Abstract
Objective
Earlier studies have suggested that a common genetic architecture underlies the clinically heterogeneous polygenic Fredrickson hyperlipoproteinemia (HLP) phenotypes defined by hypertriglyceridemia (HTG). Here, we comprehensively analyzed 504 HLP-HTG patients and 1213 normotriglyceridemic controls and confirmed that a spectrum of common and rare lipid-associated variants underlies this heterogeneity.
Methods and Results
First, we demonstrated that genetic determinants of plasma lipids and lipoproteins, including common variants associated with plasma triglyceride (TG), high-density lipoprotein cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C) from the Global Lipids Genetics Consortium were associated with multiple HLP-HTG phenotypes. Second, we demonstrated that weighted risk scores composed of common TG-associated variants were distinctly increased across all HLP-HTG phenotypes compared with controls; weighted HDL-C and LDL-C risk scores were also increased, although to a less pronounced degree with some HLP-HTG phenotypes. Interestingly, decomposition of HDL-C and LDL-C risk scores revealed that pleiotropic variants (those jointly associated with TG) accounted for the greatest difference in HDL-C and LDL-C risk scores. The APOE E2/E2 genotype was significantly overrepresented in HLP type 3 versus other phenotypes. Finally, rare variants in 4 genes accumulated equally across HLP-HTG phenotypes.
Conclusion
HTG susceptibility and phenotypic heterogeneity are both influenced by accumulation of common and rare TG-associated variants.
Keywords: lipoproteins, genetic risk scores, genetic variation, hypertriglyceridemia, pleiotropy
Plasma triglyceride (TG) concentration is reemerging as an independent risk factor for cardiovascular disease (CVD). Both fasting and postprandial plasma TG concentrations are determinants of CVD risk,1–3 increasing risk of myocardial infarction and ischemic stroke.4,5 A Mendelian randomization study using the common −1131T>C promoter variant in the APOA5 locus as a surrogate exposure to increased plasma TG concentration also supported a causal relationship with CVD risk.6 Together, this body of evidence indicates that elevated concentrations of plasma TG-rich lipoproteins increase CVD susceptibility independent of other lipoproteins.
Hypertriglyceridemia (HTG) is often defined by plasma TG concentration >95th percentile. Patients with HTG have elevated CVD risk7,8 and frequently have concomitant proatherogenic comorbidities, such as obesity, metabolic syndrome, and type 2 diabetes.8 Phenotypic heterogeneity among HTG patients is defined by qualitative and quantitative biochemical differences in plasma lipoproteins that form the basis of the World Health Organization or Fredrickson hyperlipoproteinemia (HLP) phenotypes. The HLP phenotypes defined by HTG (HLP-HTG) include 1 monogenic pediatric phenotype, called chylomicronemia (HLP type 1), and 4 polygenic familial phenotypes, called combined hyperlipidemia (HLP type 2B), dysbetalipoproteinemia (HLP type 3), primary HTG (HLP type 4), and mixed hyperlipidemia (HLP type 5). HLP type 1 is usually caused by homozygous loss-of-function mutations in genes such as LPL,9 APOC2,10 APOA5,11 LMF1,12 and GPIHBP1.13,14 However, the remaining genetic architecture underlying predisposition to HTG and phenotypic heterogeneity among the HLP-HTG phenotypes is less well defined.15
Candidate gene studies and genome-wide analyses have established that genetic variants associated with unfavorable plasma TG concentration in populations are also associated with susceptibility to clinical HTG.16–20 Both common and rare TG-associated variants accumulate in HTG patients, and they account for a large proportion of explained variation among HTG patients.20,21 However, corresponding analyses have not yet been performed on individual HLP-HTG phenotypes.
Our earlier surveys of a few isolated genetic determinants suggested to a first approximation that a shared genetic architecture underlies the phenotypic spectrum of HTG.16,18 Here, we provide a more complete analysis of the genetic etiology of the HLP-HTG phenotypes in a much larger sample of patients and controls. Further, we include all Genome-Wide Association Study (GWAS)-identified genetic determinants of lipid metabolism recently identified by the Global Lipids Genetics Consortium (GLGC)21 to assess the contribution of common variants associated with concentrations of TG, high-density lipoprotein cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C) to both HTG susceptibility and HLP-HTG phenotypic heterogeneity. Finally, we have integrated rare variants into the analyses of HLP phenotypes. We demonstrate a shared genetic architecture of common and rare TG-associated variants as the genetic foundation across HTG patients, and we provide evidence that accumulation of pleiotropic TG-associated, HDL-C-associated, and LDL-C-associated variants may together dictate susceptibility to the polygenic HLP phenotypes.
Methods
Study Subjects
This study was approved by the University of Western Ontario Institutional Review Board (protocol #07920E) and ethics boards at collaborating institutions. Study subjects provided informed consent for collection of clinical data and DNA extraction and analysis. In all, 504 HTG patients and 1213 population-based controls were studied. HTG patients were European subjects ascertained through tertiary referral lipid clinics in London, Ontario, Canada, and Amsterdam, the Netherlands. HTG patients were clinically diagnosed with 1 of 4 familial polygenic HLP-HTG phenotypes before ascertainment for this study and without genetic information (Supplemental Table I, available online at http://atvb.ahajournals.org); these patients are now referred to as having HLP-HTG case status. Population-based controls were European subjects ascertained through the Study of Health Assessment and Risk in Ethnic Groups22 and the Myocardial Infarction Genetics Consortium,23 as previously described. Control subjects with fasting plasma TG concentration >2.3 mmol/L were excluded from analyses because of potentially undiagnosed HTG. Biochemical analyses of lipoprotein traits were conducted separately in each cohort, as previously described.17,22,23
Genotyping and Sequencing
Genotyping was conducted using Affymetrix Genome-Wide Human SNP Array 6.0, as previously described.20 Imputation was conducted using HapMap2 European phased haplotypes,24 only imputing single-nucleotide polymorphisms (SNPs) with r2>0.4. Genotypes were subsequently extracted for lipid-associated loci identified by the GLGC.21 Functional variants in APOE (rs7412 and rs429358) were genotyped using validated TaqMan assays on an ABI Prism 7900HT Sequence Detection System and automated software (Applied Biosystems, Foster City, CA). Rare variants in APOA5, GCKR, LPL, and exons 26 and 29 of APOB were defined as nonsense or missense mutations with allele frequency <1% in control subjects, as previously described.20 The list of variants identified by this study are located in the supplemental material of Johansen et al.20
Statistical Analysis
Data management, linkage disequilibrium calculations, association testing, and risk score calculations were conducted as implemented in PLINK.25 Association was tested using an additive multiple logistic regression model with clinical covariates age, sex, body mass index, type 2 diabetes, and 10 principal components of ancestry.26,27 Statistical significance for association was defined as concordant direction of effect and P<0.05, as all SNPs have a priori evidence for association with lipid metabolism.
Each subject’s genetic risk index (or genetic risk score) was determined by summing risk (TG-raising, LDL-raising, and HDL-lowering) alleles at 32 TG-associated SNPs, 47 HDL-C-associated SNPs, or 37 LDL-C-associated SNPs, as identified by the GLGC.21 Genetic risk indices were measured using either unweighted scores or weighted scores, where indicated. Unweighted scores were created by summing the raw number of risk alleles carried by each subject. Weighted risk scores were created by summing risk alleles after being multiplied by their population-based effect size calculated by the GLGC.21 Units were initially reported as mg/dL, but they are converted here to mmol/L; conversion factors for TG and HDL-C/LDL-C were 88.6 and 38.7, respectively. All SNPs were included in the risk score regardless of statistical significance in the replication phase. Where SNPs were associated with multiple lipid phenotypes, they were included in each risk score, although weighted by the appropriate lipid-specific effect estimate. Pleiotropy was assessed by comparing risk scores constructed using SNPs exclusively associated with HDL-C or LDL-C, or jointly associated with HDL-C or LDL-C plus plasma TG concentration.21 Frequency distributions were plotted by dividing the range of risk scores into equal bins and plotting the relative number of HTG patients or controls in each bin. Forest plots were generated by comparing the number of HTG patients in each of 7 risk score bins to the median risk score bin or the lowest risk score bin, as indicated. Summary statistics, risk score comparisons, rare variant accumulation and explained variation calculations were conducted in SAS version 9.2 (SAS Institute, Cary, NJ). Summary statistics are displayed as mean±SD. Significance of increasing risk score in forest plots was determined using Cochrane-Armitage test for trend. Comparison of risk scores among HLP-HTG phenotypes was assessed using 1-way ANOVA and post hoc pairwise comparisons using the Tukey test. In the figures, values are mean±SE; means sharing letters are not statistically different.
