Abstract
The Australian continent holds some of the earliest archaeological evidence for the expansion of modern humans out of Africa, with initial occupation at least 40,000 y ago. It is commonly assumed that Australia remained largely isolated following initial colonization, but the genetic history of Australians has not been explored in detail to address this issue. Here, we analyze large-scale genotyping data from aboriginal Australians, New Guineans, island Southeast Asians and Indians. We find an ancient association between Australia, New Guinea, and the Mamanwa (a Negrito group from the Philippines), with divergence times for these groups estimated at 36,000 y ago, and supporting the view that these populations represent the descendants of an early “southern route” migration out of Africa, whereas other populations in the region arrived later by a separate dispersal. We also detect a signal indicative of substantial gene flow between the Indian populations and Australia well before European contact, contrary to the prevailing view that there was no contact between Australia and the rest of the world. We estimate this gene flow to have occurred during the Holocene, 4,230 y ago. This is also approximately when changes in tool technology, food processing, and the dingo appear in the Australian archaeological record, suggesting that these may be related to the migration from India.
Keywords: admixture time, population history, human evolution
Genetic and archaeological evidence suggests that anatomically modern humans expanded from Africa (1, 2) and colonized all corners of the world, replacing with limited gene flow local archaic Homo populations, such as Neanderthals (3) and the Denisovans (4, 5). The expansion of modern humans apparently proceeded via two routes: the northern dispersal that gave rise to modern Asians 23,000–38,000 y ago (6, 7) and an earlier southern dispersal, which followed the coast around the Arabian Peninsula and India, to the Australian continent (5, 7). It has been suggested that the ancestors of aboriginal Australians and Papua New Guineans diverged from the ancestral Eurasian population 62,000–75,000 y ago (7) and, based on archaeological evidence, reached Sahul (the joint Australia–New Guinea landmass) by at least 45,000 y ago (8–10). Whereas coastal New Guinea (but not the highlands) subsequently experienced additional gene flow from Asia (associated with the Austronesian expansion) (9), the extent of isolation of Aboriginal Australians following initial colonization is still debated. The prevailing view is that until the arrival of the Europeans late in the 18th century, there was little, if any, contact between Australia and the rest of the world (7, 11, 12), although some mtDNA and Y chromosomal studies suggested some gene flow to Australia from the Indian subcontinent during the Holocene (13–15). Here, we analyze genome-wide SNP data and find a significant signature of gene flow from India to Australia, which we date to about 4,230 y ago.
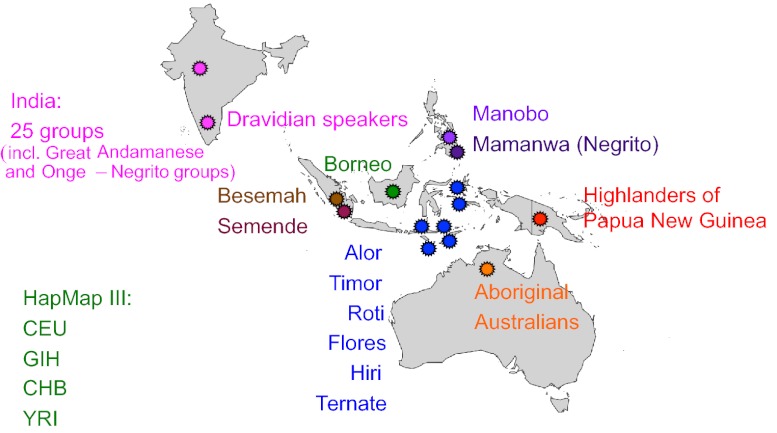
We assembled genome-wide SNP data from aboriginal Australian samples from the Northern Territories (AUA) (5, 13), highlanders of Papua New Guinea (NGH) (16), 11 populations from island Southeast (SE) Asia (5), and 26 populations from India (17), including Dravidian speakers from South India (5, 18). We also included data from the Yorubans from Ibadan; Nigeria (YRI); individuals of northern and western European ancestry living in Utah (CEU); Han Chinese individuals from Beijing, China (CHB); and Gujarati Indians from Houston, TX (GIH) (19). The final dataset comprised 344 individuals (Table S1 and Fig. 1); and after data cleaning and integration, we had 458,308 autosomal SNPs for the analysis.
Fig. 1.
Geographic distribution of samples used in this study.
Results
Genetic Relationships Between Populations.
First, to place aboriginal Australians into a global context, we carried out principal component analysis (PCA) (20). The first two principal axes are driven by genetic differentiation between Africans, Australians/Papua New Guineans, and Europeans/Indians/Asians (Fig. S1A). AUA are close to NGH but extend toward the European/Indian/Asian grouping, suggesting a common origin with the former and admixture with the latter. AUA and NGH are separated along PC4, after the separation of CEU and CHB along PC3 (Fig. S1B). The prior separation of CEU and CHB could suggest that AUA and NGH diverged after European and Asian populations, which, according to archaeological evidence (21) and estimates based on various genetic markers, happened between 37 and 60 kya (6, 7, 22). Alternatively, this result could suggest smaller Ne/stronger drift in AUA and NGH, or reflect ascertainment bias because most of the SNPs on the Affymetrix arrays were ascertained in individuals of European and African ancestry.
