [25]. COMT from ryegrass is abundantly expressed in stems [52]. Whether the differential expression of COMTs in different species suggests that they may serve additional function in some species in comparison to others is also not clear.
The difference in substrate specificity among COMTs from different plant species is intriguing. Several reports have demonstrated that a few or single amino acid changes can drastically alter substrate specificity among plant OMTs [35, 44]. Structure analysis of alfalfa COMT by crystallography suggests motifs or amino acids critical for enzyme activity on substrates [41]. Alignment of BdCOMT4 with COMTs from other species including alfalfa revealed high conservation on these amino acid sites; the change from I316 in MsCOMT to V313 in BdCOMT4 marks the only difference among the 13 active substrate binding sites (Figure 3). Interestingly, Arabidopsis COMT has valine at the corresponding position (Table 1) and its activity on caffeic acid is higher than that on caffeoyl aldehyde substrate [53]. In addition, this isoleucine in MsCOMT is changed to leucine with a similar amino acid structure in Van COMT in Vanilla planifolia. The substrate preference of Van COMT is also similar to MsCOMT with caffeoyl aldehyde as a more highly preferred substrate than caffeic acid [33]. The question of whether a single amino acid change at this site caused the difference in the substrate specificity toward caffeic acid and caffeoyl aldehyde among these COMTs merits further investigation. As we understand more of the effects of specific amino acids on enzyme activity, it may someday become possible to engineer COMT that can act on a particular substrate of interest.
Despite the importance of COMT in lignin biosynthesis, extensive characterization of COMTs has mainly been reported on dicot species such as Arabidopsis and alfalfa. Recent studies on the characterization of monocot COMTs using both biochemical and gene silencing approaches have greatly facilitated a better understanding of their functions in lignin biosynthesis in grass species [27, 38, 39, 49, 54]. Lignin content and composition are known to differ among plant species, contributing to the architecture difference of cell walls between dicots and monocots [8]. Several monocot species such as switchgrass and Miscanthus are targeted for development into an herbaceous biomass fuel crops. However, those dedicated biofuel crops are often not ideal for basic research due to their large physical stature and complex genome structure. The demonstration of many favorable characteristics of Brachypodium as a desirable experimental system promised to provide unique knowledge on grass biology including the lignin biosynthesis pathway. The notion that Brachypodium and Miscanthus have similar cell wall compositions further supports the use of Brachypodium as a model for investigating grass cell walls [55]. In this study, we showed that, by evolution, BdCOMT4 is an ortholog of COMTs in grasses including rice-and-sorghum-based gene collinearity analysis. It may also share similar biochemical and functional properties with COMTs in other grass species. Thus, knowledge gleaned from cell wall biology in Brachypodium could greatly benefit grass bioenergy crop research for sustainable fuel production.
4. Materials and Methods
4.1. Plant Materials
Brachypodium seeds (accession Bd21-3) were sown in a soilless mix (Supersoil, Rod McLellan Co., Marysville, OH, USA), placed in 4°C cold room in dark for a two-week vernalization period, and then transferred to a greenhouse with temperature range of 24°C in the day and 18°C at night with supplemental lighting to extend day length to 16 hours. Plant tissues (leaf blade, leaf sheath, stem node, stem internode, and root) were collected 3-4 weeks after growth in the greenhouse and frozen in liquid nitrogen for mRNA extraction.
4.2. Identification of Brachypodium COMT Genes and Collinearity Analysis
The rice and Arabidopsis COMT gene sequence were used in a BLASTN search against the Brachypodium genome database. Four hits matched annotated Brachypodium genes Bradi3g16530, Bradi2g02380, Bradi2g02390, and Bradi1g14870. These genes were designated BdCOMT4, BdCOMT2, BdCOMT3, and BdCOMT1, respectively. Collinearity analysis was performed using CoGe's GEvo tool (http://genomevolution.org/CoGe/index.pl/). Genomic regions (~100 kb in size) surrounding Brachypodium COMT genes were analyzed by comparison with orthologous regions in rice and sorghum. Collinear genes present in more than two genomes were identified based on BLASTN algorithm with an e-value of 1e − 30. Noncollinear gene sequences that are present in one genome, but absent in the other genomes, were also marked.
