Abstract
Western countries, prostate cancer is the most prevalent cancer of men, and one of the leading causes of cancer-related death in men. Several genome-wide association studies have yielded numerous common variants conferring risk of prostate cancer. In the present study we analyzed 32.5 million variants discovered by whole-genome sequencing 1,795 Icelanders. One variant was found to be associated with prostate cancer in European populations: rs188140481[A] (OR = 2.90, Pcomb = 6.2×10−34) located on 8q24, with an average risk allele control frequency of 0.54%. This variant is only very weakly correlated (r2 ≤ 0.06) with previously reported risk variants on 8q24, and remains significant after adjustment for all of them. Carriers of rs188140481[A] were diagnosed with prostate cancer 1.26 years younger than non-carriers (P = 0.0059). We also report results for the previously described HOXB13 mutation (rs138213197[T]), confirming it as prostate cancer risk variant in populations from all over Europe.
Risk of prostate cancer has been shown to have a strong genetic component1,2. Numerous genome-wide association studies (GWAS) are registered in the Catalog of published Genome-Wide Association Studies (see URLs), where well over 40 prostate cancer susceptibility loci are listed. However, these risk variants, which were discovered using commercial genotyping chips, are common and each confers a moderate to low risk. New high-throughput whole-genome sequencing techniques allow for systematic search for rarer risk variants, that possibly also confer greater risk. An example is the recently published missense mutation in HOXB13 conferring high risk of prostate cancer3. Here, we report one additional independent variant located on 8q24 that is rarer and confers greater risk than previously published common prostate cancer risk variants.
To search for prostate cancer risk variants, we analyzed data generated by deCODE’s ongoing whole-genome sequencing project. In short, the goal of this project is to sequence 2,500 Icelanders, to at least 10-fold coverage of the genome for each individual. The variants (SNPs and INDELs) discovered in the sequencing phase are then propagated into chip-genotyped individual as well as into individuals without genotypic information, making use of the extensive genealogical information available in Iceland as well as phasing- and imputation methods previously described4. The association analysis in our study was done at a time point when 1,795 Icelanders had been sequenced to at least a ten-fold coverage. The approximately 32.5 million variants discovered through whole-genome sequencing were imputed into 71,743 Icelanders who had all been genotyped using commercial Illumina SNP-chips as well as into 296,496 Icelanders without direct chip-genotypes but with family-based imputation genotype information4,5 (see Online Methods and Supplementary Note). Our list of prostate cancer patients contains 5,141 men diagnosed from 1955 until end of 2010; based on the nationwide Icelandic Cancer Registry (ICR; see URLs). Of those, 2,315 patients had been genotyped using one of the Illumina chips, and 2,222 additional patients had at least partial data from family based imputation; leaving 604 of the total patient list with no genotypic information. As controls we used 54,444 males (27,780 had variants imputed based on chip-genotypes and 26,664 had variants imputed with a family based methods; see Supplementary Note) not diagnosed with prostate cancer according to the nationwide ICR.
According to our analysis 252 variants were significantly associated with prostate cancer at P < 1.5×10−9 (a GWAS significance threshold applicable according to a Bonferroni correction of 32 million tests). All these variants were located at the previously described prostate cancer risk loci on 8q24 and 17q12. In order to search for novel risk variants at these two loci we used multivariate logistic regression.
When analyzing the 17q12 locus we adjusted for the variant rs4430796, the SNP originally reported at this locus6 and also the SNP most significantly associated with prostate cancer at this locus, in our present GWAS. Based on the conditional analysis no other variant on 17q12 remained significant at our GWAS P-value threshold.
