Abstract
T cells from patients with systemic lupus erythematosus (SLE) produce insufficient amounts of the vital cytokine IL-2. We previously showed that SLE T cells express decreased levels of the T-cell receptor–CD3ζ chain and forced expression of CD3ζ into SLE T cells restores IL-2 production. We recently showed that the serine arginine protein splicing factor 2/alternative splicing factor (SF2/ASF) enhances the expression of CD3ζ chain by limiting the production of an unstable splice variant. Here we demonstrate that SF2/ASF levels are decreased in patients with SLE and more so in those with active disease. More importantly, we reveal a function of SF2/ASF, independent of T-cell receptor/CD3 signaling, whereby it is recruited to the IL-2 promoter, increases transcriptional activity, and enhances IL-2 production in SLE T cells. Our results demonstrate that SF2/ASF regulates IL-2 production and that decreased SF2/ASF expression in SLE T cells contributes to deficient IL-2 production.
Keywords: autoimmunity, SFRS1, T lymphocyte, interleukin-2
Systemic lupus erythematosus (SLE) is an autoimmune disease, which mainly afflicts women in their reproductive years, causing significant morbidity and mortality. The disease is characterized by multiorgan damage mediated by immune complex deposition, lymphocytic infiltration, and inflammation. T lymphocytes are important in disease pathogenesis because they not only provide cognate help to autoreactive antibody-producing B cells, but also infiltrate target organs and contribute to tissue damage (1). SLE T cells produce decreased amounts of the vital cytokine IL-2 (2). Reduced IL-2 production is linked to reduced cytotoxicity, impaired regulatory T cells, and impaired activation-induced cell death, leading to persistence of autoreactive T cells (3–5). Thus, understanding the factors that contribute to the decreased IL-2 production is important in understanding the pathophysiology of SLE.
SLE T cells exhibit numerous signaling defects (6, 7) one of which is a reduced expression of the T-cell receptor (TCR)-associated CD3ζ chain (8, 9). Moreover, upon activation, SLE T cells display aberrantly increased tyrosine phosphorylation of signaling intermediates and excessive calcium flux (10, 11). We previously showed that forced expression of the CD3ζ chain into SLE T cells restored these signaling defects and, more importantly, enhanced IL-2 production (12). We recently found that the serine arginine (SR) protein splicing factor 2/alternative splicing factor (SF2/ASF, aka SFRS1 or SRSF1) binds to the 3′UTR of CD3ζ and limits production of an unstable splice variant of the CD3ζ mRNA, thus enhancing the expression of CD3ζ chain in human T cells (13). Based on these observations, we hypothesized that SF2/ASF expression levels may be altered in SLE T cells and may influence T-cell function, specifically IL-2 production.
IL-2 is a vital cytokine for T-cell proliferation and effector function (4). IL-2 production is regulated mainly at the level of transcription and mRNA stability (14). Following TCR activation, the CD3ζ chain is phosphorylated and recruits the ζ-associated protein-70 kinase, which phosphorylates and recruits adaptor proteins linker adaptor of T cells and SLP-76 (15). Three distinct TCR-induced signaling pathways are triggered downstream, which activate the transcription factors NF-κB, NFAT, and AP-1. The three transcription factors bind to cognate sites within the IL-2 promoter and activate IL-2 gene transcription. Besides transcriptional activation, an important mechanism of cytokine regulation is at the posttranscriptional level through mRNA stabilization and degradation (16). The exponential increase in IL-2 secretion is achieved by stabilization of the mRNA through CD28 costimulation and the JNK pathway (17, 18).
SF2/ASF is a prototype member of the SR protein family of splicing factors. The N-terminal RNA binding domain bears two RNA recognition motifs whereas the C-terminal domain contains SR repeats and is important for protein–protein interactions. Although SF2/ASF is widely known for its role in alternative splicing, it also regulates other aspects of mRNA metabolism such as mRNA decay (19, 20) and translation (21). Recently, a role for splicing factors in transcription regulation has been proposed. Interestingly, knocking down SF2/ASF or SC35, a closely related SR family member, in the murine embryonic fibroblast cell line led to a dramatic decrease in nascent mRNA, suggesting a role for these splicing regulators in gene transcription. Indeed, depletion of SC35 led to attenuated transcriptional elongation, which correlated with defective recruitment of positive transcription elongation factor b (22). More recently it was shown that AUF1, an RNA destabilizing protein, activates transcription of the telomerase gene in telomere maintenance and normal aging (23). Although SF2/ASF has been extensively studied by using model systems, very few endogenous target genes or biological pathways regulated by SF2/ASF are known. Alternative splicing of the CD45 isoforms was shown to be regulated by SF2/ASF using a minigene model system (24). We recently showed that SF2/ASF regulates the alternative splicing of the CD3ζ 3′UTR in primary human T cells (13). However, the role of SF2/ASF in human T lymphocyte function and specifically in autoimmune disease has never been studied.
We show here that SF2/ASF expression levels are decreased in T cells from patients with SLE and correlate inversely with disease activity. Importantly, SF2/ASF enhances IL-2 production in human T cells and restores IL-2 production in T cells from patients with SLE. Finally, we show that SF2/ASF regulates IL-2 at the transcriptional level.
Results
SF2/ASF Expression Levels Are Decreased in T Cells from Patients with SLE.