Rare variant accumulation was compared between HLP-HTG phenotypes and controls with the Fisher exact test, using a 2-sided P<0.05 as the threshold for statistical significance. Differences in number of rare variants in each HLP-HTG phenotype was assessed using a 2×4 contingency table (number of variants in carriers versus noncarriers for each phenotype); differences in distribution of rare variants among genes in each HLP-HTG phenotype was assessed using a 4×4 contingency table (number of variants in each gene for each phenotype).
A logistic regression model using case or control status as the dependent variable was used to calculate the proportion of variation explained by clinical and genetic variables. The proportion of variation explained by the coefficient of determination (R2) in linear regression does not have a straightforward analog in logistic regression. Therefore, we used a likelihood-based pseudo-R2 metric for logistic regression as implemented in SAS version 9.2, which compares the increase in information provided by a model fitted with predictors over a null model. We used incremental changes in this metric to assess how well each variable improves model fit, thus comparing the relative contribution of variables between models. Here, to facilitate comprehension, we describe successive increases in model fit (R2) with progressive introduction of each variable as the proportion of variation explained by each variable. These analyses only included subjects for whom both genome-wide genotyping and resequencing data were available, which comprised 346 HTG patients and 205 controls. Forward modeling was used to systematically enter clinical variables (age, sex, body mass index, and type 2 diabetes), common variants at TG-associated loci (0, 1, or 2 TG-raising alleles as each locus), the number of rare variants identified in HTG-associated genes (APOA5, GCKR, LPL, and APOB as a composite score),20 and APOE genotype (copies of each ε2/ε3/ε4 isoform) into the model. These analyses were conducted including all HTG patients and subsequently using HLP phenotype subgroups in comparison with the same group of healthy controls. This analysis served only as a relative comparison of variables contributing to the model within our cohort, and it should be considered only within the context of our study.
Results
Study Subjects
Baseline clinical characteristics of the 504 HTG patients and 1213 population-based controls are shown in Table 1. Generally, HTG patients had less favorable clinical profiles than controls. HLP type 5 had the least favorable clinical profile across all traits, including fasting plasma TG concentration >99th percentile.
Table 1.
Baseline Clinical Characteristics of HLP-HTG Patients and Controls
| All HTG | HLP Type 5 | HLP Type 4 | HLP Type 3 | HLP Type 2B | Controls | |
|---|---|---|---|---|---|---|
| No. | 504 | 180 | 128 | 37 | 159 | 1213 |
| Females, % | 32.4 | 29.4 | 22.7 | 37.8 | 42.1 | 42.0 |
| Diabetes, % | 26.4 | 31.7 | 29.7 | 24.3 | 18.2 | 0.3 |
| Age, y | 51.7 ± 12.7 | 49.6 ± 11.7 | 53.0 ± 13.4 | 51.4 ± 12.7 | 53.2 ± 13.1 | 48.2 ± 10.8 |
| BMI, kg/m2 | 30.0 ± 4.9 | 30.6 ± 4.9 | 30.5 ± 5.9 | 28.9 ± 2.5 | 29.3 ± 4.3 | 26.4 ± 4.4 |
| TC, mmol/L | 8.2 ± 3.8 | 10.8 ± 5.1 | 5.1 ± 1 | 9.4 ± 1.8 | 7.9 ± 1.5 | 5.1 ± 0.9 |
| HDL-C, mmol/L | 0.9 ± 0.3 | 0.8 ± 0.3 | 0.8 ± 0.3 | 1.0 ± 0.3 | 1.1 ± 0.3 | 1.4 ± 0.4 |
| TG, mmol/L | 13.6 ± 18.2 | 28.3 ± 24.3 | 5.4 ± 1.9 | 7.0 ± 3.9 | 5.3 ± 1.9 | 1.1 ± 0.4 |
HTG blood sampling was performed in a fasting state (12 h); control blood sampling was performed in a fasting or semifasting (4 h) state. Values are mean ± SD. BMI indicates body mass index; TC, total cholesterol.
Replication of Lipid-Associated Loci
First, we sought to replicate the genetic determinants of plasma lipoprotein metabolism from the GLGC21 in our HTG cohort. For TG-associated loci (Table 2), we replicated many loci previously associated with HTG,20 and additionally replicated recently identified loci KLHL8 (odds ratio = 1.36; P = 1.5 × 10−3) and CYP26A1 (odds ratio = 1.29; P = 5.9 × 10−3). Generally, there was a trend toward significance at additional loci, as 29 of 32 loci had directions of effect concordant with estimates from the GLGC, significantly more than would be expected by chance (P = 1.15 × 10−6). Most TG-associated loci showed different patterns of association with the HLP-HTG phenotypes (Table 2); however, there were again more concordant directions of effect than would be expected by chance (HLP 5, P = 0.007; HLP 4, P = 0.01; HLP 3, P = 0.007; HLP 2B, P = 0.0002). Taken together, these data extend the concept that common variants in TG-associated loci increase susceptibility to HTG and the HLP phenotypes.
Table 2.
