Abstract
Here we examined the involvement of Notch signaling in the endochondral ossification process, which is crucial for osteoarthritis (OA) development. Intracellular domains of Notch1 and -2 were translocated into the nucleus of chondrocytes with their differentiation in mouse limb cartilage and in mouse and human OA articular cartilage. A tissue-specific inactivation of the Notch transcriptional effector recombination signal binding protein for Ig kappa J (RBPjκ) in chondroprogenitor cells of SRY-box containing gene 9 (Sox9)-Cre;Rbpjfl/fl mouse embryos caused an impaired terminal stage of endochondral ossification in the limb cartilage. The RBPjκ inactivation in adult articular cartilage after normal skeletal growth using type II collagen (Col2a1)-CreERT;Rbpjfl/fl mice by tamoxifen injection caused resistance to OA development in the knee joint. Notch intracellular domain with the effector RBPjκ stimulated endochondral ossification through induction of the target gene Hes1 in chondrocytes. Among the Notch ligands, Jagged1 was strongly induced during OA development. Finally, intraarticular injection of N-[N-(3,5-diflurophenylacetate)-l-alanyl]-(S)-phenylglycine t-butyl ester (DAPT), a small compound Notch inhibitor, to the mouse knee joint prevented OA development. The RBPjκ-dependent Notch signaling in chondrocytes modulates the terminal stage of endochondral ossification and OA development, representing an extracellular therapeutic target of OA.
Keywords: skeletal development, cartilage degradation
Endochondral ossification is an essential process not only for physiological skeletal growth (1) but also for development of osteoarthritis (OA), which is the most prevalent form of arthritis with articular cartilage degradation (2, 3). In the process, after chondrocytes undergo hypertrophic differentiation characterized by secretion of type X collagen (COL10A1), the avascular cartilage tissue is converted into highly vascularized bone tissue via degradation of the cartilage matrix and vascular invasion (1, 4). The matrix degradation requires proteinases, among which matrix metalloproteinase-13 (MMP13) plays a major role (4, 5), and the vascular invasion depends on an angiogenic switch by vascular endothelial growth factor A (VEGFA) (6).
Notch is a single-pass transmembrane cell surface receptor, which plays a crucial role in cell fate assignment by regulating differentiation and apoptosis during embryogenesis as well as various developmental systems like neurogenesis and hematopoiesis (7, 8). In mammals, the signaling is initiated by binding of a membrane ligand (Delta-like 1, 3, 4, or Jagged1 and -2) to the Notch receptor (Notch1–4) on the adjacent cell (9). Upon binding, the Notch receptor is cleaved by proteinases like a disintegrin and metalloproteinase domain-containing proteins, and subsequently by the γ-secretase complex, and the Notch intracellular domain (ICD) is released in the cytoplasm. Then, the Notch ICD translocates to the nucleus and binds to the transcriptional effector recombination signal binding protein for Ig kappa J (RBPjκ), converting it into an activator and ultimately inducing the expression of downstream target genes like members of the hairy and enhancer of split (Hes) / hairy/enhancer-of-split related with YRPW motif (Hey) family of basic helix–loop–helix transcription factors: Hes1, Hes5, Hes7, Hey1, Hey2, and HeyL (10, 11). Accumulating evidence has shown that these Notch family members are highly expressed in chondrocytes of developmental skeletal cartilage (12–14). Furthermore, a recent report disclosed that chondrocyte differentiation is regulated via the RBPjκ-dependent Notch signaling in chondrocytes (15). The Notch family members are also expressed in adult articular cartilage (16, 17), and over 70% of chondrocytes on the surface zone of articular cartilage express Notch1 receptor (18), suggesting a significant role of the Notch signaling in regulating articular cartilage homeostasis during adult life (16, 19). A recent study demonstrated that the Notch pathway is activated in OA cartilage because several Notch signaling molecules are much more abundant than in healthy cartilage (14, 16). Here we examined the involvement of the RBPjκ-dependent Notch signaling in the endochondral ossification process during skeletal growth and OA development. We further investigated the underlying mechanism and the possibility of clinical application of the signaling as a therapeutic target of OA.
Results
Notch Signaling Molecules Are Expressed and Activated During Chondrocyte Differentiation.
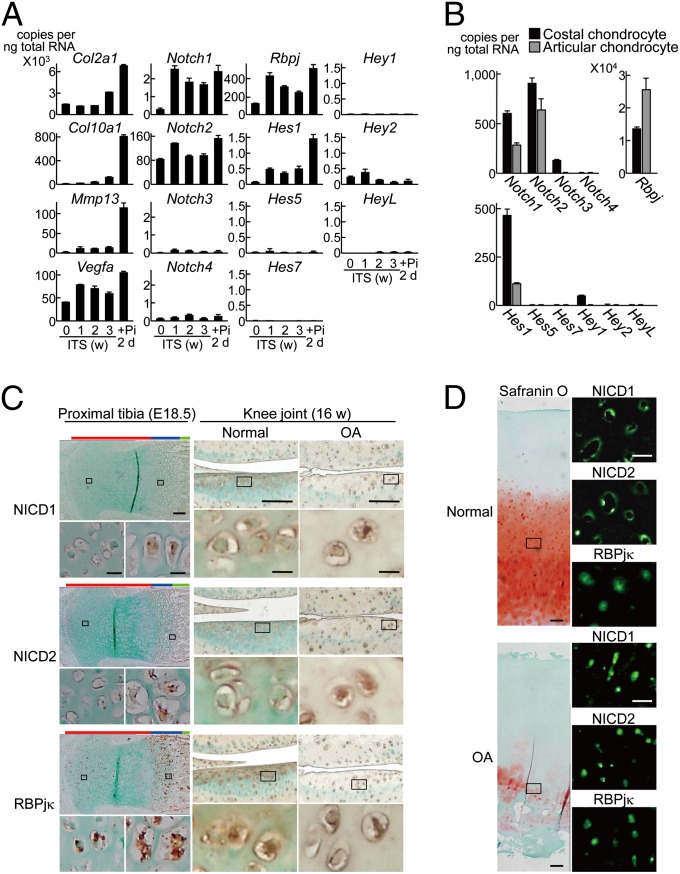
We initially looked at the in vitro and in vivo expression patterns of the Notch family members during chondrocyte differentiation. In cultures of mouse chondrogenic cell line ATDC5 and primary chondrocytes, Notch1, -2, Rbpj, and Hes1 were strongly expressed during the differentiation, whereas Notch3 and -4 and other Hes/Hey family members were little expressed (Fig. 1 A and B). In the mouse limb cartilage and knee articular cartilage, ICDs of Notch1 and Notch2 remained in the plasma membrane of less differentiated chondrocytes of the proliferative zone and undegraded normal cartilage; however, both were translocated into the nucleus of highly differentiated chondrocytes in the hypertrophic zone and in the degraded OA cartilage (Fig. 1C). The translocation of the ICDs into the nucleus of chondrocytes during OA development was reproducible in the human knee OA cartilage (Fig. 1D). Contrarily, RBPjκ remained in the nucleus without being affected by the differentiation stages or OA induction in both mouse and human samples (Fig. 1 C and D).