At the 8q24 locus, nine independent prostate cancer risk variants have been previously published7–14 (Supplementary Table 1a). These variants are distributed over five neighboring LD-regions/blocks (see Fig.1). When performing the multivariate logistic regression analysis for the variants at 8q24 in our current GWAS, we first adjusted for the SNP most significantly associated with prostate cancer, which is rs11995378 located in region-1 (r2 = 0.95 between rs11995378 and rs1447295 which is the SNP originally reported7 in region-1). In the second step we conditioned on two SNPs, including the SNP used in step-1 as well as the SNP most significant after step-1, which is rs77541621 located in region-2 (r2 = 0.64 between rs77541621 and rs16901979, which is the SNP initially reported8,10 in region-2). Based on the results from step-2, four SNPs and one INDEL, all correlated with each other (r2 ≥ 0.74; Supplementary Table 1b), remained significant at the GWAS P-value threshold. Of those, based on the unconditional family based imputed results, the most significant association with prostate cancer was observed for the minor allele (allele G) of rs183373024 (P = 1.50×10−23, OR = 2.69) and the minor allele (allele A) of rs188140481 (P = 1.5×10−22, OR = 2.88). In the third step of the logistic regression, after adding rs183373024 to the list of SNPs conditioned on, no variant remained significant at the applicable P-value threshold of P < 1.5×10−9 (Fig. 1). Hence, no further step-wise conditional analysis was done.
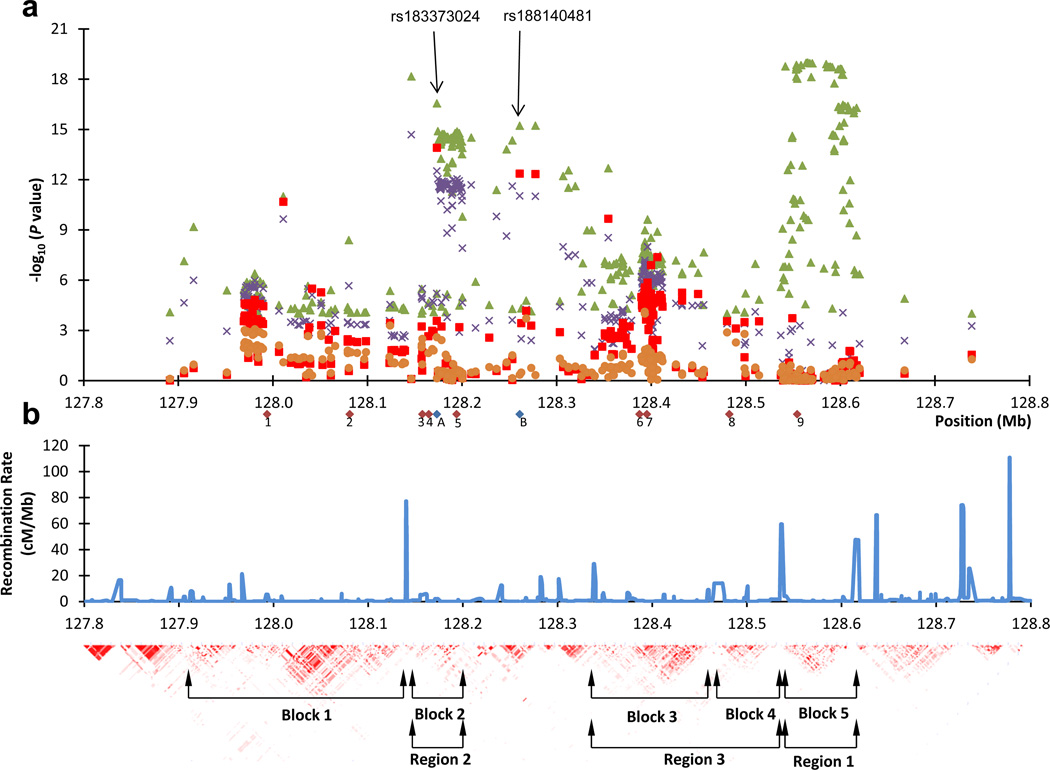
Figure 1.