Replication of TG-Associated Loci in Patients With Polygenic HTG and HLP-HTG Phenotypes
| CHR | Gene | SNP | Minor Allele/Modeled Allele | All HTG (n= 504)
|
HLP Type 5 (n = 180)
|
HLP Type 4 (n = 128)
|
HLP Type 3 (n = 37)
|
HLP Type 2B (n = 159)
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | ||||
| 11 | APOA5 | rs964184 | G/G | 3.43 (2.72 to 4.31) | 1.12 × 10−25 | 5.65 (3.92 to 8.14) | 1.81× 10−20 | 3.71 (2.56 to 5.39) | 4.9 × 10−12 | 5.02 (2.51 to 10.1) | 5.3 × 10−6 | 2.57 (1.82 to 3.63) | 8.2 × 10−8 |
| 2 | GCKR | rs1260326 | T/T | 1.64 (1.36 to 1.97) | 1.97 × 10−7 | 1.60 (1.19 to 2.15) | 1.9 × 10−3 | 1.79 (1.29 to 2.48) | 5.3 × 10−4 | 0.66 (0.35 to 1.22) | 0.18 | 1.87 (1.41 to 2.47) | 1.3 × 10−5 |
| 8 | LPL | rs12678919 | G/A | 2.21 (1.52 to 3.22) | 3.5 × 10−5 | 2.79 (1.43 to 5.44) | 2.7 × 10−3 | 5.78 (2.10 to 15.9) | 7.0 × 10−4 | 1.34 (0.45 to 3.99) | 0.60 | 1.48 (0.90 to 2.43) | 0.12 |
| 8 | TRIB1 | rs2954029 | T/A | 1.50 (1.24 to 1.81) | 3.8 × 10−5 | 1.43 (1.06 to 1.92) | 0.019 | 1.37 (0.99 to 1.89) | 0.059 | 1.60 (0.89 to 2.85) | 0.12 | 1.60 (1.20 to 2.12) | 1.2 × 10−3 |
| 1 | ANGPTL3 | rs2131925 | G/T | 1.51 (1.23 to 1.85) | 1.0 × 10−4 | 1.85 (1.31 to 2.61) | 4.7 × 10−4 | 0.98 (0.70 to 1.39) | 0.93 | 1.16 (0.63 to 2.17) | 0.63 | 1.63 (1.19 to 2.24) | 2.3 × 10−3 |
| 7 | MLXIPL | rs7811265 | G/A | 1.63 (1.25 to 2.13) | 3.3 × 10−4 | 1.70 (1.10 to 2.62) | 0.017 | 2.09 (1.25 to 3.49) | 4.8 × 10−3 | 0.82 (0.40 to 1.68) | 0.59 | 1.42 (0.97 to 2.08) | 0.073 |
| 4 | KLHL8 | rs442177 | G/T | 1.36 (1.13 to 1.64) | 1.5 × 10−3 | 1.36 (1.02 to 1.82) | 0.038 | 1.19 (0.86 to 1.64) | 0.29 | 1.86 (0.99 to 3.49) | 0.052 | 1.47 (1.11 to 1.96) | 7.6 × 10−3 |
| 10 | CYP26A1 | rs2068888 | A/G | 1.29 (1.08 to 1.55) | 5.9 × 10−3 | 1.44 (1.08 to 1.92) | 0.013 | 1.02 (0.75 to 1.40) | 0.88 | 0.96 (0.53 to 1.74) | 0.89 | 1.27 (0.96 to 1.68) | 0.093 |
| 19 | CILP2 | rs10401969 | C/T | 1.72 (1.16 to 2.54) | 6.8 × 10−3 | 2.29 (1.18 to 4.43) | 0.014 | 1.53 (0.79 to 2.95) | 0.20 | 2.94 (0.78 to 11.1) | 0.11 | 2.07 (1.13 to 3.79) | 0.018 |
| 2 | APOB | rs1042034 | C/T | 1.28 (1.02 to 1.61) | 0.032 | 1.19 (0.84 to 1.70) | 0.32 | 0.88 (0.61 to 1.28) | 0.50 | 1.92 (0.83 to 4.46) | 0.13 | 1.60 (1.11 to 2.31) | 0.01 |
| 5 | TIMD4 | rs1553318 | G/C | 1.21 (1.00 to 1.46) | 0.054 | 1.14 (0.85 to 1.54) | 0.39 | 1.22 (0.88 to 1.69) | 0.24 | 1.07 (0.60 to 1.92) | 0.82 | 1.38 (1.03 to 1.84) | 0.032 |
| 11 | FADS1 | rs174546 | T/T | 1.20 (1.00 to 1.45) | 0.054 | 1.19 (0.88 to 1.59) | 0.25 | 1.33 (0.96 to 1.84) | 0.088 | 1.08 (0.59 to 1.98) | 0.80 | 1.16 (0.88 to 1.53) | 0.29 |
| 16 | CETP | rs7205804 | A/G | 1.20 (1.00 to 1.45) | 0.056 | 1.37 (1.01 to 1.85) | 0.046 | 1.19 (0.85 to 1.65) | 0.31 | 1.04 (0.56 to 1.90) | 0.91 | 1.17 (0.88 to 1.55) | 0.28 |
| 2 | IRS1 | rs2943645 | C/T | 1.20 (0.99 to 1.46) | 0.061 | 1.20 (0.89 to 1.61) | 0.24 | 1.42 (1.00 to 2.00) | 0.049 | 1.07 (0.59 to 1.95) | 0.83 | 1.16 (0.87 to 1.53) | 0.32 |
| 6 | HLA | rs2247056 | T/C | 1.21 (0.98 to 1.50) | 0.076 | 1.29 (0.92 to 1.80) | 0.13 | 1.12 (0.77 to 1.62) | 0.55 | 0.86 (0.45 to 1.66) | 0.66 | 1.30 (0.95 to 1.77) | 0.099 |
| 1 | GALNT2 | rs1321257 | G/G | 1.16 (0.96 to 1.40) | 0.12 | 1.27 (0.95 to 1.70) | 0.11 | 1.03 (0.74 to 1.43) | 0.87 | 1.07 (0.59 to 1.93) | 0.82 | 1.07 (0.81 to 1.42) | 0.63 |
| 5 | MAP3K1 | rs9686661 | T/T | 1.19 (0.95 to 1.48) | 0.12 | 1.04 (0.73 to 1.47) | 0.84 | 0.97 (0.66 to 1.44) | 0.90 | 1.68 (0.88 to 3.21) | 0.12 | 1.40 (1.01 to 1.93) | 0.041 |
| 15 | LIPC | rs261342 | G/G | 0.84 (0.68 to 1.05) | 0.13 | 0.91 (0.64 to 1.28) | 0.58 | 0.86 (0.59 to 1.27) | 0.45 | 1.26 (0.68 to 2.33) | 0.46 | 0.8 (0.57 to 1.12) | 0.19 |
| 15 | CAPN3 | rs2412710 | A/A | 1.50 (0.87 to 2.59) | 0.14 | 1.66 (0.74 to 3.72) | 0.22 | 0.90 (0.32 to 2.55) | 0.84 | 0.83 (0.11 to 6.27) | 0.86 | 1.37 (0.60 to 3.10) | 0.46 |
| 2 | COBLL1 | rs10195252 | C/T | 1.13 (0.94 to 1.36) | 0.19 | 1.00 (0.75 to 1.33) | 0.98 | 1.42 (1.02 to 1.99) | 0.039 | 1.89 (1.00 to 3.56) | 0.049 | 0.95 (0.72 to 1.26) | 0.73 |
| 12 | LRP1 | rs11613352 | T/C | 1.11 (0.89 to 1.40) | 0.35 | 1.51 (1.04 to 2.19) | 0.030 | 1.02 (0.69 to 1.50) | 0.92 | 0.97 (0.49 to 1.92) | 0.93 | 1.09 (0.78 to 1.53) | 0.60 |
| 7 | TYW1B | rs13238203 | T/C | 1.30 (0.65 to 2.60) | 0.47 | 0.75 (0.30 to 1.9) | 0.54 | 1.64 (0.40 to 6.69) | 0.49 | NC* | NC* | 1.22 (0.41 to 3.62) | 0.72 |
| 8 | NAT2 | rs1495743 | G/G | 1.07 (0.87 to 1.33) | 0.52 | 0.92 (0.65 to 1.31) | 0.65 | 1.11 (0.76 to 1.62) | 0.61 | 2.16 (1.18 to 3.93) | 0.012 | 0.93 (0.67 to 1.29) | 0.65 |
| 3 | MSL2L1 | rs645040 | G/T | 1.06 (0.85 to 1.33) | 0.58 | 0.92 (0.66 to 1.27) | 0.61 | 1.31 (0.88 to 1.96) | 0.18 | 1.22 (0.62 to 2.42) | 0.56 | 1.23 (0.87 to 1.73) | 0.24 |
| 16 | CTF1 | rs11649653 | G/C | 1.05 (0.87 to 1.27) | 0.59 | 0.86 (0.64 to 1.14) | 0.29 | 1.33 (0.95 to 1.86) | 0.097 | 1.47 (0.81 to 2.64) | 0.20 | 1.01 (0.77 to 1.33) | 0.95 |
| 20 | PLTP | rs4810479 | C/C | 1.06 (0.85 to 1.32) | 0.60 | 1.27 (0.91 to 1.79) | 0.16 | 0.96 (0.65 to 1.42) | 0.85 | 1.15 (0.57 to 2.32) | 0.70 | 0.95 (0.67 to 1.34) | 0.77 |
| 19 | APOE | rs439401 | T/C | 0.95 (0.74 to 1.21) | 0.68 | 0.91 (0.62 to 1.34) | 0.64 | 1.00 (0.66 to 1.54) | 0.98 | 2.07 (0.96 to 4.43) | 0.062 | 0.87 (0.60 to 1.24) | 0.44 |
| 22 | PLA2G6 | rs5756931 | C/T | 1.04 (0.86 to 1.25) | 0.70 | 0.97 (0.73 to 1.30) | 0.85 | 1.06 (0.77 to 1.47) | 0.71 | 0.77 (0.44 to 1.38) | 0.38 | 1.00 (0.76 to 1.31) | 0.98 |
| 12 | ZNF664 | rs12310367 | G/A | 0.97 (0.81 to 1.17) | 0.77 | 1.17 (0.86 to 1.59) | 0.31 | 0.91 (0.66 to 1.26) | 0.58 | 1.45 (0.80 to 2.62) | 0.22 | 0.85 (0.64 to 1.12) | 0.24 |
| 15 | FRMD5 | rs2929282 | T/T | 1.06 (0.69 to 1.64) | 0.79 | 0.93 (0.46 to 1.89) | 0.84 | 0.68 (0.29 to 1.55) | 0.35 | NC* | NC* | 1.38 (0.77 to 2.47) | 0.28 |
| 10 | JMJD1C | rs10761731 | T/A | 1.00 (0.83 to 1.20) | 1.00 | 1.01 (0.76 to 1.35) | 0.94 | 0.98 (0.71 to 1.35) | 0.89 | 1.23 (0.67 to 2.24) | 0.51 | 1.13 (0.85 to 1.49) | 0.40 |
| 8 | PINX1 | rs11776767 | C/C | 1.00 (0.83 to 1.21) | 1.00 | 0.98 (0.73 to 1.33) | 0.92 | 0.95 (0.68 to 1.33) | 0.77 | 1.38 (0.77 to 2.48) | 0.28 | 1.14 (0.86 to 1.52) | 0.35 |
The modeled allele is the TG-raising allele in the combined HTG cohort. CHR indicates chromosome; Mb, megabases; NC, not calculated; OR, odds ratio.
