Abstract
BACKGROUND
Recent whole genome association studies have independently identified multiple prostate cancer (PC) risk variants on 8q24. We have evaluated association of common variants in this region with PC susceptibility and tumor aggressiveness in a sample of European American men.
METHODS
Forty-nine tagging SNPs including three previously reported significant variants (rs1447295, rs6983267, rs16901979) and seven variants in the 5′ upstream region of the MYC proto-oncogene were tested for association with susceptibility to PC and tumor aggressiveness in 596 histologically verified PC cases and 567 ethnically matched controls.
RESULTS
Significant associations with susceptibility to PC were found at 17 SNPs, four of which (rs1016342, rs1378897, rs871135 and rs6470517) remained significant after adjusting for multiple corrections. One of the associated SNPs, rs871135, is located in the putative gene POU5F1P1 within the 8q24 region. An in slico analysis showed that the associated variant of this SNP alters a transcription factor implicating a plausible regulatory role. Additionally, one of the significantly associated SNPs, rs6470517, with PC susceptibility showed a significant over-representation of the G allele in cases with aggressive tumor.
CONCLUSIONS
Although this study does not directly confirm associations of the three specific SNPs (cited above), it corroborates reported signals of association in 8q24 reaffirming that genetic variation on 8q24 influences susceptibility to PC in men of European ancestry. Although our study did not confirm the allelic association of rs1447295, meta-analysis of this SNP provided support to previous reported associations. Further, this study implicates the 8q24 region with aggressive forms of PC.
Keywords: Genetic susceptibility, association study, tagging SNPs, chromosome 8q24, MYC proto-oncogene
INTRODUCTION
Chromosome 8q24 has emerged as a potentially important region in prostate cancer (PC) genetics. Several complementary studies involving linkage, admixture mapping, and whole genome associations have identified multiple risk variants in this region associated with susceptibility to PC [1 – 6]. These results have been corroborated in multiple follow-up studies involving populations of diverse origins [7 – 14]. Importantly this region is also implicated in colorectal, breast and ovarian cancer [8, 13, 15 – 17]. These findings underscore that genetic variation in 8q24 is involved in multiple cancer types.
Based on the first genome wide association studies, three contiguous regions (Region 1, 3 and 2) spanning ~600-kb of DNA (128.10Mb to 128.70Mb on 8q24) were defined harboring variants associated with PC susceptibility [3 – 6]. Three single nucleotide polymorphisms (SNPs), rs1447295 (Region 1), rs6983267 (Region 3), rs16901979 (Region 2) were of considerable interest because of their remarkably strong independent associations with PC and most of the follow-up studies included these SNPs, particularly rs1447295. However, screening only these SNPs could have constituted an inadequate assessment of the overall spectrum of genetic association for the entire 8q24 region. To overcome this limitation, we employed a tagging approach for a comprehensive evaluation of association of common genetic variants in 8q24 with PC in a sample of European-American men. With the presence of one putative gene POU5F1P1 of unknown function, the 8q24 region is gene-poor. However, the MYC proto-oncogene, a biologically plausible candidate for cancers is located ~240-kb telomeric to the associated region. It is possible that 8q24 contains as yet unknown regulatory sequences for MYC, which may lead to the initiation of a carcinogenic cascade. We tested for association of 49 SNPs spanning 8q24 (including the three previously reported significant variants, rs1447295, rs6983267, rs16901979) and the 5′ upstream region of MYC. We found significant associations at 17 SNPs, four of which remained significant after adjusting for multiple corrections (P ~10−3-10−4) – two in Region 2 (rs1016342, rs1378897), one each in Region 3 (rs871135) and 1 (rs6470517). These results reaffirm that genetic variation on 8q24 influences susceptibility to PC in men of European ancestry.
Variants at 8q24 have been evaluated for risks of aggressiveness of PC based on age at onset, familial aggregation and tumor grades and stages [1 – 4, 7, 8, 11, 12]. While most studies have reported that the aggressive forms of the disease are influenced by the same variants associated with susceptibility to PC, few have not. We have explored this further by testing for association of all 49 tagging SNPs with PC grades defined by Gleason score and tumor stage as indices of biologic aggressiveness. We found the G allele at rs6470517, a Region 1 SNP, is significantly over-represented in cases with aggressive tumor. This corroborates previous findings that 8q24 variants are involved in poorly-differentiated forms of PC.
MATERIALS AND METHODS
Study subjects
The details of the case enrollment were reported previously [18 – 21]. Briefly, 596 men of European descent with histologically verified PC were recruited from 304 families, of which, sixty-two were singletons, three first cousins, six half-sibs, and the remainder were derived from multiplex affected full sibships. These cases were ascertained from patients seen at the Washington University School of Medicine from 1989 to 2001 by staff urologists. The age at diagnosis ranged from 40 to 91 years. Control subjects, consisting of 567 unrelated Caucasian men, were recruited from the same area of residence as the cases and during the same time frame, and were followed for several years as part of a long-term prostate cancer screening study in which PSA blood test and digital rectal examination (DRE) were performed at 6 to 12 month intervals [22] and met the following criteria: (a) at least 65 years old, (b) never had registered a PSA level >2.5 ng/ml, (c) never had DRE suspicious for prostate cancer, and (d) had no known family history of prostate cancer. PSA values used for association were the highest values prior to diagnosis. The protocol for this study was approved by the Human Studies Committee of Washington University and the Institutional Review Board of the University of Cincinnati. Written informed consent was obtained from all participants.