Fig. 1.
In vitro and in vivo expression patterns of the Notch signaling molecules during chondrocyte differentiation and OA development. (A) Time course of mRNA levels of Notch signaling molecules, and Col2a1, Col10a1, Mmp13, and Vegfa during differentiation of mouse chondrogenic ATDC5 cells cultured with insulin, transferrin, and sodium selenite (ITS) for 3 wk and for 2 d more with inorganic phosphate (Pi). Data are expressed as means ± SD. (B) mRNA levels of the Notch signaling molecules above in mouse primary costal chondrocytes and articular chondrocytes cultured for 5 and 7 d, respectively. (C) Immunostaining with antibodies to ICDs of Notch1 and Notch2 (NICD1 and NICD2), and RBPjκ in the mouse proximal tibia (E18.5) (Left; red, blue, and green bars to the Top indicate layers of proliferative, hypertrophic zones, and bone area, respectively) and in the mouse knee articular cartilage with or without surgical OA induction for 8 wk (16 wk old) (Right). Insets indicate the regions shown in the enlarged images immediately below. (Scale bars, 100 μm and 10 μm for low and high magnification images, respectively.) (D) Safranin-O staining and immunofluorescence with antibodies to NICD1, NICD2, and RBPjκ in the normal and OA cartilage of human knee joints. Insets in the safranin O staining indicate the regions shown in the enlarged immunofluorescence images. (Scale bars, 200 μm and 50 μm for low and high magnification images, respectively.)
RBPjκ-Dependent Notch Signaling Modulates Physiological Endochondral Ossification.
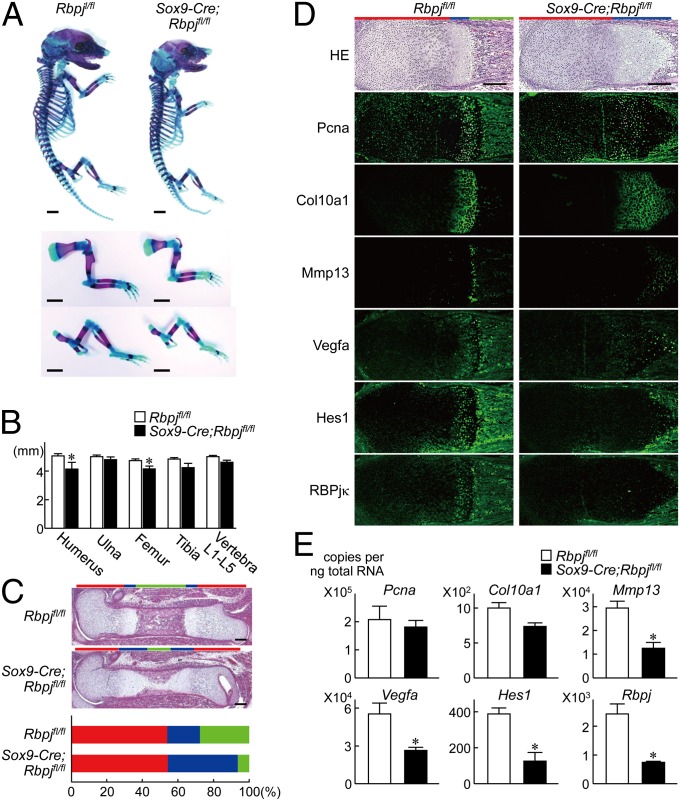
To examine the physiological role of the Notch signaling in endochondral ossification and skeletal development, we conditionally inactivated RBPjκ in chondroprogenitor cells by generating tissue-specific knockout mice by mating mice in which an internal ribosome entry site and a Cre recombinase gene were inserted into the 3′ untranslated region of the SRY-box containing gene 9 (Sox9) gene (Sox9-Cre) (20) with mice homozygous for a floxed Rbpj allele (Rbpjfl/fl) (21). Although the conditional knockout (Sox9-Cre;Rbpjfl/fl) mice died shortly after birth, they showed a slight dwarfism during the embryonic periods compared with the Rbpjfl/fl littermates (Fig. 2A): the limbs and vertebrae were about 5–10% shorter in Sox9-Cre;Rbpjfl/fl mice than in these littermates (Fig. 2B). Histological examination revealed that the percentage of the proliferative zone relative to the total limb length was comparable between the two genotypes, suggesting unaffected proliferation and hypertrophic differentiation of chondrocytes by the RBPjκ knockout (Fig. 2C). However, the percentage of the hypertrophic zone relative to the limb length was much increased, whereas that of the bone area was considerably decreased in the Sox9-Cre;Rbpjfl/fl limbs, indicating that the RBPjκ knockout impaired the terminal differentiation stage in such aspects as matrix degradation and vascular invasion (Fig. 2C). Immunofluorescence showed that intensities of the proliferating cell nuclear antigen (Pcna) and Col10a1 were comparable, whereas those of Mmp13 and Vegfa, as well as Hes1, were decreased by the RBPjκ knockout (Fig. 2D). These were confirmed by the expressions of these factors in the pellet cultures of primary chondrocytes derived from the two genotypes (Fig. 2E).
Fig. 2.