To better understand the relationships among AUA, NGH, and neighboring populations from Island SE Asia, we carried out PCA on these populations only (Fig. S1C). PC1 separates NGH and AUA from the other groups, whereas, interestingly, PC2 separates AUA and the Mamanwa (MWA) (a Negrito group from the Philippines) from NGH and the other SE Asian groups. PC3 groups the MWA with NGH but separates the Australians (Fig. S1D). The almost-identical eigenvalues for PC2 and PC3 suggest that the Mamanwa are equidistant from AUA and NGH (Fig. S1 C and D); overall, these results are consistent with previous indications of shared ancient ancestry among Australians, NGH, and the Mamanwa (5, 23).
Divergence-Time Estimation.
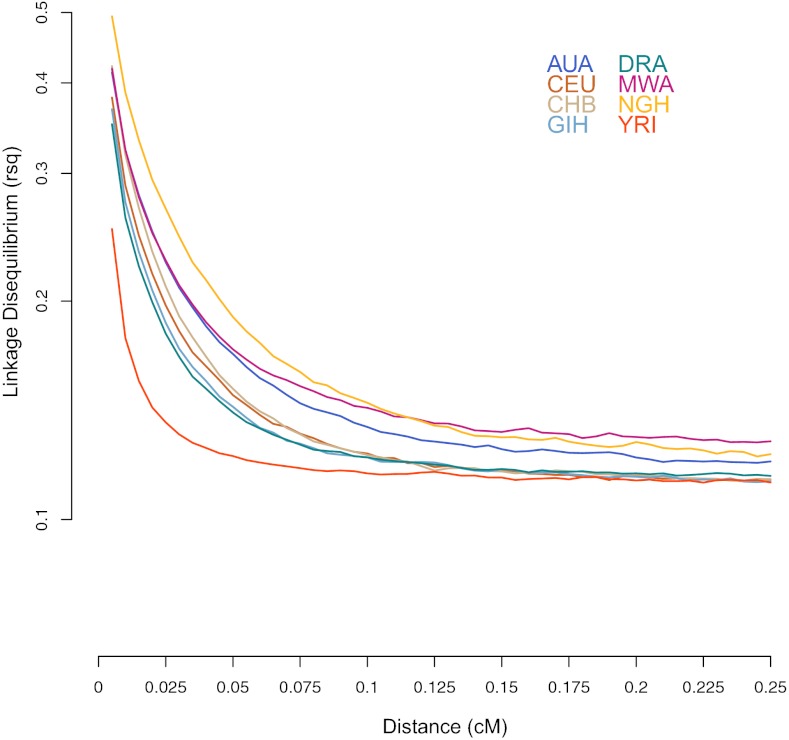
We next examined genome-wide patterns of linkage disequilibrium (LD) to estimate the divergence times among populations and investigate past population size changes (24, 25). Because LD is a property of genomic regions and not of individual SNPs, it is not expected to be strongly affected by ascertainment bias (25, 26). For this analysis, we binned the genome-wide data (588,335 SNPs) into 50 evenly spaced recombination distance categories (0.005–0.25 cM) from AUA, NGH, MWA, Dravidian speakers from South India, and the CEU, CHB, GIH, and YRI populations. Genetic distances were interpolated from genome-wide recombination rates estimated as part of the HapMap project (27). For each population and for every pair of SNPs within each distance category, we calculated the squared correlation (rLD2) in allele frequencies (25) by randomly selecting 10 individuals from each population and adjusting the measurement for each pair of SNPs by sample size (to account for missing data) (25); in total, around 150 million pairwise LD observations were made. The results (Fig. 2) show that LD increases with increasing geographic distance from Africa, as found previously (25, 26). The most extreme LD values over the shortest genomic distances (up to 0.075 cM) are seen in NGH, followed by MWA, whereas the most extreme LD values over the longest genomic distances (0.075–0.25 cM) are seen in the MWA, followed by the NGH. LD between SNPs separated by short genomic distances (short-range LD) is informative about older population size, relative to LD observed between SNPs separated by greater genomic distances (long-range LD) (12, 24, 28). Because decay of LD is inversely related to changes in effective population size (Ne) over time, our results suggest serial bottlenecks associated with the expansion of modern humans out of Africa, with the strongest ancient bottleneck being observed in the NGH. Furthermore, the MWA seem to have experienced a more recent bottleneck, possibly associated with the Austronesian expansion, as suggested previously from analyses of mtDNA sequences (29). In comparison with the NGH and the MWA, Australians exhibit the least extreme LD values, suggesting either a weaker bottleneck or less isolation experienced by this population.