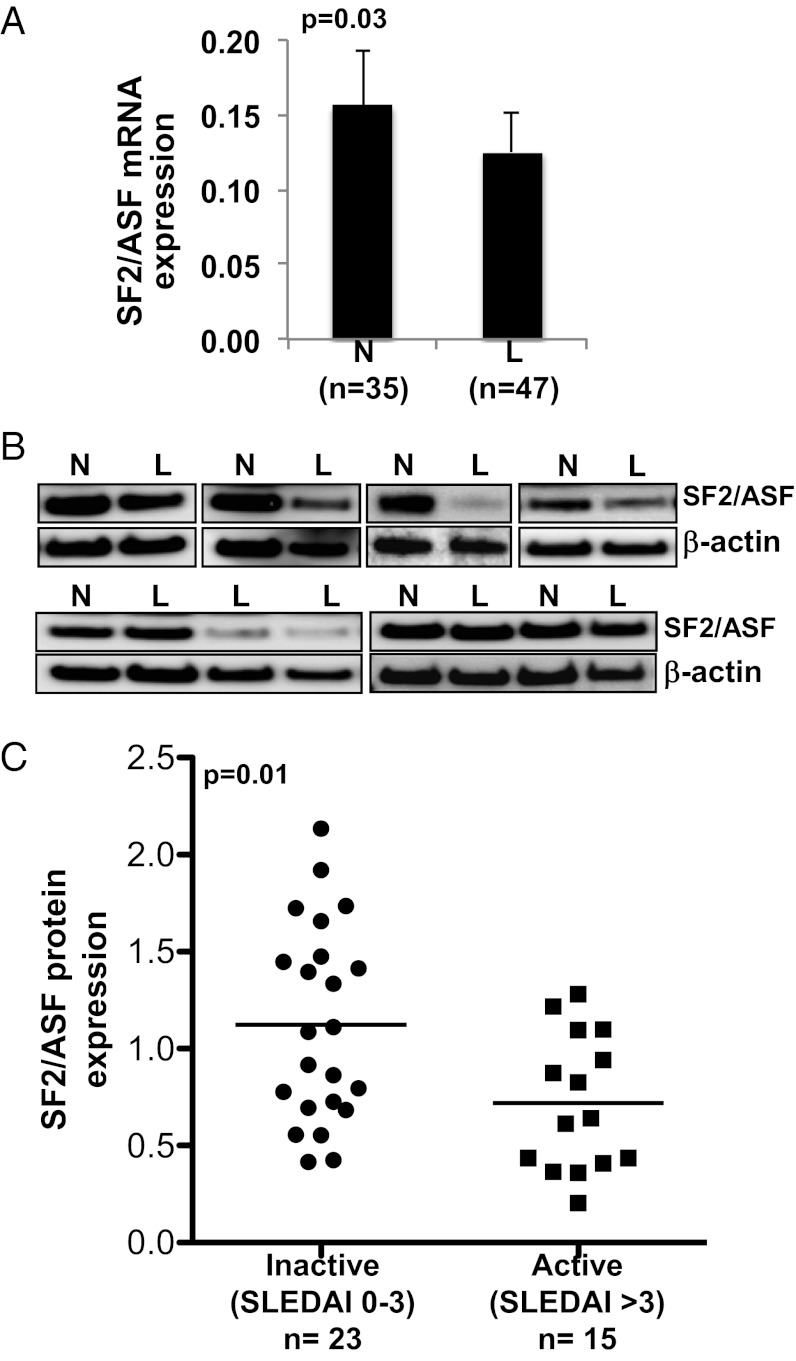
The TCR-associated CD3ζ chain expression is decreased in T cells from patients with SLE (8). Moreover, we previously showed that SF2/ASF regulates the expression of the CD3ζ chain in human T cells (13). Here we asked whether SF2/ASF expression is altered in patients with SLE. We examined the mRNA and protein expression levels of SF2/ASF in T cells from patients with SLE compared with age-, race-, and sex-matched healthy control individuals. We found that the average SF2/ASF mRNA expression is decreased in patients compared with the average expression in healthy controls (Fig. 1A). We found that protein expression is decreased in a subset of patients (Fig. 1B). Interestingly, the SF2/ASF protein expression levels segregate with the disease severity of patients as assessed by the standardized SLE disease activity index (SLEDAI) scoring system (25). Specifically, patients with moderate to severely active disease (i.e., SLEDAI score >3) show reduced expression levels compared with patients with inactive or mild disease (i.e., SLEDAI score 0–3; Fig. 1C).
Fig. 1.
SF2/ASF expression in T cells from patients with SLE and control subjects. T cells were purified from peripheral blood of patients with SLE (marked with “L”) and matched healthy donors (“N”). (A) Total RNA was isolated and reverse transcribed. SF2/ASF expression was measured by quantitative PCR and normalized to the average of GAPDH and CD3ɛ housekeeping genes. Graph shows average values, and error bars represent SEM. (B) Total protein was isolated from T cells and immunoblotted for SF2/ASF and β-actin. Representative blots are shown. (C) Densitometric quantitation of SF2/ASF from immunoblots was performed and normalized to β-actin. Graph shows relative SF2/ASF expression in patients with SLE normalized to expression from healthy control subjects, and grouped by SLEDAI score.
Overexpression of SF2/ASF in SLE T Cells Enhances IL-2 Production.
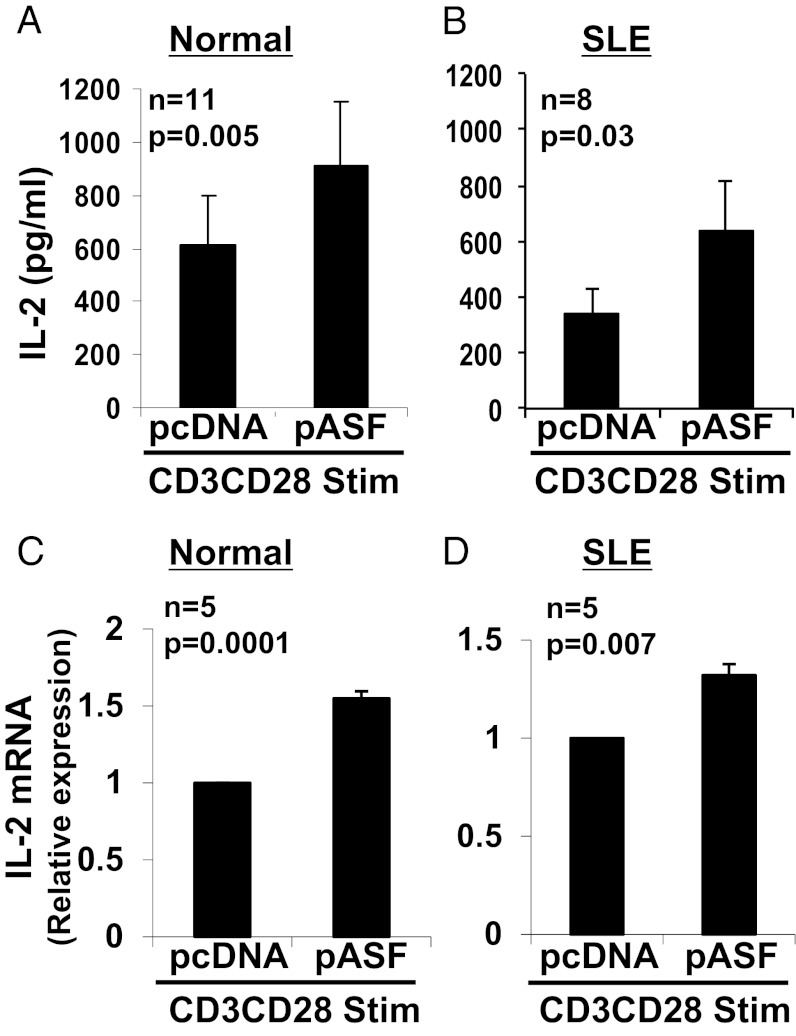
T cells from patients with SLE produce decreased amounts of the critical cytokine IL-2 (2). We previously showed that increasing the expression of CD3ζ chain in SLE T cells restored their IL-2 production (12), and that SF2/ASF increases the expression of the CD3ζ chain (13). We therefore asked whether overexpression of SF2/ASF into SLE T cells can enhance IL-2 production. We used an SF2/ASF expression vector (pASF) and an empty vector (pcDNA) as control, and transfected T cells from healthy individuals (Fig. 2A) and patients with SLE (Fig. 2B). After transfection, cells were activated with anti-CD3 and anti-CD28 antibodies, and IL-2 production was measured in the culture supernatants by ELISA. As expected, the average IL-2 secretion produced by SLE T cells (343.3 ± 94.2 pg/mL; Fig. 2B, pcDNA) was lower than the average levels produced by normal T cells (611.0 ± 184.8 pg/mL; Fig. 2A, pcDNA). Interestingly, overexpression of SF2/ASF rescued IL-2 production in the SLE T cells (640.3 ± 193.2 pg/mL; Fig. 2B, pASF), and restored levels