Schematic view of a region on chromosome 8q24 containing several prostate cancer risk variants. a) Shown are imputed association results, using only results based on chip-genotyped subjects, for variants with P < 1E-4 in the Icelandic prostate cancer GWAS and located between 127.8 Mb and 128.8 Mb (Build 36) on 8q24. Green triangles denote imputed association results (-log(P-value) of the unconditional analysis. Purple crosses denote association results after step-1 of the conditional analysis (the logistic regression), red boxed denote association results after step-2, and orange circles denote association result after step-3. The red diamonds below the X-axis denote the previously reported prostate cancer risk SNPs on 8q24 in populations of European descent (1, rs1254366314; 2, rs1008690811,14; 3, rs101634314; 4, rs1325229814; 5, rs169019798; 6, rs1690209412; 7, rs44511412 / rs62086113,14; 8, rs69832679; 9, rs14472957) and the two blue diamonds denote the two SNPs (A, rs183373024 and B, rs188140481) discussed in the main text. b) Shown are the CEU HapMap population recombination rate and the pairwise correlation coefficient (r2) for SNPs in the 1 megabase (Mb) region depicted. The annotations of blocks/regions is as previously described 7–9,14,21.
Of the previously published prostate cancer risk SNPs located on 8q24 that are not included or not strongly correlated with the SNPs conditioned on in the logistic regression analysis described above, four SNPs remain marginally significant (P < 0.05): rs13252298, rs16902094, rs445114, rs6983267 (Supplementary Table 2).
For validating the Icelandic association results we genotyped rs183373024 and rs188140481 in five prostate cancer study groups of European descent. These groups come from The Netherlands, Spain, Romania, the United Kingdom (UK) and the United States (US). The combined results of the five study groups were significant for both SNPs and combining them with the imputed results from Iceland gave an estimated OR of 2.59 for rs183373024[G] (Pcomb = 4.1×10−33) and an OR of 2.90 for rs188140481[A] (Pcomb = 6.2×10−34). The control frequency of the risk alleles of these two variants ranges from 0.1% to 1.1%, being lowest in southern Europe and higher in northern Europe. A test of heterogeneity among all six study groups showed no significant difference between the ORs of the two variants (Phet > 0.32; Table 1 and Supplementary Table 3). We also genotyped both SNPs in 498 Asian prostate cancer patients from Hong Kong and 467 African American controls from Chicago but the SNPs were not polymorphic in these study groups. Hence, if the underlying causative risk variant is present in these two ethnic groups it is either not tagged by these SNPs or very rare.
Table 1.
Summary association results for rs188140481[A] and prostate cancer
| Frequency |
||||||
|---|---|---|---|---|---|---|
| Study population |
Cases (n) |
Controls (n) |
Cases | Controls | OR (95% CI) | P value |
| Iceland | 4,537 | 54,444 | 0.032 | 0.011 | 2.88 (2.44, 3.44) | 1.5×10−22 |
| Chicago | 1,944 | 1,260 | 0.013 | 0.0052 | 2.61 (1.46, 4.69) | 0.0012 |
| Spain | 726 | 1,625 | 0.0041 | 0.0012 | 3.37 (0.87, 13.03) | 0.079 |
| The Netherlands | 1,531 | 1,956 | 0.019 | 0.0072 | 2.63 (1.68, 4.11) | 2.1×10−5 |
| Romania | 731 | 917 | 0.0068 | 0.0027 | 2.52 (0.80, 7.98) | 0.12 |
| UK | 538 | 1,825 | 0.020 | 0.0049 | 4.21 (2.23, 7.95) | 9.2×10−6 |
| All excl. Iceland | 5,470 | 7,583 | - | 0.0042 | 2.93 (2.19, 3.92) | 5.6×10−13 |
| All combined | 10,007 | 62,027 | - | 0.0054 | 2.90 (2.44, 3.44) | 6.2×10−34 |
| Phet = 0.88 | I2 = 0.0 | |||||
All P values shown are two-sided. Shown are the corresponding numbers of cases and controls (n), allelic frequencies of variants in affected and control individuals, the allelic odds-ratio (OR) with 95% confidence interval (95% CI) and P value. Also shown are the P-values for the heterogeneity of the ORs (Phet) for all study groups as well as I2 which lies between 0% and 100% and describes the proportion of total variation in study estimates that is due to heterogeneity. For the combined study populations, the reported control frequency was the average, unweighted control frequency of the individual populations, while the OR and the P value were estimated using the Mantel-Haenszel model. Of the Icelandic cases, 2,315 patients had been genotyped using one of the Illumina chips, and 2,222 additional patients had at least partial data based on family based imputation. Of the Icelandic controls 27,780 were imputed based on chip-genotypes and 26,664 were family based imputed. All non-Icelandic replication samples are directly genotyped.
