Since the discovery in 2005 of the essential role of the liver-specific microRNA, miR-122, in HCV replication (1), the mechanism by which it stimulates this process has proved elusive. In PNAS, Li et al. demonstrate that miR-122 acts to shield the HCV genome against degradation by the cytosolic RNA exonuclease, Xrn1 (2). Although this may be one way in which this microRNA promotes HCV replication, the authors also show that loss of Xrn1 is not enough to promote miR-122 independent HCV replication, indicating that miR-122 exerts yet another function in the HCV lifecycle.
Most microRNAs function to suppress gene expression, either through transcript cleavage and degradation, when complete complementarity is present between the microRNA and target, or suppression of translation, when mismatches exist between these species (3). This scenario is completely reversed for HCV and miR-122. Here, the binding of miR-122 to two sites at the far 5′ end of the HCV positive-strand RNA genome does not result in RNA degradation or in translational arrest, but instead miR-122 is essential for HCV replication (Fig. 1). MicroRNAs usually bind to specific RNAs by complementarity between the so-called seed sequence of the microRNA and the target RNA sequence. Enhancement of HCV RNA replication requires both miR-122 seed sequence interactions, as well as interactions outside of their seed sequences (4). The HCV genome does not contain a cap structure at the 5′ end, which functions on cellular RNAs to promote translation, by recruiting ribosomal components, and stability, by shielding the normally single-stranded 5′ end from exonuclease digestion (5). Indeed, it was recently shown that miR-122 increases HCV RNA stability in much the same manner as a cap (6). However, the manner of degradation that miR-122 prevents and the extent to which this increase in stability plays into the overall impact of miR-122 on HCV replication was not determined. These are the questions that Li et al. set out to answer in their PNAS publication (2).
Fig. 1.
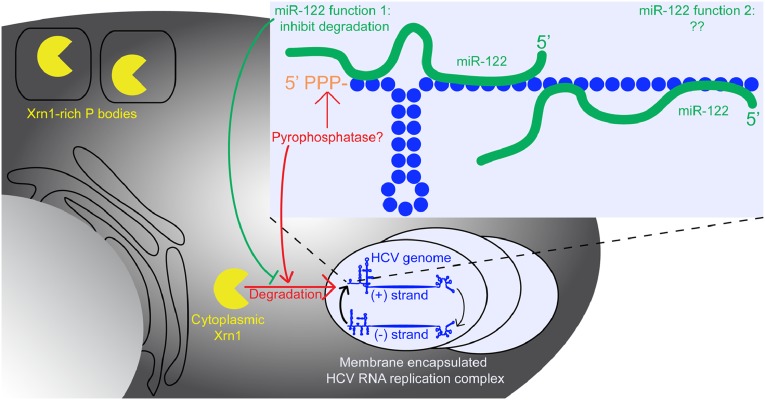
miR-122 impacts HCV RNA stability and HCV RNA replication. Illustration of HCV RNA replication within membrane bound vesicles in the host cell cytoplasm. Also shown is a zoomed-in view of the 5′ end of the HCV genomic RNA with two bound miR-122 molecules. Cytoplasmic, but not P body-associated, Xrn1 degrades the HCV genomic RNA, perhaps after the 5′ triphosphate is modified by a pyrophosphatase. Although binding of miR-122 to the HCV RNA inhibits Xrn1 degradation, miR-122 also exerts another effect to enhance HCV RNA replication.
Li et al. first tackle the question how HCV RNA is degraded by host cells (2). In eukaryotes, the exosome and Xrn1 represent two potential mRNA decay pathways (5). The exosome degrades transcripts in a 3′ to 5′ direction following deadenylation. Xrn1 is a cytoplasmic 5′ to 3′ exonuclease that degrades transcripts after the cap is removed by a decapping complex. In initial experiments, the authors find that both pathways impact the stability of HCV RNA when transfected into host cells, and HCV RNA was particularly sensitive to exosomal degradation in cell lysates. However, when examined in cells bearing actively replicating HCV RNA, only Xrn1 impacted viral RNA stability. The authors then show that miR-122 acts to protect the HCV RNA from Xrn1 degradation, as addition of extra miR-122 to cells increased HCV RNA stability to the same extent as Xrn1 silencing, and the combination of both did not exhibit an additive effect.
Based on this finding, Li et al. then proceed to test the impact of Xrn1 on HCV replication and how this may be modulated by the presence of miR-122 (2). Indeed, silencing of Xrn1, but not components of the exosome, enhanced HCV RNA replication about twofold, indicating that the Xrn1 degradation pathways negatively regulate HCV RNA replication. However, supplementing cells with additional miR-122 enhanced HCV replication even when Xrn1 was silenced. Furthermore, a mutant HCV genome bearing changes to both miR-122 binding sites, which can be efficiently rescued by expression of a mutant miR-122 with complementary changes, was not rendered replication competent by Xrn1 silencing. The authors conclude from these results that miR-122 acts to shield the HCV RNA from Xrn1 degradation and to perform another Xrn1-independent function in enhancing HCV replication (2).