similar to those produced by the normal T cells (611.0 ± 184.8 pg/mL; Fig. 2A, pcDNA). SF2/ASF also increased IL-2 secretion in T cells from healthy individuals (Fig. 2A, pASF). To determine whether SF2/ASF regulates IL-2 production at the mRNA level, we overexpressed SF2/ASF in normal and SLE T cells, followed by anti-CD3, anti-CD28 activation for 2 to 6 h, and assessed IL-2 mRNA expression by quantitative PCR. We found that SF2/ASF increased IL-2 mRNA levels in T cells from healthy individuals (Fig. 2C) and patients with SLE (Fig. 2D). These results indicate that SF2/ASF enhances IL-2 production at the mRNA and protein levels in normal and SLE T cells.
Fig. 2.
SF2/ASF enhances IL-2 cytokine production and mRNA expression in normal and SLE T cells. T cells were isolated from peripheral blood of healthy donors (A and C) or patients with SLE (B and D) and transfected with pcDNA or pASF. At 3 to 5 h after transfection, cells were stimulated with anti-CD3, anti-CD28, and cross-linker antibodies. At 18 h after activation (A and B), supernatants were collected, and IL-2 cytokine was measured by ELISA. At 2 to 6 h after activation (C and D), cells were collected, and total RNA was isolated and reverse-transcribed. IL-2 expression was measured by quantitative PCR and normalized to the cyclophilin A housekeeping gene. Graphs show average values, and error bars represent SEM.
SF2/ASF-Mediated Increase in IL-2 Production Is Independent of Proximal TCR Signaling.
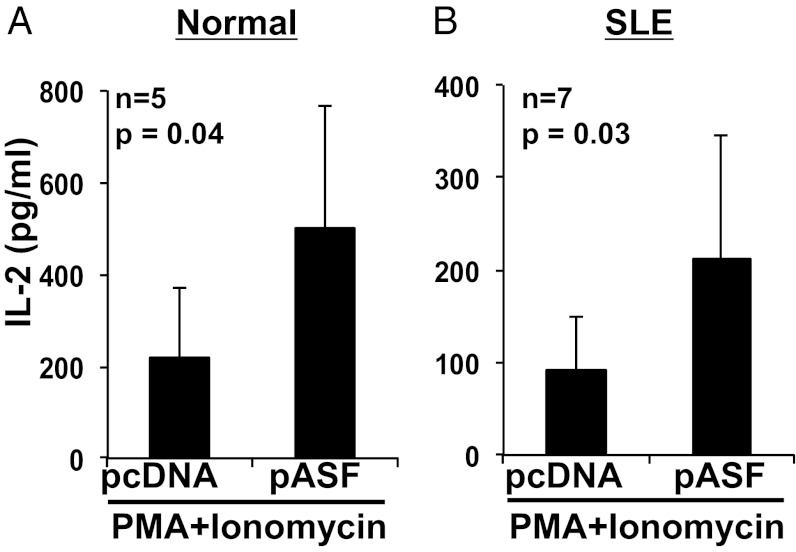
Because we had shown that SF2/ASF enhances the CD3ζ expression levels in T cells (13), and we showed here that SF2/ASF increases IL-2 production (Fig. 2), we postulated that the SF2/ASF-mediated increase in IL-2 production may result from increased CD3ζ-chain levels followed by amplification of the downstream TCR signaling cascade. To determine whether SF2/ASF mediates its effect on IL-2 via this mechanism or whether SF2/ASF may act distally, we used phorbol myristate acetate (PMA) and ionomycin for the T-cell activations. PMA activates diacylglycerol, and ionomycin is a calcium ionophore, thus triggering the downstream signaling cascade bypassing the TCR/CD3 complex entirely. Accordingly, we overexpressed SF2/ASF in T cells from healthy subjects (Fig. 3A) and patients with SLE (Fig. 3B), activated cells with PMA plus ionomycin, and measured IL-2 in the culture supernatants by ELISA. Surprisingly, SF2/ASF was able to mediate increased IL-2 production from T cells even with this stimulation, indicating that SF2/ASF can regulate IL-2 expression independent of the proximal TCR signaling.
Fig. 3.
SF2/ASF-mediated increase in IL-2 is independent of the proximal TCR signaling. T cells were isolated from peripheral blood of healthy donors (A) or patients with SLE (B) and transfected with pcDNA or pASF. At 3 to 5 h after transfection, cells were stimulated with PMA and Ionomycin. At 18 h after stimulation, supernatants were collected and IL-2 levels measured by ELISA. Graphs show average values, and error bars represent SEM.