SNP selection and DNA analysis
The markers were selected from the three defined regions, including the three previously reported significant SNPs, rs16901979 in Region 2, rs6983267 in Region 3, and rs1447295 in Region 1. Region 2 was further saturated by 13 additional SNPs that constitute HapC [3]. Twenty-eight haplotype tagging SNPs, nine from Region 3 and 19 from Region 1 were identified. Seven pairwise tagging SNPs were selected spanning ~40-kb from the 5′ upstream region of MYC. The SNPs were selected by a tagging approach [23, 24] using the Caucasian HapMap database (www.hapmap.org) based on pairwise and/or haplotype r2 (≥0.8) among all common SNPs with minor allele frequency (MAF ≥0.05).
All of the SNPs except rs1447295 were genotyped on the Applied Biosystems (ABI) SNPlex platform. TaqMan™ assay was used to genotype rs1447295, which could not be designed in the SNPlex primer pool. SNPlex and TaqMan primers and probes were obtained from ABI. Details of the genotyping protocols are described previously [20, 21].
Statistical analysis
Allele frequencies in the related PC cases were estimated by a maximum likelihood method using the USERM13 subroutine of MENDEL which is specifically designed to take into account the relatedness of the samples [25, 26]. Allele frequencies in unrelated controls were computed by gene-counting. Conformity of genotype proportions to Hardy–Weinberg equilibrium (HWE) was performed by the exact test [27].
To test for association, allele frequencies in cases were compared with controls by a likelihood ratio test as described in Suarez et al. [19]. Unconditional logistic regression was performed using Helixtree ver 6.0.2 to estimate the age-adjusted odds ratios (OR) and their respective 95% confidence intervals (CI). As our cases were related, we used a permutation-based randomization method to select one sib from each sibship to obtain a representative unrelated case population for logistic regression analysis. Bonferroni correction was used to adjust for multiple testing.
Association with tumor aggressiveness and tumor staging
We tested for association of the 8q24 variants with PC grade, defined by Gleason score and tumor stage (clinico-pathological TNM staging) as indices for biologic aggressiveness. It has been reported previously that patients with Gleason score 7 and above have a higher risk of adverse outcomes from prostate cancer [28, 29]. Therefore, we stratified our cases into two groups with Gleason scores 2–6 and 7–10 defined as low- and high-risk prostate cancer cases, respectively. To detect association with tumor stages, we categorized the cases as low-risk (T1 and T2 tumors with no involvement of lymph nodes – N0 and/or no metastasis – M0) and high-risk (T3, T4, N1 or M1) groups [30]. As noted above, controls were ascertained from a long-term prostate cancer screening study with a minimal risk for susceptibility to prostate cancer, and thus constitute a well represented low-risk population. We compared allele frequencies in controls with the high- and low-risk case groups to test for association with PC aggressiveness.
RESULTS
Allelic association with PC susceptibility
Genomic locations of the 49 SNPs along with their minor allele frequencies in cases and controls are given in Table 1. All of the SNPs are in HWE (data not shown) and polymorphic (MAF ≥0.05) in both cases and controls except for rs16901979 (MAF 0.04 and 0.03, in cases and controls, respectively). We found significant allele frequency differences (likelihood ratio χ2) between cases and controls at 17 SNPs, of which five are located in Regions 2 and 3 each, six in Region 1 and one in the upstream MYC region. However, after adjusting for multiple testing using Bonferroni correction, four SNPs (rs1016342, rs1378897, rs871135 and rs6470517) remained significant (P = 0.01 to 0.00001).
TABLE 1.
Allelic association of 49 SNPs with prostate cancer. Significant SNPs (adjusted for p-values and/or significant OR) are highlighted in italics.
