Abstract
Human leukocyte antigen-G (HLA-G) is a nonclassical major histocompatibility complex (MHC) class I molecule involved in immune tolerance processes, playing an important role in the maintenance of the semi-allogeneic fetus. Although HLA-G expression is restricted in normal tissues, it is broadly expressed in malignant tumors and may favor tumor immune escape. We analyzed HLA-G protein and mRNA expression in tumor samples from patients with glioblastoma collected in France, Denmark, and Brazil. We found HLA-G protein expression in 65 of 108 samples and mRNA in 20 of 21 samples. The absence of HLA-G protein expression was associated with a better long-term survival rate. The mechanisms underlying HLA-G gene expression were investigated in glioma cell lines U251MG, D247MG, and U138MG. Induction of HLA-G transcriptional activity was dependent of 5-aza-2′-deoxycytidine treatment and enhanced by interferon-γ. HLA-G protein expression was observed in U251MG cells only. These cells exhibited a permissive chromatin state at the HLA-G gene promoter and the highest levels of induced HLA-G transcriptional activity following 5-aza-2′-deoxycytidine treatment. Several antigen-presenting machinery components were up-regulated in U251MG cells after demethylating and IFN-γ treatments, suggesting an effect on the up-regulation of HLA-G cell surface expression. Therefore, because of its role in tumor tolerance, HLA-G found to be expressed in glioblastoma samples should be taken into consideration in clinical studies on the pathology and in the design of therapeutic strategies to prevent its expression in HLA-G–negative tumors.
Glioblastomas are very aggressive brain tumors, exhibiting poor response to therapeutic agents. Most of the evasion mechanisms developed by tumors are related to impairment of immune system cell function, which should be taken into account regarding the development of novel immunotherapeutic approaches. In this respect, human leukocyte antigen-G (HLA-G) is a pertinent molecule that is involved in immune tolerance processes and is expressed in numerous tumors, but has been poorly analyzed in glioblastoma.
HLA-G is a nonclassical HLA class I antigen characterized by restricted tissue expression in normal conditions,1 low polymorphism in the HLA-G gene coding region,2 and alternative splicing of primary transcript generating membrane-bound (HLA-G1-G4) and soluble (HLA-G5-G7) isoforms.3,4 HLA-G can inhibit natural killer (NK) and T-cell cytotoxicity as well as allogeneic proliferation.5–7 These functions are mediated through the direct binding of HLA-G to the inhibitory receptors ILT-2 (LILRB1/CD85j) and ILT-4 (LILRB2/CD85d)8,9 and KIR2DL4 (CD158d). The latter is expressed only by NK cells,10 and the KIR2DL4:HLA-G interaction is still debated.11,12
HLA-G is predominantly produced during pregnancy by invasive cytotrophoblasts13 and has also been detected in a few healthy adult and fetal tissues. On the other hand, HLA-G expression is induced in several pathological situations, such as inflammatory and autoimmune diseases, transplantation, and cancer.14 HLA-G expression in tumor lesions was first demonstrated with melanoma. In particular, HLA-G was expressed in nodular areas of primary melanoma and lymph node metastases from one patient, but it was not detected in the healthy skin and regressive area of the primary tumor in the same patient.15 To date, HLA-G expression has been detected in more than 1000 tumor lesions from at least 26 distinct tissue origins varying from approximately 20% to more than 80% of the lesions.14 HLA-G can be detected in tumor cells, in tumor-infiltrating cells, or both, being expressed on the cell surface, secreted, or incorporated into tumor-derived exosomes.15 HLA-G aberrant expression in tumors has thus been suggested to be part of the strategies that tumors use to escape from the host’s immunosurveillance. In agreement with that, HLA-G–mediated protection of tumoral cells against NK and T-cell cytotoxicity was demonstrated in vitro and could involve trogocytosis.15 Moreover, a correlation between poor clinical outcomes and HLA-G expression was reported in several tumoral diseases such as melanoma,16 B cell chronic lymphocytic leukemia,17 nasopharyngeal carcinoma,18 breast cancer,19 esophageal squamous cell carcinoma,20 non small cell lung cancer,21 colorectal tumors,22 and neuroblastoma.23 More recently, the demonstration has been made by means of a xenotumor model in mice that the HLA-G–positive tumor cell develops and tolerizes the host antitumor immune response in vivo.24 Collectively, these findings strongly argue for HLA-G as a prognostic biomarker.
There are only a few studies investigating HLA-G expression in glioma, which account for more than 70% of all brain tumors. Of these, glioblastoma is the most frequent and malignant histologic type (World Health Organization grade IV). Despite optimal treatment, the prognosis of glioma patients is still very poor, as only 9.8% of glioblastoma patients are still alive at 5 years after diagnosis.25 The expression and functional activity of HLA-G in glioblastoma cells was first reported by Wiendl et al.26 They found HLA-G expression in four of five tumor biopsy samples and in some cultured glioblastoma cell lines; moreover, the expression was enhanced after INF-γ stimulation. Notably, Wiendl et al26 observed that a few HLA-G–positive cells were sufficient to inhibit alloreactive lysis of HLA-G–negative glioma cells and that the expression of HLA-G rendered glioma cells less susceptible to alloreactive cytolytic killing. The investigators thus proposed that HLA-G–dependent suppression of T-cell responses represents a novel immune escape pathway of human glioblastoma. More recently, Kren et al27,28 reported two immunohistochemistry analyses using MEM-G/02 anti–HLA-G monoclonal antibody, one analysis performed in 39 cases of glioblastoma and the other analysis in 26 cases. Despite the small case number analyzed, HLA-G expression was observed in 64% of cases in neoplastic astrocytes and in 73% of cases in tumor-infiltrating ameboid microglia/macrophages. These data again reinforce the importance of investigating HLA-G in a high number of glioblastoma, as well as the mechanisms of its induction and up-regulation.
Interestingly, by comparing HLA-G expression in cultured cell lines of different tumor types with surgically removed tumor samples, HLA-G protein expression appeared more frequent in lesions than in cell lines.29 Therefore, HLA-G protein expression in vitro may require additional stimulators that are present in the tumoral microenvironment only. This is strongly supported by data showing that a primary culture of the melanoma cell line FON exhibited a high level of HLA-G1 cell-surface expression that was maintained until passage 40, started to decrease from passage 66, and then become completely negative at passage 70.30 In agreement with this observation, loss of HLA-G1 cell-surface expression was reported in in vitro culture of renal carcinoma cell lines31 and short-term ovarian carcinoma cell lines.32 Viral and environmental factors, such as cytokines (granulocyte macrophage-colony stimulating factor, interferons, IL-10, and leukemia inhibitory factor), hormones (progesterone, hydrocortisone), stress factors (arsenic, hypoxia), nutrient deprivation, and increased acidity have been associated with HLA-G expression in tumors.1 Nevertheless, most of them have no direct effect on the induction of HLA-G gene transcription in tumor cells when HLA-G gene is repressed. Indeed, we demonstrated that epigenetics is crucial in HLA-G repression/activation, as DNA demethylating agents and histone deacetylase inhibitors can reactivate HLA-G gene and protein expression in malignant cells in vitro.33,34 Finally, peptide loading and antigen-processing machinery (APM) components such as low molecular proteasome (LMP) subunits, LMP2, LMP7, LMP10, transporter associated with antigen processing (TAP)1, TAP2, calnexin, calreticulin, and tapasin, as well as β2-microglobulin (β2-m) are involved both in classical HLA class I and HLA-G cell-surface expression.35,36 APM expression may be modulated in the tumoral context37 and after epigenetic treatments in vitro and thus may also affect the magnitude of HLA-G expression on the cell surface.35,36
In the present study, we revaluated the extent of HLA-G expression in glioblastoma samples and its impact in the survival rate of patients. We also investigated the impact of epigenetic processes on the HLA-G gene and APM component expression in glioma cell lines exposed or not exposed to IFN-γ treatment.
Materials and Methods
Tissues and Patients
This retrospective study included initial specimen surgically removed from 122 patients with glioblastoma (World Health Organization grade IV) and no prior treatment to the craniotomy (Table 1). We used collected archival material from three different countries: i) Seventy-one glioblastomas were obtained from patients between 2001 and 2005 in Odense University Hospital (Denmark); ii) Twenty-seven glioblastomas were obtained from patients between 2004 and 2008 in Clinics Hospital of Ribeirão Preto of the University of São Paulo (Brazil); and iii) Ten glioblastomas were obtained from patients between 2004 and 2008 in Centre Hospitalier Universitaire Côte de Nacre (Caen, France). Fresh tissue biopsy samples were fixed in 4% neutral buffered formalin and paraffin embedded. Sections (3 to 4 μmol/L) were cut on a microtome, stained routinely with H&E to define representative tumor regions, and used for immunohistochemistry (IHC). We also used total RNAs extracted from 21 glioblastoma biopsy samples obtained from patients in Caen between 2003 and 2005; seven of these biopsies were included in the IHC analysis.
Table 1.
Immunohistochemical Analysis of HLA-G Expression in Glioblastoma Biopsy Samples
| France | Denmark | Brazil | Total | |||||
|---|---|---|---|---|---|---|---|---|
| Glioblastomas | ||||||||
| HLA-G status | G+ | G− | G+ | G− | G+ | G− | G+ | G− |
| No. of samples (n = 108) | 6 | 4 | 40 | 31 | 19 | 8 | 65 | 43 |
| (60.0%) | (40.0%) | (56.3%) | (43.7%) | (70.4%) | (29.6%) | (60.2%) | (39.8%) | |
| Patients | ||||||||
| No., male | 5 | 3 | 28 | 19 | 10 | 6 | 43 | 28 |
| Age, mean | 60.4 | 68.7 | 60.3 | 59.8 | 52.2 | 48.5 | 59.7 | 58.3 |
| Range | 47–68 | 63–73 | 21–78 | 37–74 | 31–67 | 26–70 | 21–78 | 31–74 |
| No., female | 1 | 1 | 12 | 12 | 8 | 2 | 21 | 15 |
| Age, mean | 70 | 67 | 60.2 | 64.5 | 62.7 | 67 | 61.3 | 65 |
| Range | 70 | 67 | 43–75 | 48–78 | 33–77 | 65–69 | 33–77 | 48–78 |
Immunohistochemistry
Sections from paraffin-embedded specimens were immunostained with MEM-G/02 monoclonal antibody (mAb) (1/50) that recognizes the free heavy chain of all HLA-G isoforms (Exbio, Praha, Czech Republic). An isotype-matched antibody was used under similar conditions to control nonspecific staining. Invasive cytotrophoblast from third trimester human placenta served as a positive HLA-G protein control.