4.3. Protein Sequence Alignment
COMT protein sequences of alfalfa, Arabidopsis, maize, and rice were collected from the NCBI database. The four Brachypodium COMT protein sequences annotated in the genome database were confirmed with the full-length cDNA sequences amplified from Brachypodium total mRNA (see below). A total of eight COMT protein sequences were aligned with ClustalW2 (multiple sequence alignment tool) (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Predicted substrate binding sites, methyl donor binding sites, and catalytic sites were inferred according to alfalfa COMT crystal structure analysis [41].
4.4. Phylogeny Analysis
The coding sequences of the four Brachypodium COMT genes were used to construct a phylogeny tree with available COMT sequences from different plant species by MEGA5 program with the neighbor-joining method. The evolutionary history was inferred using the neighbor-joining method [56]. The bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history of the taxa analyzed [57]. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates were collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) were shown next to the branches [57]. All positions containing gaps and missing data were eliminated from the dataset for analysis.
4.5. RT-PCR
Plant tissues were ground in liquid nitrogen and extracted with TRIzol Reagent (Invitrogen, Inc.). Total mRNA was quantified by ND-1000 Spectrophotometer (NanoDrop Technology, Inc.). cDNA was synthesized by the SuperScript III kit (Invitrogen, Inc.). PCR primers specific for each Brachypodium COMT gene were designed as below: BdCOMT1: forward, 5′-GAGGTCGAGGAGGGGCAGGAG-3′, reverse, 5′-CGCCAACATCCACGAGGGTCTT-3′. BdCOMT2: forward, 5′-TGAGCGCCGGCAGGGAAGC-3′, reverse, 5′-T GGCCTCGTTGTACATGCGGTTGA-3′; BdCOMT3: forward, 5′-CGTCTCCATGGCCC CCTTCTG-3′, reverse, 5′-CCGGCGACATCCACGAGGAC-3′; BdCOMT4: forward, 5′-CCGCCCTCGCGCTCATGA A-3′ reverse, 5′-ACGTGGGGGAGGTCG AAGTTGAT-3′; The Brachypodium actin gene served as an expression control for RT-PCR. The primer set for amplifying its transcript is as follow: forward: 5′-AAGTACCCTATTGAGCATGG-3′; reverse: 5′-CGTAGTCAAGAGCCACATATG-3′.
4.6. cDNA Isolation and Expression Vector Cloning
Full-length cDNA of each BdCOMT gene was amplified by gene-specific primers. To facilitate cloning, NdeI and XhoI restriction sites were added at the beginning of 5′ primers and in the end of 3′ primers, respectively. Amplified fragments were cloned in frame at the NdeI and XhoI sites into pQE-T7-1 vector (Qiagen, Inc.). Cloned plasmids were named as pQE-BdCOMT1, pQE-BdCOMT2, pQE-BdCOMT3, and pQE-BdCOMT4. Each plasmid was sequenced to confirm the sequence accuracy and then retransformed into E. coli Bd21 (DES) strains (Invitrogen, Inc.) for protein expression.
The cloning primers for each COMT gene are listed below: BdCOMT4: forward, 5′-GGAATTCCA TATGAAACAGATGGGTTCCACGGCGGCGGA-3′; reverse, 5′-CCGCTCGAGTCTTACTACTACTTG GTGAACTCGATGG-3′; BdCOMT2: forward, 5′-GGAATTCCATATGAAACAGATGGCCGAGGA GGAGGCGTG-3′; reverse, 5′-CCGCTCGAGTCTTACTACTACTTGGTGTACTCGATGGCCCAGA A-3′; BdCOMT3: forward, 5′-GGAATTCCATATGAAACAGATGGCGGAAGAGGAAGCCGGGTG-3′; reverse, 5′-CCGCTCGAGTCTTACTACTACTTGGTGTACTCGATGGCCCAGAA-3′; BdCOMT1: forward, 5′-GGAATTCCATATGAAACAGATGGGCTCTGCCCGCACCG-3′; reverse, 5′-CCGCTC GAGTCTTACTACTATTTGGTGTACTCAATGGCCCAGGAG-3′.
4.7. Protein Expression, Purification, and Quantification
A single E. coli clone carrying the expression construct was inoculated into 6 mL Luria broth (LB) medium with 50 μg/mL kanamycin and incubated in 37°C shaker overnight. The overnight culture (1 mL) was added to 500 mL (1 : 500 ratio) LB medium with 50 μg/mL kanamycin. When OD 600 of the culture reached 0.7, IPTG was added to the final concentration of 0.2 mM to induce protein expression and incubated at room temperature for 16 hours with shaking.