| dbSNP ID (rs) | Region | Position | Variation and Associated Allele | MAF | LR P | Adjusted P | OR Log-additive (95% CI) | ||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Control | ||||||||
| N = 596 | N = 567 | ||||||||
| rs1456314 | 2 | 128144872 | A→G | G | 0.28 | 0.32 | 0.071 | - | 1.0 (0.76–1.33) |
| rs17831626 | 2 | 128149605 | G→T | T | 0.49 | 0.45 | 0.025 | - | 1.15 (0.88–1.50) |
| rs6993569 | 2 | 128153279 | G→A | G | 0.86 | 0.84 | 0.160 | - | 1.09 (0.76–1.56) |
| rs6994316 | 2 | 128153721 | A→G | G | 0.27 | 0.26 | 0.718 | - | 1.07 (0.80–1.43) |
| rs6470494 | 2 | 128157086 | C→T | T | 0.27 | 0.27 | 0.954 | - | 1.0 (0.75–1.35) |
| RS1016342 | 2 | 128161637 | C→T | C | 0.65 | 0.59 | 9.05×10−6 | 4.3×10−4 | 1.26 (1.01–1.65) |
| RS1016343 | 2 | 128162479 | C→T | T | 0.24 | 0.19 | 1.0×10−3 | 0.076 | 1.55 (1.13–2.13) |
| rs1456306 | 2 | 128185682 | C→T | C | 0.73 | 0.71 | 0.493 | - | 1.32 (0.97–1.79) |
| RS1378897 | 2 | 128191841 | G→A | G | 0.96 | 0.93 | 8.0×10−4 | 0.038 | 1.26 (0.91–2.25) |
| rs16901979 | 2 | 128194098 | C→A | A | 0.04 | 0.03 | 0.285 | - | 1.29 (0.83–2.01) |
| rs10505482 | 2 | 128195031 | T→C | C | 0.29 | 0.28 | 0.678 | - | 1.01 (0.75–1.35) |
| RS1456305 | 2 | 128196434 | T→C | T | 0.93 | 0.90 | 4.8×10−3 | 0.23 | 0.656 (0.48–0.895) |
| rs17184796 | 2 | 128197641 | G→T | T | 0.38 | 0.37 | 0.691 | - | 1.07 (0.81–1.234) |
| rs7816535 | 2 | 128206850 | G→A | A | 0.22 | 0.21 | 0.178 | - | 1.12 (0.86–1.68) |
| rs750816 | 3 | 128460719 | C→G | G | 0.24 | 0.21 | 0.068 | - | 1.22 (0.99–1.49) |
| rs1949808 | 3 | 128463720 | G→C | C | 0.27 | 0.26 | 0.729 | - | 1.04 (0.86–1.26) |
| RS6986543 | 3 | 128465498 | C→A | A | 0.41 | 0.36 | 0.024 | - | 1.27 (1.03–1.46) |
| rs1562871 | 3 | 128470954 | A→G | G | 0.16 | 0.17 | 0.618 | - | 0.94 (0.75–1.18) |
| rs10441525 | 3 | 128472135 | T→C | C | 0.13 | 0.11 | 0.208 | - | 1.17 (0.90–1.52) |
| RS10956365 | 3 | 128473069 | G→A | G | 0.85 | 0.82 | 0.032 | - | 1.02 (0.73–1.44) |
| RS3847136 | 3 | 128476372 | G→A | A | 0.33 | 0.28 | 6.1×10−3 | 0.293 | 1.24 (1.07–1.54) |
| rs6983267 | 3 | 128482487 | G→T | G | 0.54 | 0.49 | 0.044 | - | 1.03 (0.79–1.34) |
| RS871135 | 3 POU5F1P1 |
128495575 | G→T | G | 0.49 | 0.41 | 3.0×10−4 | 0.014 | 1.39 (1.17–1.65) |
| rs10090421 | 1 | 128522947 | A→G | G | 0.21 | 0.20 | 0.469 | - | 1.05 (0.86–1.29) |
| rs12334695 | 1 | 128523110 | T→C | C | 0.39 | 0.38 | 0.142 | - | 1.09 (0.92–1.29) |
| rs7012462 | 1 | 128526872 | C→T | T | 0.42 | 0.40 | 0.215 | - | 1.11 (0.93–1.32) |
| rs4871791 | 1 | 128527826 | C→T | C | 0.53 | 0.50 | 0.274 | - | 1.26 (0.97–1.63) |
| RS6470517 | 1 | 128529586 | A→G | G | 0.22 | 0.16 | 1.6×10−5 | 7.6×10−4 | 1.58 (1.28–1.96) |
| RS10094059 | 1 | 128530789 | G→C | G | 0.22 | 0.26 | 0.109 | - | 0.81 (0.67–0.98) |
| rs7841264 | 1 | 128535996 | C→T | T | 0.19 | 0.18 | 0.731 | - | 1.04 (0.82–1.32) |
| RS10099905 | 1 | 128537116 | C→A | C | 0.17 | 0.21 | 0.023 | - | 0.79 (0.64–0.98) |
| rs10094871 | 1 | 128541151 | A→G | G | 0.21 | 0.21 | 0.409 | - | 1.04 (0.85–1.274) |
| RS1447293 | 1 | 128541502 | A→G | G | 0.43 | 0.39 | 0.021 | - | 1.29 (1.0–1.68) |
| rs921146 | 1 | 128544367 | A→C | C | 0.13 | 0.15 | 0.350 | - | 0.88 (0.67–1.15) |
| rs1447295 | 1 | 128554220 | C→A | A | 0.13 | 0.11 | 0.332 | - | 1.43 (0.96–2.14) |
| rs9297758 | 1 | 128555770 | G→A | G | 0.62 | 0.61 | 0.532 | - | 1.27 (0.95–1.68) |
| RS13260378 | 1 | 128557932 | G→T | T | 0.42 | 0.48 | 0.011 | 0.518 | 0.78 (0.66–0.92) |
| rs10956373 | 1 | 128559758 | T→C | C | 0.25 | 0.21 | 0.019 | 0.912 | 1.24 (1.02–1.51) |
| RS6985504 | 1 | 128565958 | G→A | A | 0.31 | 0.27 | 0.011 | 0.518 | 1.39 (1.04–1.84) |
| rs13258548 | 1 | 128566029 | A→G | A | 0.67 | 0.65 | 0.928 | - | 1.03 (0.79–1.35) |