The reaction product was visualized using the Ultratech HRP Streptavidin-Biotin Universal Detection System (Immunotech-Coulter, Villepinte, France) according to the manufacturer’s recommendations. Briefly, after rinsing the sections in phosphate-buffered saline solution with 0.1% saponin, endogenous peroxidases were inhibited using 3% hydrogen peroxide. Samples were initially incubated with specific or irrelevant antibodies for 1 hour at room temperature and subsequently with a solution containing a peroxidase-labeled polymer conjugated with a goat anti-mouse immunoglobulin for 30 minutes. The diaminobenzidine plus substrate-chromogen was finally used to develop antibody fixation (Dako EnVision-System Peroxidase; Dako, Trappes, France). All tissues were counterstained with hematoxylin (Dako). Two independent pathologists (E.L.-Z. and B.W.K.) interpreted HLA-G staining results.
Human Cell Lines and Culture
U251MG and U138MG human glioblastoma cell lines were provided by Dr. Heinz Wiendl (University of Wuerzburg, Wuerzburg, Germany) and were initially obtained from Dr. Nicolas de Tribolet’s laboratory (Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland).26 D247MG glioblastoma cell line was from the laboratory of S.Fe. All cells were cultured and analyzed in the CEA/Saint-Louis Hospital laboratory. JEG-3 human choriocarcinoma cell line (ATCC, Manassas, VA) and M8 melanoma cell line33 were used as positive and negative controls for HLA-G expression, respectively. Glioma and choriocarcinoma cell lines were maintained in Dulbecco’s modified Eagle’s medium medium supplemented with Glutamax-I (Invitrogen, Cergy Pontoise, France), M8 cells were maintained in RPMI-1640 medium (Sigma-Aldrich, St. Quentin Fallavier, France) supplemented with l-glutamine (Sigma-Aldrich). All cultures were supplemented with 10% heat-inactivated fetal calf serum, 250 μg/L fungizone (Invitrogen), and 10 mg/L gentamicin (Invitrogen). Cells were routinely tested for absence of contamination with mycoplasma (plasmoTest, InvivoGen, San Diego CA).
HLA-G Gene Sequencing and CpG Methylation Analysis
The HLA-G coding region in U251MG, D247MG and U138MG cell lines was analyzed for the presence of nonsense mutations by direct sequencing of PCR products using an ABI310 Genetic Analyzer (Applied Biosystems, Foster City, CA) as previously described.38 Methylation status of CpG sites was performed using a sodium bisulfite modification of DNA as previously described,34 with a DNA methylation detection kit (Biochain, Hayward, CA) according to the manufacturer’s recommendations. Methylation status of HLA-G region spanning 443 bp upstream of initiation codon was analyzed by PCR amplication using the primers GP328 (5′-AAGAGTATAGGAGGATAGGTAAGG-3′) and GP776 (5′-AACACCATAACCACCATCCTTAAC-3′) for bisulfite-treated DNA and GP327N (5′-GAAGAGTAC-AGGAGGACAGGC-3′) and GP717N (5′-AGAATGAGT-CCGGGTGGGTGA-3′) for non–bisulfite-treated DNA.34
Cell Treatments
The U251MG, D247MG, and U138MG cell lines were cultured at the initial concentration of 2 × 106 cells for 24 hours. The culture medium was then changed, and cells were further cultured in medium (untreated), in medium supplemented with either 10 μmol/L or 100 μmol/L demethylating drug 5-aza-2′-deoxycytidine (5-aza-dC; Sigma-Aldrich), with 500 U/mL IFN-γ (PeproTech, Rocky Hill, NJ) or with 5-aza-dC and IFN-γ combined. Conditions were maintained until day 5 in most experiments.
Flow Cytometry Analysis
Cells were harvested and washed with PBS containing 0.1% bovine serum albumin and blocked with human immunoglobulins (Alphaglobin; Grifols, Langen, Germany) for 10 minutes at 4°C. After one washing step, the unlabeled first antibody was added at the final concentration. HLA-G cell-surface expression was evaluated with MEM-G/9 mAb, which recognizes HLA-G molecules associated with β2-m (Exbio, Prague, Czech Republic). Isotype control IgG1 was used at the same concentration as the primary antibody. Incubation was done on ice for 30 minutes, followed by two washes. Goat anti-mouse IgG PE-conjugated F(ab′)2 fragment (5 μg/mL; Sigma-Aldrich) or dichlorotriazinyl-fluorescein-conjugated F(ab′)2 fragment (10 μg/mL; Dianova, Hamburg, Germany) were used as a secondary antibody. Cells were analyzed using a flow Epics XL cytometer using Expo32 ADC 1.1C software (Beckman Coulter, Rossy, France). Specific fluorescence indexes (SFIs) were calculated by dividing the mean fluorescence obtained with specific antibody by mean fluorescence obtained with isotype control antibody. An SFI value of 1.5 was considered positive.
Quantitative Real-Time RT-PCR analysis
RNAs were withdrawn using RNeasy Mini Kit (Qiagen, Courtaboeuf, France) according to the manufacturer’s protocol. A 1 μg quantity of total RNA from each tumor sample was reverse transcribed at 42°C for 1 hour using the Promega RT system (Promega, Charbonnieres, France). Total RNA from cell lines was reverse transcribed using oligo-(dT)12-18 priming and M-MLV Reverse Transcriptase (Invitrogen, Cergy Pontoise, France). Real-time RT-PCR was performed in triplicate as previously described33 with ABI Prism 7000 SDS (Applied Biosystems, Courtaboeuf, France) in a duplex PCR with 40 amplification rounds, using TaqMan Universal PCR Mix, predeveloped TaqMan assay reagent glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an endogenous control, and an HLA-G–specific probe located in exon 5 that targets all HLA-G mRNAs. Quantification was performed relative to amounts of HLA-G transcripts in HLA-G–positive JEG-3 using the comparative CT method33: ΔCT = CT HLA-G − CT GAPDH; ΔΔCT = ΔCT sample − ΔCT JEG-3; relative HLA-G expression = 2 −ΔΔCT. We analyzed at least two independent experiments including treated (5-aza-dC, IFN-γ) and untreated glioma cells.
Western Blot Analysis
Untreated and treated cells with 5-aza-dC and/or IFN-γ were collected (5 × 106) and lysed in lysis buffer (1% NP40, 50 mmol/L Tris HCl, 150 mmol/L NaCl, 2 mmol/L EDTA, and 10% glycerol). After centrifugation at 16,000 × g at 4°C for 30 minutes, supernatants were transferred to Eppendorf tubes (Dutscher, Brumath, France). Equal amounts of cell lysates (50 μg) were normalized by the micro-BCA assay (Pierce, Rockford, IL) and separated in 10% SDS-PAGE. All samples were electroblotted onto Hybond-C extra membrane (Amersham, Les Ulis, France) and blocked by incubation with Tris-buffered saline (TBS) 5% nonfat dry milk for 30 minutes. After blocking, membranes were washed three times in TBS solution containing 0.2% Tween-20, then probed with the corresponding mAb overnight at 4°C and washed three times in TBS containing 0.2% Tween-20. The membranes were incubated for 30 minutes at room temperature with peroxidase-conjugated goat anti-mouse IgG directed against heavy and light chains (H&L) and washed three times in TBS containing 0.2% Tween-20. The membranes were treated with West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL) for 1 minute and exposed to Kodak Biomax MR film. The cell line JEG-3 was used as a positive HLA-G control.
The following mAbs were used in Western blotting: an antitubulin antibody for internal control (Sigma-Aldrich); the anti-pan HLA-G 4H84 (mouse IgG1; provided by Michael McMaster, University of California, San Francisco, CA); the β2m-specific mAb L36839; the LMP2-specific mAb SY-1; the LMP7-specific mAb HB-2; the LMP10-specific mAb TO-6; the TAP1-specific mAb NOB-1; the TAP2-specific mAb NOB-2; the calnexin-specific mAb TO-5; the calreticulin-specific mAb TO-11; the ERp57-specific mAb TO-2, and the tapasin-specific mAb TO-3 were developed and characterized as described elsewhere.40–42 The mAbs were purified from ascitic fluid by sequential precipitation with ammonium sulphate and caprylic acid.43 The purity of mAb preparations was assessed by SDS-PAGE. The activity of the mAb preparations was monitored by testing with a lymphoid cell lysate in Western blotting. Intensities of the bands were analyzed using AlphaEaseFC software version 3.1.2 (FluorChem 8800 Digital Imaging; Alpha Innotech., San Leandro, CA).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as previously described34 with anti–phospho(S10)-acetyl(K14)-histone H3 (07-081; Upstate, Lake Placid, NY), anti–acetyl(K18)-histone H3 (07-354, Upstate), and anti–RNA polymerase II (sc-899 X; Santa Cruz Biotechnology, Santa Cruz, CA). Briefly, cell line cultures were exposed to formaldehyde for cross-linking proteins and DNA together, followed by chromatin sonication into pieces of approximately 300 to 500 bp. Cross-linked chromatin was immunoprecipitated with either no antibody (−) or the indicated antibody. The antibody-protein-DNA complexes were purified using beads coupled to protein A–sepharose (Sigma-Aldrich). The DNA was isolated from the complexes using a combination of heat to reverse cross-linking, RNase, and proteases and was then purified using phenol extraction and ethanol precipitation. The final DNAs from ChIP and input chromatin (PCR control) were then used as templates in PCR using primer set −187 forward (5′-CCCGCGTTGGGGATTCTCTC-3′) / +4 reverse (5′-AGAGGGTTCGGGGCGCCATGAC-3′) specific for the HLA-G gene proximal promoter and primer set 1089 F (5′-CCCTTTGTGACTTCAAGAAC-3′), and 1252 R (5′-AAGTTATAGCTCAGTGGACC-3′) specific for the HLA-G 3′ untranslated region (3′UTR).
Statistical Analysis
We used Prism version 5.0 (GraphPad Software, San Diego, CA) and the nonparametric U-test for comparison of values obtained by real-time PCR analysis. Data are presented as means ± SEM for at least three independent experiments. Survival analysis was performed using the log-rank test and graphical representation was performed by Kaplan-Meier. The analysis took into account patients who died of the tumor excluding other reasons (data were validated from the hospital charts and death certificate). Significance was defined as a P < 0.05 at a confidence interval of 95%.