Fig. 2.
LD measured for each population and each pair of SNPs within the 50 evenly spaced recombination distance categories. Shortest genetic distances between the SNPs are represented on the left and progress toward the largest genetic distances on the right.
The AUA, the NGH, and the MWA have all experienced ancient admixture with the Denisova hominins (5), and although admixture in general is known to decrease genome-wide LD (30), ancient admixture has been shown to increase long-range LD (30, 31). It is possible that long-range LD values in these three populations are inflated because of this ancient gene flow from the Denisovans. However, because the population bottlenecks, associated with the expansion of modern humans out of Africa increase genome-wide LD (25, 26), including the long-range LD, and we do expect to observe a strong effect of these bottlenecks in the Australian, the NGH and the MWA populations, it is hard to distinguish here the signature of bottlenecks from the possible signal of ancient admixture. We, therefore, can only conclude that the Denisova gene flow might have contributed to the increase in the long-range LD we observe in these three populations. Importantly, however, because the Denisova gene flow occurred into the common ancestor of these three populations (5), the differences in LD we observe between them cannot be attributable to this ancient admixture.
The correlation in LD patterns between populations can be used to estimate their time of divergence (25). The rationale behind this calculation is that immediately after two populations diverge, genome-wide LD in the two daughter populations should be perfectly correlated, but the correlation will decay exponentially over time, with the rate of decay dependent only on the recombination distance between the markers, not on Ne (25). The correlation in LD between populations is independent of Ne, because recombination events essentially behave like new neutral mutations: there will be more of them in a big population but fewer of them will fix via drift than in a small population, and as these two processes cancel each other out exactly, the rate of LD decay is not influenced by the population size. We computed the correlation between the LD values for each pair of populations and for each recombination distance category and estimated the time of divergence from the rate of decay of the correlation in LD values with recombination distance (25). To be able to compare our results to previous studies (25), and to exclude the effect of potential later admixture (32), for this analysis, we used only the first 20 recombination distance categories, i.e., only SNP pairs located at distances of up to 0.1 cM from each other. We estimate the average time of divergence for the main continental groups as follows: European (CEU) and Asian (CHB) populations and populations of greater Australia (AUA and NGH) have diverged from the African populations (YRI) 66 kya, and the split between CEU and CHB is estimated to have occurred 43 kya. These dates are in good agreement with previous studies, based on different types of data and using different methods (6, 22, 25). The divergence times among the AUA, NGH, and MWA (the putative descendants of the early southern route migration) were 36 kya, roughly in concordance with the date of divergence estimated based on the distribution of the bacterium Helicobacter pylori (33) but too recent given the purported date of the dispersal into Sahul at 45 kya (8–10). Despite LD being a measure expected to be relatively unaffected by ascertainment bias (25, 26), this may reflect some effect of this bias on the estimation; because a smaller number of SNPs included into the genotyping platform is expected to be polymorphic in these populations relative to the populations in which these SNPs were discovered, a smaller number of pairwise LD observations could be made. This will make the observed correlation in LD measurements between any two populations appear higher, reducing the rate of decay of the correlation in LD values and resulting in the time of divergence being underestimated. A previous study that estimated the time of divergence for the YRI, CEU, and CHB populations, using the same LD measure but half the number of markers, also reported low divergence dates (25). Because the Denisova gene flow occurred into the common ancestor of the AUS, NGH, and MWA (5), which is before the divergence time, it should not have any effect on the time of divergence estimation. In sum, these results confirm a common origin but an ancient split (at least 36 kya) for the Mamanwa, Australians, and NGH, supporting the view that these populations represent the descendants of an early southern route migration out of Africa and that Australians and New Guineans diverged early in the history of Sahul, when they were still one landmass, and not when the lands were separated by rising sea waters around 8,000 y ago.
Admixture with India.