Regardless of the mode of action of miR-122 in the HCV lifecycle, dissection of the way HCV RNA decays is an important advance toward understanding how HCV interacts with its host. Li et al. (2) speculate that the distinct role that Xrn1 plays in the turnover of HCV RNA in replicating cells may reflect the specific location where viral RNA replication takes place, i.e., membrane compartments that encapsulate the HCV RNA replication complex. Nucleases may not have access to this compartment, and therefore viral RNA may only be degraded when these compartments break down or when RNA is released for other events such as packaging into virions. Xrn1 is specifically concentrated in P bodies, but the authors show that HCV RNA is not present in these structures, and instead only colocalizes with Xrn1 in the cytoplasm of infected cells (2). Another interesting consideration discussed by the authors is that Xrn1 is active against only the 5′ end of RNA molecules bearing a monophosphate. The HCV RNA, which has a triphosphate at its 5′ end, would first need to be modified by a pyrophosphatase to be susceptible to Xrn1 mediated decay. Although miR-122 may also play a role in inhibiting this modification, as this would be in the same pathway as Xrn1 degradation, it is unlikely to account for the additional role of miR-122 in the HCV lifecycle the authors observe.
One might also speculate about whether the negative, or complementary, HCV RNA strand is also affected by Xrn1. The negative sense HCV RNA is not known to bind miR-122 or any other microRNA. As this strand is known only to function as a replication intermediate, it may never be actively released from the membrane bound replication compartment and therefore be protected from Xrn1 degradation. Additionally, the 5′ stem structure present at the end of the HCV negative RNA may serve to increase stability. Indeed, the HCV-related pestiviruses contain double-stranded stem structures at the 5′ ends of their viral RNAs that may enhance RNA stability. Why HCV would not have evolved a similar mechanism to stabilize its genome is unclear. Perhaps this difference is reflective of the additional undefined role miR-122 plays in HCV replication—as this microRNA is recruited to the HCV genome for an Xrn1-independent reason, using it to enhance stability may have been merely a function that the miR-122 conveniently took on secondary to another role.
One interesting outstanding question concerns the roles of the two separate miR-122 binding sites in HCV replication. Although it has not been conclusively shown that both are simultaneously occupied by miR-122 molecules, both sites are important to HCV replication. If the primary mechanism by which miR-122 stabilizes the HCV genome is by base pairing with the single-stranded sequence at the very distal viral RNA 5′ end, one would expect
Li et al. demonstrate that miR-122 acts to shield the HCV genome against degradation by the cytosolic RNA exonuclease, Xrn1.
only the microRNA that binds to the first site to exert this effect. However, it is possible that proteins miR-122 brings to the HCV genome, such as the RISC complex, may be responsible for the shielding effect from Xrn1, and such an effect may be mediated through miR-122 binding to either site. On the contrary, if the second site does not participate in regulating HCV genome stability, perhaps it is more important for the additional uncharacterized mechanism by which this microRNA stimulates HCV replication. Such possibilities could be explored by mutating each site individually and assaying the impact on HCV replication and stability in the absence and presence of microRNA mutants with complementary changes.
Of course, another intriguing question that is highlighted by the study of Li et al. (2) regards the identity of the still undefined mode of action of miR-122 in the HCV life cycle. The authors rule out a role for miR-122 in viral translation and instead suggest that miR-122 may be important in initiating RNA synthesis or establishing/assembling a functional RNA replication complex. In such a model, miR-122 may act as a bridge to bring factors involved in RNA replication to the HCV genome, or act to correctly position such factors at particular regions in the genome to enhance RNA replication. Alternatively, miR-122 may act to bring the HCV RNA to distinct intracellular locations required for the life cycle of the virus. One would expect that, by modulating the functions of miR-122 that are required for HCV replication, one could render HCV replication independent of this microRNA, as attempted by Li et al. (2). Although miR-122 appears to be universally required by all HCV genomes, one recent study found that the HCV replication could be rendered miR-122–independent by replacement of the first miR-122 binding site and a stem-loop structure at the 5′ end of the genome with a stem-loop from the U3 small nucleolar RNA (7). It would be interesting to examine how degradation of this mutant is affected by Xrn1 or whether it can be used to uncover non-Xrn1 miR-122 HCV replication mechanisms.
Ultimately, the findings by Li et al. provide clear-cut mechanistic details by which miR-122 acts to enhance HCV RNA stability and thus provide essential insights into its role in the viral life cycle, as well as provide exciting new directions to further define this as yet unique use of a microRNA by a viral pathogen. The study of viruses often yields insights into normal cellular processes that would be difficult to observe otherwise. The finding that HCV replication requires miR-122 revealed that a microRNA can be used for functions other than suppressing gene expression. Although the use of a microRNA to promote RNA stability is startling, we expect that further research in this area is likely to give clues into yet additional previously unrecognized functions of microRNAs.
Footnotes
The authors declare no conflict of interest.
See companion article on page 1881.
References
- 1.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309(5740):1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, et al. Competing and noncompeting activities of miR-122 and the 5′ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc Natl Acad Sci USA. 2012;110:1881–1886. doi: 10.1073/pnas.1213515110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: A look under the hood of miRISC. Nat Struct Mol Biol. 2012;19(6):586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 4.Machlin ES, Sarnow P, Sagan SM. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci USA. 2011;108(8):3193–3198. doi: 10.1073/pnas.1012464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8(2):113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 6.Shimakami T, et al. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci USA. 2012;109(3):941–946. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li YP, Gottwein JM, Scheel TK, Jensen TB, Bukh J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1-6 and reduced efficacy by host RNA insertion or mutations in the HCV 5′ UTR. Proc Natl Acad Sci USA. 2011;108(12):4991–4996. doi: 10.1073/pnas.1016606108. [DOI] [PMC free article] [PubMed] [Google Scholar]