Variant is monomorphic in HTG cases.
For HDL-C-associated loci (Supplemental Table II), we replicated associations at 4 loci with HTG. The replicated loci were predominantly jointly associated with plasma TG as the lead trait, including MLXIPL, TRIB1, and FADS1, but we also replicated the HDL-C associated IRS1 locus. Generally, there was a trend toward association at additional loci, as 31 of 47 loci had directions of effect concordant with estimates from GLGC (P = 0.01). Few loci were replicated among individual HLP-HTG types, although concordant directions of effect were overrepresented among HLP types 4 and 5 (P = 0.005 and P = 0.05, respectively), but not HLP types 2B and 3 (P = 0.11 and P = 0.11, respectively). Together, these data suggest that common variants in some HDL-C-associated loci contribute to the phenotypic spectrum within HTG.
For LDL-C-associated loci (Supplemental Table III), we replicated associations at 9 loci with HTG. The strongest association was at the DNAH11 locus, whereas remaining loci had similarly large effects (odds ratio = 1.2 to 1.4) but with marginal statistical significance. Generally, there was a trend toward association at additional loci, as 25 of 37 loci had directions of effect concordant with estimates from GLGC (P=0.01). Among HLP-HTG phenotypes, few loci were replicated in phenotypes other than HLP type 2B; however, concordant directions of effect were overrepresented among HLP types 2B, 4, and 5 (P=0.0009, P=0.04, and P=0.02, respectively). Together, these data suggest that common variants in some LDL-associated loci also contribute to the phenotypic spectrum within HTG.
Genetic Risk Scores in HTG and the HLP-HTG Phenotypes
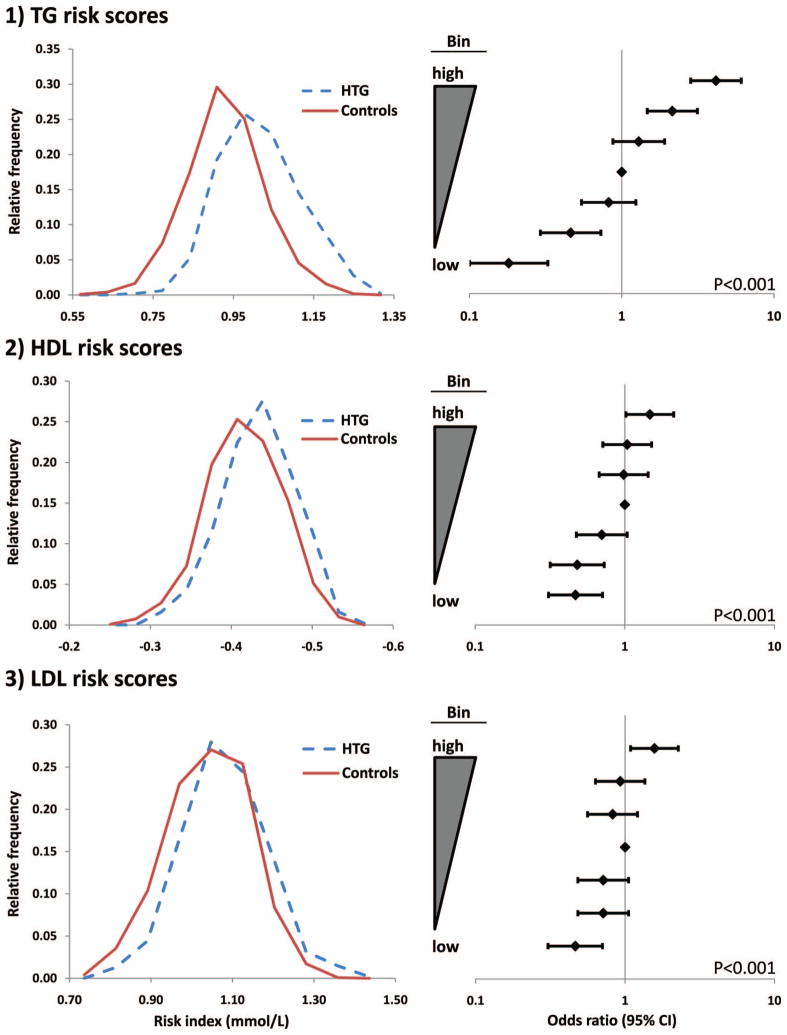
Next, we constructed weighted allelic risk scores to assess the accumulation of lipid-associated risk alleles in the study sample (Figure 1). Consistent with our previous report in a smaller sample,21 weighted TG risk scores were significantly increased in HTG patients versus controls (Figure 1, top). The relative frequency distribution was clearly shifted toward increased scores in HTG patients versus controls: 0.982±0.004 mmol/L versus 0.896±0.003 mmol/L (P=1.6×10−53); unweighted risk scores were similarly increased in HTG patients (Supplemental Figure I). Weighted TG risk scores were associated with increased HTG susceptibility (Ptrend<0.001), as subjects in the highest risk score bin were 4.15 (95% CI: 2.84 to 6.09) times more likely to be HTG cases than healthy controls compared with the median risk score bin (P=8.56×10−14). Subjects in the highest risk score bin were 23.0 (95% CI: 12.86 to 41.14) times more likely to be HTG cases than healthy controls compared with subjects in the lowest risk score bin as the reference group (P=3.51×10−40).
Figure 1.
Weighted allelic risk scores are increased in polygenic HTG patients vs controls. Left: Relative frequency distributions of weighted risk scores in HTG patients and controls. Right: Forest plots demonstrating incremental increases in HTG susceptibility or protection as weighted risk scores deviate from the median. Probability values represent Cochrane-Armitage test for trend.
Weighted HDL-C risk scores were also significantly increased in HTG patients versus controls (Figure 1, middle). The relative frequency distribution of HDL-C risk scores was modestly shifted toward less favorable scores in HTG patients versus controls, corresponding to a highly significant difference between HTG cases and controls of −0.449 ± 0.002 mmol/L versus −0.431 ± 0.001 mmol/L, respectively (P = 1.3 × 10−12). Incremental increases in HDL-C risk score also corresponded to increased HTG susceptibility (Ptrend < 0.001), as subjects in the highest risk score bin were 1.50 (95% CI: 1.02 to 2.12) times more likely to be HTG cases than healthy controls compared with the median risk score bin (P = 0.05). Subjects in the highest risk score bin were 3.14 (95% CI: 2.09 to 4.72) times more likely to be HTG cases than healthy controls compared with the lowest risk score bin as the reference group (P = 2.87 × 10−8).