Since, the association effect of the two variants cannot be distinguished from each other, we elected to focus on rs188140481[A] in subsequent investigations, based on its slightly stronger combined replication results than in the case of rs183373024. Neither rs188140481 nor any of the highly correlated (r2 > 0.7; Supplementary Table 1b) variants are present in the latest HapMap release (data release 28). Therefore, association analysis based on HapMap data did not yield the association signal reported here. The SNP rs188140481 is however, in the 1000-genome data set registered into the dbSNP-135 database in July 2011.
The SNP, rs188140481, is located telomeric to the previously published region-2 on 8q24; originally reported to contain the prostate cancer risk SNP rs169019798 and later also rs1016343 and rs1325229814 (Fig. 1). rs188140481 has D’ = 1 with those three SNPs but it is only very weakly correlated with them (r2 ≤ 0.005). Furthermore, the correlation (the r-coefficient) between rs188140481 and rs16901979 is in a negative direction. Of the previously published risk SNPs in populations of European descent and located on 8q24, rs16901979 has the closest resemblance in terms of conferred risk and allele frequency (risk allele control frequency in Iceland is ~3% and ~1% for rs16901979[A] and rs188140481[A], respectively). This indicates that the two risk variants, are never together on the same haplotype. Consequently, the association results for rs16901979 and rs188140481 remain significant after being adjusted for each other. Furthermore, rs188140481 is only very weakly correlated (r2 < 0.06) with the previously published leukemia15, prostate7–10,14, colon16–18, breast19, or bladder cancer20 risk variants on 8q24 (Supplementary Table 1a), and the association results for rs188140481 remain significant after being adjusted for all nine previously published prostate cancer risk SNPs at 8q24 (Table 2).
Table 2.
Adjusted and unadjusted association results for rs188140481[A] and prostate cancer
| Unadjusted |
Adjusted |
|||
|---|---|---|---|---|
| Study group | OR (95% CI) | P value | OR (95% CI) | P value |
| Iceland | 3.01 (2.23, 4.06) | 5.9×10−13 | 2.04 (1.42, 2.92) | 1.0×10−4 |
| Chicago | 2.63 (1.50, 4.60) | 7.0×10−4 | 1.95 (1.07, 3.56) | 0.029 |
| The Netherlands | 2.67 (1.73, 4.13) | 1.0×10−5 | 1.93 (1.21, 3.08) | 0.006 |
| Romania | 2.52 (0.90, 7.09) | 0.08 | 2.06 (0.71, 5.97) | 0.18 |
| Spain | 3.3 (0.95, 11.45) | 0.06 | 3.31 (0.92, 11.86) | 0.066 |
| UK | 4.22 (2.24, 7.96) | 8.8×10−6 | 3.20 (1.66, 6.15) | 4.9×10−4 |
| All combined | 2.97 (2.42, 3.65) | 3.4×10−25 | 2.15 (1.71, 2.70) | 7.0×10−11 |
| Phet = 0.89 | I2 = 0.0 | Phet = 0.81 | I2 = 0.0 | |
Shown are association results (the allelic odds-ratio (OR) with 95% confidence interval (95% CI) and P value) for rs188140481 adjusted and unadjusted for the 9 previously published anchor prostate cancer risk SNPs in populations of European descent and located on 8q24 (rs1254366314, rs1008690811,14, rs101634314, rs1325229814, rs169019798, rs1690209412, rs44511412/rs62086113,14, rs69832679, rs14472957). Only individuals directly genotyped for rs188140481 were used in this analysis.