SF2/ASF Does Not Regulate Stability of IL-2 mRNA.
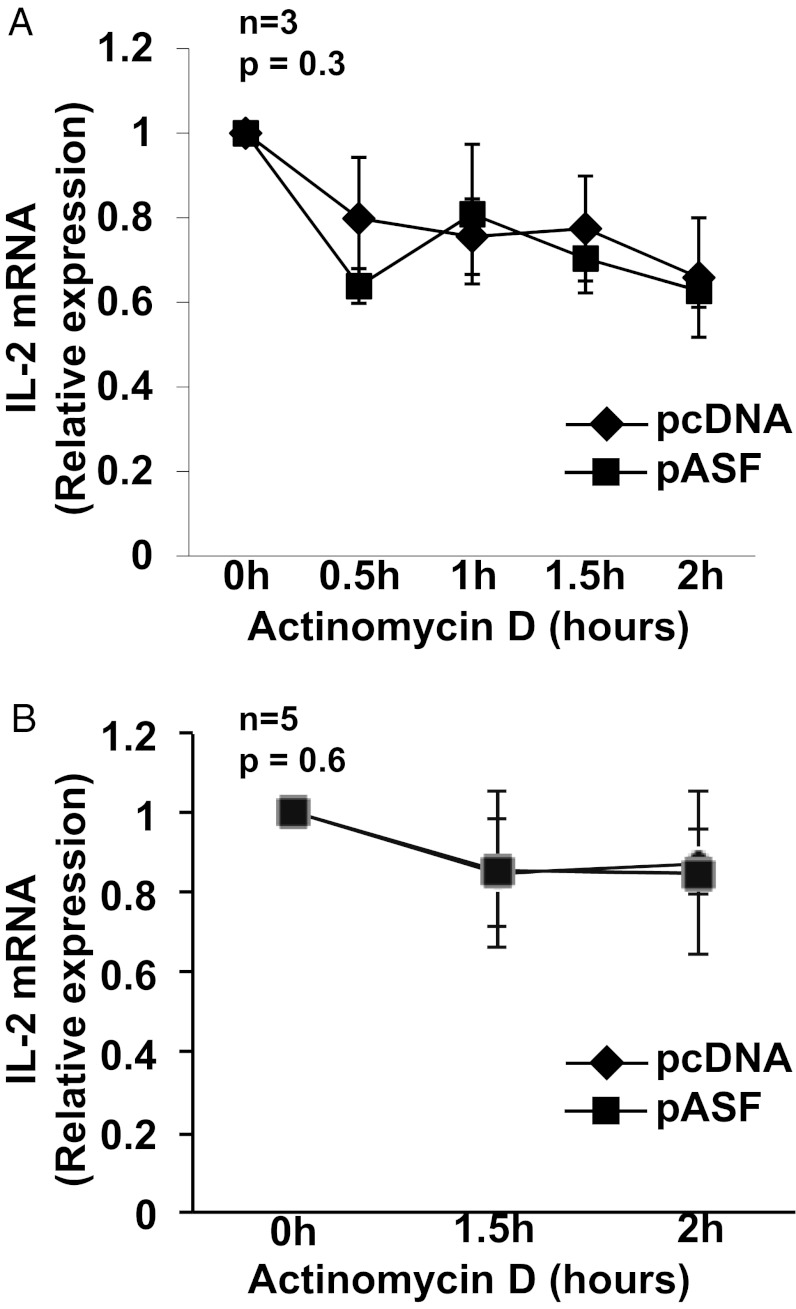
IL-2 is one of the earliest cytokines to be up-regulated in T cells following TCR stimulation, and its expression is mainly regulated at two levels—transcription and mRNA stabilization. In fact, mRNA stabilization accounts for the exponential increase in IL-2 secretion within hours after TCR activation. SF2/ASF was shown to regulate mRNA stability of the PKCIR (19) and CXCL2 (20) genes. We therefore asked whether SF2/ASF regulates the mRNA stability of IL-2. We overexpressed SF2/ASF in T cells, activated the cells, and then blocked transcription by adding actinomycin D to the cell cultures. IL-2 mRNA expression was measured by quantitative PCR. We found no differences in IL-2 mRNA decay with or without SF2/ASF overexpression in T cells from healthy individuals (Fig. 4A) or patients with SLE (Fig. 4B).
Fig. 4.
SF2/ASF does not regulate stability of IL-2 mRNA. T cells were isolated from peripheral blood of healthy donors (A) or patients with SLE (B) and transfected with pcDNA or pASF. At 16 to 18 h after transfection, cells were stimulated with anti-CD3, anti-CD28, and cross-linker antibodies. At 2 to 4 h after activation, actinomycin D was added to cell cultures, and cells were collected at indicated time points. Total RNA was isolated and reverse-transcribed. IL-2 expression was measured by quantitative PCR and normalized to the cyclophilin A housekeeping gene. Graphs show average values, and error bars represent SEM.
Overexpression of SF2/ASF Leads to Transcriptional Activation of IL-2.