Skeletal abnormality in Sox9-Cre;Rbpjfl/fl mouse embryos. (A) Double staining with Alizarin red and Alcian blue of the whole skeleton (Top), upper extremities (Middle), and lower extremities (Bottom) of Rbpjfl/fl and Sox9-Cre;Rbpjfl/fl littermate embryos (E17.5). (Scale bars, 1 mm.) (B) Length of long bones and vertebra (first to fifth lumbar spines) of Rbpjfl/fl and Sox9-Cre;Rbpjfl/fl littermate embryos. Data are expressed as means ± SD of six mice per group. *P < 0.05 versus Rbpjfl/fl. (C) H&E staining of whole femurs of the Rbpjfl/fl and Sox9-Cre;Rbpjfl/fl littermate embryos. (Scale bars, 200 μm.) Lower graph indicates percentage of the length of proliferative zone (red), hypertrophic zone (blue), and bone area (green) over the total femoral length of the Rbpjfl/fl and Sox9-Cre;Rbpjfl/fl littermate embryos. (D) H&E staining and immunofluorescence with antibodies to Pcna, Col10a1, Mmp13, Vegfa, Hes1, and RBPjκ of distal femurs of the Rbpjfl/fl and Sox9-Cre;Rbpjfl/fl littermate embryos. Red, blue, and green bars (Upper) indicate layers of proliferative, hypertrophic zones, and bone area, respectively. (Scale bars, 200 μm.) (E) mRNA levels of Pcna, Col10a1, Mmp13, Vegfa, Hes1, and Rbpj in the pellet cultures of primary costal chondrocytes derived from Rbpjfl/fl and Sox9-Cre;Rbpjfl/fl littermate embryos. Data are expressed as means ± SD *P < 0.01 versus Rbpjfl/fl.
RBPjκ-Dependent Notch Signaling Modulates OA Development.
We next examined the contribution of RBPjκ in cartilage to OA development in which pathological endochondral ossification in the adult articular cartilage is known to play a crucial role (2). Because the Sox9-Cre;Rbpjfl/fl mice died shortly after birth, we sought to create RBPjκ knockout mice that would undergo normal skeletal growth and joint formation under physiological conditions. To inactivate RBPjκ in later stages of chondrocyte differentiation than Sox9-Cre;Rbpjfl/fl mice, we generated conditional knockout mice by mating type II collagen (Col2a1) promoter-driven Cre-transgenic mice (Col2a1-Cre) (22) with the Rbpjfl/fl mice. Among three lines of the Col2a1-Cre;Rbpjfl/fl mice obtained, two were perinatal lethal as previously reported (23); however, one line survived even after birth and grew normally without skeletal abnormality (Fig. S1A). This may be because the RBPjκ inactivation in cartilage was not complete but partial in this line (Fig. S1B). Our further analysis of the knee joints showed that joint morphology as well as phenotypes of articular cartilage, meniscus, and subchondral bone were comparable to the Rbpjfl/fl littermates under physiological conditions (Fig. S1C). However, when we created the surgical OA model through induction of instability to the knee joints (24), the cartilage degradation of the Col2a1-Cre;Rbpjfl/fl joints after 8 wk was suppressed compared with the Rbpjfl/fl littermate joints (Fig. S1D). This suppression was associated with the decreases in expressions of Mmp13, Vegfa, and Hes1, but not Col10a1, indicating prevention of the terminal stage of endochondral ossification by the RBPjκ insufficiency, similar to the Sox9-Cre;Rbpjfl/fl limb cartilage. Quantification by grading systems (25) confirmed that the RBPjκ insufficiency caused significant resistance to OA development (Fig. S1E).
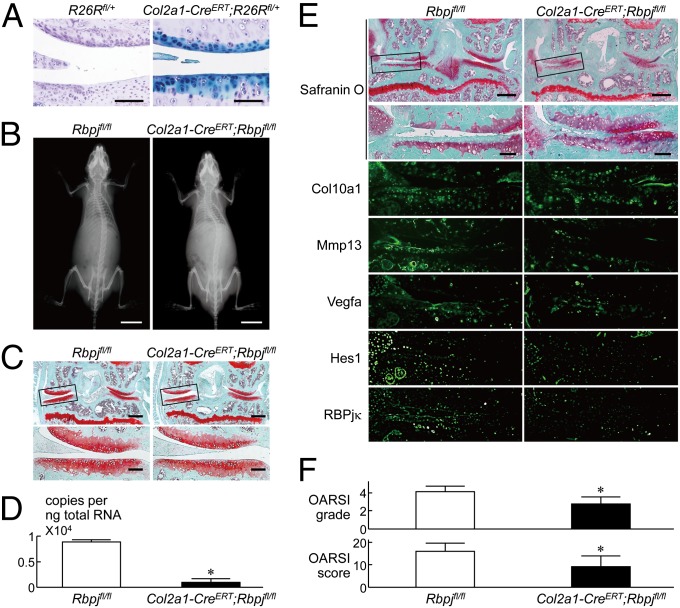
To further learn the effect of RBPjκ inactivation in adult articular cartilage after skeletal growth, we used transgenic mice in which the Cre recombinase was fused to a mutated ligand binding domain of the human estrogen receptor (ER) driven by the Col2a1 promoter (Col2a1-CreERT), so that the fusion protein was translocated into nuclei causing gene targeting by administration of the estrogen antagonist tamoxifen (26). We generated inducible conditional knockout mice by mating the Col2a1-CreERT mice with the Rbpjfl/fl mice (Col2a1-CreERT;Rbpjfl/fl), injected tamoxifen to the 7-wk-old Col2a1-CreERT;Rbpjfl/fl mice and the Rbpjfl/fl littermates daily for 5 d, and created the surgical OA model 2 d after the last injection at 8 wk. Initially, we confirmed that Cre-recombination was successfully achieved in adult articular chondrocytes after tamoxifen induction by LacZ staining (Fig. 3A). The Col2a1-CreERT;Rbpjfl/fl mice developed and grew normally without abnormality in the skeleton, articular cartilage, or their joint phenotypes under physiological conditions (Fig. 3 B and C), although RBPjκ was confirmed to be efficiently inactivated in the articular chondrocytes (Fig. 3D). Under the surgical OA induction (24), however, the cartilage degradation as well as expressions of Mmp13, Vegfa, and Hes1 were suppressed in the Col2a1-CreERT;Rbpjfl/fl knee joints, compared with the Rbpjfl/fl joints (Fig. 3 E and F), confirming the resistance to OA development by the RBPjκ insufficiency in adult articular cartilage. Because histomorphometric analyses of subchondral bones in the knee joints revealed no difference between the two genotypes, the OA resistance by the RBPjκ insufficiency was proved not to be due to alteration in bone morphology or joint biomechanics (Table S1).
Fig. 3.