We examined rs188140481 and the four correlated (r2 > 0.7) variations for known functional features from The Encyclopedia of DNA Elements (ENCODE) project. We noted that rs183373024 is located at a site of DNaseI hypersensitivity in prostate adenocarcinoma and normal prostate cell lines (LNCaP and PrEC; Supplementary Fig.1). Whether that has any biological significance remains to be studied further. Other correlated variants were not found at sites predicted to have a biological effect.
Recently, the rare missense mutation G84E (rs138213197[T]) in HOXB13 was found to confer risk of prostate cancer in a US study population3. In order to assess the HOXB13 association signal in study populations independent of the original report, we genotyped rs138213197 in five study groups of European descent. Combination of the imputed Icelandic results and the results from the five directly genotyped replication study groups gave an estimated OR of 7.06 (Pcomb. = 1.5×10−19; Table 3). This effect is comparable but somewhat lower than the effect reported in the initial study3. We also genotyped rs138213197 in 498 Asian prostate cancer patients from Hong Kong and 467 African American controls from Chicago but we did not detect the risk allele in these individuals, suggesting that either is the risk variant very rare or not present in these two ethnic groups.
Table 3.
Summary association results for rs138213197[T] in HOXB13 and prostate cancer
| Frequency |
||||||
|---|---|---|---|---|---|---|
| Study population |
Cases (n) |
Control s (n) |
Cases | Control s |
OR (95% CI) | P value |
| Iceland | 4,537 | 54,444 | 0.0029 | 0.00080 | 3.67 (1.71, 7.90) | 8.8×10−4 |
| Chicago | 1,982 | 1,260 | 0.0058 | 0.00040 | 14.70 (3.59, 60.14) | 1.8×10−4 |
| Spain | 716 | 1,692 | 0.00070 | 0.0 | inf | 0.30 |
| The Netherlands | 1,520 | 1,916 | 0.015 | 0.0021 | 7.51 (3.99, 14.11) | 3.9×10−10 |
| Romania | 722 | 857 | 0.0014 | 0.0012 | 1.19 (0.00, inf) | 1.0 |
| UK | 511 | 1,825 | 0.012 | 0.00082 | 14.44 (4.74, 44.03) | 2.7×10−6 |
| All excl. Iceland | 5,451 | 7,550 | - | 0.00089 | 9.41 (5.66, 15.64) | 5.1×10−18 |
| All combined | 9,988 | 61,994 | - | 0.00088 | 7.06 (4.62, 10.78) | 1.5×10−19 |
| Phet = 0.36 | I2 = 8.6 | |||||
All P values shown are two-sided. Shown are the corresponding numbers of cases and controls (n), allelic frequencies of variants in affected and control individuals, the allelic odds-ratio (OR) with 95% confidence interval (95% CI) and P value. Also shown are the P-values for the heterogeneity of the ORs (Phet) for all study groups as well as I2 which lies between 0% and 100% and describes the proportion of total variation in study estimates that is due to heterogeneity. For the combined study populations, the reported control frequency was the average, unweighted control frequency of the individual populations, while the OR and the P value were estimated using the Mantel-Haenszel model. Of the Icelandic cases, 2,315 patients had been genotyped using one of the Illumina chips, and 2,222 additional patients had at least partial data based on family based imputation. Of the Icelandic controls 27,780 were imputed based on chip-genotypes and 26,664 were family based imputed. All non-Icelandic replication samples are directly genotyped.
In order to analyze further the new variant on 8q24 and the HOXB13 mutation we stratified the patients from the six European study groups according to age at diagnosis and aggressiveness of the disease. For rs188140481[A] at 8q24, when comparing the group of patients with a more aggressive phenotype (Gleason ≥7 and/or T3 or higher and/or node positive and/or metastatic disease) to the group with less aggressive tumors (Gleason <7 and T2 or lower), a non-significant trend towards being diagnosed with an advanced disease was observed (OR = 1.30, Pcomb. = 0.08; Supplementary Table 4a). The same type of analysis for the HOXB13 mutation resulted in an OR = 1.05 for the more aggressive phenotype (Pcomb. = 0.90; Supplementary Table 4b). When stratifying according to age at diagnosis, carriers of allele A of rs188140481on 8q24 were found to be diagnosed 1.26 years younger than non-carriers (Pcomb = 0.0059, Supplementary Table 5a), and carriers of the HOXB13 mutation (rs138213197[T]) were found to be diagnosed 1.16 years younger than non-carriers (Pcomb =0.098; Supplementary Table 5b).