Results
HLA-G Is Frequently Expressed in Glioblastoma Biopsy Samples
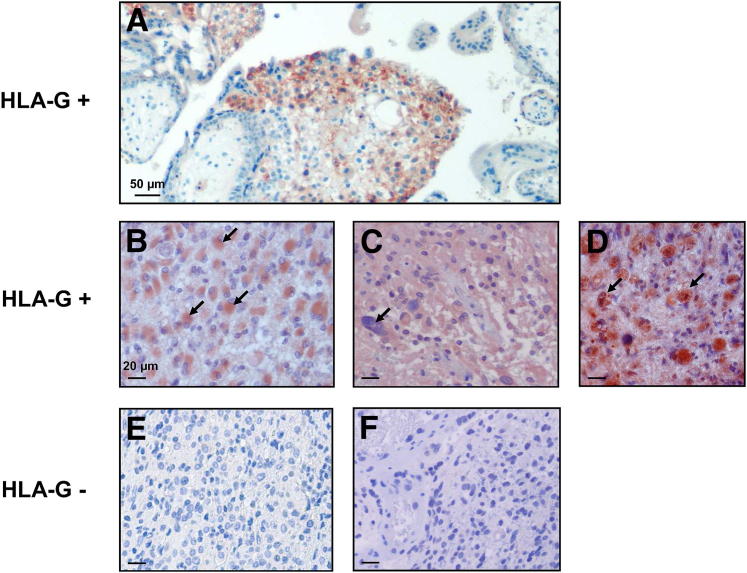
To evaluate the extent of HLA-G protein expression in glioblastoma lesions, we performed a multicentric IHC analysis of paraffin-embedded biopsy sections of 108 glioblastoma lesions (World Health Organization grade IV) from French, Danish, and Brazilian patients using the anti–HLA-G monoclonal antibody MEM-G/02, which recognize all HLA-G isoforms. Results in Table 1 show that HLA-G was detected in 60.2% of cases, ranging from 56.3% in Danish patients to 70.4% in Brazilian patients. Therefore, the majority of glioblastoma specimens are HLA-G positive, whatever the geographic origin of patients. No correlation between age or sex and HLA-G expression could be established. In most cases, HLA-G expression was observed in astrocyte gemistocytics but also was observed in inflammatory infiltrating cells (Figure 1).
Figure 1.
HLA-G expression in paraffin-embedded sections of glioblastoma. A: Invasive cytotrophoblast from third trimester human placenta served as a HLA-G–positive control immunostained with the anti–HLA-G–specific MEM-G/02 monoclonal antibody (mAb) (all isoforms are recognized). B–D: Representative HLA-G–positive glioblastoma specimens immunostained with MEM-G/02 mAb. Arrows indicate gemistocytic tumor cells in B, giant tumor cells in C, and macrophages in D. E and F: Representative HLA-G–negative glioblastoma specimens immunostained with MEM-G/02 mAb. Scale bars: 50 μm (A), 20 μm. (B–F). Original magnification, ×40.
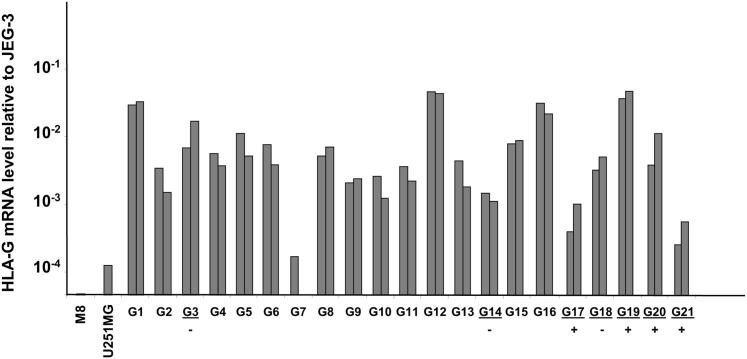
HLA-G transcriptional activity was investigated on 21 glioblastoma biopsy samples from French patients using quantitative real-time RT-PCR; seven of the samples (G3, G14, G17, G18, G19, G20, and G21) were also included in the IHC analysis. By comparing glioblastoma HLA-G transcriptional activity with that of HLA-G–positive cell line JEG-3 and HLA-G–negative cell lines U251 and M8, we observed that with the exception of the G7 biopsy sample, levels of HLA-G gene transcription were clearly higher (at least ninefold) in glioblastoma biopsy samples than in HLA-G–negative cells (Figure 2). The highest amounts of HLA-G mRNA were found in G1, G12, G16, and G19 samples corresponding to ∼ 1 in 50 of JEG-3 HLA-G mRNA. It is of note that HLA-G protein expression analyzed by IHC was detected only in G17, G19, G20, and G21, whereas G3, G14, and G18 were HLA-G negative, suggesting that post-transcriptional mechanisms participate in the observed HLA-G protein expression.
Figure 2.
HLA-G transcriptional activity in 21 glioblastoma biopsy samples (G1 to G21). Results of real-time RT-PCR performed twice (each in triplicate) are presented, with two distinct sticks (mean of triplicates) for each sample, except G7 (only one experiment). Results are compared to the HLA-G mRNA expression in HLA-G–positive choriocarcinoma JEG-3 cells (assigned a value of 1) and HLA-G–negative cell lines melanoma M8 and glioblastoma U251MG. Glioblastoma specimens studied in IHC are underlined. Plus (+) and minus (−) signs indicate positive and negative HLA-G protein expression (IHC), respectively.
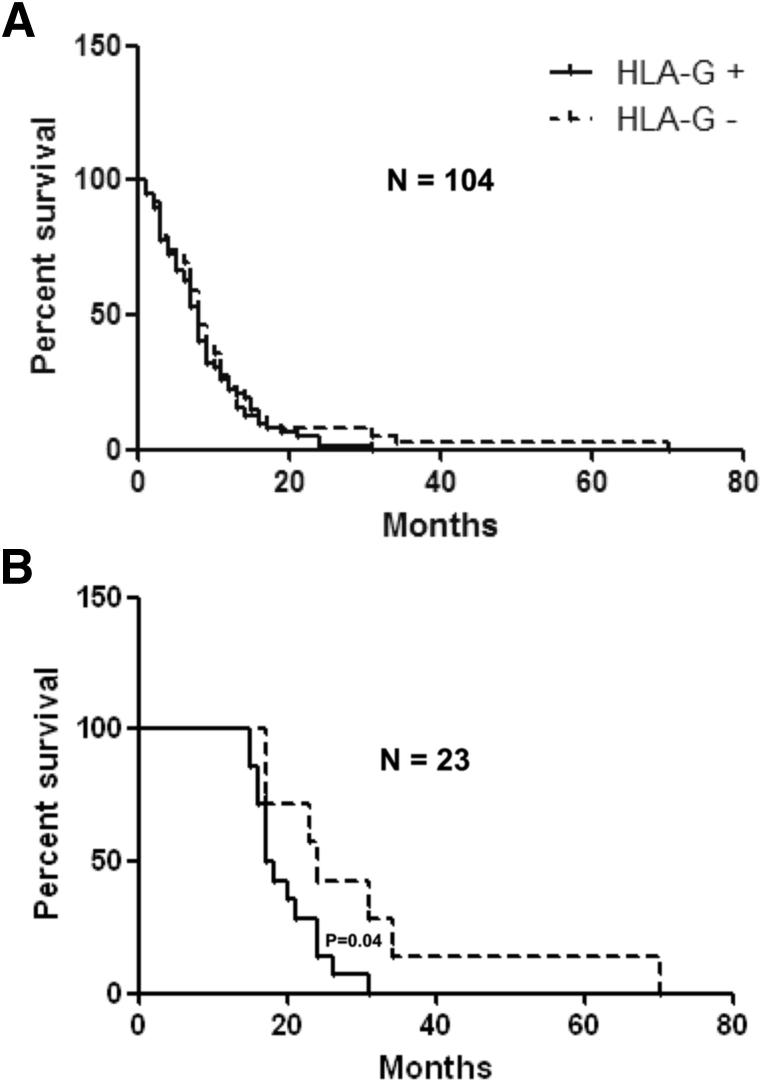
Lack of HLA-G Expression May Be a Pronostic Indicator of Long-Term Survival Rate
In an effort to assess the impact of HLA-G protein expression on the survival rate of patients with glioblastoma, we generated Kaplan-Meier survival curves using available data in 106 patients (26 Brazilian, 9 French, and 71 Danish patients). We excluded one patient (HLA-G–negative) who was still alive at the time of the study, and one patient (HLA-G–positive, 6-month survival) who died of pulmonary embolism rather than glioblastoma. The survival time of 104 patients from the date of craniotomy varied between 1 and 70 months (one patient with 70-month survival). HLA-G expression was not significantly correlated with a lower survival rate of an HLA-G–positive patient compared with HLA-G–negative patients (mean survival rate, 8 months) (Figure 3A). To further analyze the impact of HLA-G expression in patients with long-term survival, we gradually analyzed patients having a survival of more than 8 months and found no significant association before 15 months. After 15 months, it was possible to observe that HLA-G–negative patients were still alive 7 months longer than HLA-G–positive patients (P = 0.04) (Figure 3B).
Figure 3.
Survival analysis of 104 patients with glioblastoma either expressing or not expressing HLA-G protein at the tumor site (Kaplan-Meier survival curves). A: Survival time of 104 patients from the date of craniotomy. The median survival rate is 8 months for HLA-G–positive and HLA-G–negative patients. B: Survival time of 23 patients after 15 months from the date of craniotomy. HLA-G–negative patients are still alive 7 months longer than HLA-G–positive patients (P = 0.04).