| rs723555 | 1 | 128569281 | A→G | G | 0.19 | 0.19 | 0.831 | - | 1.01 (0.82–1.26) |
| rs16902173 | 1 | 128573181 | A→G | G | 0.15 | 0.16 | 0.839 | - | 0.97 (0.765–1.22) |
| rs6984323 | upstream MYC | 128776090 | T→C | T | 0.53 | 0.49 | 0.140 | - | 1.49 (1.11–2.00) |
| rs12547643 | MYC | 128782355 | G→A | A | 0.35 | 0.33 | 0.439 | - | 1.18 (0.88–1.58) |
| rs9642880 | MYC | 128787250 | G→T | T | 0.47 | 0.44 | 0.150 | - | 1.157 (0.96–1.39) |
| RS17186926 | MYC | 128787625 | A→G | G | 0.22 | 0.17 | 6.0×10−3 | 0.288 | 1.34 (1.08–1.67) |
| rs16902359 | MYC | 128812033 | C→T | T | 0.09 | 0.09 | 0.523 | - | 0.96 (0.72–1.27) |
| rs3891248 | intron 1 MYC | 128819321 | T→A | A | 0.13 | 0.13 | 0.907 | - | 1.04 (0.79–1.35) |
| rs4645959 | nsSNP MYC | 128819722 | A→G | G | 0.05 | 0.06 | 0.542 | - | 0.89 (0.63–1.27) |
We did not find significant association with any of the three previously reported strongly significant SNPs (rs16901979 in Region 2, rs6983267 in Region 3, and rs1447295 in Region 1). Minor allele frequencies at rs16901979 are <0.05 in the cases and controls, and our sample size is not large enough with adequate power to capture signals of association. However, we found significant association with two other Region 2 SNPs, rs1378897 (adjusted P = 0.04) and rs1016342 (adjusted P = 4.3×10−4), which are 2.3kb and 32.5kb upstream of rs16901979, respectively. Marginal association was found with rs6983267 (unadjusted P = 0.04); however, after Bonferroni correction, the significance did not survive and also, the OR was not significant. However, the immediate downstream SNP (13kb telomeric), rs871135 showed significant association (adjusted P = 0.01; OR = 1.39, 95% CI: 1.17–1.65). Interestingly, this SNP is located in the putative gene POU5F1P1. Also, rs3847136, which is 6kb upstream of rs6983267, showed a modest level of association (unadjusted P = 6.1×10−3; OR = 1.24, 95% CI: 1.07–1.54), although the corrected P value is not significant. With respect to association of Region 1 variants, rs6470517 showed significant association (adjusted P = 7.6×10−4; OR = 1.58, 95% CI: 1.28–1.96), which is located 24.6kb upstream of rs1447295. Taken together, although our study did not replicate the significance of the three specific SNPs reported previously, it reaffirms that genetic variation at 8q24 influences susceptibility to prostate cancer. Of the seven tagSNPs in the upstream region of MYC, rs17186926 showed an unadjusted significant P value (6.0×10−3) and OR of 1.34, 95% CI: 1.08–1.67. However, after correction for multiple testing, the statistical significance did not remain (P = 0.29).
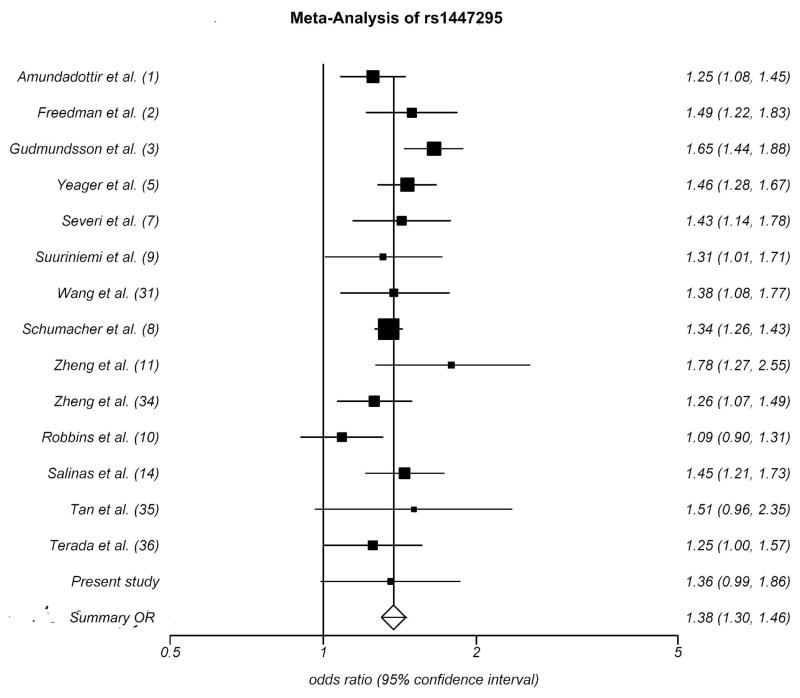
Meta-analysis of rs1447295
Among the 8q24 variants, rs1447295 is the most commonly studied marker and its association with PC susceptibility has been consistently strong. Although we did not find significant association, we performed a meta-analysis of this SNP using the published data to date (Fig 1). The R package “meta” ver 0.8–2 (http://cran.r-project.org/web/packages/meta/index.html) was used for this analysis. Allelic odds were computed for each of the published studies, and estimated odds ratios were weighted by the inverse variance random-effect model approach. The analysis showed a significant association of rs1447295 with PC susceptibility (two-tailed P ≤10−4) with a summary OR of 1.38 (95% CI: 1.30–1.46). The meta-analysis was expectedly driven by the largest follow-up study by Schumacher et al. [8]. The estimated OR for our study sample was 1.36 (95% CI: 0.99–1.84), which is similar to the summary OR yielded by the meta-analysis (1.37; 95% CI: 1.3–1.48).