When examining the genotypic status of all subjects in our study, one subject was found to be homozygous for the HOXB13 mutation; a Dutch prostate cancer patient diagnosed at the age of 69 and with a positive family history of prostate cancer (two brothers diagnosed at the age of 67 and 74, but no DNA samples were available). In the Icelandic study group, we identified 13 Icelandic male carriers of the HOXB13 mutation who have been diagnosed with prostate cancer. Thereof, four patients have a positive family history of prostate cancer, two belong to a brother pair, and two belong to a father-offspring pair. The remaining nine patients do not have any 1st or 2nd degree male relatives diagnosed with prostate cancer, based on deCODE’s extensive genealogy database and the nationwide ICR database.
We identified 16 homozygous carriers of the 8q24 variant: a Dutch prostate cancer patient diagnosed at the age of 69 and not known to have a positive family history of prostate cancer; 8 Icelandic females and 7 Icelandic males. Of the 7 Icelandic males 5 have been diagnosed with prostate cancer (age at diagnosis ranges between 55 and 87 years) and the remaining two are both born in 1953 (current age is 59 years) and not known to have been diagnosed with prostate cancer. The 5 Icelandic homozygous prostate cancer patients (of whom four belong to two brother pairs) all have one or more close relatives within the nuclear family diagnosed with prostate cancer (Supplementary Fig. 2). This demonstrates a high prevalence of the disease among homozygous male carriers over 60 years of age. Furthermore, using directly generated genotypes only, the carrier frequency among Icelandic patients with a positive family history was estimated to be 10.9% compared to 4.2% among patients without a family history (OR = 2.70, P = 1.8×10−5).
In summary, we have discovered a new independent prostate cancer risk variant at the 8q24 locus. The risk profile of this new variant differs from other GWAS-based discovered risk variants in being rare and conferring greater risk of the disease. The replication results for the HOXB13 coding variant presented above, confirm it as a prostate cancer risk variant in populations from across Europe.
Online Methods
Study populations
For a description of the study populations see Supplementary Note.
Genotyping Methods
Illumina genome-wide genotyping
The Icelandic chip-typed samples were assayed with the Illumina Human Hap300, Hap CNV370, Hap 610, 1M or Omni-1 Quad bead chips at deCODE genetics. SNPs were excluded if they had (i) yield less than 95%, (ii) minor allele frequency less than 1% in the population or (iii) significant deviation from Hardy-Weinberg equilibrium in the controls (P < 0.001), (iv) if they produced an excessive inheritance error rate (over 0.001), (v) if there was substantial difference in allele frequency between chip types (from just a single chip if that resolved all differences, but from all chips otherwise). All samples with a call rate below 97% were excluded from the analysis. The final set of SNPs used for long-range phasing was composed of 785,863 SNPs.
Single track assay SNP genotyping
Genotyping of the SNP reported in Tables 1–3 of the main text for the five case-control groups from the Netherlands, Spain, Romania, the UK and the US was carried out by deCODE Genetics in Reykjavik, Iceland, applying the Centaurus22 (Nanogen) platform. Using the Centaurus single-track assay, we genotyped all cases and controls available from these study populations. For confirming the imputed Icelandic association results for rs188140481on 8q24 and rs138213197 in HOXB13, we directly genotyped over 3.900 Icelandic study subjects using Centaurus single-track assay for these SNPs. The correlation (r2) between the results of two genotyping methods for these two SNPs was >0.94.