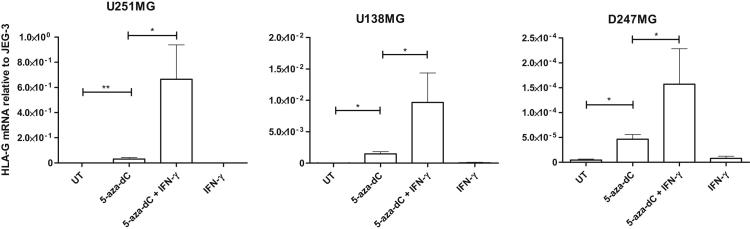
HLA-G Induction in Glioma Cells Requires DNA Demethylation and Is Enhanced by Incubation with IFN-γ
To investigate the mechanisms underlying the induction of HLA-G expression in glioblastoma cells, we used the human glioma cell lines U251MG, U138MG, and D247MG as models for in vitro studies. We did not detect any significant HLA-G mRNA amounts using quantitative real-time RT-PCR or protein expression using Western blot and flow cytometry, even after incubation with IFN-γ (Figures 4 and 5). Interestingly, sodium bisulfite genomic sequencing performed with U251MG cells showed that 18 CpG sites are methylated among the 19 CpG sites located within the HLA-G gene fragment spanning 443 bp upstream to ATG (Supplemental Figure S1). Therefore, having previously demonstrated that DNA demethylation at the HLA-G locus induced HLA-G gene expression, we cultured the three cell lines in medium supplemented with the demethylating agent 5-aza-dC (100 μmol/L) and/or IFN-γ (500 U/mL). As expected, HLA-G mRNA was up-regulated in all cell lines treated with 5-aza-dC and 5-aza-dC/IFN-γ compared with cells in medium alone (untreated). In addition, levels of HLA-G transcripts were increased in the three cell lines after combined treatments with 5-aza-dC/IFN-γ compared with 5-aza-dC alone (Figure 4). It is noteworthy that the highest amounts of HLA-G mRNA were observed in the U251MG cell line after 5-aza-dC treatment (U251MG × U138MG: P = 0.04; U251MG × D247MG: P = 0.01; U-test; n = 3) and after 5-aza-dC/INF-γ treatment (U251MG × U138MG, P = 0.05; U251MG × D247MG, P = 0.05; U-test; n = 3). We performed an additional experiment with U251MG cells in which 5-aza-dC and IFN-γ were removed from the growth medium after 5 days of treatment. We analyzed HLA-G transcription at day 5 after 5-aza-dC and IFN-γ treatments and at days 6, 7, and 8 of cell culture without treatment. We observed that the induced HLA-G transcriptional activity was still maintained, even in the absence of treatment (Supplemental Figure S2).
Figure 4.
Induction of HLA-G gene transcription in HLA-G–negative glioma cells after treatments with 100 μmol/L 5-aza-dC and 5-aza-dC/IFN-γ (500 U/mL). Real-time RT-PCRs (performed in triplicate for each experiment) targeting all HLA-G mRNA forms are shown for each glioma cell line (U251MG, U138MG, and D247MG), either untreated (UT) or treated with 5-aza-dC alone, 5-aza-dC and IFN-γ combined, and IFN-γ alone, for 5 days. The y axis indicates amounts of HLA-G mRNA levels relative to mRNA expression in HLA-G–positive JEG-3 choriocarcinoma cells (assigned a value of 1). Data are presented as mean ± SEM for at least three independent experiments. Statistical analysis was performed using U-tests; *P < 0.05 and **P < 0.01.
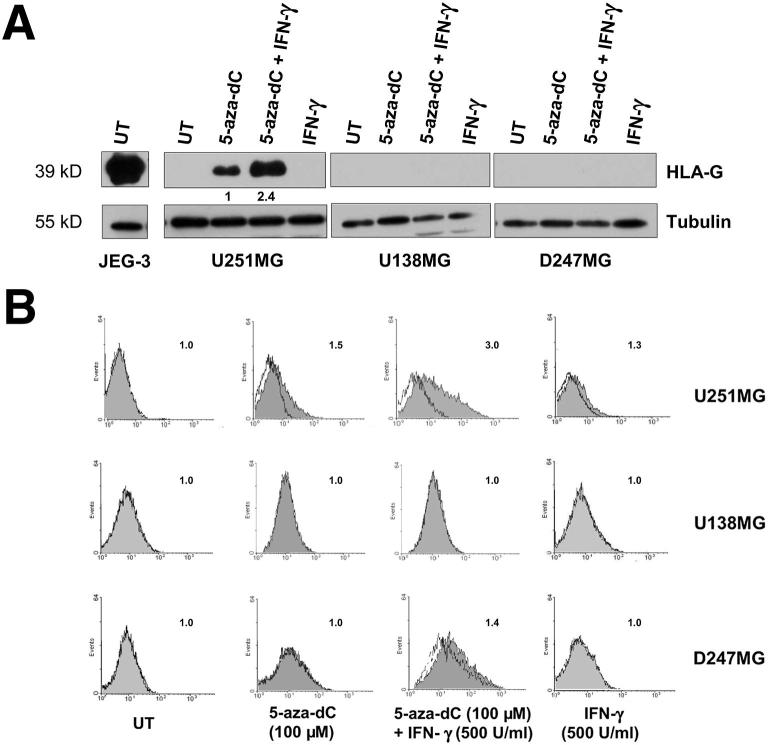
Figure 5.
Induction and up-regulation of HLA-G protein expression in U251MG glioma cells upon 5-aza-dC and 5-aza-dC/IFN-γ treatments (day 5). U251MG, D247MG, and U138MG cells were either untreated (UT) or treated with 5-aza-dC alone, 5-aza-dC and IFN-γ combined, or IFN-γ alone. A: Representative Western blots performed with the anti–HLA-G 4H84 and anti-tubulin antibodies (four independent experiments). A value of 1 is assigned to the HLA-G protein level in 5-aza-dC–treated cells. B: Representative flow cytometry analysis was performed with the anti–HLA-G MEM-G/9 antibody (dark) and IgG1 isotype control (three independent experiments with similar results). Numbers in the upper right corner of each box indicate SFI values. An SFI value of 1.5 was considered positive.
Then, to test whether the induced HLA-G transcripts were translated into proteins, we performed Western blot assays using anti–HLA-G 4H84 mAb. Figure 5A presents a representative result from four independent experiments, showing that the incubation with IFN-γ alone had no detectable effect on the HLA-G protein expression in the three cell lines analyzed. Cell treatment with 100 μmol/L 5-aza-dC alone induced HLA-G protein expression in U251MG, but not in D247MG and U138MG cell lines. In U251MG cells, a 2.4-fold increase in HLA-G expression was observed after incubation with 5-aza-dC and IFN-γ combined (mean 1.95-fold increase; SEM = 0.185; n = 4), and was still not detectable on D247MG and U138MG cells. The complete coding sequence analysis of the HLA-G alleles in the three cell lines did not reveal any nonsense mutation (data not shown).
On the other hand, flow cytometry analysis of the cell lines stained with the anti–HLA-G mAb MEM/G9 revealed a weak staining of the cell membrane of U251MG cells after a 24-hour incubation (data not shown); the staining progressively increased by prolonging the incubation to 120 hours (SFI = 1.5 was observed in two of three experiments) (Figure 5B). In agreement with the results of QRT-PCR and western blot experiments, the incubation with 5-aza-dC and IFN-γ increased the level of HLA-G expression on the cell membrane of U251MG cells (SFI = 3; mean SFI = 2.9 of three independent experiments) (Figure 4B) compared to cells incubated with 5-aza-dC alone. Moreover, we observed that the HLA-G expression was maintained in U251 treated cultures for at least 15 days (data not shown).
HLA-G Expression in Glioma Cells Is Associated with Chromatin Remodeling at the HLA-G Promoter and the Modulation of APM Components upon Treatment
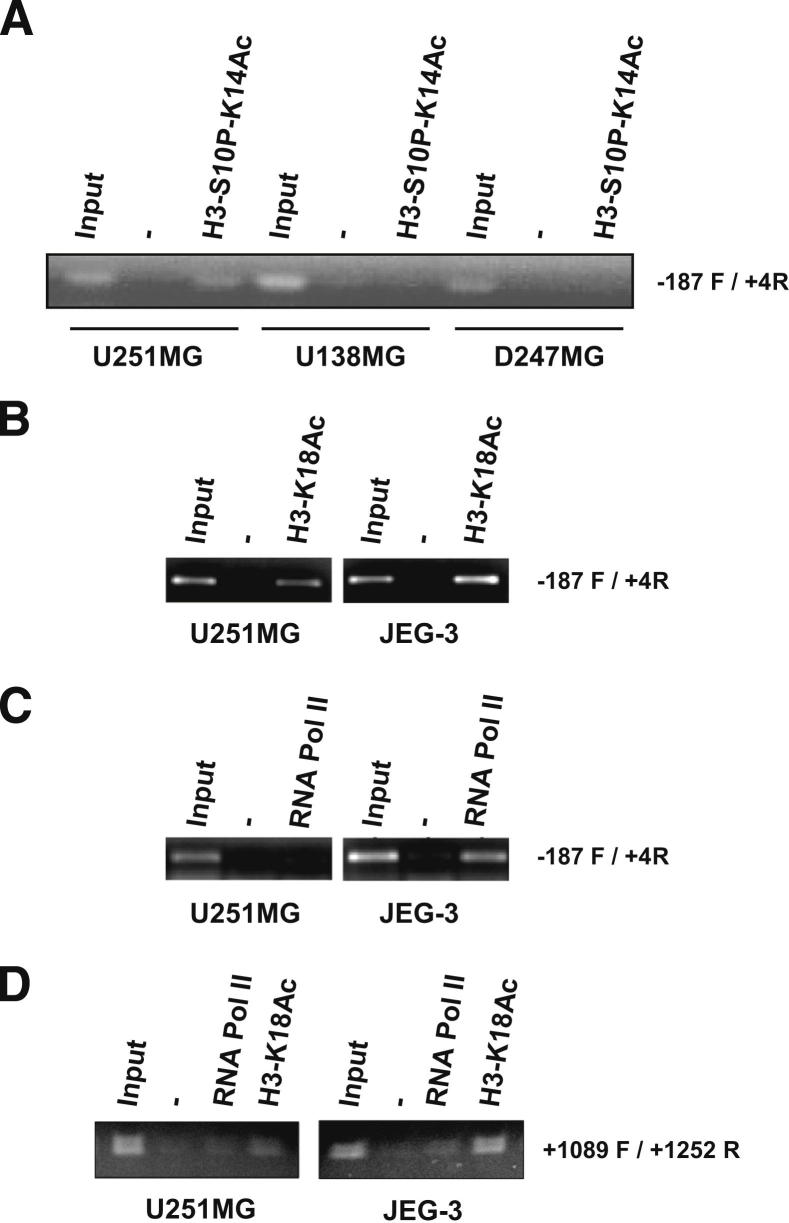
To identify elements influencing HLA-G gene induction and cell surface expression, we first examined chromatin modifications at the proximal HLA-G promoter region in untreated U251MG, D247MG, and U138MG cell lines. By ChIP, we observed that S10 phosphorylated K14-acetylated H3 histone, a pattern which is often associated with gene activation, is found only in U251MG cells (Figure 6A). Next, we used ChIP assays to explore the H3-K18 acetylation, another hallmark of gene activation, at the HLA-G gene promoter and 3′ untranslated region in U251MG and HLA-G–positive JEG-3 cells, and found H3 acetylation in both cell lines (Figure 6, B and D). On the other hand, ChIP assays targeting RNA pol II was positive only at the HLA-G gene promoter in JEG-3 (Figure 6, C and D), a result in agreement with the very low presence or the absence of HLA-G transcriptional activity observed in untreated U251MG cells (Figure 4). Therefore, the chromatin structure at the HLA-G promoter in U251MG cells exhibits a pattern that is compatible with HLA-G transcription, but the structure is still not accessible to RNA pol II when cells are cultured in medium alone.