FIGURE 1.
Meta-Analysis of rs1447295. The x-axis represents the per-allele odds ratio, each row represents one published study. The area of the square for each study is proportional to the inverse of variance of odds ratio estimates, and the corresponding horizontal lines represent 95% confidence intervals. The diamond at the bottom row represents summary odds ratio (1.38) and the width of the diamond represent 95% confidence intervals (1.30–1.46).
Association of the 8q24 variants with tumor aggressiveness
As described in the Methods, the cases were divided into groups based upon Gleason scores (low: 2–6 and high: 7–10) and TNM staging (localized: T1, T2, N0 or M0 and advanced: T3, T4, N1 or M1). We compared allele frequency differences at all 49 SNPs between each of these groups and the controls to test for association with tumor aggressiveness (Fig 2). We found a significant association of rs6470517, a Region 1 SNP, with aggressive phenotypes defined by both Gleason score and TNM staging. The minor allele (G) of this SNP was significantly over-represented in the aggressive cases (P = 10−4-10−5). As noted above, this SNP also showed significant association with PC susceptibility.
FIGURE 2.
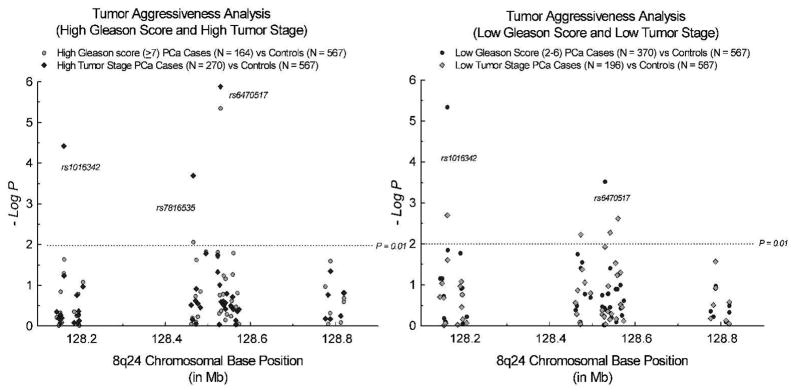
Association of the 8q24 variants with prostate cancer tumor grade and stage.
DISCUSSION
We have analyzed 49 SNPs spanning the 8q24 region in which linkage and association with PC susceptibility and aggressiveness has been reported. We observed significant allele frequency differences at 17 SNPs between PC cases and controls. However, after correcting for multiple tests, statistical significance remained only at four SNPs (rs1016342, rs1378897, rs871135, rs6470517). Using a similar approach, Salinas et al. (14) reported significant association with rs1016343 with PC susceptibility. Although in our study, the same SNP did not attain statistical significance after the Bonferroni multiple test correction (unadjusted P = 0.001; adjusted P = 0.07), we observed association with rs1016342 (unadjusted P = 9.05×10−6; adjusted P = 4.3×10−4), which is located 824 kb upstream of rs1016343. We did not find association with the three previously reported SNPs (rs16901979, rs6983267, rs1447295) that had shown strong association with PC susceptibility in several studies [3 – 5]. However, we found two significantly associated Region 2 SNPs, one of which (rs1016342) is located very close (2.3kb upstream) to rs16901979. The Region 3 SNP, rs871135 showed significant association, which is located 13kb downstream of rs6983267.
It is somewhat intriguing that we did not find association with rs1447295. This is the most strongly associated and consistently replicated SNP in studies involving samples of European descent. Among the African Americans, however, the association is not consistent [1 – 3, 10, 14]. It is likely that we failed to find significant association due to the relatively small sample size. Note that, as described in methods using permutation-based randomization to select one sib from each sibship to obtain a representative unrelated case sample for logistic regression analysis resulted in a reduced sample size. However, meta-analysis of rs1447295 showed that the OR in our sample was significant. Also, we found strong evidence of association of an upstream Region 3 SNP, rs6470517 (adjusted P = 7.6×10−4; OR = 1.58, 95% CI: 1.28–1.96). Taken together, these results indicate that the 8q24 region harbors multiple risk associated variants for prostate cancer. Failure to find association with previously reported specific variants could be attributed to several factors. First, these SNPs are not likely the causal variants rather these are in linkage disequilibrium (LD) with other risk associated variants; owing to population heterogeneity the nature of LD may vary from population to population. Second, the sample size of our study may lack statistical power to capture significant association at some markers. Nonetheless, use of tagging SNPs not only provided a comprehensive assessment of the region of interest, it guarded against the potential limitation of the power based on single or a small number of previously identified significant variants.
Previous studies have assessed the association of the 8q24 variants with age at onset, family history and clinical stage of PC. These investigations mostly confined to the variants that were strongly associated with risk of PC susceptibility. The results show that, in general, 8q24 variants are associated with earlier onset cases, positive family history and aggressive forms of PC classified by tumor grade and stage [1, 3, 4, 12, 31]. Few studies, however, did not indicate strong association of the risk variants with tumor stage or age at diagnosis [7, 8]. We performed a comprehensive analysis taking into consideration of all 49 markers spanning the entire 8q24 region. Our study showed a significant association of rs6470517 in Region 1 with aggressive phenotypes defined by both Gleason score and TNM staging. The minor allele (G) of this SNP was significantly over-represented in the aggressive cases (P = 10−4-10−5).