OA development in Col2a1-CreERT;Rbpjfl/fl mice. (A) Beta-galactosidase (LacZ) staining of knee joints of Col2a1-CreERT;R26Rf/+ mice and the Cre negative control littermates (R26Rf/+) 8 wk after tamoxifen induction (16 wk old). (Scale bars, 100 μm.) Haematoxylin staining was performed as a counterstain. (B) Plain radiographs of the entire bodies of Rbpjfl/fl and Col2a1-CreERT;Rbpjfl/fl littermates (8 wk old) after tamoxifen injection for 5 d. (Scale bars, 10 mm.) (C) Safranin O staining of mouse knee joints in 8-wk-old Rbpjfl/fl and Col2a1-Cre;Rbpjfl/fl littermates above under physiological conditions. Insets in the Upper safranin O-stained image indicate the regions shown in the enlarged images (Lower). (Scale bars, 400 μm and 100 μm for low and high magnification images, respectively.) (D) mRNA levels of Rbpj in articular chondrocytes from Rbpjfl/fl and Col2a1-CreERT;Rbpjfl/fl littermates above (8 wk old). Data are expressed as means ± SD *P < 0.01 versus Rbpjfl/fl. (E) Cartilage degradation assessed by safranin O staining and immunofluorescence with antibodies to Col10a1, Mmp13, Vegfa, Hes1, and RBPjκ in mouse knee joints 8 wk after creating a surgical OA model in 8-wk-old Rbpjfl/fl and Col2a1-CreERT;Rbpjfl/fl littermates above. Insets in the Upper safranin O-stained images indicate the regions shown in the enlarged safranin O-stained or immunofluorescence images (Lower). (Scale bars, 400 μm and 100 μm for low and high magnification images, respectively.) (F) Quantification of OA development by Osteoarthritis Research Society International (OARSI) grading systems. Data are expressed as means ± SD of nine mice per group. *P < 0.05 versus Rbpjfl/fl.
Notch Signaling Enhances the Terminal Stage of Endochondral Ossification Through Hes1 Induction.
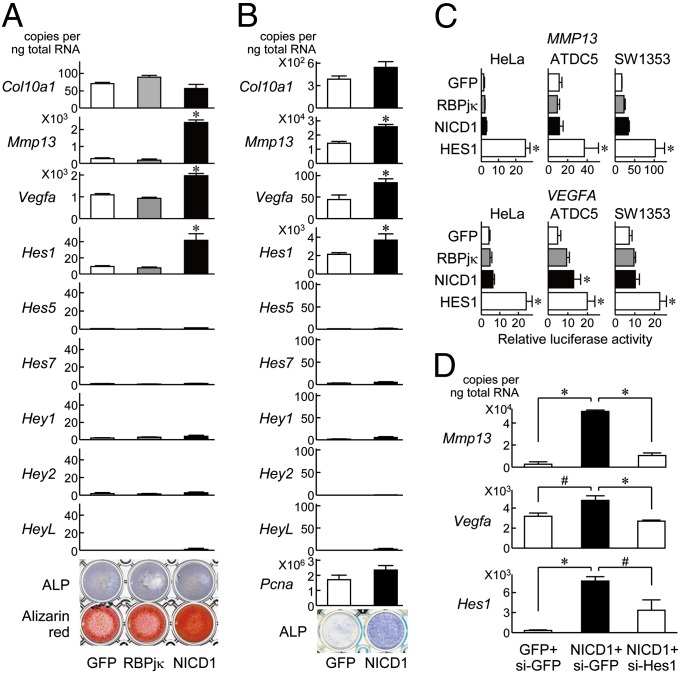
We then examined the mechanism underlying regulation of endochondral ossification by the Notch signaling. In cultures of ATDC5 cells (Fig. 4A) and primary articular chondrocytes (Fig. 4B), Mmp13, Vegfa, and Hes1 expressions, as well as the alkaline phosphatase (ALP) and Alizarin red staining were increased by overexpression of Notch1 ICD, although Col10a1, Pcna, and other Hes/Hey family members were unaffected (Fig. 4A). Contrarily, none of the expression or staining was altered by the RBPjκ overexpression (Fig. 4A). Taken together with the in vivo loss-of-function analyses (Figs. 2 and 3 and Fig. S1), endogenous RBPjκ is likely to be essential to maintain the Notch function as the transcriptional effector of Notch ICD for endochondral ossification under physiological and pathological conditions. Luciferase analyses revealed that the promoter activities of MMP13 and VEGFA were enhanced slightly by Notch1 ICD and notably by Hes1 transfection, whereas not by RBPjκ (Fig. 4C), implicating a transcriptional regulation of these factors by Hes1. In fact, the Notch1–ICD-induced Mmp13 and Vegfa expressions were suppressed by the Hes1 knockdown through the specific siRNA transfection (Fig. 4D). These lines of results indicate that the RBPjκ-dependent Notch signaling enhances the terminal stage of endochondral ossification through induction of the target gene Hes1, which transactivates Mmp13 and Vegfa.
Fig. 4.
Regulation of Col10a1, Mmp13, Vegfa, and Hes/Hey family members in cultures of chondrocytes. (A) mRNA levels of the factors above, and alkaline phosphatase (ALP) and Alizarin red stainings in stable lines of ATDC5 cells retrovirally transfected with GFP, RBPjκ, and Notch1–ICD (NICD1) after culture for 3 wk with ITS and 2 d more with Pi. *P < 0.01 versus GFP. (B) mRNA levels of the factors above, Pcna, and ALP staining in primary articular chondrocytes adenovirally transfected with GFP and NICD1 after culture for 5 d with differentiation medium. *P < 0.01 versus GFP. (C) Luciferase activities by the transfections of GFP, RBPjκ, NICD1, and HES1 into HeLa, ATDC5, and human chondrogenic SW1353 cells with a reporter construct containing a fragment of the MMP13 gene (Upper) or the VEGFA gene (Lower) (−1000 bp to transcriptional start site). *P < 0.01 versus GFP. (D) mRNA levels of Mmp13, Vegfa, and Hes1 in ATDC5 cells that were transfected with GFP and NICD1, and further cotransfected with siRNA for GFP (si-GFP) or Hes1 (si-Hes1). Data are expressed as means ± SD #P < 0.05, *P < 0.01.
Jagged1 Expression Is Most Strongly Enhanced During OA Development Among Notch Ligands.
Next, we looked at the upstream signals regulating the Notch signaling in chondrocytes during endochondral ossification and OA development. Among the five canonical membrane ligands Delta-like 1, 3, 4, and Jagged1 and -2, Jagged1 was most strongly expressed in cultures of mouse primary costal chondrocytes and articular chondrocytes (Fig. S2A). Furthermore, only Jagged1 expression was enhanced during OA development in the mouse knee articular cartilage, whereas four other ligands were unaffected or somewhat decreased (Fig. S2B). In addition to the canonical ligands, several noncanonical ligands have been reported to bind to the Notch receptors and activate the signaling, like CCN3, a member of the connective tissue growth factor/cysteine-rich 61/nephroblastoma overexpressed gene family, and microfibril-associated glycoproteins (MAGPs), components of extracellular microfibrils (9). Among them, CCN3 was most strongly expressed in mouse articular chondrocytes (Fig. S2C); however, none of the expression was associated with the OA development (Fig. S2D). Taken together, Jagged1 might be the most likely ligand regulating the Notch signaling during OA development.