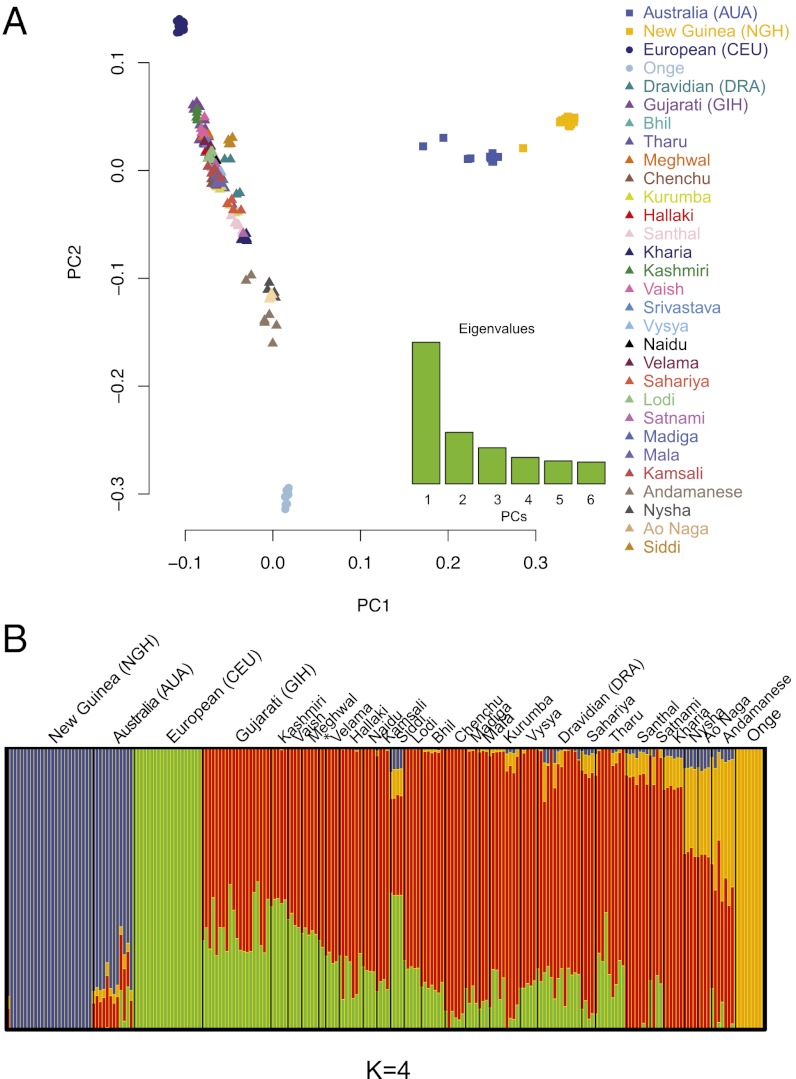
The PCA results clearly indicate some signal of admixture in the Australians (Fig. S1A). This could be attributable to recent European admixture, as reported previously (12, 34). To investigate this signal of admixture, we first carried out a PCA of AUA, NGH, Europe, and India (Fig. 3A). PC1 separates AUA and NGH from the other groups, whereas PC2 separates the Andamanese Onge at one end and CEU at the other, with mainland Indian populations spread roughly along a north-to-south cline, as observed previously (17). Apart from two outliers, the Australians are distributed toward the middle of the Indian cline and not toward Europe. Thus, these results do not indicate that Europeans are the source population for the signal of admixture and suggest, instead, that the signal comes from the Indian subcontinent.
Fig. 3.
Results of the PCA and ADMIXTURE analyses. (A) PCA of AUA, NGH, CEU, and 26 Indian populations. PC1 is driven by differences between the populations of Sahul and Eurasia. PC2 reflects a north-to-south gradient of European ancestry observed in Indian groups, with the southernmost group being the Onge, a Negrito population from the Andaman islands. (B) Population structure estimated using ADMIXTURE for K = 4. Each vertical bar represents an individual and each color describes the proportion of each individual’s genome that comes from one of the four hypothetical ancestral populations (K). The asterisk indicates the two individuals from the Srivastava group.
It has been shown previously that uneven sampling has a strong influence on the results of PCA (35). Although the sample sizes of populations used in this analysis were unequal, by making them equal, we would introduce a bias in that the analysis will cease to be blind to population labels (i.e., we have to know how to group individuals into populations to make population sizes equal). Therefore, to test the robustness of these results to uneven sampling, we repeated the analysis 10 times, each time randomly sampling 70% of the samples. Although slight differences in the results were present, the overall results and conclusions remain unchanged (Fig. S2).
To further investigate this result, we then analyzed genetic ancestry using the maximum-likelihood–based clustering algorithm ADMIXTURE (36). Briefly, this method considers each person’s genome as having originated from a specified number (K) of hypothetical ancestral populations and then describes the proportion of each individual’s genome that comes from each of these ancestral populations. To avoid potential problems caused by existing LD between markers, we first used the PLINK tool to thin the dataset by excluding from the analysis SNPs in strong LD. Our initial experiments showed that LD pruning, based either on correlations between SNPs or on correlations between linear combinations of SNPs, did not have any noticeable effect on the results of the ADMIXTURE analysis. Nevertheless, to save computational time, we used the pruned dataset, comprising nearly 170,000 markers, for all of the subsequent runs of ADMIXTURE. We tested K = 2 through K = 10 and performed 10 independent runs for each value of K. We monitored consistency between the runs and used ADMIXTUREs cross-validation procedure to establish the value of K that fits the data best (Fig. S3). Although the lowest cross-validation error is exhibited by K = 3 (Fig. S3), the Indian component we are interested in is identified only at K = 4; because the difference between the CV error for K = 3 and K = 4 is quite small, at least four times smaller than the difference between K = 3 and any other value of K (Fig. S3), we report here the results for K = 4. At K = 4 (Fig. 3B), Australians are assigned a component that is present at high frequency in mainland India and is shared exclusively between Australia and India [with the exception of one NGH individual, who is an outlier relative to the other NGH samples and, according to PCA results (Fig. S1A), is closer to AUA than to other individuals in the NGH population]. Moreover, this component is observed in similar proportions in all of the Australians, suggesting that it is uniform throughout the genome. By contrast, the “European” ancestry component is present in only a few Australians and in varying amounts, as expected, for very recent admixture such as observed in African Americans (20, 37). These AUA individuals showing evidence of recent European ancestry were excluded from further analyses. Thus, the Indian admixture signal revealed in AUA by this analysis does not exhibit the same characteristics as recent European admixture. Identical results were obtained using another maximum-likelihood–based software frappe (38) with the full set of 460,000 markers (Fig. S4).