The 8q24 region is relatively gene-poor. The only known pseudogene, POU5F1P1, is located in the adjoining area of Regions 3 and 1. One of the significantly associated SNPs in our study, rs871135, is located in Intron 1 of POU5F1P1. We conducted an in silico analysis to search for consensus sequences for transcriptional factors (TF) in 8q24 (128.14Mb to 128.82Mb). Interestingly, we found that the ‘G’ allele at rs871135 alters the TF binding domain sequence, cyclic AMP response element binding protein (CRE-BP) (Table 2). Another TF binding domain, NKx-2.5 is also located 2bp 3′ to this SNP. This observation is noteworthy as recent studies suggested significantly increased CpG promoter methylation of NKx-2.5 transcription factor in prostate tumors [32]. Dysregulation of CRE-BP and activation of cyclic AMP response element binding was linked with progression to aggressive and possibly predispose to metastatic tumors [33]. These findings suggest a regulatory role of 8q24, particularly the region in and around POU5F1P1, in prostate carcinogenesis.
TABLE 2.
In silico sequence analysis on rs871135.
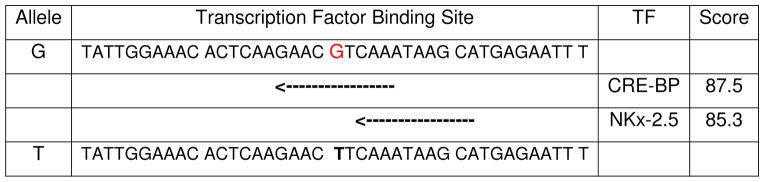
|
The MYC proto-oncogene is located ~230Kb telomeric to Region 1. We analyzed seven tagSNPs in the 5′ untranslated region of MYC. One of the SNPs, rs17186926, located ~30kb centromeric from the transcriptional start site had an unadjusted P value of 6.0×10−3. Although, the significance did not hold after multiple correction, the estimated OR (1.34, 95% CI: 1.08–1.67) marginally indicated the role of MYC in PC risk independently if not in conjunction with neighboring 8q24 variants. Previous studies have reported inconsistent associations of MYC variants with PC risk; three of these reported no association [1, 5, 11], while Salinas et al. [14] found association of a SNP, rs3891248 in intron 1 of MYC, with protective effect in Caucasian men with prostate cancer, but not among African Americans. Analysis of this SNP in our sample did not show association with PC susceptibility. The frequency of the A allele in both our cases and controls was 13%; the frequencies of this allele in cases and controls in Salinas et al.’s study were 13% and 15%, respectively. Overall these results suggest further work to evaluate the role of MYC in PC susceptibility.
Acknowledgments
This study was supported by a grant from the Urological Research Foundation.
References
- 1.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, Jakobsdottir M, Kostic J, Magnusdottir DN, Ghosh S, Agnarsson K, Birgisdottir B, Le Roux L, Olafsdottir A, Blondal T, Andresdottir M, Gretarsdottir OS, Bergthorsson JT, Gudbjartsson D, Gylfason A, Thorleifsson G, Manolescu A, Kristjansson K, Geirsson G, Isaksson H, Douglas J, Johansson JE, Bälter K, Wiklund F, Montie JE, Yu X, Suarez BK, Ober C, Cooney KA, Gronberg H, Catalona WJ, Einarsson GV, Barkardottir RB, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. A common variant associated with prostate cancer in African and European populations. Nature Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 2.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, Penney K, Steen RG, Ardlie K, John EM, Oakley-Girvan I, Whittemore AS, Cooney KA, Ingles SA, Altshuler D, Henderson BE, Reich D. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-Americans. Proc Natl Acad Sci USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Xu J, Blondal T, Kostic J, Sun J, Ghosh S, Stacey SN, Mouy M, Saemundsdottir J, Backman VM, Kristjansson K, Tres A, Partin AW, Albers-Akkers MT, Godino-Ivan Marcos J, Walsh PC, Swinkels DW, Navarrete S, Isaacs SD, Aben KK, Graif T, Cashy J, Ruiz-Echarri M, Wiley KE, Suarez BK, Witjes JA, Frigge M, Ober C, Jonsson E, Einarsson GV, Mayordomo JI, Kiemeney LA, Isaacs WB, Catalona WJ, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nature Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 4.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ, Greenway SC, Stram DO, Le Marchand L, Kolonel LN, Frasco M, Wong D, Pooler LC, Ardlie K, Oakley-Girvan I, Whittemore AS, Cooney KA, John EM, Ingles SA, Altshuler D, Henderson BE, Reich D. Multiple regions within 8q24 independently affect risk for prostate cancer. Nature Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hunter DJ, Chanock SJ, Thomas G. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nature Genet. 2007;39:646–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 6.Witte JS. Multiple prostate cancer risk variants on 8q24. Nature Genet. 2007;39:579–580. doi: 10.1038/ng0507-579. [DOI] [PubMed] [Google Scholar]