Weighted LDL-C risk scores were less dramatically increased in HTG patients versus controls (Figure 1, bottom). A modest shift toward higher weighted risk scores was observed in HTG patients, corresponding to a mean risk score of 1.04 ± 0.005 mmol/L in HTG patients versus 1.00 ± 0.003 mmol/L in controls (P = 6.9 × 10−12). Incremental increases in LDL-C risk score were marginally associated with increased HTG susceptibility (Ptrend< 0.001), as subjects in the highest risk score bin were 1.57 (95% CI: 1.09 to 2.27) times more likely to be HTG cases than healthy controls compared with the median risk score bin (P = 0.02). Subjects in the highest risk score bin were 3.39 (95% CI: 2.25 to 5.11) times more likely to be HTG cases than healthy controls compared with the lowest risk score bin as the reference group (P = 3.03 × 10−9).
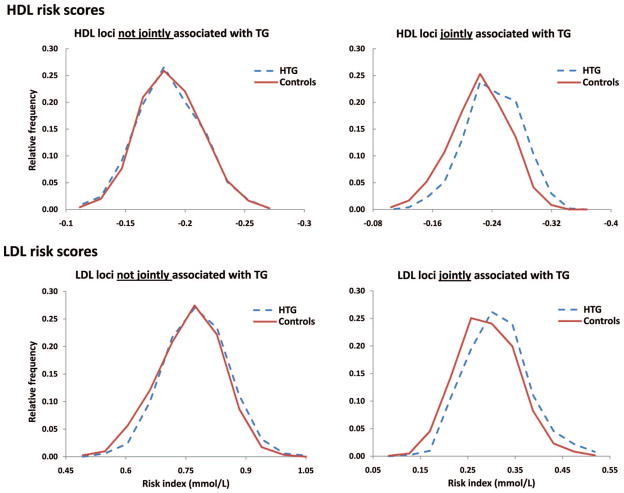
We hypothesized that the increased HDL-C and LDL-C risk scores observed in HTG patients were mediated by the pleiotropic effect of 15 of 47 (32%) HDL-C or 10 of 37 (27%) LDL-C variants that were jointly associated with plasma TG concentration.21 Accordingly, we constructed HDL-C and LDL-C risk scores composed of either (1) variants associated exclusively with HDL-C or LDL-C, or (2) pleiotropic variants jointly associated with plasma TG concentration in addition to HDL-C or LDL-C (Figure 2). For HDL-C, risk scores composed exclusively of HDL-C-associated variants were not different between HTG patients and healthy controls (P = 0.72); however, risk scores composed of pleiotropic HDL-C-associated variants were increased in HTG patients versus controls (P = 1.7 × 10−18) (Figure 2, top). For LDL-C, risk scores composed exclusively of LDL-C-associated variants were slightly increased in HTG patients versus controls (P = 0.0001); however, risk scores composed of pleiotropic LDL-C-associated variants were greatly increased in HTG patients versus controls (P = 8.8 × 10−11) (Figure 2, bottom). We considered the possibility that the large effect sizes of many pleiotropic variants could have inflated the observed variant accumulation; however, this is unlikely because parallel analyses using unweighted risk scores were similarly increased in HTG patients (Supplemental Figure II).
Figure 2.
Increased weighted allelic HDL-C and LDL-C risk scores in HTG patients are driven primarily by loci jointly associated with plasma TG concentration. Weighted risk scores from Figure 1 were separated by loci associated with only HDL-C (top left) or LDL-C (bottom left) or loci jointly associated with plasma TG in addition to HDL-C (top right) or LDL-C (bottom right). Weighted allelic risk scores were constructed from the sum of HDL-C or LDL-C effect estimates at each locus, not effect estimates for associations with plasma TG.
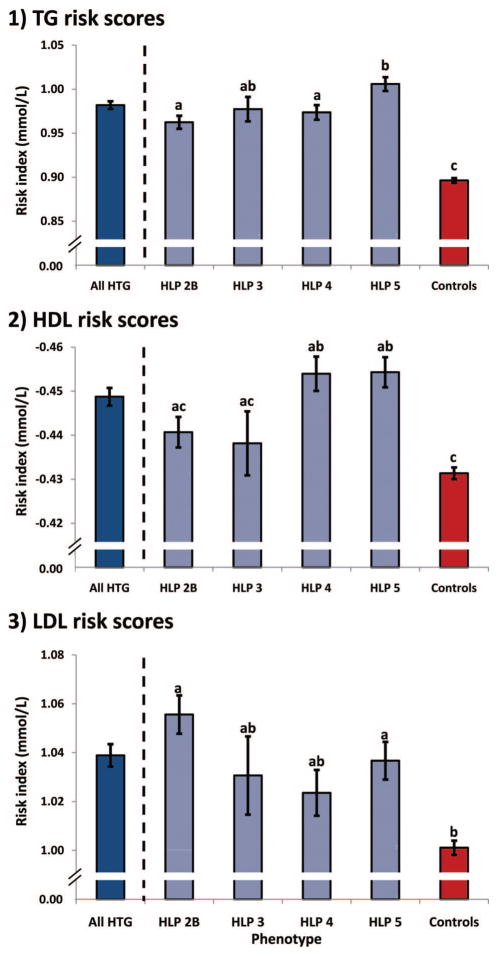
Finally, we tested whether the accumulation of lipid-associated risk alleles observed in HTG patients was consistent across the different HLP-HTG phenotypes (Figure 3). We hypothesized that weighted TG risk scores would be equal among HLP-HTG phenotypes, whereas weighted HDL-C and LDL-C risk scores might differ among HTG patients and phenotypes. For TG risk scores, all HLP-HTG phenotypes had risk scores elevated above controls, with HLP type 5 patients having risk scores further increased over other phenotypes, suggesting an accumulation of common variants with larger effects in these more severely affected patients. For HDL-C risk scores, only HLP types 4 and 5 had scores increased above controls, consistent with the excess of HDL-C-associated variants with directions of effect concordant with GLGC estimates. For LDL-C risk scores, HLP phenotypes 2B and 5 had risk scores increased above controls, also consistent with the excess of LDL-C-associated variants with directions of effect concordant with GLGC estimates. Although the greatest differences in risk scores between HLP phenotypes and controls were attributed to pleiotropic variants, the contribution of both pleiotropic and lipid-specific variants likely contribute to the overall phenotypic heterogeneity among HLP-HTG phenotypes (Supplemental Figure III). These data collectively suggest that common variants associated with plasma lipid concentrations accumulate in HTG patients: TG-associated variants accumulate in all HLP-HTG phenotypes, whereas HDL-C and LDL-C associated variants preferentially accumulate in an HLP-HTG phenotype-dependent manner.
Figure 3.
Weighted allelic risk scores differ among polygenic HLP-HTG phenotypes. The mean risk score in all HTG patients is provided as a reference only; it was not included in statistical comparisons. Values are mean ± standard error; means sharing letters are not statistically different.