Cells were harvested and suspended in 30 mL lysis buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole). A lysozyme was added to the final concentration of 1 mg/mL and the mixture was placed on ice for 30 min before sonication. E. coli soluble fraction was collected after centrifugation and incubated with 1 mL Ni-NTA agarose beads (Qiagen) by shaking at room temperature for 1 hour. Beads were washed three times in a column each with 50 mL lysis buffer containing imidazole with step increase of concentration at 20 mM, 30 mM, and 50 mM. Proteins were eluted by elution buffer (50 mM NaH2PO4, 300 mM NaCl, 200 mM imidazole, and 0.05% Tween 20) and stored at −20°C following addition of glycerol to a final concentration of 30%. Proteins were quantified by the BCA assay kit (Thermal Scientific, Inc.).
4.8. Enzyme Activity Assay
Assay reaction mixture contained 5 mM substrate, 3 μg COMT protein, 0.5 mM S-[methyl-14C]-adenosyl-L-methionine (SAM) (47 mCi mmol−1), 0.1 M Tris-HCl (pH = 7.5) in a final volume of 50 μL reaction system. After 20-minute incubation at 30°C, the reaction was stopped by adding 50 μL 0.2 M HCl and 200 μL hexane : ethyl acetate (1 : 1 ratio) mixture to extract methylated substrate. 20 μL of the supernatant layer containing 35S-labeled methylated products from the reaction mixture was moved into 2 mL cocktail solution for liquid scintillation counting by LS6500 Scintillation System (Beckman Coulter, Inc.). Each assay was performed with three replicates and repeated three times. Control reaction was performed in the same manner without addition of COMT protein.
4.9. Determination of K m and V max Value of BdCOMT1 Protein
Steady kinetic curves were determined using different caffeic acid substrate concentrations under the same reaction conditions as described in the enzyme activity assay except 3 μL [14C]SAM and 10 μL of unlabeled S-adenosyl-L-methionine (SAM) (Sigma, Inc.) were used. The substrate concentrations in this array were 0.05, 0.1, 0.25, 0.5, 1.0, and 2.0 mM. The K m and V max were calculated from the nonlinear Michaelis-Menten plot enzyme activity assay by Prism 5 for Windows (GraphPad Software, Inc.). The counts per second were calculated to nanomoles of product produced per second (nkat), based on the specific activity (SA) of the 14C-labeled substrate and the disintegrations per Minute (DPMs) read by the scintillation counter.
Supplementary Material
Supplemental Figure 1: Phylogenetic analysis of COMT and CCoAOMT genes among species
Supplemental Figure 2: Phylogenetic analysis of BdCOMT and barley COMT ESTs
Authors' Contribution
X. Wu and J. Wu contributed equally to the work.
Conflict of Interests
The authors do not have a direct financial relation with the commercial identity mentioned in this work that might lead to a conflict of interests.
Acknowledgments
The authors thank Xiaohua He for the critical reading of this paper. This work was supported in part by the United States Department of Agriculture, Agriculture Research Service CRIS 5325-21000-015 and 532521000-17 and by the office of Science (DER), US Department of Energy, Interagency Agreement no. DE-AI02-07ER64452.
Abbreviations
- CCoAOMT:
Caffeoyl CoA 3-o-methyltransferase
- COMT:
Caffeic acid o-methyltransferase
- PCR:
Polymerase chain reaction.