- 7.Severi G, Hayes VM, Padilla EJ, English DR, Southey MC, Sutherland RL, Hopper JL, Giles GG. The common variant rs1447295 on chromosome 8q24 and prostate cancer risk: results from an Australian population-based case-control study. Cancer Epidemiol Biomarkers Prev. 2007;16:610–612. doi: 10.1158/1055-9965.EPI-06-0872. [DOI] [PubMed] [Google Scholar]
- 8.Schumacher FR, Feigelson HS, Cox DG, Haiman CA, Albanes D, Buring J, Calle EE, Chanock SJ, Colditz GA, Diver WR, Dunning AM, Freedman ML, Gaziano JM, Giovannucci E, Hankinson SE, Hayes RB, Henderson BE, Hoover RN, Kaaks R, Key T, Kolonel LN, Kraft P, Le Marchand L, Ma J, Pike MC, Riboli E, Stampfer MJ, Stram DO, Thomas G, Thun MJ, Travis R, Virtamo J, Andriole G, Gelmann E, Willett WC, Hunter DJ, et al. A common 8q24 variant in prostate and breast cancer from a large nested case-control study. Cancer Res. 2007;67:2951–2956. doi: 10.1158/0008-5472.CAN-06-3591. [DOI] [PubMed] [Google Scholar]
- 9.Suuriniemi M, Agalliu I, Schaid DJ, Johanneson B, McDonnell SK, Iwasaki L, Stanford JL, Ostrander EA. Confirmation of a positive association between prostate cancer risk and a locus at chromosome 8q24. Cancer Epidemiol Biomarkers Prev. 2007;16:809–814. doi: 10.1158/1055-9965.EPI-06-1049. [DOI] [PubMed] [Google Scholar]
- 10.Robbins C, Torres JB, Hooker S, Bonilla C, Hernandez W, Candreva A, Ahaghotu C, Kittles R, Carpten J. Confirmation study of prostate cancer risk variants at 8q24 in African Americans identifies a novel risk locus. Genome Res. 2007;17:1717–1722. doi: 10.1101/gr.6782707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng SL, Sun J, Cheng Y, Li G, Hsu FC, Zhu Y, Chang BL, Liu W, Kim JW, Turner AR, Gielzak M, Yan G, Isaacs SD, Wiley KE, Sauvageot J, Chen HS, Gurganus R, Mangold LA, Trock BJ, Gronberg H, Duggan D, Carpten JD, Partin AW, Walsh PC, Xu J, Isaacs WB. Association between two unlinked loci at 8q24 and prostate cancer risk among European Americans. J Natl Cancer Inst. 2007;99:1525–1533. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]
- 12.Beebe-Dimmer JL, Levin AM, Ray AM, Zuhlke KA, Machiela MJ, Halstead-Nussloch BA, Johnson GR, Cooney KA, Douglas JA. Chromosome 8q24 markers: risk of early-onset and familial prostate cancer. Int J Cancer. 2008;122:2876–2879. doi: 10.1002/ijc.23471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, Pooley KA, Ramus SJ, Kjaer SK, Hogdall E, DiCioccio RA, Whittemore AS, Gayther SA, Giles GG, Guy M, Edwards SM, Morrison J, Donovan JL, Hamdy FC, Dearnaley DP, Ardern-Jones AT, Hall AL, O’Brien LT, Gehr-Swain BN, Wilkinson RA, Brown PM, Hopper JL, Neal DE, Pharoah PD, Ponder BA, Eeles RA, Easton DF, Dunning AM. UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology; UK ProtecT Study Collaborators. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salinas CA, Kwon E, Carlson CS, Koopmeiners JS, Feng Z, Karyadi DM, Ostrander EA, Stanford JL. Multiple independent genetic variants in the 8q24 region are associated with prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:1203–1213. doi: 10.1158/1055-9965.EPI-07-2811. [DOI] [PubMed] [Google Scholar]
- 15.Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, Kolonel LN, Wu AH, Reich D, Henderson BE. A common genetic risk factor for colorectal and prostate cancer. Nature Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E, Ferretti V, Laflamme P, Sundararajan S, Roumy S, Olivier JF, Robidoux F, Sladek R, Montpetit A, Campbell P, Bezieau S, O’Shea AM, Zogopoulos G, Cotterchio M, Newcomb P, McLaughlin J, Younghusband B, Green R, Green J, Porteous ME, Campbell H, Blanche H, Sahbatou M, Tubacher E, Bonaiti-Pellié C, Buecher B, Riboli E, Kury S, Chanock SJ, Potter J, Thomas G, Gallinger S, Hudson TJ, Dunlop MG. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nature Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 17.Fletcher O, Johnson N, Gibson L, Coupland B, Fraser A, Leonard A, Silva IDS, Ashworth A, Houlston R, Peto J. Association of genetic variants at 8q24 with breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:702–705. doi: 10.1158/1055-9965.EPI-07-2564. [DOI] [PubMed] [Google Scholar]
- 18.Burmester JK, Suarez BK, Lin JH, Jin CH, Miller RD, Zhang KQ, Salzman SA, Reding DJ, Catalona WJ. Analysis of candidate genes for prostate cancer. Hum Hered. 2004;57:172–178. doi: 10.1159/000081443. [DOI] [PubMed] [Google Scholar]