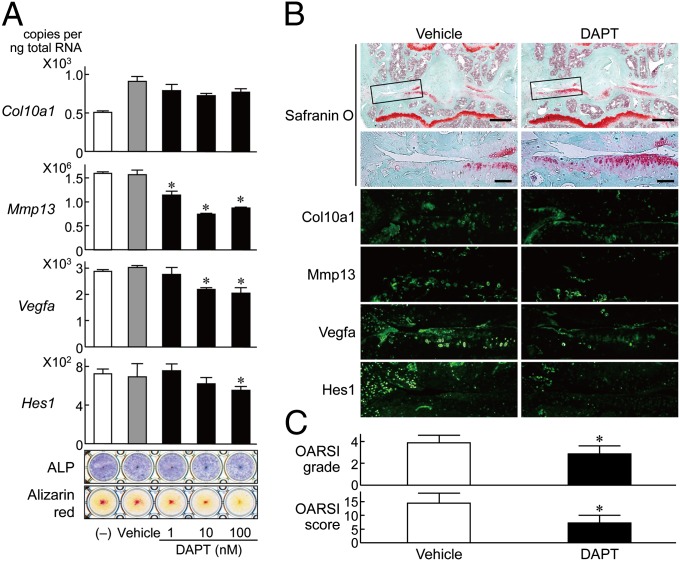
Small Compound of Notch Inhibitor Suppresses OA Development.
Finally, to examine the possibility of the Notch signaling being a clinical therapeutic target of OA, we used DAPT (N-[N-(3,5-diflurophenylacetate)-l-alanyl]-(S)-phenylglycine t-butyl ester), a pharmacological inhibitor of the Notch signaling. In the cultures of articular chondrocytes, treatment with DAPT inhibited Mmp13, Vegfa, and Hes1 expressions and also alkaline phosphatase and Alizarin red stainings (Fig. 5A). We then performed intraarticular injection (two times per week) of DAPT to the mouse knee joints of the surgical OA model for 10 wk. The cartilage degradation was suppressed by this injection compared with the control vehicle-injected knee joints (Fig. 5B). This suppression was associated with the decreases in expressions of Mmp13, Vegfa, and Hes1, but not Col10a1 (Fig. 5B), similar to the RBPjκ insufficiency. Quantification by grading systems (25) confirmed that the DAPT injection significantly suppressed OA development (Fig. 5C).
Fig. 5.
Effects of a small compound DAPT, a Notch inhibitor, on endochondral ossification and OA development. (A) mRNA levels of Col10a1, Mmp13, Vegfa, Hes1, and ALP and Alizarin red stainings in mouse articular chondrocytes cultured for 10 d with and without DAPT (1, 10, 100 nM in DMSO) or DMSO alone (vehicle). Data are expressed as means ± SD *P < 0.05 versus vehicle. (B) Cartilage degradation assessed by safranin O staining and immunofluorescence with antibodies to Col10a1, Mmp13, Vegfa, and Hes1 in the mouse surgical knee OA joints with intraarticular injection (two times per week) of 10 μL of 2.5 μM DAPT solution (25 mM DAPT/DMSO in PBS, 1:10,000) or DMSO alone in PBS (1:10,000; vehicle) for 10 wk. Insets in the Upper safranin O-stained image indicate the regions shown in the enlarged safranin O-stained or immunofluorescence images (Lower). (Scale bars, 400 μm and 100 μm for low and high magnification images, respectively.) (C) Quantification of OA development by OARSI grading systems. Data are expressed as means ± SD of seven mice per group. *P < 0.05 versus vehicle.
Discussion
The present study showed that the RBPjκ-dependent Notch signaling in chondrocytes plays a role in the maintenance of the terminal stage of endochondral ossification like matrix degradation and vascular invasion. There are, however, several cell culture studies showing that Notch signaling functions as an inhibitor of the early stage of chondrocyte differentiation (13, 27, 28). In vivo studies on genetic gain- and loss of function of the signaling using paired-related homeobox gene 1 (Prx1) promoter-driven Cre-transgenic (Prx1-Cre) mice also showed that activation of Notch receptors and RBPjκ in mesenchymal progenitor cells inhibited their differentiation into chondrocytes and osteoblasts, and thereby suppressed cartilage and bone formation, respectively (29, 30). This action in the early stage is likely to be mediated by inhibition of Sox9 expression (23, 29). Hence, we initially used Sox9-Cre;Rbpjfl/fl mice to know the role of the Notch signaling in later stages of differentiation by excluding the influence of the early Notch/Sox9 signal, and found that it was not inhibitory but stimulatory at the terminal stage. The role of Notch signaling in the process of chondrocyte differentiation and endochondral ossification thus seems complicated. It is inhibitory at the initial stage although stimulatory at the terminal stage, implicating that endogenous Notch signaling may be required for appropriate timing and localization with a delicate balance to maintain the cartilage homeostasis.
Here we propose Hes1 as the most probable downstream molecule for the Notch signaling in chondrocytes. In fact, Hes1 is known to be expressed strongly in chondrocyte progenitor cells and plays important roles in the progenitor maintenance via the suppression of chondrogenesis in the early differentiation stage (29). The present study shows that Hes1 induces MMP13 and VEGFA expressions in the late stage of chondrocyte differentiation. Because the 1-kb MMP13 promoter and VEGFA promoter used in this study contain three and two E-boxes, the representative motifs of Hes1, respectively, Hes1 might directly transactivate MMP13 and VEGFA. Otherwise, Hes1 might indirectly function possibly through the mediation of other transcription factors, e.g., runt-related transcription factor 2 (Runx2), a major signal for endochondral ossification (31), because Hes1 is known to enhance the transcriptional activity of Runx2 by binding to (32) or increasing the protein stability of Runx2 (33). Also, we previously reported that Runx2 heteroknockout mice showed resistance to OA development with decreased Mmp13 expression (34), similar to Col2a1-CreERT;Rbpjfl/fl mice and Col2a1-Cre;Rbpjfl/fl mice in the present study, implicating a possible mediation of the Runx pathway in the Notch pathway during the OA development as well. Further studies will be necessary to examine the direct binding of Hes1 to the promoters of MMP13 and VEGFA.