References
- 1.Carpita NC, McCann MC. Maize and sorghum: genetic resources for bioenergy grasses. Trends in Plant Science. 2008;13(8):415–420. doi: 10.1016/j.tplants.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Chakrabarti SK, Roychoudhury PK, Bajpai PK. Biphasic biomethanation of wood-hydrolysate effluent. Artificial Cells, Blood Substitutes, and Immobilization Biotechnology. 1999;27(5-6):461–467. doi: 10.3109/10731199909117720. [DOI] [PubMed] [Google Scholar]
- 3.Missaoui AM, Paterson AH, Bouton JH. Investigation of genomic organization in switchgrass (Panicum virgatum L.) using DNA markers. Theoretical and Applied Genetics. 2005;110(8):1372–1383. doi: 10.1007/s00122-005-1935-6. [DOI] [PubMed] [Google Scholar]
- 4.Somleva MN, Snell KD, Beaulieu JJ, Peoples OP, Garrison BR, Patterson NA. Production of polyhydroxybutyrate in switchgrass, a value-added cO-product in an important lignocellulosic biomass crop. Plant Biotechnology Journal. 2008;6(7):663–678. doi: 10.1111/j.1467-7652.2008.00350.x. [DOI] [PubMed] [Google Scholar]
- 5.Brkljacic J, Grotewold E, Scholl R, et al. Brachypodium as a model for the grasses: today and the future. Plant Physiology. 2011;157:3–13. doi: 10.1104/pp.111.179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogel JP, Garvin DF, Mockler TC, et al. Genome sequencing and analysis of the model grass Brachypodium distachyon . Nature. 2010;463(7282):763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- 7.Wolny E, Hasterok R. Comparative cytogenetic analysis of the genomes of the model grass Brachypodium distachyon and its close relatives. Annals of Botany. 2009;104(5):873–881. doi: 10.1093/aob/mcp179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogel J. Unique aspects of the grass cell wall. Current Opinion in Plant Biology. 2008;11(3):301–307. doi: 10.1016/j.pbi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordon RE, Craig SJ, Foong FC. Molecular engineering of the cellulosome complex for affinity and bioenergy applications. Biotechnology Letters. 2009;31(4):465–476. doi: 10.1007/s10529-008-9899-7. [DOI] [PubMed] [Google Scholar]
- 11.Porter J, Costanza R, Sandhu H, Sigsgaard L, Wratten S. The value of producing food, energy, and ecosystem services within an agrO-ecosystem. Ambio. 2009;38(4):186–193. doi: 10.1579/0044-7447-38.4.186. [DOI] [PubMed] [Google Scholar]
- 12.Schmitz PM, Kavallari A. Crop plants versus energy plants—on the international food crisis. Bioorganic and Medicinal Chemistry. 2009;17(12):4020–4021. doi: 10.1016/j.bmc.2008.11.041. [DOI] [PubMed] [Google Scholar]
- 13.Suh MC, Kim MJ, Hur CG, et al. Comparative analysis of expressed sequence tags from Sesamum indicum and Arabidopsis thaliana developing seeds. Plant Molecular Biology. 2003;52(6):1107–1123. doi: 10.1023/b:plan.0000004304.22770.e9. [DOI] [PubMed] [Google Scholar]
- 14.Yuan JS, Tiller KH, Al-Ahmad H, Stewart NR, Stewart CN., Jr. Plants to power: bioenergy to fuel the future. Trends in Plant Science. 2008;13(8):421–429. doi: 10.1016/j.tplants.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Chen F, Dixon RA. Lignin modification improves fermentable sugar yields for biofuel production. Nature Biotechnology. 2007;25(7):759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- 16.Chiang VL. From rags to riches. Nature Biotechnology. 2002;20(6):557–558. doi: 10.1038/nbt0602-557. [DOI] [PubMed] [Google Scholar]
- 17.Fu C, Mielenz JR, Xiao X, et al. Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(9):3803–3808. doi: 10.1073/pnas.1100310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCann MC, Carpita NC. Designing the deconstruction of plant cell walls. Current Opinion in Plant Biology. 2008;11(3):314–320. doi: 10.1016/j.pbi.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Weber C, Farwick A, Benisch F, et al. Trends and challenges in the microbial production of lignocellulosic bioalcohol fuels. Applied Microbiology and Biotechnology. 2010;87(4):1303–1315. doi: 10.1007/s00253-010-2707-z. [DOI] [PubMed] [Google Scholar]
- 20.Weng JK, Li X, Bonawitz ND, Chapple C. Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Current Opinion in Biotechnology. 2008;19(2):166–172. doi: 10.1016/j.copbio.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Pauly M, Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant Journal. 2008;54(4):559–568. doi: 10.1111/j.1365-313X.2008.03463.x. [DOI] [PubMed] [Google Scholar]