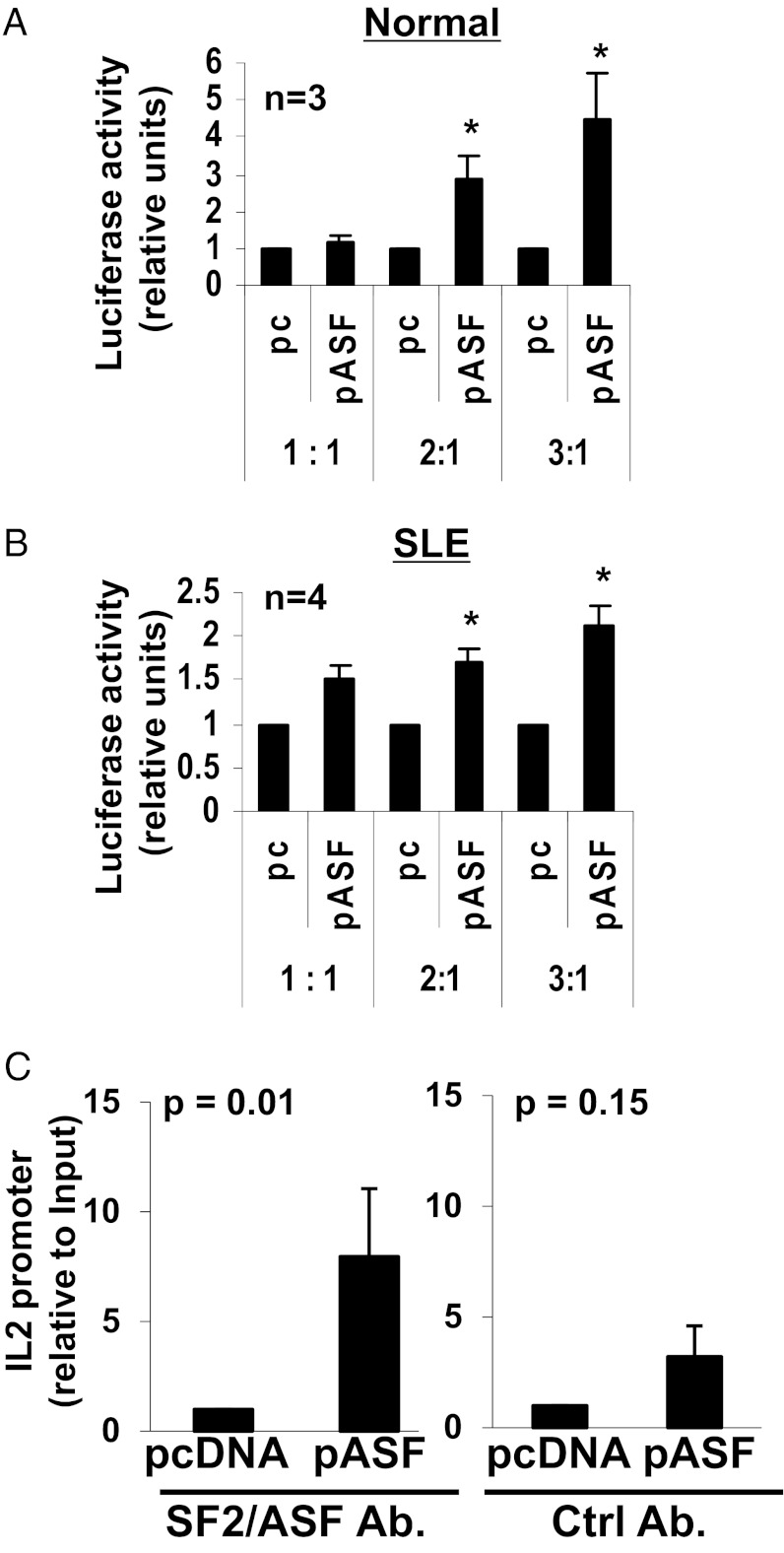
Having ruled out a role for SF2/ASF in the stabilization of the IL-2 mRNA, and because of its recently recognized role in transcription regulation (22), we asked whether SF2/ASF regulates IL-2 expression at the transcription level. We transfected normal (Fig. 5A) or SLE T cells (Fig. 5B) with an IL-2 promoter–luciferase “reporter” construct and cotransfected the cells with increasing amounts of the “effector” construct SF2/ASF (i.e., pASF), or an empty vector as control (i.e., pc). Thus, the relative amounts of the cotransfected effector:reporter constructs were 1:1, 2:1, and 3:1, respectively. We observed a dose-dependent increase in luciferase activity with increasing amounts of SF2/ASF in normal and SLE T cells (Fig. 5 A and B), indicating that SF2/ASF increases transcriptional activity of the IL-2 promoter. We further determined whether SF2/ASF is recruited to the transcriptional complex binding to the IL-2 promoter, by using reporter-ChIP (R-ChIP) assays. In this assay, cells were transfected with the IL-2 promoter–luciferase reporter vector and cotransfected with the SF2/ASF vector (i.e., pASF) or empty vector (i.e., pcDNA), then activated, followed by crosslinking of the DNA–protein complexes, cell lysis, chromatin shearing, and immunoprecipitation with an SF2/ASF-specific antibody (Fig. 5C, Left) or a control antibody (Fig. 5C, Right). After washes and reverse crosslinking, DNA was purified and the presence of the IL-2 promoter was measured by quantitative PCR using primers spanning the IL-2 promoter and the beginning of the luciferase construct, and normalized to that obtained from the input control DNA. We observed an increased recruitment of SF2/ASF to the IL-2 promoter in the SF2/ASF-transfected cells compared with control-transfected cells (Fig. 5C). These results indicate that SF2/ASF is recruited to the IL-2 promoter binding complex and increases transcriptional activity of IL-2.
Fig. 5.
SF2/ASF induces transcriptional activity of IL-2. T cells from healthy donors (A) or patients with SLE (B) were cotransfected with an IL-2 promoter–luciferase vector and empty vector (pc) or pASF in increasing concentrations, such that the relative ratio of the pcDNA or pASF (i.e., effector) vector to the luciferase (i.e., reporter) vector was 1:1, 2:1, or 3:1. The pRL-TK plasmid was cotransfected into all cells as an internal transfection control. At 2 to 4 h after transfection, cells were stimulated with anti-CD3, anti-CD28, and cross-linker antibodies. At 2 h after activation, cells were collected and lysed, and luciferase activity was measured by using the dual-luciferase assay system. Graphs show average values, and error bars represent SEM (*P < 0.05). (C) T cells from healthy donors were cotransfected with an IL-2 promoter–luciferase reporter vector and pcDNA or pASF. At 2 to 4 h after transfection, cells were stimulated with anti-CD3, anti-CD28, and cross-linker antibodies. At 2 h after activation, cells were collected, and R-ChIP assays were performed by using an SF2/ASF-specific antibody (Left) or control antibody (Right). The IL-2 promoter was amplified by quantitative PCR and normalized to the values obtained from the input DNA. Graph shows average values from six independent experiments. Error bars represent SEM.
Discussion
In this study, we present a number of interesting findings. First, we show that SF2/ASF expression levels are reduced in T cells from patients with SLE and correlate inversely with their disease activity (Fig. 1); second, we show that SF2/ASF is a regulator of T-cell function because it enhances IL-2 production in human T cells (Fig. 2 A and C); third, we show that SF2/ASF can rescue IL-2 secretion in T lymphocytes of patients with SLE (Fig. 2B); and finally, we show that SF2/ASF is a regulator of IL-2 transcription (Fig. 5).