Figure 6.
Differential pattern of H3 phospho-acetylation at the HLA-G locus in glioma cell lines U251MG, U138MG, and D247MG. Representative ChIP experiments are performed with no antibody (−), antibodies directed against H3-S10P-K14Ac (A), H3-K18Ac (B–D), and anti-RNA polymerase II (C and D). B–D: HLA-G promoter and 3′ untranslated region (3′UTR) modifications in U251MG are compared with those of choriocarcinoma cell line JEG-3, which exhibits high levels of HLA-G transcripts. −187 F / +4R and + 1089F / + 1252R indicate primer set used for PCR targeting proximal HLA-G promoter and HLA-G 3′ UTR, respectively; input is input chromatin used as PCR control.
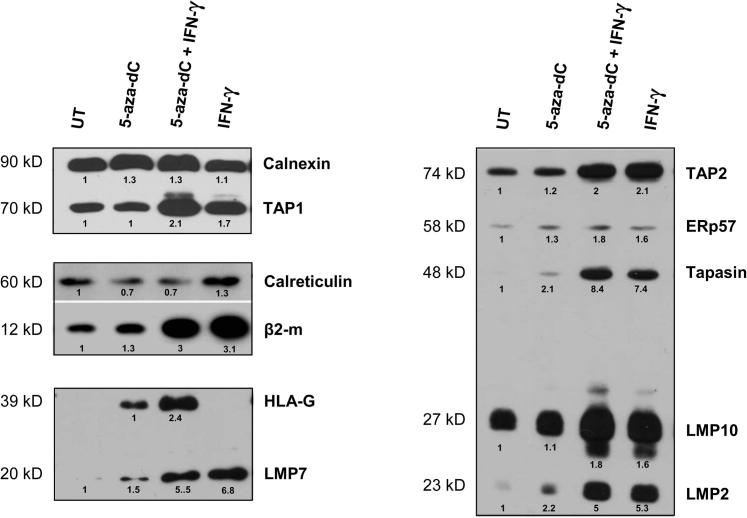
Next, we evaluated the effect of 5-aza-dC and IFN-γ treatments on the modulation of accessory proteins that are involved both in classical HLA class I and HLA-G cell surface expression. Calnexin, calreticulin, TAP-1, TAP-2, tapasin, Erp57, proteasome subunits LMP-2, -7, -10, and β2-m expressions were analyzed by Western blots of protein extracts of U251MG cells either exposed or not exposed to 5-aza-dC and IFN-γ for 4 or 5 days (Figure 7). The demethylating agent, either alone or combined with IFN-γ, had a slight effect on the up-regulation of APM components and β2-m except calreticulin. On the other hand, although IFN-γ alone had no effect on the HLA-G protein expression, IFN-γ increased almost all APM components and β2-m, with stronger effect in comparison with 5-aza-dC. In particular, mean values from two independent experiments revealed a 1.8-fold increase in β2-m expression, a twofold increase in TAP-1 expression, and 3.9- and 4.7-fold increases in LMP-2 and LMP-7 expression, respectively, in U251MG cells treated with IFN-γ compared with cells treated with 5-aza-dC alone (data not shown). A combination of treatments had no strong additive effect on APM component expression (Figure 7).
Figure 7.
APM components and β2-m up-regulation in U251MG glioma cell line upon 5-aza-dC and IFN-γ treatments. Representative Western blots are presented (at least two independent experiments). Protein extracts (50 μg) are from U251MG cells either untreated (UT) or treated with 5-aza-dC alone, 5-aza-dC/IFN-γ combined, and IFN-γ alone. Results obtained from the same gel are framed. Quantitative variations are indicated; a value of 1 is assigned to the amount of APM component in untreated cells or to the amount of HLA-G expression in cells treated with 5-aza-dC alone.
Discussion
Despite better knowledge of the molecular mechanisms involved in glioma phenotype and introduction of the new treatment with radiation and temozolomide in 2005, the median survival time for patients continues to be very poor. Glioblastoma appears to benefit from the central nervous system, which has been considered an immunologically privileged site. In fact, T lymphocytes and antigen presenting cells pass through the central nervous system during pathologic conditions such as neurodegenerative diseases44 and glioma.45 Microglia, an important portion of resident central nervous system cells, exhibits the ability to generate significant innate and adaptive immune responses after infectious and traumatic stimuli. Indeed, under pathological conditions, microglia express the molecular machinery required to present antigen and acquire the potential to interact with T cells in an antigen-specific manner.46 Therefore, the brain should be viewed as an immunologically quiescent site, and the poor prognosis for glioblastoma may be due in part to mechanisms that promote tumor tolerance and immune suppression.47
Previous studies47 have highlighted that glioblastoma may use multiple immune evasion strategies such as secretion of immunosuppressive cytokines and factors (prostaglandin E2, transforming growth factor β, vascular endothelial growth factor, and IL-10), and impairment of adhesive effectors to extracellular matrix proteins. Other mechanisms, including glioblastoma up-regulation of B7 homologue 148 and STAT-3,49 have been proposed in inhibiting allogenic T-cell activation and/or modifying cytokine profile expression toward Th2. With regard to classical HLA class I molecules, a defect was previously reported in only ∼ 50% of glioblastomas tested and was significantly correlated with astrocytic tumor grade, although, no correlation between HLA class I expression and survival was found.37 Interestingly, an observation was made that APM alterations may occur in tumor cells expressing normal levels of HLA class I molecules. Therefore, it was proposed that production of peptide-free HLA molecules would affect immune recognition of tumors cells by T cells and promote HLA class I inhibition of NK cells.50 Indeed, LMP2, TAP1, and β2-m were previously demonstrated to be down-regulated in high-grade astrocytomas,50 a result that we confirmed in the present study (Figure 7). In addition, tapasin down-regulation was observed associated with HLA class I antigen down-regulation as well as tumor grade, although not significant (possibly due to sample size).37 The present work shows that nonclassical HLA-G is frequently transcribed in glioblastoma and the protein expressed in ∼ 60.2% of cases (n = 108), and may be induced in HLA class I–positive cell lines. Despite having studied smaller patient cohorts, several reports strongly support our finding : i) HLA-G mRNA expression was detected in four of 12 tested glioma cell lines26; ii) transcriptomic microarray performed with total mRNA from 20 glioblastoma specimens detected 12 tumors with significant HLA-G mRNA expression compared with a pool of non-neoplastic brain samples51; and iii) immunohistochemical analyses performed with different anti–HLA-G mAb revealed that HLA-G protein is expressed in 75% (n = 4),26 73% (n = 26),28 and 64% (n = 39),27 of the tested glioblastoma samples, both in tumor cells and in infiltrating microglia/macrophages. HLA-G expression in the latter cells thus makes them behave as regulatory cells involved in immune suppression. Therefore, even if the mechanism involving classical HLA class I molecules is likely to participate in glioma escape from the immune system, HLA-G would exert its immunosuppressive properties both in the absence and presence of classical HLA class-I molecules.15 Interestingly, recent reports52,53 have shown that ILT-4 (LILRB2/CD85d) can recognize the β2M-free HLA-G, but ILT-2 (LILRB1/CD85j) cannot. Therefore, despite down-regulation of β2-m in glioblastoma, HLA-G would be capable of conferring an inhibitory effect selectively on ILT-4–expressing cells, such as antigen-presenting cells. A previous study27 performed with 39 cases reported that the expression of HLA-G in glioblastoma cells was prognostically neutral. By studying a larger number of cases, we also found that HLA-G expression in tumor samples with no prior treatments was prognostically neutral. However, we cannot exclude the possibility that therapeutic treatments could have modulated HLA-G expression. One possibility is that HLA-G has been induced in some permissive tumors, thus reducing time survival of some patients who were diagnosed as HLA-G negative before treatment. Interestingly, we found that the absence of HLA-G expression in glioblastoma at the time of craniotomy was associated with a favorable outcome in patients after 15 months of survival. This is in agreement with several studies describing a positive correlation between HLA-G expression in cancer and shortened survival of patients,54 and suggests a potential role of HLA-G participation in the down-regulation of anti-glioblastoma immune responses.55
Focusing on the HLA-G expression in glioblastoma and consequently the possible effect in tumor escape, we thus investigated mechanisms that could induce it. Considering the literature data in the field, the up-regulation of HLA-G in glioblastoma is likely to involve the interaction of tumor microenvironment such as hypoxia, immunosuppressive molecules, and therapeutic treatments, with HLA-G alleles of the patients.1,56 Indeed, the HLA-G gene polymorphism is frequent in 5′ and 3′ untranslated regions and may affect the binding of transcription factors and miRNAs,57,58 thus providing a tangible explanation for its participation in the absence or in the low level of HLA-G protein expression in almost 40% of the tested biopsy samples. Moreover, by analyzing HLA-G mRNA levels in 21 glioblastoma biopsy samples, we observed quantitative variations, even though almost all samples exhibited HLA-G transcriptional activity. However, amounts of HLA-G transcripts were lower than those of choriocarcinoma JEG-3 cell line, probably due to tumoral tissue heterogeneity. Finally, regarding the HLA-G expressed in infiltrating microglia/macrophages, it is tempting to speculate that it might reflect a tumor-derived HLA-G acquisition through trogocytosis, a mechanism that has been previously observed in vitro.15
Here, we investigated the impact of 5-aza-dC and IFN-γ treatments. There is evidence that DNA hypermethylation is detected in 87% of glioblastoma lesions.59 Accordingly, to develop tumor antigen-specific active immunotherapy, it has been proposed to up-regulate melanoma associated antigen (MAGE)1 by 5-aza-dC treatment combined with a strategy to up-regulate major histocompatibility complex levels by IFN-γ to augment tumor cell recognition by tumor antigen-specific T cells.60 5-aza-dC treatment was also proposed as a drug candidate to up-regulate human telomerase reverse transcriptase in glioma to reduce telomerase expression.61 In agreement with previous observations in several tumor cell types,33,34 here we demonstrate that HLA-G promoter region is methylated in U251MG cells and that HLA-G transcriptional expression in glioma cells is always induced by 5-aza-dC treatment. Methylation/demethylation processes are thus key events in the observed HLA-G transcriptional up-regulation in glioma cells. Interestingly, levels of HLA-G mRNAs were enhanced by combining 5-aza-dC with IFN-γ treatment, whereas as previously reported for NK and NK-like cells,62 IFN-γ alone had only a slight effect on the induction of HLA-G gene transcriptional activity, suggesting that IFN-γ probably required HLA-G gene demethylation before having an effect. The cell lines in the present study were studied elsewhere,26 showing a constitutive HLA-G transcription only in U251MG and D247MG cells, whereas HLA-G transcription became detectable in U138MG cells after IFN-γ treatment. In addition, HLA-G was not detectable on the cell membrane and was induced with IFN-γ at the cell surface of D247MG and U138MG, suggesting that HLA-G stability is not affected in these cells. It is noteworthy that, by studying these cell lines in the CEA/Saint-Louis Hospital laboratory, we did not find such HLA-G expression supporting that environmental conditions are of crucial importance in the induction of HLA-G expression and its maintenance in culture. Besides, our findings did not support the induction of high levels of HLA-G mRNA in HLA-G–negative glioma cells after IFN-γ treatment alone.26 Nonetheless, HLA-G mRNA levels were assessed by Northern blot analysis, a less sensitive methodology that would not allow detection of low level HLA-G mRNA in untreated cells, and thus could explain such apparently contradictory results.