Next, to be more confident that the Indian component we observe in AUA is indeed Indian and does not reflect some unsampled ancestry, we repeated the ADMIXTURE analysis with individuals of African, European, Asian, and SE Asian ancestry, including the Mamanwa. For the Indian group, we used genotypes from the Chenchu and Kurumba (tribal Dravidian-speaking populations) and from the nontribal Dravidian speakers from south India, because these groups are closest to the axis of admixture in the PCA and have the highest frequencies of the shared Australia–India ancestry component in the previous ADMIXTURE analysis. After the dataset was thinned for SNPs in LD, we had 187,470 SNPs remaining for this analysis. This time, the lowest cross-validation error is exhibited by K = 5, whereas the Indian component is identified at K = 7 (Fig. S5). At K = 5, the proportion of Australian ancestry not shared with the New Guineans most closely resembles the ancestry profile of the three Indian populations at this value of K. Additionally, at K = 7, six runs with the highest log-likelihood scores ascribe 11% of Australian ancestry to India, whereas an additional 9% is shared with the Mamanwa (Fig. S5).
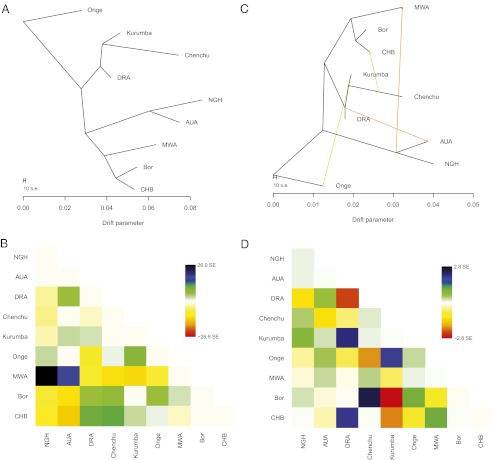
To further verify the signal of Indian admixture, we used TreeMix (39) to find a population graph that best describes the relationship between populations in the dataset by testing for gene flow between them. This method uses the genome-wide allele frequency data to first find the maximum-likelihood tree of populations and then infer migration events by identifying populations that poorly fit this tree. Because it has been shown previously that migrations inferred for Oceanian populations differ depending on whether the SNPs involved in the analysis were ascertained in a Yoruban or a French individual (39), we have excluded YRI and CEU individuals from this analysis. (For the results of the analysis that included these populations, see Fig. S6.] After removal of SNPs in LD, the resulting dataset comprised 150,000 markers. We first inferred the maximum likelihood tree of the nine populations included in the analysis (Fig. 4A) and then analyzed the residuals (Fig. 4B) to identify pairs of populations that are more related to each other than is captured by this tree. We then sequentially added migration events to the tree, until we found a graph with the smallest residuals (Fig. 4 C and D). The graph that best fits the data has four inferred migration edges: Chenchu to CHB (weight, 4%), Onge to India (17) (weight, 6%); one of the edges captures shared ancestry between NGH, AUA, and MWA (5, 23) (weight, 15%); and one of the edges provides evidence for the gene flow from India to Australia. The weight for this migration edge is estimated to be 11%, in agreement with the admixture proportion obtained in the ADMIXTURE analysis. The P value (which here describes how much a particular inferred migration improves the fit to the data) for all migrations is estimated to be at least 1 × 10−5.
Fig. 4.
Results of the TreeMix analysis. (A) The maximum-likelihood tree of nine populations included in the analysis. (B) Residual fit from the tree. Residuals above zero indicate pairs of populations that are candidates for admixture events. (C) Population graph that best fits the data, based on the smallest residuals (D).