Sanger Sequencing
For confirming the genotypes from the Centaurus single-track assay genotyping, using standard procedure, we Sanger sequenced all carriers of the HOXB13 mutation identified in all the study populations as well as all homozygous carriers and 25 heterozygous Icelandic carriers of the 8q24 variant rs188140481. Concordance rate between the different genotyping platforms was above 98%.
Whole-genome sequencing and SNP imputations
Of the 1,795 individuals whole-genome sequence and used in the current study, 53 have been diagnosed with prostate cancer according to the nationwide list maintained by the Icelandic Cancer Registry. For a more detailed description of the key steps of the whole-genome sequencing and imputation see Supplementary Note.
Bioinformatics
For the list of SNPs correlated with rs188140481 we carried out a search for overlaps between SNP position and known bioinformatic features. We retrieved data from UCSC test browser (HG19 build 37). We accessed all feature tracks relevant to the prostate tissue as well as containing genome positional information, and identified those features that overlapped with the SNPs. These are recognized draft quality data and were used as is without quality filtering.
Statistical analysis
Long range phasing
Long range phasing of all chip-genotyped individuals was performed with methods described previously4,23–26. In brief, phasing is achieved using an iterative algorithm which phases a single proband at a time given the available phasing information about everyone else that shares a long haplotype identically by state with the proband. Given the large fraction of the Icelandic population that has been chip-typed, accurate long range phasing is available genome-wide for all chip-typed Icelanders.
Genotype imputation
We imputed the SNPs identified and genotyped through sequencing into all Icelanders who had been phased with long range phasing using the same model as used by IMPUTE27 (see Supplementary Note for a more detailed description).
In-silico genotyping
In addition to imputing sequence variants from the whole genome sequencing effort into chip genotyped individuals, we also performed a second imputation step where genotypes were imputed into relatives of chip genotyped individuals, creating in-silico genotypes. The inputs into the second imputation step are the fully phased (in particular every allele has been assigned a parent of origin) imputed and chip type genotypes of the available chip typed individuals (see Supplementary Note for a detailed description).
Genotype imputation information
The informativeness of genotype imputation was estimated by the ratio of the variance of imputed expected allele counts and the variance of the actual allele counts. For the present study, when imputed genotypes are used, the information value for all SNPs is between 0.92 and 0.99.
Case control association testing
Logistic regression was used to test for association between SNPs and disease, treating disease status as the response and expected genotype counts from imputation or allele counts from direct genotyping as covariates. Testing was performed using the likelihood ratio statistic (for a detailed description of the genotyping, sequencing and statistical methods see Supplementary Note and Ref.24–26).
Multivariate logistic regression
For this analysis we only used cases and controls which have been genotyped using the Illumina chip-genotyping platform. The multivariate logistic regression analysis was performed conditioning for a given marker by adjusting for the estimated allele count based on imputation of this marker. The genomic control correction factor is the same as used for the unadjusted association analysis. A forward selection multiple logistic regression model was used to further define the extent of the genetic association. Briefly, all imputed SNPs located within the interval 127.8 Mb to 128.8 on 8q24 were tested for possible incorporation into a multiple regression model. In stepwise fashion, a SNP was added to the model if it had the smallest P-value among all SNPs not yet included in the model and if it had a P-value below 1.5×10−9 (a GWAS significance threshold applicable according to a Bonferroni correction of 32 million tests). In the last step none of the SNPs remained significant at this threshold.
Inflation factor adjustment
In order to account for the relatedness and stratification within our case and control sample sets we applied the method of genomic control based on chip markers. For prostate cancer GWAS the correction factor based on the genomic control is 1.33.
Supplementary Material
ACKNOWLEDGMENTS
We thank the individuals who participated in the study and whose contribution made this work possible. This project was funded in part by contract number 202059 (PROMARK) from the 7th Framework Program of the European Union, and in part by the Urological Research Foundation, U01 CA089600, P50 CA90386, P30 CA60553. The UK ProtecT study is ongoing and is funded by the Health Technology Assessment Programme (projects 96/20/06, 96/20/99). The ProtecT trial is supported by Department of Health, England; Cancer Research UK grant number C522/A8649; Medical Research Council of England grant number G0500966; ID 75466; the National Cancer Research Institute (NCRI), UK; and the Southwest National Health Service Research and Development. The bio-repository from ProtecT is supported by the NCRI (ProMPT) study and the Cambridge and Oxford British Medical Research Council grants from National Institute for Health Research.