Furthermore, we showed that U251MG cells exhibited better efficiency in HLA-G gene induction upon 5-aza-dC treatment than U138MG and D247MG cells. It is noteworthy that doubly modified H3-S10PK14Ac was observed only at the proximal promoter of U251MG. This dual modification is known to allow interaction of 14-3-3 proteins, which are important factors for mediating the switch from transcriptional repressive to active chromatin in vivo, and thus induction of many genes. Interestingly, previous studies found that strong 14-3-3 protein expression was observed and associated with tumor genesis and progression in glioma.63 In addition, we revealed H3-K18Ac post-transcriptional modification at the HLA-G promoter in U251MG cells, a chromatin pattern again associated with gene activation. Altogether, these chromatin modifications strongly suggest that the amplitude of HLA-G gene activation in glioblastoma cells after 5-aza-dC treatment is dependent on the permissive (or nonpermissive) statute of the chromatin at the HLA-G locus. Accordingly, further exploration in glioblastoma biopsy samples either expressing or not expressing HLA-G is justified and currently under investigation.
As previously demonstrated for JAR, Raji, and LCL 721.221 cell lines,33 5-aza-dC treatment of U251MG cells also directly induced the expression of HLA-G protein and its expression at the cell surface (Figure 4B). Conversely, HLA-G protein induction was not observed in U138MG and D247MG cells. Because we did not find any nonsense mutations in the coding region of the HLA-G alleles, we can hypothesize this result to be due either to lower levels of induced HLA-G mRNA compared to U251MG cells or to other regulatory mechanisms acting at the post-transcriptional level, such as miRNAs.58,64 In agreement, by investigating RNA and protein on the same tissue specimen (Figure 2), we observed that although HLA-G transcriptional activity was observed in all samples, HLA-G protein was detected only in four of seven samples.
It is noteworthy that the modulation of the expression of APM components showed in the present study is also a pertinent parameter to consider in the induction of HLA-G cell-surface expression. The effect of 5-aza-dC treatment on APM components was previously investigated on the melanoma cell line OGM-1A, showing TAP1, TAP2, LMP2, and tapasin increase.36 In the present work, we observed the up-regulation of some APM components but the increase was small compared with the effect of IFN-γ. In particular, reduced expression of the TAP1 molecule has been reported in glioma cell lines and, in agreement with our data, was up-regulated upon IFN-γ treatment.65 In addition, we noted that calreticulin expression was not up-regulated with both treatments. Interestingly, it has been reported that in neither the untreated nor treated OCM-1A cells, calreticulin expression was detected. Moreover HLA-G cell surface expression in 5-aza-dC–treated OCM-1A melanoma cells was much less than that on the HLA-G–positive JEG-3 cell, whereas a similar amount of total HLA-G was observed. Calreticulin was thus hypothesized to be critical for levels of HLA-G cell surface expression in OCM-1A,36 and therefore could be involved in the level of HLA-G cell surface expression in U251MG cells.
In conclusion, taken together, our data strongly suggest that new therapeutic strategies such us immunotherapy for glioma need to take into account antigens constitutively expressed by cells in the tumor lesion and their impact on glioma-associated immune suppression and invasion. Indeed, the down-regulation of some APM components in glioblastoma might not affect the expression of all HLA-G isoforms and HLA-G cell surface expression could be induced and/or up-regulated after treatments in permissive cells. The lack of HLA-G expression in some glioblastomas and the variability of HLA-G induction in vitro among cell lines also suggest that the HLA-G gene polymorphism56 should be taken into consideration, as it can affect the activity of transcription factors66 and miRNA binding.58,67 Therefore, it could be very useful to decide HLA-G typing in favor of or against therapeutic treatments using 5-aza-dC and IFN-γ.
Acknowledgments
We thank Prof. Heinz Wiendl for glioma cell lines, Deborah Sangrouber for preliminary experiments, Gustavo Martelli Palomino for HLA-G gene sequencing, and Nathalie Rouas-Freiss and Irene Krawice-Radanne for helpful advice in immunohistochemistry analyses. This manuscript is dedicated to Laurent Bachelard.
Footnotes
Supported by the Commissariat à l’Energie Atomique et aux Energies Alternatives (CEA), the binational research cooperation program CAPES/Brazil-COFECUB/France project Me 653/09, the French Embassy in Denmark (scientific and academic cooperation), PHS grants PO1CA109688 (S.Fe.), RO1CA104947 (S.Fe.), and RO1CA110249 (S.Fe.) awarded by the National Cancer Institute; a post-doctoral fellowship from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (I.J.W.), a post-doctoral fellowship from CAPES (Process no. 3260/06-2) (R.T.S.), an IRTELIS fellowship from CEA and Association pour la Recherche sur le Cancer (ARC No. DOC20110603234) (L.Y.), and a doctoral fellowship from CAPES-COFECUB (J.T.P.).
I.J.W. and R.T.S. contributed equally to this work.
Current address of I.J.W., Pontificial Catholic University of Goiás, Goiás, Brasil; of R.T.S., Núcleo de Pós-graduação e Pesquisa do Hospital da Santa Casa de Belo Horizonte, Brazil; of J.T.P., Departamento de Ciências da Saúde, Universidade Federal do Espírito Santo, São Mateus, Brazil; of S.Fl. INSERM, UMR1011, Université Lille Nord de France, Lille, France.
Supplemental Data
CpG methylation status of the 443 bp-HLA-G region upstream the ATG initiation codon in U251MG cells. Upper nucleotide sequence is from non-bisulfite-treated DNA compared with lower nucleotide sequence from bisulfite treated DNA. Methylated CpG (18) are in bold, the unique unmethylated CpG is underlined. The box indicates the ATG initiation codon. Efficiency of sodium bisulfite treatment was >98 %.
Persistence of the induced HLA-G expression in the absence of 5-aza-dC and IFN-γ treatment. U251MG cells were cultured during 5 days in the presence of 5-aza-dC and IFN-γ (5-aza-dC+IFN-γ) or absence (UT) and analyzed by real-time RT-PCR in triplicate for HLA-G transcriptional activity. Culture medium was changed at day 5 and cells were then cultured without any treatment and analyzed at day (D) 6 (UT D1), 7 (UT D2), and 8 (UT D3). The y axis indicates amounts of HLA-G mRNA levels relative to mRNA expression in HLA-G positive JEG-3 choriocarcinoma cells (assigned a value 1).
Supplemental Data
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2012.10.021.