Finally, to further test the robustness of this inference, we used the 4 Population Test statistic f4 (17). The four populations considered in this analysis were AUA, NGH, India, and YRI. For the Indian group, we again used genotypes from the Chenchu and Kurumba, as well as from the nontribal Dravidian speakers from south India. YRI were chosen as an outgroup that is equally distant from the other three groups, and the allele frequencies in the YRI were used for normalization, where we weighted each SNP by a quantity proportional to its expected genetic drift in the ancestral group (YRI) (17). We calculated allele frequency differences at each SNP between all pairs of populations, restricting the analysis to SNPs that were polymorphic in all of the groups to minimize any effect of the ascertainment bias. This reduced the dataset to 250,000 SNPs. The expectation is that if there was no gene flow from India into Australia, then the allele frequency differences observed between YRI and India should be uncorrelated with the allele frequency differences observed between AUAs and NGH. However, if this correlation deviates from zero, then this suggests that there was gene flow from India into either AUA, NGH, or both. A Weighted Block Jackknife approach, where the genome was divided into nonoverlapping 5-cM blocks and each block was dropped sequentially (17, 40), was used to correct for nonindependence of SNPs and to assess statistical significance via a Z score (17). In our analysis, the f4 statistic has a Z = −1.93 (P = 0.026), allowing us to reject the simple tree (YRI(India(AUA,NGH))) and suggesting, instead, that the data are best described by a mixture of two trees: (YRI(India(AUA,NGH))) and (YRI(NGH(AUA, India))). The fact that the Z score has a negative sign is important here, because it indicates gene flow between India and AUA (or NGH and Yoruba) and not between India and NGH. We repeated this analysis, substituting an Asian population (CHB) and a Negrito population of the Andaman Islands (Onge) for India; for both analyses, the resulting f4 statistic had much higher P values (Z = −0.11, P value = 0.45; and Z = −0.28, P value = 0.38, respectively). Thus, the f4 statistics indicate a signal of gene flow from India to Australia and, furthermore, that the source population is more closely related to present-day Dravidian-speaking Indian groups than to Onge.
In sum, four analyses (PCA, ADMIXTURE, TreeMix, and f4 statistics) all indicate gene flow from India to Australia. Although previous analyses based on a limited number of markers (41) or uniparental data (13, 14) also suggested genetic relationships between Australia and India, neither a previous study of genome-wide SNP data from Australians (12) nor the analysis of a genome sequence of an aboriginal Australian (7) reported any such gene flow. However, the genome-wide SNP study (12) did not include any populations from India, and although the analysis of the Australian genome sequence did find indications of genetic relationships with groups from India, they concluded that this represented some genetic ancestry in the Australian genome sequence that could not be assigned to any existing population (7). Based on the results above, it is likely that the signal of Indian genetic ancestry in the Australian genome sequence does, in fact, reflect the same gene flow from India that we detect in our analyses.
Admixture-Time Estimation.
We next analyzed the genome-wide admixture pattern to estimate the time of admixture. We first used StepPCO (20) to obtain the block-like admixture signal across each chromosome for each Australian (excluding two individuals with evidence of European admixture). The NGH and India (represented, again, by Chenchu and Kurumba and the nontribal Dravidian speakers from South India) were used as proxies for the parental populations (Fig. S7). We then applied wavelet-transform analysis to the StepPCO signal and used the wavelet transform coefficients to infer time since admixture (20). Briefly, this wavelet transform represents the admixture signal as the sum of simple waves, each characterized by its frequency (width) and position within the signal. The dominant frequency present in the signal is an indirect measure of an average width of the admixture blocks, and from this, the time of admixture is estimated by comparing this observed dominant frequency to that obtained for simulated data generated using the admixture rate observed in the empirical data (20). The spectral analysis of the StepPCO signal revealed that the estimated average dominant frequency for the Australians was 3.9, which corresponds to an abundance of high-frequency wavelets (that is, narrow ancestry blocks). Based on simulations, this estimate corresponds to an admixture time of 141 generations ago. Assuming a generation time of 30 y (42), our results indicate that the gene flow from India into Australia occurred around 4,230 y ago, consistent with a previous estimate based on a small number of Y-STR (short tandem repeats on the Y-chromosome) loci (14).
Interestingly, at around this time, several changes take place in the archaeological record of Australia. There is a sudden change in stone tool technologies, with microliths appearing for the first time (43), and people start processing plants differently (14, 44). It has been a matter of controversy as to whether these changes occurred in situ (45) or reflect contact with people from outside Australia or some combination of both factors. However, the dingo also first appears in the fossil record at this time and must have come from outside Australia (46). Although dingo mtDNA appears to have a SE Asian origin (47), morphologically, the dingo most closely resembles Indian dogs (46). The fact that we detect a substantial inflow of genes from India into Australia at about this same time does suggest that all of these changes in Australia may be related to this migration.
Discussion
In conclusion, our results suggest an ancient association between Australia, New Guinea, and the Mamanwa (a Negrito group from the Philippines), with a time of divergence of at least 35,000 y ago, implying a common origin but an early separation for these groups, and supporting the view that these populations represent the descendants of an early “southern migration route” (5, 7). Strikingly, we also detect a signal of substantial gene flow between Indian and Australia populations before European contact. We estimate the date of this admixture to be 141 generations ago and suggest that this gene flow may be associated with the changes documented in the Australian archaeological record at about this time.