Jagged1 seemed to be the most probable Notch ligand in the present study; however, the upstream of the Notch signaling in chondrocytes has been rather complicated. Delta-like negatively regulates the transition from prehypertrophic to hypertrophic chondrocytes (27). Contrarily, Jagged maintains the mesenchymal progenitor state through suppression of further differentiation (35). Although little is known about the function of these ligands in the late stage of chondrocyte differentiation, the enhanced expression of Jagged1 during OA development in the present study is consistent with a previous report using human knee OA samples (16). From a therapeutic aspect, Jagged1 is an extracellular protein that may be easier to target than intracellular molecules like RBPjκ and Hes1; however, even for cell surface/extracellular targets, the therapeutic agent must traverse the cartilage matrix lacking in vasculature and reach the chondrocytes, which is difficult for large macromolecules like antibodies. Because we successfully used i.p. injection of tamoxifen in Col2a1-CreERT;Rbpjfl/fl mice, a small molecule may reach the chondrocytes and efficiently function intracellularly even if it is delivered systemically. We have actually shown that intraarticular injection of DAPT, a small compound Notch inhibitor, in the surgical mouse OA model caused suppression of cartilage degradation (Fig. 5B). Although the effects of DAPT in the chondrocyte culture system suggest that the underlying mechanism may be inhibition of the terminal stage of endochondral ossification (Fig. 5A), we cannot deny the possibility that DAPT has effects on other processes that require Notch signaling. In fact, DAPT is known to inhibit the hypoxia-induced angiogenesis mediated by interactions between Notch1 and hypoxia-inducible factor-1α in synovial cells of inflammatory arthritis (36). Also, inhibition of Notch signaling with DAPT is reported to cause delayed dedifferentiation in chondrocytes and maintain Col2a1 expression (14). As the Notch signaling is implicated in several human diseases (37), its modulation by the DAPT administration may clinically offer an optimal treatment of OA in either mechanism.
Considering that the Notch signaling system is believed to be activated via cell–cell contact, there remains a question of how an intercellular signaling mechanism could be active in the cartilage where chondrocytes are distributed in substantial extracellular matrix. One hypothesis involves the role of soluble ligands. In mammals, the Delta-like and Jagged ligands mature as a result of cleavages by tumor necrosis factor α-converting enzyme-like protease and γ-secretase, and could be released as extracellular soluble forms (38). Furthermore, Notch ligands may also be released extracellularly via secreted vesicles (39). Besides Delta and Jagged family members, CCN3 and MAGPs can function as the Notch ligand, causing dissociation of the Notch1 extracellular domain and activation of the receptor (9). The next major challenge may be to identify the novel activator of the Notch signaling in cartilage including the noncanonical ligands, because this will considerably advance our understanding of the Notch signaling in the pathogenesis of OA.
Materials and Methods
Cell Cultures.
We cultured HeLa cells (RIKEN BioResource Center, RIKEN BRC) and SW1353 cells (American Type Culture Collection) in DMEM with 10% (vol/vol) FBS, and ATDC5 cells (RIKEN BRC) in DMEM/F12 (1:1) with 5% (vol/vol) FBS. To induce chondrocyte differentiation, we cultured ATDC5 cells in the presence of insulin, transferring, and sodium selenite (ITS) supplement (Sigma) for 3 wk and replaced it with αMEM/5% (vol/vol) FBS with 4 mM inorganic phosphate (Pi) for 2 d (40), or cultured as pellets in DMEM/F12 (1:1) with 5% (vol/vol) FBS for 1 wk. For generation of the stable cell lines, we cultured ATDC5 cells with retrovirus supernatant with polybrene (8 μg/mL) for 2 d and then with 3 μg/mL puromycin until confluency (41). We performed ALP staining with a solution containing 0.01% naphthol AS-MX phosphate disodium salt, 1% N,N-dimethyl-formamide and 0.06% fast blue BB (Sigma). In addition, we stained the cells with 2% (g/vol) Alizarin red S solution (Sigma). For primary cell cultures, we isolated costal and articular chondrocytes from wild-type mice [embryonic day E18.5 (E18.5) and 6 d old, respectively] (42). To confirm the RBPjκ inactivation, we isolated articular chondrocytes from wild-type and Col2a1-Cre;Rbpjfl/fl (6 d old) or Col2a1-CreERT;Rbpjfl/fl mice (8 wk old). We performed pellet culture of primary costal chondrocytes in DMEM with 10% FBS for 1 wk. We performed monolayer culture of primary articular chondrocytes with or without DAPT (Calbiochem) in DMEM containing 10% (vol/vol) FBS, 10 mM b-glycerophosphate, and 10 μg/mL ascorbic acid for 2 wk (43).
Construction of Expression Vectors.
We amplified full-length cDNAs of RBPjκ and HES1, and NOTCH1–ICD sequence (coding sequence, 5266–7668) by PCR and cloned them into pCMV-HA (Clontech). We cloned RBPjκ and NICD into pMX-puro for retrovirus vectors. We constructed adenovirus vectors by the AdenoX expression system (Clontech). We constructed an siRNA vector for the mouse Hes1 gene (coding sequence, 222–242) with piGENEmU6 vector (iGENE Therapeutics). An expression vector for GFP and an siRNA vector for GFP were constructed as previously described (41).
Real-Time RT-PCR Analysis.
We isolated total RNA with an RNeasy mini kit, and reverse transcribed 1 μg of total RNA with MultiScribe reverse transcriptase (Applied Biosystems) to generate single-stranded cDNA. We performed real-time RT-PCR with an Mx3000P Real-time PCR system (Stratagene). Each PCR consisted of 1× FullVelocity SYBR Green QPCR Master mix (Stratagene), 0.3 μM specific primers, and 500 ng of cDNA. We calculated the mRNA copy number of a specific gene in total RNA using a standard curve generated by serially diluted plasmids containing PCR amplicon sequences and normalized to the human or rodent total RNA (Applied Biosystems) with the mouse glyceraldehyde-3-phosphate dehydrogenase (Gapdh) or the β-actin (Actb) as an internal control. We ran all reactions in triplicate. Primer sequence information is available upon request.
Luciferase Assay.
We constructed the reporter vectors of the human MMP13 and VEGFA promoter regions as previously described (44). We performed the luciferase assay with a dual luciferase-reporter assay system (Promega), and showed the data as the ratio of the firefly activities to the renilla activities.
Animals.