References
- 1.Moreau P., Flajollet S., Carosella E.D. Non-classical transcriptional regulation of HLA-G: an update. J Cell Mol Med. 2009;13:2973–2989. doi: 10.1111/j.1582-4934.2009.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donadi E.A., Castelli E.C., Arnaiz-Villena A., Roger M., Rey D., Moreau P. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell Mol Life Sci. 2011;68:369–395. doi: 10.1007/s00018-010-0580-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishitani A., Geraghty D.E. Alternative splicing of HLA-G transcripts yields proteins with primary structures resembling both class I and class II antigens. Proc Natl Acad Sci U S A. 1992;89:3947–3951. doi: 10.1073/pnas.89.9.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirszenbaum M., Moreau P., Gluckman E., Dausset J., Carosella E. An alternatively spliced form of HLA-G mRNA in human trophoblasts and evidence for the presence of HLA-G transcript in adult lymphocytes. Proc Natl Acad Sci USA. 1994;91:4209–4213. doi: 10.1073/pnas.91.10.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rouas-Freiss N., Goncalves R.M., Menier C., Dausset J., Carosella E.D. Direct evidence to support the role of HLA-G in protecting the fetus from maternal uterine natural killer cytolysis. Proc Natl Acad Sci U S A. 1997;94:11520–11525. doi: 10.1073/pnas.94.21.11520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Gal F.A., Riteau B., Sedlik C., Khalil-Daher I., Menier C., Dausset J., Guillet J.G., Carosella E.D., Rouas-Freiss N. HLA-G-mediated inhibition of antigen-specific cytotoxic T lymphocytes. Int Immunol. 1999;11:1351–1356. doi: 10.1093/intimm/11.8.1351. [DOI] [PubMed] [Google Scholar]
- 7.Riteau B., Menier C., Khalil-Daher I., Sedlik C., Dausset J., Rouas-Freiss N., Carosella E.D. HLA-G inhibits the allogeneic proliferative response. J Reprod Immunol. 1999;43:203–211. doi: 10.1016/s0165-0378(99)00034-0. [DOI] [PubMed] [Google Scholar]
- 8.Colonna M., Navarro F., Bellon T., Llano M., Garcia P., Samaridis J., Angman L., Cella M., Lopez-Botet M. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J Exp Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colonna M., Samaridis J., Cella M., Angman L., Allen R.L., O'Callaghan C.A., Dunbar R., Ogg G.S., Cerundolo V., Rolink A. Human myelomonocytic cells express an inhibitory receptor for classical and nonclassical MHC class I molecules. J Immunol. 1998;160:3096–3100. [PubMed] [Google Scholar]
- 10.Rajagopalan S., Long E.O. A human histocompatibility leucocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med. 1999;189:1093–1099. doi: 10.1084/jem.189.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez-Lozano N., de Pablo R., Puente S., Vilches C. Recognition of HLA-G by the NK cell receptor KIR2DL4 is not essential for human reproduction. Eur J Immunol. 2003;33:639–644. doi: 10.1002/eji.200323741. [DOI] [PubMed] [Google Scholar]
- 12.Rajagopalan S. Endosomal signaling and a novel pathway defined by the natural killer receptor KIR2DL4 (CD158d) Traffic. 2010;11:1381–1390. doi: 10.1111/j.1600-0854.2010.01112.x. [DOI] [PubMed] [Google Scholar]
- 13.McMaster M.T., Librach C.L., Zhou Y., Lim K.H., Janatpour M.J., DeMars R., Kovats S., Damsky C., Fisher S.J. Human placental HLA-G expression is restricted to differentiated cytotrophoblasts. J Immunol. 1995;154:3771–3778. [PubMed] [Google Scholar]
- 14.Carosella E.D., Moreau P., Lemaoult J., Rouas-Freiss N. HLA-G: from biology to clinical benefits. Trends Immunol. 2008;29:125–132. doi: 10.1016/j.it.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Rouas-Freiss N., Moreau P., Menier C., LeMaoult J., Carosella E.D. Expression of tolerogenic HLA-G molecules in cancer prevents antitumor responses. Semin Cancer Biol. 2007;17:413–421. doi: 10.1016/j.semcancer.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Wagner S.N., Rebmann V., Willers C.P., Grosse-Wilde H., Goos M. Expression analysis of classic and non-classic HLA molecules before interferon alfa-2b treatment of melanoma. Lancet. 2000;356:220–221. doi: 10.1016/S0140-6736(00)02486-7. [DOI] [PubMed] [Google Scholar]
- 17.Nuckel H., Rebmann V., Durig J., Duhrsen U., Grosse-Wilde H. HLA-G expression is associated with an unfavorable outcome and immunodeficiency in chronic lymphocytic leukemia. Blood. 2005;105:1694–1698. doi: 10.1182/blood-2004-08-3335. [DOI] [PubMed] [Google Scholar]
- 18.Ghandri N, Gabbouj S, Farhat K, Bouaouina N, Abdelaziz H, Nouri A, Chouchane L, Hassen E: Association of HLA-G polymorphisms with nasopharyngeal carcinoma risk and clinical outcome. Hum Immunol 72:150–158 [DOI] [PubMed]
- 19.de Kruijf E.M., Sajet A., van Nes J.G., Natanov R., Putter H., Smit V.T., Liefers G.J., van den Elsen P.J., van de Velde C.J., Kuppen P.J. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J Immunol. 2010;185:7452–7459. doi: 10.4049/jimmunol.1002629. [DOI] [PubMed] [Google Scholar]
- 20.Lin A., Zhang X., Zhou W.J., Ruan Y.Y., Xu D.P., Wang Q., Yan W.H. HLA-G expression is associated with a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Cancer. 2011;129:1382–1390. doi: 10.1002/ijc.25807. [DOI] [PubMed] [Google Scholar]
- 21.Yie S.M., Yang H., Ye S.R., Li K., Dong D.D., Lin X.M. Expression of human leucocyte antigen G (HLA-G) is associated with prognosis in non-small cell lung cancer. Lung Cancer. 2007;58:267–274. doi: 10.1016/j.lungcan.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 22.Zhu C.B., Wang C.X., Zhang X., Zhang J., Li W. Serum sHLA-G levels: A useful indicator in distinguishing colorectal cancer from benign colorectal diseases. Int J Cancer. 2010;128:617–622. doi: 10.1002/ijc.25372. [DOI] [PubMed] [Google Scholar]
- 23.Morandi F., Levreri I., Bocca P., Galleni B., Raffaghello L., Ferrone S., Prigione I., Pistoia V. Human neuroblastoma cells trigger an immunosuppressive program in monocytes by stimulating soluble HLA-G release. Cancer Res. 2007;67:6433–6441. doi: 10.1158/0008-5472.CAN-06-4588. [DOI] [PubMed] [Google Scholar]
- 24.Agaugue S., Carosella E.D., Rouas-Freiss N. Role of HLA-G in tumor escape through expansion of myeloid-derived suppressor cells and cytokinic balance favoring Th2 vs Th1/Th17. Blood. 2011;117:7021–7031. doi: 10.1182/blood-2010-07-294389. [DOI] [PubMed] [Google Scholar]
- 25.Stupp R., Hegi M.E., Mason W.P., van den Bent M.J., Taphoorn M.J., Janzer R.C., Ludwin S.K., Allgeier A., Fisher B., Belanger K., Hau P., Brandes A.A., Gijtenbeek J., Marosi C., Vecht C.J., Mokhtari K., Wesseling P., Villa S., Eisenhauer E., Gorlia T., Weller M., Lacombe D., Cairncross J.G., Mirimanoff R.O. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 26.Wiendl H., Mitsdoerffer M., Hofmeister V., Wischhusen J., Bornemann A., Meyermann R., Weiss E.H., Melms A., Weller M. A functional role of HLA-G expression in human gliomas: an alternative strategy of immune escape. J Immunol. 2002;168:4772–4780. doi: 10.4049/jimmunol.168.9.4772. [DOI] [PubMed] [Google Scholar]
- 27.Kren L., Slaby O., Muckova K., Lzicarova E., Sova M., Vybihal V., Svoboda T., Fadrus P., Lakomy R., Vanhara P., Krenova Z., Sterba J., Smrcka M., Michalek J. Expression of immune-modulatory molecules HLA-G and HLA-E by tumor cells in glioblastomas: an unexpected prognostic significance? Neuropathology. 2011;31:129–134. doi: 10.1111/j.1440-1789.2010.01149.x. [DOI] [PubMed] [Google Scholar]
- 28.Kren L., Muckova K., Lzicarova E., Sova M., Vybihal V., Svoboda T., Fadrus P., Smrcka M., Slaby O., Lakomy R., Vanhara P., Krenova Z., Michalek J. Production of immune-modulatory nonclassical molecules HLA-G and HLA-E by tumor infiltrating ameboid microglia/macrophages in glioblastomas: a role in innate immunity? J Neuroimmunol. 2010;220:131–135. doi: 10.1016/j.jneuroim.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 29.Frumento G., Franchello S., Geraghty E., Ferrara G.B. Analysis of HLA-G expression in tumor cell lines. Transplant Proc. 1999;31:1847–1848. doi: 10.1016/s0041-1345(99)00185-2. [DOI] [PubMed] [Google Scholar]
- 30.Rouas-Freiss N., Moreau P., Ferrone S., Carosella E.D. HLA-G proteins in cancer: do they provide tumor cells with an escape mechanism? Cancer Res. 2005;65:10139–10144. doi: 10.1158/0008-5472.CAN-05-0097. [DOI] [PubMed] [Google Scholar]
- 31.Bukur J., Malenica B., Huber C., Seliger B. Altered expression of nonclassical HLA class Ib antigens in human renal cell carcinoma and its association with impaired immune response. Hum Immunol. 2003;64:1081–1092. doi: 10.1016/j.humimm.2003.08.350. [DOI] [PubMed] [Google Scholar]
- 32.Malmberg K.J., Levitsky V., Norell H., de Matos C.T., Carlsten M., Schedvins K., Rabbani H., Moretta A., Soderstrom K., Levitskaya J., Kiessling R. IFN-gamma protects short- term ovarian carcinoma cell lines from CTL lysis via a CD94/NKG2A-dependent mechanism. J Clin Invest. 2002;110:1515–1523. doi: 10.1172/JCI15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreau P., Mouillot G., Rousseau P., Marcou C., Dausset J., Carosella E.D. HLA-G gene repression is reversed by demethylation. Proc Natl Acad Sci U S A. 2003;100:1191–1196. doi: 10.1073/pnas.0337539100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mouillot G., Marcou C., Rousseau P., Rouas-Freiss N., Carosella E.D., Moreau P. HLA-G gene activation in tumor cells involves cis-acting epigenetic changes. Int J Cancer. 2005;113:928–936. doi: 10.1002/ijc.20682. [DOI] [PubMed] [Google Scholar]
- 35.Park B., Ahn K. An essential function of tapasin in quality control of HLA-G molecules. J Biol Chem. 2003;278:14337–14345. doi: 10.1074/jbc.M212882200. [DOI] [PubMed] [Google Scholar]
- 36.Yan W.H., Lin A.F., Chang C.C., Ferrone S. Induction of HLA-G expression in a melanoma cell line OCM-1A following the treatment with 5-aza-2′-deoxycytidine. Cell Res. 2005;15:523–531. doi: 10.1038/sj.cr.7290376. [DOI] [PubMed] [Google Scholar]