Although SF2/ASF has been widely studied as a splicing factor, its role in all aspects of gene expression, including mRNA stability, translation, and, more recently, transcription, is now being uncovered. Because IL-2 is regulated mainly at the transcription and mRNA stabilization levels, we primarily focused evaluating the role of SF2/ASF in these two mechanisms. We found that SF2/ASF increases transcriptional activity of the IL-2 promoter (Fig. 5), but does not appear to regulate the stability of IL-2 mRNA (Fig. 4).
We observed that, whereas the average mRNA expression of SF2/ASF is decreased in patients with SLE compared with average mRNA expression from healthy controls, protein levels are decreased in only a subset of patients. A potential explanation for this finding is that SF2/ASF may undergo compensatory posttranscriptional regulation to maintain protein levels within the cell. This is not surprising, because SF2/ASF is an essential cell survival gene. Expression levels are tightly regulated, and knockdown of SF2/ASF leads to genomic instability and eventually cell death (26, 27). Hence, the low mRNA expression of SF2/ASF in SLE T cells may trigger compensatory posttranscriptional regulation to prevent decrease in protein levels and ensure cell survival. Indeed, SF2/ASF can regulate its own expression via mechanisms at the alternative splicing and translational levels such that overexpression of SF2/ASF leads to unproductive splicing and translational repression (28).
Interestingly, SF2/ASF protein expression levels were decreased in patients with high disease activity as measured by SLEDAI (Fig. 1C). This suggests that reduced SF2/ASF expression may contribute to the T-cell defect in patients with SLE, and/or may represent a potential molecular marker of disease activity. Indeed, we show that replenishing SF2/ASF in T cells from patients with SLE restores their IL-2 production. This suggests that the reduced SF2/ASF expression may be a contributor of the IL-2 defect. Furthermore, SF2/ASF may represent a potential therapeutic target to correct T-cell function.
We had previously shown that SF2/ASF regulates expression of the TCR CD3ζ signaling chain. Therefore, we assumed that SF2/ASF might regulate IL-2 production by increasing CD3ζ chain expression, thus enhancing downstream signaling events, and increasing IL-2 expression. However, our PMA/ionomycin activation studies revealed that SF2/ASF could enhance IL-2 production independent of the proximal TCR signaling, indicating a distal mechanism of regulation (Fig. 3). Interestingly, we found that SF2/ASF induces transcriptional activity and is recruited to the IL-2 promoter (Fig. 5). Although SF2/ASF is not a DNA-binding protein, interactions with proteins of the basal transcription machinery could explain its recruitment to the IL-2 promoter. Indeed, SC35, a closely related SR family member, induces transcription by recruiting components of the general transcription machinery such as positive transcription elongation factor b, which is suggested to relieve pausing of the RNA polymerase II within the body of the target gene (22). It is not known if SF2/ASF activates transcriptional initiation or elongation in the regulation of IL-2. A number of transcription factors such as NFAT, AP-1, and NF-κB are known to be important in IL-2 transcription control (14, 29). We previously showed that increased expression of the cAMP response element modulator α repressor (30) and reduced expression and binding of the AP-1 activator (31) are factors that contribute to the reduced IL-2 expression in T cells from patients with SLE. We also showed reduced NF-κB p65 binding activity to the IL-2 promoter in SLE T cells (32). It is not known whether SF2/ASF may interact with or regulate these molecules, in addition to its direct role in activating IL-2 transcription. Further studies are needed to elucidate these processes in detail.
Our study brings to light a role of SF2/ASF in the immune system, both in physiology as well as potentially in autoimmunity and disease. IL-2 is critical in autoimmunity, because poor IL-2 production is linked to reduced T-cell cytotoxicity; reduced activation induced cell death leading to persistence of autoreactive T cells, and, importantly, impaired regulatory T-cell function. It would be interesting to see whether SF2/ASF is involved in these processes and if it contributes to autoimmunity. Whether it has a role in autoimmunity and in lupus disease will need to be addressed by manipulating SF2/ASF expression using in vivo animal model systems.
In conclusion, we have shown that SF2/ASF is a regulator of human T-cell function, can rescue IL-2 production in T cells from patients with SLE, and may represent a molecular mechanism of the T-cell defect in SLE.
Materials and Methods
Patients and Healthy Controls.
Ninety-nine patients with SLE, all fulfilling the American College of Rheumatology classification criteria (33), were recruited at the Rheumatology clinic at the Beth Israel Deaconess Medical Center (Boston, MA). Blood samples from 70 patients were used for experiments in Fig. 1; 47 of these were used for SF2/ASF mRNA analysis and 38 were used for protein analysis (15 samples were common to both groups). Details of patients’ demographics, clinical scores, and therapy are listed in Table S1. For patients in Fig. 1, age-, race-, and sex- matched healthy individuals were recruited as controls. Samples from 29 patients were used for experiments in the remaining figures. For these experiments, deidentified peripheral blood samples from healthy donors were obtained from the Kraft Donor Center (Dana–Farber Cancer Institute, Boston, MA). Peripheral blood was drawn from patients and control subjects by venipuncture. All studies were approved by the institutional review board (Committee on Clinical Investigations) at the Beth Israel Deaconess Medical Center, and informed consent was obtained from all participants.
Cells, Antibodies, Plasmids, and Reagents.
Peripheral blood was collected by venipuncture, and total T cells were purified by using the Rosette Sep T-cell kit (StemCell Technologies). The SF2/ASF antibody was purchased from Life Technologies, β-actin antibody from Sigma-Aldrich, and HRP-conjugated secondary antibodies from Santa Cruz Biotechnology. The pcDNA3.1-SF2/ASF expression plasmid was a gift from James Manley (Columbia University, New York, NY). The pGL3-IL-2 promoter-luciferase reporter construct (spanning −996 to +40 bp of the human IL-2 promoter cloned into the pGL3-Basic luciferase vector at the MluI and BglII restriction sites) was custom synthesized by Genewiz. The pRL-TK plasmid was purchased from Promega. Actinomycin D was purchased from Sigma-Aldrich.
Plasmid Transfections.