- 22.York WS, O’Neill MA. Biochemical control of xylan biosynthesis—which end is up? Current Opinion in Plant Biology. 2008;11(3):258–265. doi: 10.1016/j.pbi.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Vanholme R, Ralph J, Akiyama T, et al. Engineering traditional monolignols out of lignin by concomitant up-regulation of F5H1 and down-regulation of COMT in Arabidopsis. Plant Journal. 2010;64(6):885–897. doi: 10.1111/j.1365-313X.2010.04353.x. [DOI] [PubMed] [Google Scholar]
- 24.Lam KC, Ibrahim RK, Behdad B, Dayanandan S. Structure, function, and evolution of plant O-methyltransferases. Genome. 2007;50(11):1001–1013. doi: 10.1139/g07-077. [DOI] [PubMed] [Google Scholar]
- 25.Collazo P, Montoliu L, Puigdomènech P, Rigau J. Structure and expression of the lignin O-methyltransferase gene from Zea mays L. Plant Molecular Biology. 1992;20(5):857–867. doi: 10.1007/BF00027157. [DOI] [PubMed] [Google Scholar]
- 26.Capellades M, Torres MA, Bastisch I, et al. The maize caffeic acid O-methyltransferase gene promoter is active in transgenic tobacco and maize plant tissues. Plant Molecular Biology. 1996;31(2):307–322. doi: 10.1007/BF00021792. [DOI] [PubMed] [Google Scholar]
- 27.Piquemal J, Chamayou S, Nadaud I, et al. Down-regulation of caffeic acid O-methyltransferase in maize revisited using a transgenic approach. Plant Physiology. 2002;130(4):1675–1685. doi: 10.1104/pp.012237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vignols F, Rigau J, Torres MA, Capellades M, Puigdomenech P. The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell. 1995;7(4):407–416. doi: 10.1105/tpc.7.4.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goujon T, Sibout R, Pollet B, et al. A new Arabidopsis thaliana mutant deficient in the expression of O-methyltransferase impacts lignins and sinapoyl esters. Plant Molecular Biology. 2003;51(6):973–989. doi: 10.1023/a:1023022825098. [DOI] [PubMed] [Google Scholar]
- 30.Guillaumie S, Goffner D, Barbier O, Martinant JP, Pichon M, Barrière Y. Expression of cell wall related genes in basal and ear internodes of silking brown-midrib-3, caffeic acid O-methyltransferase (COMT) down-regulated, and normal maize plants. BMC Plant Biology. 2008;8, article 71 doi: 10.1186/1471-2229-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo D, Chen F, Inoue K, Blount JW, Dixon RA. Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell. 2001;13(1):73–88. doi: 10.1105/tpc.13.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kota P, Guo D, Zubieta C, Noel J, Dixon RA. O-Methylation of benzaldehyde derivatives by "lignin specific" caffeic acid 3-O-methyltransferase. Phytochemistry. 2004;65(7):837–846. doi: 10.1016/j.phytochem.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 33.Li HM, Rotter D, Hartman TG, Pak FE, Havkin-Frenkel D, Belanger FC. Evolution of novel O-methyltransferases from the Vanilla planifolia caffeic acid O-methyltransferase. Plant Molecular Biology. 2006;61(3):537–552. doi: 10.1007/s11103-006-0029-4. [DOI] [PubMed] [Google Scholar]
- 34.Lu F, Marita JM, Lapierre C, et al. Sequencing around 5-hydroxyconiferyl alcohol-derived units in caffeic acid O-methyltransferase-deficient poplar lignins. Plant Physiology. 2010;153(2):569–579. doi: 10.1104/pp.110.154278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma QH, Xu Y. Characterization of a caffeic acid 3-O-methyltransferase from wheat and its function in lignin biosynthesis. Biochimie. 2008;90(3):515–524. doi: 10.1016/j.biochi.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 36.Yoshihara N, Fukuchi-Mizutani M, Okuhara H, Tanaka Y, Yabuya T. Molecular cloning and characterization of O-methyltransferases from the flower buds of Iris hollandica . Journal of Plant Physiology. 2008;165(4):415–422. doi: 10.1016/j.jplph.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Bout S, Vermerris W. A candidate-gene approach to clone the sorghum Brown midrib gene encoding caffeic acid O-methyltransferase. Molecular Genetics and Genomics. 2003;269(2):205–214. doi: 10.1007/s00438-003-0824-4. [DOI] [PubMed] [Google Scholar]
- 38.Lin F, Yamano G, Hasegawa M, Anzai H, Kawasaki S, Kodama O. Cloning and functional analysis of caffeic acid 3-O-methyltransferase from rice (Oryza sativa) Journal of Pesticide Science. 2006;31(1):47–53. [Google Scholar]