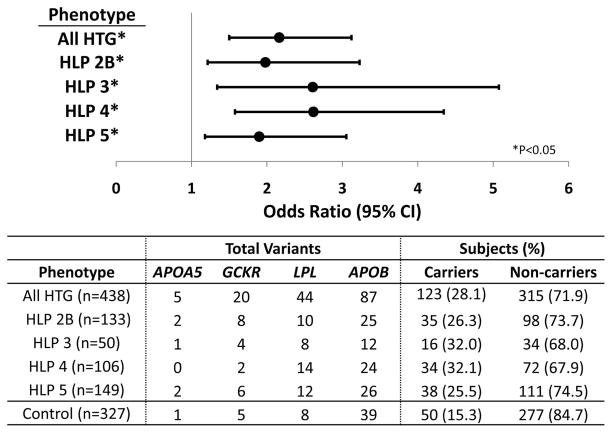
Rare Variants in TG-Associated Genes Among HLP-HTG Phenotypes
We previously demonstrated that a significant excess of rare variants is present in GWAS-identified genes associated with HTG, including APOA5, GCKR, LPL, and APOB, in HTG patients compared with controls.20 Rare variants identified in this previous study were often found in multiple subjects; however, we could not identify any trends between genes, variants, or HLP-HTG phenotypes. Here, we tested whether this excess of rare variants extended to the HLP phenotypes in subjects with available sequencing data. Carriers and noncarriers were compared in this analysis, as some subjects carried >1 rare variant. Indeed, a significant excess of rare variants was observed across all HLP-HTG phenotypes (Figure 4). Comparison of rare variants between each HLP-HTG phenotype and healthy controls using the Fisher exact test revealed that carriers were 1.8 to 2.6 times more likely to have a HLP-HTG case status than a control status. There were no differences in number of rare variant carriers or noncarriers among the 4 phenotypes (P = 0.59), nor were there differences in distribution of rare variants among genes in each phenotype (P = 0.69), as assessed using contingency table analysis. We also found no differences in TG risk scores among carriers and noncarriers of rare variants in HTG patients and controls (Supplemental Figure IV). These data suggest that the distribution of rare variants in HTG-associated genes occurs nonpreferentially across all HLP-HTG phenotypes in several TG-associated genes, on top of an accumulation of lipid-associated common variants.
Figure 4.
Excess of rare variants is similar among polygenic HLP-HTG phenotypes in terms of carrier number and gene distribution. Top: Rare variant accumulation was compared between each HLP-HTG phenotype and controls. Statistical significance was measured using the Fisher exact test using a 2-sided P < 0.05. Bottom: Distribution of rare variants across genes and subjects. Carriers are subjects with ≥1 rare variant in ≥1 genes.
Comparison of Clinical and Genetic Variable Contribution Among HLP-HTG Phenotypes
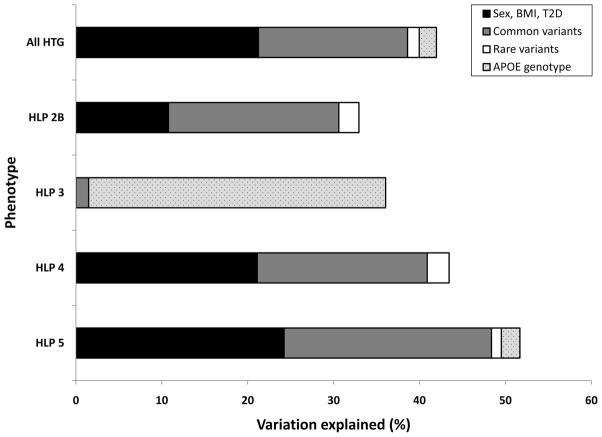
Given the limitations associated with calculating explained variation from logistic regression models, we measured changes in model goodness-of-fit (pseudo-R2) attributed to different clinical and genetic variables in a logistic regression model discriminating between HLP-HTG phenotypes and healthy controls as a surrogate metric for explained variation (Figure 5; see Methods). Within HTG patients, the total proportion of variation explained by the model was 42.0%, very similar to our previous estimates.20 Clinical variables explained 21.2%, common variants explained 17.4%, and rare variants explained 1.4%. In this analysis, APOE genotype was also included because of its historical importance in HLP type 3, which explained an additional 2% of variation among HTG cases. Among the HLP-HTG phenotypes, the proportion of variation attributable to these variables differed significantly. In HLP type 3, APOE ε2 genotype explained 34.6% of variation; common variation explained only an additional 1.5% of variation. Conversely, common genetic variants explained 20% to 24% of variation among remaining phenotypes. APOA5, GCKR, MLXIPL, and CILP2 explained the majority of this variation, although LPL explained additional variation in HLP type 4, TRIB1 explained additional variation in HLP type 2B, and ANGPTL3 explained additional variation in HLP types 2B and 5. Rare genetic variants in the 4 genes also explained 1.2% to 2.6% of variation among HLP types 2B, 4, and 5. Finally, APOE genotype explained ≈2.2% of variation among HLP type 5 patients, but it did not explain additional variation among remaining phenotypes. Thus, the contribution of common and rare variants to the model was relatively similar among HLP phenotypes 2B, 4, and 5, whereas APOE genotype was the best predictor of HLP type 3 case status.
Figure 5.
Comparison of variation attributable to clinical and genetic variables among polygenic HLP-HTG phenotypes within our cohort. The proportion of variation explained was calculated from changes in model fit (R2) from multiple logistic regression entering clinical and genetic variables using forward modeling. Common variants included risk alleles (0, 1, or 2 alleles) from multiple TG-associated loci selected by the model. BMI, body mass index; T2D, type 2 diabetes.
Discussion
We have demonstrated that TG loci were most robustly associated with HTG, although HDL-C and LDL-C loci were also associated with HTG. Common variants were often associated with multiple HLP-HTG phenotypes. Weighted TG risk scores composed of TG, HDL-C, or LDL-C variants were distinctly increased across all HLP-HTG phenotypes compared with controls. Weighted HDL-C and LDL-C risk scores were also increased, although less pronounced, in some HLP-HTG phenotypes compared with controls. Interestingly, decomposition of HDL-C and LDL-C risk scores revealed that pleiotropic variants (those jointly associated with TG) accounted for the greatest difference in HDL-C and LDL-C risk scores among HLP-HTG phenotypes. Rare variants in 4 genes also accumulated equally among HLP-HTG phenotypes. Thus, HTG susceptibility and phenotypic heterogeneity appear to be influenced by accumulation of common and rare TG-associated variants.
This comprehensive analysis yields several new insights. First, common variants in loci associated not only with TG but also with HDL-C and LDL-C are associated with HTG and the HLP-HTG phenotypes. Second, common and rare variants in TG-associated loci are overrepresented in HTG patients and across all HLP-HTG phenotypes. Third, common variants in HDL-C and LDL-C loci are overrepresented in HTG patients but accumulate preferentially in different HLP-HTG phenotypes. Finally, pleiotropic variants jointly associated with plasma TG concentration in addition to HDL-C or LDL-C account for most of the accumulation of HDL-C and LDL-C associated variants in the HLP-HTG phenotypes. Taken together, these findings provide a more complete accounting of the genetic architecture of HTG susceptibility and the basis of the phenotypic heterogeneity in HTG.
Our previous preliminary studies provided a first approximation of the genetic architecture of HTG and the HLP-HTG phenotypes,16–18 whereas the novelty of the current study lies in the comprehensive nature of our analysis, which included all recently identified lipid-associated variants from the GLGC.21 We have confirmed the hypothesis that a common genetic architecture of many common and rare TG-associated variants underlies predisposition to HTG,28 and demonstrated that pleiotropic variants and accumulation of additional lipid-associated variants may partly explain an origin for the phenotypic heterogeneity among HTG patients.
Our interpretation of these results is that a genetic burden of deleterious variants may predispose some subjects to HTG. Common and rare variants accumulate more often in HTG patients, suggesting that risk alleles may incrementally contribute to HTG susceptibility. However, it appears that no single variant or accumulation of variants is sufficient to cause HTG or any particular HLP-HTG phenotype. Despite seemingly strong phenotype-specific associations, risk alleles at associated loci are overrepresented in multiple HLP-HTG phenotypes compared with controls. Similarly, rare HTG-associated variants also accumulate comparably across the HLP-HTG phenotypes, in addition to an elevated background of common risk variants, again suggesting that rare variants are not sufficient for HTG causation. Ultimately, the significant overlap of risk alleles among HTG patients and controls suggests that—as with most complex traits— genetic variants only account for a portion of variation in HTG case status.