Transient transfections of primary human T cells were carried out using the Nucleofector system (Lonza). Briefly, 3 to 5 million cells were resuspended in 100 μL of Nucleofector solution, plasmid DNA (pcDNA or pASF, 0.5 μg/106 cells) added, and cells transfected by using the U-014 program. Cells were rescued immediately in prewarmed RPMI medium supplemented with 10% (vol/vol) FBS and 1% penicillin and streptomycin.
Cytokine Analysis.
T cells were activated with anti-CD3 (5 μg/mL), anti-CD28 (2.5 μg/mL), and crosslinker (2.5 μg/mL) antibodies or with PMA (1 ng/mL) and ionomycin (40 ng/mL; Sigma-Aldrich). At 16 to 24 h after activation, supernatants were collected and IL-2 production measured by ELISA using the human IL-2 ELISA kit (eBiosciences).
mRNA Expression.
Total RNA was isolated using the RNeasy mini kit (Qiagen). Total RNA (200 ng) was reverse-transcribed into cDNA by using the RNA to cDNA premix (Clontech). Real-time PCR amplification was carried out with SYBR Green I using a LightCycler 480 (Roche) and the following program: initial denaturation at 95 °C for 5 min; 40 cycles of amplification (denaturation at 95 °C for 15 s, annealing at 60 °C for 15 s, extension at 72 °C for 30 s); one cycle of melting curves [95 °C for 15 s, 65 °C for 2 min, and 97 °C (continuous)], and a final cooling at 37 °C. PCR reactions in Fig.1A were performed by using a probes MasterMix and the following program: initial denaturation at 95 °C for 10 min, 45 cycles of amplification (denaturation at 95 °C for 10 s, annealing at 60 °C for 30 s, extension at 72 °C for 1 s); and a final cooling at 40 °C. All PCR reactions were performed in duplicate or triplicate. Threshold cycle (i.e., Ct) values were used to calculate relative mRNA expression by the ΔCt relative quantification method. Primer sequences were as follows: IL-2, forward, 5′-CAC ACT CAC AGT AAC CTC AAC TCC T -3′; and reverse, 5′- GTG GGA AGC ACT TAA TTA TCA AGT CAG TG-3′; housekeeping genes, cyclophilin A, forward, 5′- TTC ATC TGC ACT GCC AAG AC-3′; reverse, 5′-TCG AGT TGT CCA CAG TCA GC-3′; CD3ɛ, forward, 5′-CAA GGC CAA GCC TGT GAC-3′; and reverse, 5′-TCA TAG TCT GGG TTG GGA ACA-3′; and GAPDH, forward, 5′-AGC CAC ATC GCT CAG ACA C-3′; and reverse, 5′-GCC CAA TAC GAC CAA ATC C-3′.
Western Blotting.
Cells were pelleted and lysed with radioimmunoprecipitation assay buffer (Boston Bioproducts). Lysates were resolved on 4% to 12% (wt/vol) Bis-Tris gels and transferred to PVDF membrane. Membranes were blocked with 5% (wt/vol) nonfat milk in Tris-buffered saline solution with 0.05% Tween 20 (TBS-T) for 1 h, incubated with primary antibody (1:1,000; or 1:4,000 for β-actin antibody) at room temperature for 1 h, washed three times with TBS-T, incubated with horseradish peroxidase-conjugated secondary antibody (1:2,000) for 1 h, washed three times with TBS-T, developed with ECL detection reagents (GE Healthcare), and visualized by the Fujifilm LAS-4000 imager. Densitometry was performed using Quantity One software (Bio-Rad).
Luciferase Assays.
Five million primary human T cells were cotransfected with 0.8 μg of the pGL3-IL-2 promoter–luciferase reporter construct and increasing amounts (0.8 μg, 1.6 μg, 2.4 μg) of effector plasmid (pcDNA3.1 or pcDNA3.1-SF2/ASF) to obtain effector: reporter plasmid ratios of 1:1, 2:1, and 3:1. Each transfection included 25 ng of the pRL-TK Renilla luciferase construct as an internal control. Two hours after transfection, cells were activated with anti-CD3, anti-CD28, and cross-linker antibodies. Two hours after activation, cells were collected and lysed, and luciferase activity was quantified by using the dual-luciferase assay system (Promega) according to the manufacturer’s instructions.
R-ChIP Assays.
Primary human T cells were cotransfected with the human IL-2-promoter luciferase construct (0.16 μg/106 cells) and empty vector or SF2/ASF expression vector (0.5 μg/106 cells). At 2 to 3 h after transfection, cells were activated with anti-CD3, anti-CD28 and, cross-linker antibodies. At 2 h after activation, cells were collected, and the assay was performed by using the MAGnify ChIP kit (Life Technologies) according to the manufacturer’s instructions. Briefly, cells were fixed for 10 min in 1% formaldehyde to cross-link DNA–protein and protein–protein complexes, and glycine (1.25 M) was added for 5 min to stop cross-linking. Cells were washed with cold PBS solution, resuspended in lysis buffer containing protease inhibitors and sonicated to shear DNA, and sedimented, and diluted supernatants were immunoprecipitated with the indicated antibodies overnight. Ten percent of the diluted supernatants were kept as “input” for normalization. After several washing steps, the protein was digested with proteinase K, and the cross-linking reversed at 65 °C. DNA was purified and amplified by quantitative PCR on a LightCycler 480 real-time PCR system (Roche) using specific primers flanking the proximal IL-2 promoter (forward primer, 5′-ACC TCA ACT CCT GCC ACA AT-3′) and the luciferase gene (reverse primer, 5′-GCC TTA TGC AGT TGC TCT CC-3′). Threshold cycle (i.e., Ct) values were used to calculate relative mRNA expression by the ΔΔCt relative quantification method.
Statistical Analysis.
The Student t test was used for statistical analysis. The Wilcoxon signed-rank test or Mann–Whitney test was used for nonparametric analysis.
Supplementary Material
Acknowledgments
We thank Ms. Hannah Robertson for technical assistance and Dr. Martin Flajnik for critical reading of the manuscript. The work was funded by National Institutes of Health Grants R01 AI42269 (to G.C.T.) and K01 AR060781 (to V.R.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214207110/-/DCSupplemental.