The signal of Indian gene flow might not necessarily come directly from India; it is easy to envision a scenario whereby the Indian ancestry comes to Australia indirectly, e.g., via contact with island SE Asian populations. Indeed, it is known that some pre-European trade existed between the northeastern coast of Australia and Indonesia (45). However, our study includes 11 populations from island SE Asia, but there is no signal whatsoever of recent gene flow from India into these populations or from these populations into Australia (Fig. S8), which renders this scenario of Indian ancestry via SE Asia unlikely.
It has been shown that ancient population structure could produce patterns similar to those generated by admixture (48). However, even if this substructure existed in the ancestral population of the AUA and NGH, to suspect that the gene flow we detect here might be an artifact attributable to this substructure would require the age of this ancestry to be much older, predating the colonization of the Sahul (49). The fact that the date we obtain is comparatively very recent argues against this possibility. Moreover, the amount of ancestry shared between Australians and Denisovans is approximately the same as that shared between NGH and Denisovans (5). This might seem surprising, because we do expect that later mid-Holocene gene flow into Australia (but not NGH) should diminish the proportion of the Denisovan ancestry in the AUA but not the NGH. However, given that the total Denisovan contribution into the ancestor of these populations is around 3–5% (5) and the amount of Indian contribution is estimated here to be around 11%, the expected impact of Indian genetic material would be to decrease the estimated Denisovan ancestry in the Australian genome by about 0.3–0.5%, which is too small to be detected in our data.
Lastly, although the Australian samples presented in this study come from a broad geographical area of the Northern Territories of Australia, they might not be representative of the Australian aboriginals as a whole. As others (12) have pointed out, comprehensive studies of the genetic variation in Australia would be very desirable to further understand their increasingly complex history.
Materials and Methods
Population Samples and Data.
The aboriginal Australian samples were obtained in the early 1990s by forensic scientists from individuals throughout the Northern Territory, who gave oral consent for their samples to be used in studies of population history, and have been used in previous such studies (5, 13). This study was approved by the ethical review board of the University of Leipzig Medical Faculty. All samples were genotyped on Affymetrix 6.0 arrays, and quality filtering was performed as described previously (5, 16). YRI, CEU, CHB, and GIH genotypes were downloaded from the International HapMap project home page (http://hapmap.ncbi.nlm.nih.gov/downloads/genotypes/2009-01_phaseIII/). The data were merged using PLINK (50) to include only markers that passed quality filters in all datasets.
Statistical Analyses.
PCA and time of admixture estimation was performed using the StepPCO software (20). All PCA analyses were run on 458,308 markers. Genome-wide LD calculation and divergence-time estimation were performed using custom scripts. Individual ancestry components and admixture proportions were inferred using ADMIXTURE (36). The LD pruning for the ADMIXTURE was done with PLINK tool (50), using the following settings: –indep-pairwise 200 25 0.4 (ref. 7), which reduced the dataset to 168,051 markers. Calculation of allele frequencies for the TreeMix analysis was performed using PLINK tool (50). The Onge samples were set as an outgroup, and we used the window size of 500 (-k option).
Supplementary Material
Acknowledgments
We thank Slava Matveyev, Michael Lachmann, David Reich, and Nick Patterson for valuable discussions and comments on the manuscript; Cesare de Filippo for his contribution to the final version of this manuscript; and the two anonymous reviewers for the careful and constructive reviews. This research was funded by the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211927110/-/DCSupplemental.