Our developing model of polygenic HTG is that accumulation of common and rare TG-associated risk alleles gradually increases HTG susceptibility. Furthermore, the accumulation of pleiotropic risk alleles in addition to lipid-associated risk alleles may contribute to the phenotypic heterogeneity characteristic of the classical HLP-HTG phenotypes. However, we do not exclude the possibility that HTG susceptibility in some patients can be independent of risk allele accumulation. In our sample, we observed that HLP type 2B appeared to be associated preferentially with common variants in pleiotropic and exclusively LDL-C-associated loci, HLP type 4 appeared to be associated with common variants in pleiotropic HDL-C-associated loci, and the most severe phenotype HLP type 5 was associated with large-effect common TG-associated risk alleles, with additional contributions from both pleiotropic LDL-C- and HDL-C-associated loci. The contribution of risk alleles from APOA5, GCKR, MLXIPL, and CILP2 appeared to contribute relatively equally among these phenotypes, with additional smaller contributions from other TG-associated variants in each HLP-HTG phenotype.
HLP type 3 is unique among HLP-HTG phenotypes as it is predominantly explained by the classical APOE ε2/ε2 isoform (APOE E2/E2 genotype). However, even APOE ε2/ε2 is individually insufficient to cause HLP type 3. HLP type 3 is defined by the accumulation of intermediate-density lipoproteins visible as β-very-low-density lipoprotein (or a broad β-band) on electrophoresis, resulting in a high ratio of very-low-density lipoprotein cholesterol to TG. Many HLP type 3 patients have APOE ε2/ε2, although some HLP type 5 patients that are clinically defined by fasting chylomicronemia and pancreatitis in our sample also have APOE ε2/ε2 genotype; many healthy controls in the general population also have APOE ε2/ε2 genotype. Variants such as APOA5 that are strongly associated with HLP type 3, in addition to other TG-raising risk alleles, likely contribute additionally to HLP type 3 susceptibility.
Our conclusions must be interpreted in the context of important limitations of our study. First, our sample size and statistical power were limited, although we had the largest cohort of HTG patients reported to date. Accordingly, our sample was insufficient to replicate most small effect variants identified by the GLGC.21 This was compounded by the need for subgroup analyses, which prevented replication of many variants in the HLP-HTG phenotypes, even those associated with HTG at P < 0.05, but was unavoidable because of insufficient numbers. To compensate for this, we have used additional analyses, including risk score analyses and estimates of directionality of effect to overcome this limitation. Second, the analysis of GWAS lead SNPs is likely insufficient to demonstrate the full spectrum of genetic heterogeneity within the HLP-HTG phenotypes. We genotyped tagSNPs that serve as surrogates for functional variants at each locus, not truly querying the specific functional variants that increase HTG susceptibility. Direct genotyping of functional variants, such as the APOA5 −1131T>C promoter variant and the Ser19Trp variant, which are both strongly associated with HTG16–18 but are each in relatively weak linkage disequilibrium with the APOA5 tag-SNP rs964184 (r2 = 0.29 and r2 = 0.34 in sample subsets) may improve the resolution of our risk scores and thus the ability to detect differences between phenotypes. This might be expected given that APOE genotype at functional variants rs7412 and rs429358 (encoding the APOE E2/E3/E4 isoforms) contributed significantly more information to HLP type 3 case status than the APOE tagSNP. Third, only 4 HTG-associated genes were selected for resequencing. This limited view of rare variant accumulation may preclude our ability to detect genetic heterogeneity in other genes that could mediate phenotypic heterogeneity within the HLP-HTG phenotypes. Furthermore, functional data are necessary to separate deleterious from benign variants, which may also introduce noise into this analysis preventing detection of heterogeneity. However, many variants in these genes were identified across multiple HLP-HTG phenotypes. For example, the loss-of-function variant LPL Gly188Glu,29 which completely attenuates LPL function, was identified as a heterozygous mutation in HLP types 3, 4, and 5. This suggests that rare variants in these genes have a general effect on HTG susceptibility, without predisposing subjects to a specific HLP-HTG phenotype. Ultimately, the combination of low power arising from a limited sample size with an incomplete survey of functional genetic variants in all HTG-associated genes may hide some of the genetic heterogeneity initially expected to underlie the HLP-HTG phenotypes. However, our results based on GWAS tagSNPs and 4 HTG-associated genes provide evidence for a common overlapping genetic etiology for the phenotypic spectrum of HLP-HTG phenotypes.
Future studies will require exhaustive resequencing of TG-associated genes to identify additional genetic variants involved in HTG susceptibility. These will include both common functional variants that underlie GWAS signals and large effect rare variants that mediate significant HTG predisposition in their carriers. Together, these variants will likely explain additional variation in HTG case status and may provide a better explanation for the genetic basis of the phenotypic heterogeneity among HLP-HTG patients. Evaluation of non-GWAS candidate genes, including APOC2, APOC3, APOE, GPIHBP1, and LMF1, will also be essential to fully document the genetic determinants of HTG. Consideration of these determinants in the context of multiplicative or synergistic effects between genes, variants, and metabolic and environmental exposures may contribute additionally to the unexplained variation among HTG patients.
Evaluation of remaining unexplored variables may eventually lead to improved detection of subjects with increased HTG susceptibility. For example, genetic risk scores composed of functional HTG-associated variants may facilitate presymptomatic identification of clinic patients at increased risk for development of HTG. Common variants in genes such as APOA5, LPL, TRIB1, and CILP2 are strong determinants of HTG but also of CVD, and they could be further integrated into risk prediction algorithms such as the Framing-ham risk score as individual variants or genetic risk scores to help predict subjects at increased risk of CVD. The clinical utility of HTG-associated variants in the prediction of HTG or CVD susceptibility has not been evaluated, but it could be used in the future to direct evidence-based treatments or lifestyle interventions at those with highest HTG susceptibility.
In conclusion, we demonstrate that a spectrum of common and rare genetic variants in lipid-associated loci underlies the biochemically defined HLP phenotypes characterized by HTG. We confirm that an accumulation of common TG-associated risk alleles is observed in HTG patients and provide new evidence supporting the accumulation of pleiotropic TG-, HDL-C-, and LDL-C-associated risk alleles as possible determinants of HTG phenotypic heterogeneity. The significant overlap of risk alleles among multiple HLP-HTG phenotypes indicates a shared yet complex and heterogeneous genetic HTG susceptibility. Ongoing discovery of additional TG-associated alleles and elucidation of specific functional variants contributing to HTG pathophysiology will be required in the context of normal and perturbed metabolic and environmental conditions.
Supplementary Material
Acknowledgments
Sources of Funding
Dr Johansen is supported by the Canadian Institutes of Health Research (CIHR) Banting and Best Canada Graduate Scholarship Doctoral Research Award and the University of Western Ontario MD/PhD program, and is a CIHR fellow in Vascular Research. Dr Hegele is supported by the Jacob J. Wolfe Distinguished Medical Research Chair at the University of Western Ontario; the Edith Schulich Vinet Canada Research Chair in Human Genetics (Tier I); the Martha G. Blackburn Chair in Cardiovascular Research; and operating grants from the CIHR (MOP-13430, MOP-79523, CTP-79853), the Heart and Stroke Foundation of Ontario (NA-6059, T-6018, PRG-4854), the Pfizer Jean Davignon Distinguished Cardiovascular and Metabolic Research Award, and Genome Canada through the Ontario Genomics Institute.
Footnotes
Disclosures
None.