We performed all experiments according to the protocol approved by the animal care and use committee of the University of Tokyo. In each experiment, we compared genotypes of littermates that were maintained in a C57BL/6 background with a standard diet. We purchased Col2a1-Cre mice from The Jackson Laboratory, and Rbpj-flox mice from RIKEN BRC. Col2a1-CreERT mice were provided to us by Fanxin Long (Washington University, St. Louis). The Cre-recombination efficiency was evaluated by LacZ staining in Col2a1-CreERT mice that were mated with Rosa26 reporter mice (R26Rf/+). To generate Sox9-Cre;Rbpjfl/fl mice, Col2a1-Cre;Rbpjfl/fl mice or Col2a1-CreERT;Rbpjfl/fl mice, Rbpjfl/fl mice were mated with Sox9-Cre mice, Col2a1-Cre mice, or Col2a1-CreERT mice to obtain Sox9-Cre;Rbpjfl/+ mice, Col2a1-Cre;Rbpjfl/+ mice, or Col2a1-CreERT;Rbpjfl/+ mice, respectively, which were then mated with Rbpjfl/fl mice.
Histological Analyses.
We performed double staining of skeletons of mouse embryos with a solution containing Alizarin red S and Alcian blue 8GX (Sigma) after fixation in 99.5% ethanol and acetone. H&E staining was done according to standard protocols after fixation in 4% paraformaldehyde buffered with PBS. For immunohistochemistry, we incubated the sections with antibodies to Notch1–ICD (1:100; Santa Cruz Biotechnology), Notch2–ICD (1:100; Santa Cruz Biotechnology), RBPjκ (1:100; Santa Cruz Biotechnology), Hes1 (1:100; Santa Cruz Biotechnology), Dll1 (1:100; Santa Cruz Biotechnology), Dll3 (1:100; Santa Cruz Biotechnology), Dll4 (1:100; Santa Cruz Biotechnology), Jag1 (1:100; Santa Cruz Biotechnology), Jag2 (1:100; Santa Cruz Biotechnology), Ccn3 (1:100; Santa Cruz Biotechnology), Magp1 (1:100; Santa Cruz Biotechnology), Magp2 (1:100; Santa Cruz Biotechnology), Vegfa (1:100, Santa Cruz Biotechnology), Col10a1 (1:500; LSL), Mmp13 (1:200; Chemicon) and Pcna (1:100; Cell Signaling Technology). For visualization, we used a tyramide signal amplification (TSA) Fluorescence system (PerkinElmer). For radiological analysis, plain radiographs were taken using a soft X-ray apparatus (Softex CMB-2; Softex).
OA Experiment.
Tamoxifen (Sigma; 100 μg per gram of body weight) was intraperitoneally injected to 7-wk-old Col2a1-CreERT;Rbpjfl/fl mice and the Rbpjfl/fl littermates daily for 5 d. We then performed the surgical procedure to create an experimental OA model on 8-wk-old male mice as previously reported (24), and we analyzed them 8 wk after surgery. We quantified OA severity by the OARSI system (25), which was assessed by a single observer who was blinded to the experimental group.
Human Samples.
We obtained human samples from individuals undergoing total knee arthroplasty after obtaining written informed consent as approved by the ethics committee of the University of Tokyo.
Intraarticular Administration of DAPT.
We performed intraarticular administration of DAPT to C57BL/6J mice twice a week for 10 wk after the surgical induction. For each administration, we injected 10 μL of 2.5 μM DAPT solution, which was prepared by diluting 25 mM DAPT in dimethyl sulfoxide (DMSO) with PBS at 1:10,000, and 10 μL of DMSO diluted with PBS (1:10,000) as the control.
Histomorphometric Analysis.
We performed histomorphometric analyses in eight optical fields of the subchondral bone of distal femurs, according to the American Society for Bone and Mineral Research nomenclature report (45).
Statistical Analysis.
We performed statistical analyses of experimental data with the unpaired two-tailed Student t test.
Supplementary Material
Acknowledgments
We thank Dr. Fanxin Long for generously providing an important Col2a1-CreERT mouse strain and R. Yamaguchi, H. Kawahara, and D. Mori for technical assistance. This study was supported by Grants-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology 19109007, 23390358, and 23689065.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207458110/-/DCSupplemental.
References
- 1.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 2.Kawaguchi H. Endochondral ossification signals in cartilage degradation during osteoarthritis progression in experimental mouse models. Mol Cells. 2008;25(1):1–6. [PubMed] [Google Scholar]
- 3.Sharma L, Kapoor D. In: Osteoarthritis, Diagnosis and Medical/Surgical Management. 4th Ed. Moskowits RE, Altman RD, Hochberg MC, Buckwalter JA, Goldberg VM, editors. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 3–26. [Google Scholar]
- 4.Ortega N, Behonick DJ, Werb Z. Matrix remodeling during endochondral ossification. Trends Cell Biol. 2004;14(2):86–93. doi: 10.1016/j.tcb.2003.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stickens D, et al. Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development. 2004;131(23):5883–5895. doi: 10.1242/dev.01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zelzer E, et al. VEGFA is necessary for chondrocyte survival during bone development. Development. 2004;131(9):2161–2171. doi: 10.1242/dev.01053. [DOI] [PubMed] [Google Scholar]
- 7.Calvi LM, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425(6960):841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 8.Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: Insights from mouse mutants. Nat Neurosci. 2005;8(6):709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 9.D’Souza B, Meloty-Kapella L, Weinmaster G. Canonical and non-canonical Notch ligands. Curr Top Dev Biol. 2010;92:73–129. doi: 10.1016/S0070-2153(10)92003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iso T, Kedes L, Hamamori Y. HES and HERP families: Multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194(3):237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 11.Kopan R, Ilagan MX. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayes AJ, Dowthwaite GP, Webster SV, Archer CW. The distribution of Notch receptors and their ligands during articular cartilage development. J Anat. 2003;202(6):495–502. doi: 10.1046/j.1469-7580.2003.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlsson C, et al. Notch and HES5 are regulated during human cartilage differentiation. Cell Tissue Res. 2007;327(3):539–551. doi: 10.1007/s00441-006-0307-0. [DOI] [PubMed] [Google Scholar]
- 14.Sassi N, et al. Expression of Notch family members in cultured murine articular chondrocytes. Biotech Histochem. 2009;84(6):313–320. doi: 10.3109/10520290903054382. [DOI] [PubMed] [Google Scholar]
- 15.Kohn A, et al. Cartilage-specific RBPjκ-dependent and -independent Notch signals regulate cartilage and bone development. Development. 2012;139(6):1198–1212. doi: 10.1242/dev.070649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlsson C, Brantsing C, Egell S, Lindahl A. Notch1, Jagged1, and HES5 are abundantly expressed in osteoarthritis. Cells Tissues Organs. 2008;188(3):287–298. doi: 10.1159/000121610. [DOI] [PubMed] [Google Scholar]