- 39.Ma QH. The expression of caffeic acid 3-O-methyltransferase in two wheat genotypes differing in lodging resistance. Journal of Experimental Botany. 2009;60(9):2763–2771. doi: 10.1093/jxb/erp132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou JM, Seo YW, Ibrahim RK. Biochemical characterization of a putative wheat caffeic acid O-methyltransferase. Plant Physiology and Biochemistry. 2009;47(4):322–326. doi: 10.1016/j.plaphy.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 41.Zubieta C, Kota P, Ferrer JL, Dixon RA, Noel JP. Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell. 2002;14(6):1265–1277. doi: 10.1105/tpc.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnable JC, Freeling M, Lyons E. Genome-wide analysis of syntenic gene deletion in the grasses. Genome Biology and Evolution. 2012;4(3):265–277. doi: 10.1093/gbe/evs009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rutter MT, Cross KV, Van Woert PA. Birth, death and subfunctionalization in the Arabidopsis genome. Trends in Plant Science. 2012;17(4):204–212. doi: 10.1016/j.tplants.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Gang DR, Lavid N, Zubieta C, et al. Characterization of phenylpropene O-methyltransferases from sweet basil: facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell. 2002;14(2):505–519. doi: 10.1105/tpc.010327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parvathi K, Chen F, Guo D, Blount JW, Dixon RA. Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant Journal. 2001;25(2):193–202. doi: 10.1046/j.1365-313x.2001.00956.x. [DOI] [PubMed] [Google Scholar]
- 46.Boerjan W, Ralph J, Baucher M. Lignin Biosynthesis. Annual Review of Plant Biology. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 47.Li L, Popko JL, Umezawa T, Chiang VL. 5-Hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. Journal of Biological Chemistry. 2000;275(9):6537–6545. doi: 10.1074/jbc.275.9.6537. [DOI] [PubMed] [Google Scholar]
- 48.Osakabe K, Tsao CC, Li L, et al. Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(16):8955–8960. doi: 10.1073/pnas.96.16.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Louie GV, Bowman ME, Tu Y, Mouradov A, Spangenberg G, Noel JP. Structure-function analyses of a caffeic acid O-methyltransferase from perennial ryegrass reveal the molecular basis for substrate preference. Plant Cell. 2010;22(12):4114–4127. doi: 10.1105/tpc.110.077578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmer NA, Sattler SE, Saathoff AJ, Sarath G. A continuous, quantitative fluorescent assay for plant caffeic acid O-methyltransferases. Journal of Agricultural and Food Chemistry. 2010;58(9):5220–5226. doi: 10.1021/jf904445q. [DOI] [PubMed] [Google Scholar]
- 51.Ma QH, Xu Y, Lin ZB, He P. Cloning of cDNA encoding COMT from wheat which is differentially expressed in lodging-sensitive and -resistant cultivars. Journal of Experimental Botany. 2002;53(378):2281–2282. doi: 10.1093/jxb/erf102. [DOI] [PubMed] [Google Scholar]
- 52.McAlister FM, Jenkins CLD, Watson JM. Sequence and expression of a stem-abundant caffeic acid O-methyltransferase cDNA from perennial ryegrass (Lolium perenne) Australian Journal of Plant Physiology. 1998;25(2):225–235. [Google Scholar]
- 53.Moinuddin SGA, Jourdes M, Laskar DD, et al. Insights into lignin primary structure and deconstruction from Arabidopsis thaliana COMT (caffeic acid O-methyl transferase) mutant Atomt1 . Organic and Biomolecular Chemistry. 2010;8(17):3928–3946. doi: 10.1039/c004817h. [DOI] [PubMed] [Google Scholar]
- 54.Tu Y, Rochfort S, Liu Z, et al. Functional analyses of caffeic acid O-methyltransferase and Cinnamoyl-CoA-reductase genes from perennial ryegrass (Lolium perenne) Plant Cell. 2010;22(10):3357–3373. doi: 10.1105/tpc.109.072827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gomez LD, Bristow JK, Statham ER, McQueen-Mason SJ. Analysis of saccharification in Brachypodium distachyon stems under mild conditions of hydrolysis. Biotechnology for Biofuels. 2008;1, article 15 doi: 10.1186/1754-6834-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 57.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Phylogenetic analysis of COMT and CCoAOMT genes among species
Supplemental Figure 2: Phylogenetic analysis of BdCOMT and barley COMT ESTs