- 19.Suarez BK, Pal P, Jin CH, Kaushal R, Sun G, Jin L, Pasche B, Deka R, Catalona WJ. TGFBR1*6A is not associated with prostate cancer in men of European ancestry. Prostate Cancer Prostatic Dis. 2005;8:50–53. doi: 10.1038/sj.pcan.4500765. [DOI] [PubMed] [Google Scholar]
- 20.Pal P, Xi H, Kaushal R, Sun G, Jin CH, Suarez BK, Catalona WJ, Deka R. Variants in the HEPSIN gene are associated with prostate cancer in men of European origin. Hum Genet. 2006;12:187–192. doi: 10.1007/s00439-006-0204-3. [DOI] [PubMed] [Google Scholar]
- 21.Pal P, Xi H, Sun G, Meeks JJ, Thaxton CS, Guha S, Jin CH, Suarez BK, Catalona WJ, Deka R. Tagging SNPs in the Kallikrein genes 3 and 2 on 19q13 and their association with prostate cancer in men of European Origin. Hum Genet. 2007;122:251–259. doi: 10.1007/s00439-007-0394-3. [DOI] [PubMed] [Google Scholar]
- 22.Smith DS, Humphrey PA, Catalona WJ. The early detection of prostate carcinoma with prostate specific antigen: the Washington University experience. Cancer. 1997;80:1852–1856. [PubMed] [Google Scholar]
- 23.Weale ME, Depondt C, Macdonald SJ, Smith A, Lai PS, Shorvon SD, Wood NW, Goldstein DB. Selection and evaluation of tagging SNPs in the neuronal-sodium-channel gene SCN1A: implications for linkage-disequilibrium gene mapping. Am J Hum Genet. 2003;73:551–565. doi: 10.1086/378098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lange K, Weeks D, Boehnke M. Programs for pedigree analysis: MENDEL, FISHER and dGENE. Genet Epidemiol. 1988;5:471–472. doi: 10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]
- 26.Boehnke M. Allele frequency estimation from data on relatives. Am J Hum Genet. 1991;48:22–25. [PMC free article] [PubMed] [Google Scholar]
- 27.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephan C, Yousef GM, Scorilas A, Jung K, Jung M, Kristiansen G, Hauptmann S, Kishi T, Nakamura T, Loening SA, Diamondia EP. Hepsin is highly over expressed in and a new candidate for a prognostic indicator in prostate cancer. J Urol. 2004;171:187–191. doi: 10.1097/01.ju.0000101622.74236.94. [DOI] [PubMed] [Google Scholar]
- 29.Albertsen PC, Hanley JA, Fine J. 20-year outcomes following conservative management of clinically localized prostate cancer. JAMA. 2005;293:2095–2101. doi: 10.1001/jama.293.17.2095. [DOI] [PubMed] [Google Scholar]
- 30.Stanford JL, McDonnell SK, Friedrichsen DM, Carlson EE, Kolb S, Deutsch K, Janer M, Hood L, Ostrander EA, Schaid DJ. Prostate cancer and genetic susceptibility: a genome scan incorporating disease aggressiveness. Prostate. 2006;66:317–325. doi: 10.1002/pros.20349. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, McDonnell SK, Slusser JP, Hebbring SJ, Cunningham JM, Jacobsen SJ, Cerhan JR, Blute ML, Schaid DJ, Thibodeau SN. Two common chromosome 8q24 variants are associated with increased risk for prostate cancer. Cancer Res. 2007;67:2944–2950. doi: 10.1158/0008-5472.CAN-06-3186. [DOI] [PubMed] [Google Scholar]
- 32.Kwabi-Addo B, Chung W, Shen L, Ittmann M, Wheeler T, Jelinek J, Issa JP. Age-related DNA methylation changes in normal human prostate tissues. Clinical Cancer Res. 2007;13:3796–3802. doi: 10.1158/1078-0432.CCR-07-0085. [DOI] [PubMed] [Google Scholar]
- 33.Wu D, Zhau HE, Huang W-C, Iqbal S, Habib FK, Sartor O, Cvitanovic L, Marshall FF, Xu Z, Chung LWK. cAMP-responsive element-binding protein regulates vascular endothelial growth factor expression: implication in human prostate cancer bone metastasis. Oncogene. 2007;26:5070–5077. doi: 10.1038/sj.onc.1210316. [DOI] [PubMed] [Google Scholar]
- 34.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, Adami H-O, Hsu F-C, Zhu Y, Balter K, Kader AK, Turner AR, Liu W, Bleecker ER, Meyers DA, Duggan D, Carpten JD, Chang B-L, Isaacs WB, Xu J, Gronberg H. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 35.Tan YC, Zeigler-Johnson C, Mittal RD, Mandhani A, Mital B, Rebbeck TR, Rennert H. Common 8q24 sequence variations are associated with Asian Indian advanced prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:2431–2435. doi: 10.1158/1055-9965.EPI-07-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terada N, Tsuchiya N, Ma Z, Shimizu Y, Kobayashi T, Nakamura E, Kamoto T, Habuchi T, Ogawa O. Association of genetic polymorphisms at 8q24 with the risk of prostate cancer in a Japanese population. Prostate. 2008;68:1689–1695. doi: 10.1002/pros.20831. [DOI] [PubMed] [Google Scholar]