- 17.Ustunel I, et al. The immunohistochemical localization of notch receptors and ligands in human articular cartilage, chondroprogenitor culture and ultrastructural characteristics of these progenitor cells. Acta Histochem. 2008;110(5):397–407. doi: 10.1016/j.acthis.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Grogan SP, Miyaki S, Asahara H, D’Lima DD, Lotz MK. Mesenchymal progenitor cell markers in human articular cartilage: Normal distribution and changes in osteoarthritis. Arthritis Res Ther. 2009;11(3):R85. doi: 10.1186/ar2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sassi N, et al. The role of the Notch pathway in healthy and osteoarthritic articular cartilage: From experimental models to ex vivo studies. Arthritis Res Ther. 2011;13(2):208. doi: 10.1186/ar3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akiyama H, et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci USA. 2005;102(41):14665–14670. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han H, et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14(6):637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- 22.Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26(2):145–146. [PubMed] [Google Scholar]
- 23.Mead TJ, Yutzey KE. Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development. Proc Natl Acad Sci USA. 2009;106(34):14420–14425. doi: 10.1073/pnas.0902306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamekura S, et al. Osteoarthritis development in novel experimental mouse models induced by knee joint instability. Osteoarthritis Cartilage. 2005;13(7):632–641. doi: 10.1016/j.joca.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Pritzker KP, et al. Osteoarthritis cartilage histopathology: Grading and staging. Osteoarthritis Cartilage. 2006;14(1):13–29. doi: 10.1016/j.joca.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 26.Hilton MJ, Tu X, Long F. Tamoxifen-inducible gene deletion reveals a distinct cell type associated with trabecular bone, and direct regulation of PTHrP expression and chondrocyte morphology by Ihh in growth region cartilage. Dev Biol. 2007;308(1):93–105. doi: 10.1016/j.ydbio.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crowe R, Zikherman J, Niswander L. Delta-1 negatively regulates the transition from prehypertrophic to hypertrophic chondrocytes during cartilage formation. Development. 1999;126(5):987–998. doi: 10.1242/dev.126.5.987. [DOI] [PubMed] [Google Scholar]
- 28.Fujimaki R, Toyama Y, Hozumi N, Tezuka K. Involvement of Notch signaling in initiation of prechondrogenic condensation and nodule formation in limb bud micromass cultures. J Bone Miner Metab. 2006;24(3):191–198. doi: 10.1007/s00774-005-0671-y. [DOI] [PubMed] [Google Scholar]
- 29.Dong Y, et al. RBPjkappa-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development. Development. 2010;137(9):1461–1471. doi: 10.1242/dev.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilton MJ, et al. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med. 2008;14(3):306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komori T. Signaling networks in RUNX2-dependent bone development. J Cell Biochem. 2011;112(3):750–755. doi: 10.1002/jcb.22994. [DOI] [PubMed] [Google Scholar]
- 32.McLarren KW, et al. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor Cbfa1. J Biol Chem. 2000;275(1):530–538. doi: 10.1074/jbc.275.1.530. [DOI] [PubMed] [Google Scholar]
- 33.Suh JH, Lee HW, Lee JW, Kim JB. Hes1 stimulates transcriptional activity of Runx2 by increasing protein stabilization during osteoblast differentiation. Biochem Biophys Res Commun. 2008;367(1):97–102. doi: 10.1016/j.bbrc.2007.12.100. [DOI] [PubMed] [Google Scholar]
- 34.Kamekura S, et al. Contribution of runt-related transcription factor 2 to the pathogenesis of osteoarthritis in mice after induction of knee joint instability. Arthritis Rheum. 2006;54(8):2462–2470. doi: 10.1002/art.22041. [DOI] [PubMed] [Google Scholar]
- 35.Oldershaw R, Murdoch A, Brennan K. The putative role of the Notch ligand, jaggedl, in the mediation of the early events of human mesenchymal stem cell chondrogenesis. Int J Exp Pathol. 2005;86:47–48. [Google Scholar]
- 36.Gao W, et al. Notch-1 mediates hypoxia-induced angiogenesis in rheumatoid arthritis. Arthritis Rheum. 2012;64(7):2104–2113. doi: 10.1002/art.34397. [DOI] [PubMed] [Google Scholar]
- 37.Zanotti S, Canalis E. Notch regulation of bone development and remodeling and related skeletal disorders. Calcif Tissue Int. 2012;90(2):69–75. doi: 10.1007/s00223-011-9541-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LaVoie MJ, Selkoe DJ. The Notch ligands, Jagged and Delta, are sequentially processed by alpha-secretase and presenilin/gamma-secretase and release signaling fragments. J Biol Chem. 2003;278(36):34427–34437. doi: 10.1074/jbc.M302659200. [DOI] [PubMed] [Google Scholar]
- 39.Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005;132(8):1751–1762. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- 40.Magne D, et al. Phosphate is a specific signal for ATDC5 chondrocyte maturation and apoptosis-associated mineralization: Possible implication of apoptosis in the regulation of endochondral ossification. J Bone Miner Res. 2003;18(8):1430–1442. doi: 10.1359/jbmr.2003.18.8.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saito T, Ikeda T, Nakamura K, Chung UI, Kawaguchi H. S100A1 and S100B, transcriptional targets of SOX trio, inhibit terminal differentiation of chondrocytes. EMBO Rep. 2007;8(5):504–509. doi: 10.1038/sj.embor.7400934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gosset M, Berenbaum F, Thirion S, Jacques C. Primary culture and phenotyping of murine chondrocytes. Nat Protoc. 2008;3(8):1253–1260. doi: 10.1038/nprot.2008.95. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Z, et al. Neogenin regulation of BMP-induced canonical Smad signaling and endochondral bone formation. Dev Cell. 2010;19(1):90–102. doi: 10.1016/j.devcel.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saito T, et al. Transcriptional regulation of endochondral ossification by HIF-2alpha during skeletal growth and osteoarthritis development. Nat Med. 2010;16(6):678–686. doi: 10.1038/nm.2146. [DOI] [PubMed] [Google Scholar]
- 45.Parfitt AM, et al. Report of the ASBMR Histomorphometry Nomenclature Committee Bone histomorphometry: Standardization of nomenclature, symbols, and units. J Bone Miner Res. 1987;2(6):595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.