References
- 1.Cullen P. Evidence that triglycerides are an independent coronary heart disease risk factor. Am J Cardiol. 2000;86:943–949. doi: 10.1016/s0002-9149(00)01127-9. [DOI] [PubMed] [Google Scholar]
- 2.Hokanson JE, Austin MA. Plasma triglyceride level is a risk factor for cardiovascular disease independent of high-density lipoprotein cholesterol level: a meta-analysis of population-based prospective studies. J Cardiovasc Risk. 1996;3:213–219. [PubMed] [Google Scholar]
- 3.Bansal S, Buring JE, Rifai N, Mora S, Sacks FM, Ridker PM. Fasting compared with nonfasting triglycerides and risk of cardiovascular events in women. JAMA. 2007;298:309–316. doi: 10.1001/jama.298.3.309. [DOI] [PubMed] [Google Scholar]
- 4.Nordestgaard BG, Benn M, Schnohr P, Tybjaerg-Hansen A. Nonfasting triglycerides and risk of myocardial infarction, ischemic heart disease, and death in men and women. JAMA. 2007;298:299–308. doi: 10.1001/jama.298.3.299. [DOI] [PubMed] [Google Scholar]
- 5.Freiberg JJ, Tybjaerg-Hansen A, Jensen JS, Nordestgaard BG. Nonfasting triglycerides and risk of ischemic stroke in the general population. JAMA. 2008;300:2142–2152. doi: 10.1001/jama.2008.621. [DOI] [PubMed] [Google Scholar]
- 6.Sarwar N, Sandhu MS, Ricketts SL, Butterworth AS, Di Angelantonio E, Boekholdt SM, Ouwehand W, Watkins H, Samani NJ, Saleheen D, Lawlor D, Reilly MP, Hingorani AD, Talmud PJ, Danesh J. Triglyceride-mediated pathways and coronary disease: collaborative analysis of 101 studies. Lancet. 2010;375:1634–1639. doi: 10.1016/S0140-6736(10)60545-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hegele RA, Pollex RL. Hypertriglyceridemia: phenomics and genomics. Mol Cell Biochem. 2009;326:35–43. doi: 10.1007/s11010-008-0005-1. [DOI] [PubMed] [Google Scholar]
- 8.Yuan G, Al-Shali KZ, Hegele RA. Hypertriglyceridemia: its etiology, effects and treatment. CMAJ. 2007;176:1113–1120. doi: 10.1503/cmaj.060963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Havel RJ, Gordon RS., Jr Idiopathic hyperlipemia: metabolic studies in an affected family. J Clin Invest. 1960;39:1777–1790. doi: 10.1172/JCI104202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connelly PW, Maguire GF, Little JA. Apolipoprotein CIISt. Michael: familial apolipoprotein CII deficiency associated with premature vascular disease. J Clin Invest. 1987;80:1597–1606. doi: 10.1172/JCI113246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Priore Oliva C, Pisciotta L, Li Volti G, Sambataro MP, Cantafora A, Bellocchio A, Catapano A, Tarugi P, Bertolini S, Calandra S. Inherited apolipoprotein A-V deficiency in severe hypertriglyceridemia. Arterioscler Thromb Vasc Biol. 2005;25:411–417. doi: 10.1161/01.ATV.0000153087.36428.dd. [DOI] [PubMed] [Google Scholar]
- 12.Peterfy M, Ben-Zeev O, Mao HZ, Weissglas-Volkov D, Aouizerat BE, Pullinger CR, Frost PH, Kane JP, Malloy MJ, Reue K, Pajukanta P, Doolittle MH. Mutations in LMF1 cause combined lipase deficiency and severe hypertriglyceridemia. Nat Genet. 2007;39:1483–1487. doi: 10.1038/ng.2007.24. [DOI] [PubMed] [Google Scholar]
- 13.Beigneux AP, Franssen R, Bensadoun A, Gin P, Melford K, Peter J, Walzem RL, Weinstein MM, Davies BS, Kuivenhoven JA, Kastelein JJ, Fong LG, Dallinga-Thie GM, Young SG. Chylomicronemia with a mutant GPIHBP1 (Q115P) that cannot bind lipoprotein lipase. Arterioscler Thromb Vasc Biol. 2009;29:956–962. doi: 10.1161/ATVBAHA.109.186577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Hegele RA. Homozygous missense mutation (G56R) in glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 (GPI-HBP1) in two siblings with fasting chylomicronemia (MIM 144650) Lipids Health Dis. 2007;6:1–4. doi: 10.1186/1476-511X-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegele RA. Plasma lipoproteins: genetic influences and clinical implications. Nat Rev Genet. 2009;10:109–121. doi: 10.1038/nrg2481. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Ban MR, Kennedy BA, Anand S, Yusuf S, Huff MW, Pollex RL, Hegele RA. APOA5 genetic variants are markers for classic hyperlipoproteinemia phenotypes and hypertriglyceridemia. Nat Clin Pract Cardiovasc Med. 2008;5:730–737. doi: 10.1038/ncpcardio1326. [DOI] [PubMed] [Google Scholar]
- 17.Hegele RA, Ban MR, Hsueh N, Kennedy BA, Cao H, Zou GY, Anand S, Yusuf S, Huff MW, Wang J. A polygenic basis for four classical Fredrickson hyperlipoproteinemia phenotypes that are characterized by hypertriglyceridemia. Hum Mol Genet. 2009;18:4189–4194. doi: 10.1093/hmg/ddp361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Ban MR, Zou GY, Cao H, Lin T, Kennedy BA, Anand S, Yusuf S, Huff MW, Pollex RL, Hegele RA. Polygenic determinants of severe hypertriglyceridemia. Hum Mol Genet. 2008;17:2894–2899. doi: 10.1093/hmg/ddn188. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Cao H, Ban MR, Kennedy BA, Zhu S, Anand S, Yusuf S, Pollex RL, Hegele RA. Resequencing genomic DNA of patients with severe hypertriglyceridemia (MIM 144650) Arterioscler Thromb Vasc Biol. 2007;27:2450–2455. doi: 10.1161/ATVBAHA.107.150680. [DOI] [PubMed] [Google Scholar]
- 20.Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, Ban MR, Martins RA, Kennedy BA, Hassell RG, Visser ME, Schwartz SM, Voight BF, Elosua R, Salomaa V, O’Donnell CJ, Dallinga-Thie GM, Anand SS, Yusuf S, Huff MW, Kathiresan S, Hegele RA. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anand SS, Yusuf S, Vuksan V, Devanesen S, Teo KK, Montague PA, Kelemen L, Yi C, Lonn E, Gerstein H, Hegele RA, McQueen M. Differences in risk factors, atherosclerosis, and cardiovascular disease between ethnic groups in Canada: the Study of Health Assessment and Risk in Ethnic groups (SHARE) Lancet. 2000;356:279–284. doi: 10.1016/s0140-6736(00)02502-2. [DOI] [PubMed] [Google Scholar]
- 23.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, Erdmann J, Reilly MP, Rader DJ, Morgan T, Spertus JA, Stoll M, Girelli D, McKeown PP, Patterson CC, Siscovick DS, O’Donnell CJ, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Melander O, Altshuler D, Merlini PA, Berzuini C, Bernardinelli L, Peyvandi F, Tubaro M, Celli P, Ferrario M, Fetiveau R, Marziliano N, Casari G, Galli M, Ribichini F, Rossi M, Bernardi F, Zonzin P, Piazza A, Yee J, Friedlander Y, Marrugat J, Lucas G, Subirana I, Sala J, Ramos R, Meigs JB, Williams G, Nathan DM, MacRae CA, Havulinna AS, Berglund G, Hirschhorn JN, Asselta R, Duga S, Spreafico M, Daly MJ, Nemesh J, Korn JM, McCarroll SA, Surti A, Guiducci C, Gianniny L, Mirel D, Parkin M, Burtt N, Gabriel SB, Thompson JR, Braund PS, Wright BJ, Balmforth AJ, Ball SG, Hall AS, Linsel-Nitschke P, Lieb W, Ziegler A, Konig I, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Ouwehand W, Deloukas P, Scholz M, Cambien F, Li M, Chen Z, Wilensky R, Matthai W, Qasim A, Hakonarson HH, Devaney J, Burnett MS, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Epstein SE, Scheffold T, Berger K, Huge A, Martinelli N, Olivieri O, Corrocher R, McKeown P, Erdmann E, Konig IR, Holm H, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Do R, Xie C, Siscovick D. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Abecasis GR. Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK. a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 28.Johansen CT, Kathiresan S, Hegele RA. Genetic determinants of plasma triglycerides. J Lipid Res. 2011;52:189–206. doi: 10.1194/jlr.R009720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emi M, Wilson DE, Iverius PH, Wu L, Hata A, Hegele R, Williams RR, Lalouel JM. Missense mutation (Gly-Glu188) of human lipoprotein lipase imparting functional deficiency. J Biol Chem. 1990;265:5910–5916. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.