- 37.Facoetti A., Nano R., Zelini P., Morbini P., Benericetti E., Ceroni M., Campoli M., Ferrone S. Human leukocyte antigen and antigen processing machinery component defects in astrocytic tumors. Clin Cancer Res. 2005;11:8304–8311. doi: 10.1158/1078-0432.CCR-04-2588. [DOI] [PubMed] [Google Scholar]
- 38.Castelli E.C., Mendes-Junior C.T., Veiga-Castelli L.C., Roger M., Moreau P., Donadi E.A. A comprehensive study of polymorphic sites along the HLA-G gene: implication for gene regulation and evolution. Mol Biol Evol. 2011;28:3069–3086. doi: 10.1093/molbev/msr138. [DOI] [PubMed] [Google Scholar]
- 39.Lampson L.A., Fisher C.A., Whelan J.P. Striking paucity of HLA-A, B, C and beta 2-microglobulin on human neuroblastoma cell lines. J Immunol. 1983;130:2471–2478. [PubMed] [Google Scholar]
- 40.Bandoh N., Ogino T., Cho H.S., Hur S.Y., Shen J., Wang X., Kato S., Miyokawa N., Harabuchi Y., Ferrone S. Development and characterization of human constitutive proteasome and immunoproteasome subunit-specific monoclonal antibodies. Tissue Antigens. 2005;66:185–194. doi: 10.1111/j.1399-0039.2005.00462.x. [DOI] [PubMed] [Google Scholar]
- 41.Ogino T., Wang X., Kato S., Miyokawa N., Harabuchi Y., Ferrone S. Endoplasmic reticulum chaperone-specific monoclonal antibodies for flow cytometry and immunohistochemical staining. Tissue Antigens. 2003;62:385–393. doi: 10.1034/j.1399-0039.2003.00114.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang X., Campoli M., Cho H.S., Ogino T., Bandoh N., Shen J., Hur S.Y., Kageshita T., Ferrone S. A method to generate antigen-specific mAb capable of staining formalin-fixed, paraffin-embedded tissue sections. J Immunol Methods. 2005;299:139–151. doi: 10.1016/j.jim.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 43.Temponi M., Kageshita T., Perosa F., Ono R., Okada H., Ferrone S. Purification of murine IgG monoclonal antibodies by precipitation with caprylic acid: comparison with other methods of purification. Hybridoma. 1989;8:85–95. doi: 10.1089/hyb.1989.8.85. [DOI] [PubMed] [Google Scholar]
- 44.Larochelle C., Alvarez J.I., Prat A. How do immune cells overcome the blood-brain barrier in multiple sclerosis? FEBS Lett. 2011;585:3770–3780. doi: 10.1016/j.febslet.2011.04.066. [DOI] [PubMed] [Google Scholar]
- 45.Yang I., Tihan T., Han S.J., Wrensch M.R., Wiencke J., Sughrue M.E., Parsa A.T. CD8+ T-cell infiltrate in newly diagnosed glioblastoma is associated with long-term survival. J Clin Neurosci. 2010;17:1381–1385. doi: 10.1016/j.jocn.2010.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang I., Han S.J., Kaur G., Crane C., Parsa A.T. The role of microglia in central nervous system immunity and glioma immunology. J Clin Neurosci. 2010;17:6–10. doi: 10.1016/j.jocn.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gomez G.G., Kruse C.A. Mechanisms of malignant glioma immune resistance and sources of immunosuppression. Gene Ther Mol Biol. 2006;10:133–146. [PMC free article] [PubMed] [Google Scholar]
- 48.Wintterle S., Schreiner B., Mitsdoerffer M., Schneider D., Chen L., Meyermann R., Weller M., Wiendl H. Expression of the B7-related molecule B7-H1 by glioma cells: a potential mechanism of immune paralysis. Cancer Res. 2003;63:7462–7467. [PubMed] [Google Scholar]
- 49.Brantley E.C., Nabors L.B., Gillespie G.Y., Choi Y.H., Palmer C.A., Harrison K., Roarty K., Benveniste E.N. Loss of protein inhibitors of activated STAT-3 expression in glioblastoma multiforme tumors: implications for STAT-3 activation and gene expression. Clin Cancer Res. 2008;14:4694–4704. doi: 10.1158/1078-0432.CCR-08-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mehling M., Simon P., Mittelbronn M., Meyermann R., Ferrone S., Weller M., Wiendl H. WHO grade associated downregulation of MHC class I antigen-processing machinery components in human astrocytomas: does it reflect a potential immune escape mechanism? Acta Neuropathol. 2007;114:111–119. doi: 10.1007/s00401-007-0231-8. [DOI] [PubMed] [Google Scholar]
- 51.de Tayrac M., Etcheverry A., Aubry M., Saikali S., Hamlat A., Quillien V., Le Treut A., Galibert M.D., Mosser J. Integrative genome-wide analysis reveals a robust genomic glioblastoma signature associated with copy number driving changes in gene expression. Genes Chromosomes Cancer. 2009;48:55–68. doi: 10.1002/gcc.20618. [DOI] [PubMed] [Google Scholar]
- 52.Gonen-Gross T., Achdout H., Arnon T.I., Gazit R., Stern N., Horejsi V., Goldman-Wohl D., Yagel S., Mandelboim O. The CD85J/leukocyte inhibitory receptor-1 distinguishes between conformed and beta2-microglobulin-free HLA-G molecules. J Immunol. 2005;175:4866–4874. doi: 10.4049/jimmunol.175.8.4866. [DOI] [PubMed] [Google Scholar]
- 53.Shiroishi M., Kuroki K., Rasubala L., Tsumoto K., Kumagai I., Kurimoto E., Kato K., Kohda D., Maenaka K. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d) Proc Natl Acad Sci U S A. 2006;103:16412–16417. doi: 10.1073/pnas.0605228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amiot L., Ferrone S., Grosse-Wilde H., Seliger B. Biology of HLA-G in cancer: a candidate molecule for therapeutic intervention? Cell Mol Life Sci. 2011;68:417–431. doi: 10.1007/s00018-010-0583-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiendl H., Mitsdoerffer M., Weller M. Hide-and-seek in the brain: a role for HLA-G mediating immune privilege for glioma cells. Semin Cancer Biol. 2003;13:343–351. doi: 10.1016/s1044-579x(03)00025-7. [DOI] [PubMed] [Google Scholar]
- 56.Donadi E.A., Castelli E.C., Arnaiz-Villena A., Roger M., Rey D., Moreau P. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell Mol Life Sci. 2011;68:369–395. doi: 10.1007/s00018-010-0580-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan Z., Shon A.M., Ober C. Evidence of balancing selection at the HLA-G promoter region. Hum Mol Genet. 2005;14:3619–3628. doi: 10.1093/hmg/ddi389. [DOI] [PubMed] [Google Scholar]
- 58.Tan Z., Randall G., Fan J., Camoretti-Mercado B., Brockman-Schneider R., Pan L., Solway J., Gern J.E., Lemanske R.F., Nicolae D., Ober C. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waha A., Guntner S., Huang T.H., Yan P.S., Arslan B., Pietsch T., Wiestler O.D. Epigenetic silencing of the protocadherin family member PCDH-gamma-A11 in astrocytomas. Neoplasia. 2005;7:193–199. doi: 10.1593/neo.04490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu G., Ying H., Zeng G., Wheeler C.J., Black K.L., Yu J.S. HER-2, gp100, and MAGE-1 are expressed in human glioblastoma and recognized by cytotoxic T cells. Cancer Res. 2004;64:4980–4986. doi: 10.1158/0008-5472.CAN-03-3504. [DOI] [PubMed] [Google Scholar]
- 61.Patel R., Shervington L., Lea R., Shervington A. Epigenetic silencing of telomerase and a non-alkylating agent as a novel therapeutic approach for glioma. Brain Res. 2008;1188:173–181. doi: 10.1016/j.brainres.2007.10.043. [DOI] [PubMed] [Google Scholar]
- 62.Teyssier M., Bensussan A., Kirszenbaum M., Moreau P., Gluckman E., Dausset J., Carosella E. Natural killer cells are the unique lymphocyte cell subset which do not express HLA-G. Nat Immun. 1995;14:262–270. [PubMed] [Google Scholar]
- 63.Cao W., Yang X., Zhou J., Teng Z., Cao L., Zhang X., Fei Z. Targeting 14-3-3 protein, difopein induces apoptosis of human glioma cells and suppresses tumor growth in mice. Apoptosis. 2010;15:230–241. doi: 10.1007/s10495-009-0437-4. [DOI] [PubMed] [Google Scholar]
- 64.Castelli E.C., Moreau P., Oya e Chiromatzo A., Mendes-Junior C.T., Veiga-Castelli L.C., Yaghi L., Giuliatti S., Carosella E.D., Donadi E.A. In silico analysis of microRNAS targeting the HLA-G 3′ untranslated region alleles and haplotypes. Hum Immunol. 2009;70:1020–1025. doi: 10.1016/j.humimm.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 65.Satoh E., Mabuchi T., Satoh H., Asahara T., Nukui H., Naganuma H. Reduced expression of the transporter associated with antigen processing 1 molecule in malignant glioma cells, and its restoration by interferon-gamma and -beta. J Neurosurg. 2006;104:264–271. doi: 10.3171/jns.2006.104.2.264. [DOI] [PubMed] [Google Scholar]
- 66.Ober C., Billstrand C., Kuldanek S., Tan Z. The miscarriage-associated HLA-G -725G allele influences transcription rates in JEG-3 cells. Hum Reprod. 2006;21:1743–1748. doi: 10.1093/humrep/del036. [DOI] [PubMed] [Google Scholar]
- 67.Castelli E.C., Mendes-Junior C.T., Deghaide N.H., de Albuquerque R.S., Muniz Y.C., Simoes R.T., Carosella E.D., Moreau P., Donadi E.A. The genetic structure of 3′untranslated region of the HLA-G gene: polymorphisms and haplotypes. Genes Immun. 2010;11:134–141. doi: 10.1038/gene.2009.74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CpG methylation status of the 443 bp-HLA-G region upstream the ATG initiation codon in U251MG cells. Upper nucleotide sequence is from non-bisulfite-treated DNA compared with lower nucleotide sequence from bisulfite treated DNA. Methylated CpG (18) are in bold, the unique unmethylated CpG is underlined. The box indicates the ATG initiation codon. Efficiency of sodium bisulfite treatment was >98 %.
Persistence of the induced HLA-G expression in the absence of 5-aza-dC and IFN-γ treatment. U251MG cells were cultured during 5 days in the presence of 5-aza-dC and IFN-γ (5-aza-dC+IFN-γ) or absence (UT) and analyzed by real-time RT-PCR in triplicate for HLA-G transcriptional activity. Culture medium was changed at day 5 and cells were then cultured without any treatment and analyzed at day (D) 6 (UT D1), 7 (UT D2), and 8 (UT D3). The y axis indicates amounts of HLA-G mRNA levels relative to mRNA expression in HLA-G positive JEG-3 choriocarcinoma cells (assigned a value 1).