Footnotes
AUTHOR CONTRIBUTION
The study was designed and results were interpreted by J.G., P.S., A.K., U.T., T.R., and K.S. Statistical analysis was carried out by P.S., D.F.G., G.T., J.G., and A.K. Subject recruitment, biological material collection and handling along with genotyping was supervised and carried out by J.G., B.A., K.R.B., S.N.S., A.S., S.A.G, H.J., H.Th.H., A.J., S.B.O., J.G.J., L.T., S.N., F.F., B.T.H., Q.H., I.E.C., I.N.M., V.J., K.K.H.A., I.M.vO., S.H.V., J.L.D., F.C.H., C-F.N., P.K.F.C., K-M.L., M.C.Y.N., J.R.G., G.M., W.J.C., J.I.M., G.V.E., R.B.B., E.J., D.M., D.A.N., L.A.K., U.T. and T.R. Authors J.G., P.S., T.R. and K.S. drafted the manuscript. All authors contributed to the final version of the paper. Principal investigators and corresponding authors for the respective replication study populations are: The Netherlands, L.A.K.; Spain, J.I.M.; Romania, D.M.; Chicago, W.J.C; Hong Kong, C-F.N., and M.C.Y.N., the UK, F.C.H., J.L.D., D.E.N.
COMPETING INTERESTS STATEMENT
The authors from deCODE genetics declare competing financial interests.
URLs
Catalog of published Genome-Wide Association Studies (http://www.genome.gov/gwastudies/) The Icelandic Cancer Registry (http://www.krabbameinsskra.is/indexen.jsp?icd=C61)
References
- 1.Lichtenstein P, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 2.Amundadottir LT, et al. Cancer as a Complex Phenotype: Pattern of Cancer Distribution within and beyond the Nuclear Family. PLoS Med. 2004;1:e65. doi: 10.1371/journal.pmed.0010065. Epub 2004 Dec 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ewing CM, et al. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366:141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong A, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat Genet. 2008;40:1068–1075. doi: 10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gudmundsson J, et al. Discovery of common variants associated with low TSH levels and thyroid cancer risk. Nat Genet. 2012;44:319–322. doi: 10.1038/ng.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gudmundsson J, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 7.Amundadottir LT, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 8.Gudmundsson J, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 9.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 10.Haiman CA, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng SL, et al. Association between two unlinked loci at 8q24 and prostate cancer risk among European Americans. J Natl Cancer Inst. 2007;99:1525–1533. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]
- 12.Gudmundsson J, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeager M, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet. 2009;41:1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al Olama AA, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41:1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 15.Crowther-Swanepoel D, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat Genet. 2010;42:132–136. doi: 10.1038/ng.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomlinson I, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 17.Zanke BW, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 18.Haiman CA, et al. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiemeney LA, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Witte JS. Multiple prostate cancer risk variants on 8q24. Nat Genet. 2007;39:579–580. doi: 10.1038/ng0507-579. [DOI] [PubMed] [Google Scholar]
- 22.Kutyavin IV, et al. A novel endonuclease IV post-PCR genotyping system. Nucleic Acids Research. 2006;34:e128. doi: 10.1093/nar/gkl679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong A, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 24.Sulem P, et al. Identification of low-frequency variants associated with gout and serum uric acid levels. Nat Genet. 2011;43:1127–1130. doi: 10.1038/ng.972. [DOI] [PubMed] [Google Scholar]
- 25.Rafnar T, et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nat Genet. 2011;43:1104–1107. doi: 10.1038/ng.955. [DOI] [PubMed] [Google Scholar]
- 26.Stacey SN, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011;43:1098–1103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.