References
- 1.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365(22):2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Linker-Israeli M, et al. Defective production of interleukin 1 and interleukin 2 in patients with systemic lupus erythematosus (SLE) J Immunol. 1983;130(6):2651–2655. [PubMed] [Google Scholar]
- 3.Bachmann MF, Oxenius A. Interleukin 2: from immunostimulation to immunoregulation and back again. EMBO Rep. 2007;8(12):1142–1148. doi: 10.1038/sj.embor.7401099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malek TR. The biology of interleukin-2. Annu Rev Immunol. 2008;26:453–479. doi: 10.1146/annurev.immunol.26.021607.090357. [DOI] [PubMed] [Google Scholar]
- 5.Pipkin ME, et al. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32(1):79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsokos GC, Nambiar MP, Tenbrock K, Juang YT. Rewiring the T-cell: Signaling defects and novel prospects for the treatment of SLE. Trends Immunol. 2003;24(5):259–263. doi: 10.1016/s1471-4906(03)00100-5. [DOI] [PubMed] [Google Scholar]
- 7.Moulton VR, Tsokos GC. Abnormalities of T cell signaling in systemic lupus erythematosus. Arthritis Res Ther. 2011;13(2):207. doi: 10.1186/ar3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nambiar MP, Mitchell JP, Ceruti RP, Malloy MA, Tsokos GC. Prevalence of T cell receptor zeta chain deficiency in systemic lupus erythematosus. Lupus. 2003;12(1):46–51. doi: 10.1191/0961203303lu281oa. [DOI] [PubMed] [Google Scholar]
- 9.Brundula V, et al. Diminished levels of T cell receptor zeta chains in peripheral blood T lymphocytes from patients with systemic lupus erythematosus. Arthritis Rheum. 1999;42(9):1908–1916. doi: 10.1002/1529-0131(199909)42:9<1908::AID-ANR17>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 10.Liossis SN, Ding XZ, Dennis GJ, Tsokos GC. Altered pattern of TCR/CD3-mediated protein-tyrosyl phosphorylation in T cells from patients with systemic lupus erythematosus. Deficient expression of the T cell receptor zeta chain. J Clin Invest. 1998;101(7):1448–1457. doi: 10.1172/JCI1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez D, Perl A. mTOR signaling: a central pathway to pathogenesis in systemic lupus erythematosus? Discov Med. 2010;9(46):173–178. [PMC free article] [PubMed] [Google Scholar]
- 12.Nambiar MP, et al. Reconstitution of deficient T cell receptor zeta chain restores T cell signaling and augments T cell receptor/CD3-induced interleukin-2 production in patients with systemic lupus erythematosus. Arthritis Rheum. 2003;48(7):1948–1955. doi: 10.1002/art.11072. [DOI] [PubMed] [Google Scholar]
- 13.Moulton VR, Tsokos GC. Alternative splicing factor/splicing factor 2 regulates the expression of the zeta subunit of the human T cell receptor-associated CD3 complex. J Biol Chem. 2010;285(17):12490–12496. doi: 10.1074/jbc.M109.091660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain J, Loh C, Rao A. Transcriptional regulation of the IL-2 gene. Curr Opin Immunol. 1995;7(3):333–342. doi: 10.1016/0952-7915(95)80107-3. [DOI] [PubMed] [Google Scholar]
- 15.Weiss A, Littman DR. Signal transduction by lymphocyte antigen receptors. Cell. 1994;76(2):263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 16.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9(4):353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 17.Chen CY, et al. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000;14(10):1236–1248. [PMC free article] [PubMed] [Google Scholar]
- 18.Lindstein T, June CH, Ledbetter JA, Stella G, Thompson CB. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science. 1989;244(4902):339–343. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- 19.Lemaire R, et al. Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: A novel function for SR proteins. Genes Dev. 2002;16(5):594–607. doi: 10.1101/gad.939502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun D, et al. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing-regulatory factor SF2 (ASF) Nat Immunol. 2011;12(9):853–860. doi: 10.1038/ni.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michlewski G, Sanford JR, Cáceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30(2):179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15(8):819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pont AR, Sadri N, Hsiao SJ, Smith S, Schneider RJ. mRNA decay factor AUF1 maintains normal aging, telomere maintenance, and suppression of senescence by activation of telomerase transcription. Mol Cell. 2012;47(1):5–15. doi: 10.1016/j.molcel.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemaire R, Winne A, Sarkissian M, Lafyatis R. SF2 and SRp55 regulation of CD45 exon 4 skipping during T cell activation. Eur J Immunol. 1999;29(3):823–837. doi: 10.1002/(SICI)1521-4141(199903)29:03<823::AID-IMMU823>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 25.Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH. The Committee on Prognosis Studies in SLE Derivation of the SLEDAI. A disease activity index for lupus patients. Arthritis Rheum. 1992;35(6):630–640. doi: 10.1002/art.1780350606. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122(3):365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Wang J, Manley JL. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 2005;19(22):2705–2714. doi: 10.1101/gad.1359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun S, Zhang Z, Sinha R, Karni R, Krainer AR. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat Struct Mol Biol. 2010;17(3):306–312. doi: 10.1038/nsmb.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hench VK, Su L. Regulation of IL-2 gene expression by Siva and FOXP3 in human T cells. BMC Immunol. 2011;12:54. doi: 10.1186/1471-2172-12-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenbrock K, Juang YT, Gourley MF, Nambiar MP, Tsokos GC. Antisense cyclic adenosine 5′-monophosphate response element modulator up-regulates IL-2 in T cells from patients with systemic lupus erythematosus. J Immunol. 2002;169(8):4147–4152. doi: 10.4049/jimmunol.169.8.4147. [DOI] [PubMed] [Google Scholar]
- 31.Kyttaris VC, Juang YT, Tenbrock K, Weinstein A, Tsokos GC. Cyclic adenosine 5′-monophosphate response element modulator is responsible for the decreased expression of c-fos and activator protein-1 binding in T cells from patients with systemic lupus erythematosus. J Immunol. 2004;173(5):3557–3563. doi: 10.4049/jimmunol.173.5.3557. [DOI] [PubMed] [Google Scholar]
- 32.Wong HK, Kammer GM, Dennis G, Tsokos GC. Abnormal NF-kappa B activity in T lymphocytes from patients with systemic lupus erythematosus is associated with decreased p65-RelA protein expression. J Immunol. 1999;163(3):1682–1689. [PubMed] [Google Scholar]
- 33.Tan EM, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25(11):1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.