References
- 1.Ramachandran S, et al. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102(44):15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu H, Prugnolle F, Manica A, Balloux F. A geographically explicit genetic model of worldwide human-settlement history. Am J Hum Genet. 2006;79(2):230–237. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328(5979):710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468(7327):1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reich D, et al. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am J Hum Genet. 2011;89(4):516–528. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5(10):e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen M, et al. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334(6052):94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Connell J, Allen J. Dating the colonization of Sahul (Pleistocene Australia–New Guinea): A review of recent research. J Archaeol Sci. 2004;31(6):835–853. [Google Scholar]
- 9.Kayser M. The human genetic history of Oceania: Near and remote views of dispersal. Curr Biol. 2010;20(4):R194–R201. doi: 10.1016/j.cub.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Summerhayes GR, et al. Human adaptation and plant use in highland New Guinea 49,000 to 44,000 years ago. Science. 2010;330(6000):78–81. doi: 10.1126/science.1193130. [DOI] [PubMed] [Google Scholar]
- 11.Hudjashov G, et al. Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis. Proc Natl Acad Sci USA. 2007;104(21):8726–8730. doi: 10.1073/pnas.0702928104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McEvoy BP, et al. Whole-genome genetic diversity in a sample of Australians with deep Aboriginal ancestry. Am J Hum Genet. 2010;87(2):297–305. doi: 10.1016/j.ajhg.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redd AJ, Stoneking M. Peopling of Sahul: mtDNA variation in aboriginal Australian and Papua New Guinean populations. Am J Hum Genet. 1999;65(3):808–828. doi: 10.1086/302533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Redd AJ, et al. Gene flow from the Indian subcontinent to Australia: Evidence from the Y chromosome. Curr Biol. 2002;12(8):673–677. doi: 10.1016/s0960-9822(02)00789-3. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S, et al. Reconstructing Indian-Australian phylogenetic link. BMC Evol Biol. 2009;9:173. doi: 10.1186/1471-2148-9-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wollstein A, et al. Demographic history of Oceania inferred from genome-wide data. Curr Biol. 2010;20(22):1983–1992. doi: 10.1016/j.cub.2010.10.040. [DOI] [PubMed] [Google Scholar]
- 17.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461(7263):489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cordaux R, et al. Mitochondrial DNA analysis reveals diverse histories of tribal populations from India. Eur J Hum Genet. 2003;11(3):253–264. doi: 10.1038/sj.ejhg.5200949. [DOI] [PubMed] [Google Scholar]
- 19. Altshuler DM, et al. (2010) Integrating common and rare genetic variation in diverse human populations. Nature 467(7311):52–58. [DOI] [PMC free article] [PubMed]
- 20.Pugach I, Matveyev R, Wollstein A, Kayser M, Stoneking M. Dating the age of admixture via wavelet transform analysis of genome-wide data. Genome Biol. 2011;12(2):R19. doi: 10.1186/gb-2011-12-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stringer C. Modern human origins: Progress and prospects. Philos Trans R Soc Lond B Biol Sci. 2002;357(1420):563–579. doi: 10.1098/rstb.2001.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhivotovsky LA, Rosenberg NA, Feldman MW. Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am J Hum Genet. 2003;72(5):1171–1186. doi: 10.1086/375120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delfin F, et al. The Y-chromosome landscape of the Philippines: Extensive heterogeneity and varying genetic affinities of Negrito and non-Negrito groups. Eur J Hum Genet. 2011;19(2):224–230. doi: 10.1038/ejhg.2010.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenesa A, et al. Recent human effective population size estimated from linkage disequilibrium. Genome Res. 2007;17(4):520–526. doi: 10.1101/gr.6023607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McEvoy BP, Powell JE, Goddard ME, Visscher PM. Human population dispersal “Out of Africa” estimated from linkage disequilibrium and allele frequencies of SNPs. Genome Res. 2011;21(6):821–829. doi: 10.1101/gr.119636.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jakobsson M, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451(7181):998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 27.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayes BJ, Visscher PM, McPartlan HC, Goddard ME. Novel multilocus measure of linkage disequilibrium to estimate past effective population size. Genome Res. 2003;13(4):635–643. doi: 10.1101/gr.387103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gunnarsdóttir ED, Li M, Bauchet M, Finstermeier K, Stoneking M. High-throughput sequencing of complete human mtDNA genomes from the Philippines. Genome Res. 2011;21(1):1–11. doi: 10.1101/gr.107615.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeGiorgio M, Jakobsson M, Rosenberg NA. Out of Africa: Modern human origins special feature: Explaining worldwide patterns of human genetic variation using a coalescent-based serial founder model of migration outward from Africa. Proc Natl Acad Sci USA. 2009;106(38):16057–16062. doi: 10.1073/pnas.0903341106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plagnol V, Wall JD. Possible ancestral structure in human populations. PLoS Genet. 2006;2(7):e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moorjani P, et al. The history of African gene flow into Southern Europeans, Levantines, and Jews. PLoS Genet. 2011;7(4):e1001373. doi: 10.1371/journal.pgen.1001373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moodley Y, et al. The peopling of the Pacific from a bacterial perspective. Science. 2009;323(5913):527–530. doi: 10.1126/science.1166083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ballantyne KN, et al. MtDNA SNP multiplexes for efficient inference of matrilineal genetic ancestry within Oceania. Forensic Sci Int Genet. 2012;6(4):425–436. doi: 10.1016/j.fsigen.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 35.McVean G. A genealogical interpretation of principal components analysis. PLoS Genet. 2009;5(10):e1000686. doi: 10.1371/journal.pgen.1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryc K, et al. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci USA. 2010;107(2):786–791. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: Analytical and study design considerations. Genet Epidemiol. 2005;28(4):289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 39.Pickrell JK, Pritchard JK. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet 8(11):e1002967.
- 40.Kunsch HR. The jackknife and the bootstrap for general stationary observations. Ann Stat. 1989;17(3):1217–1241. [Google Scholar]
- 41.Stoneking M, et al. Alu insertion polymorphisms and human evolution: Evidence for a larger population size in Africa. Genome Res. 1997;7(11):1061–1071. doi: 10.1101/gr.7.11.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fenner JN. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol. 2005;128(2):415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- 43.Glover I, Presland G. 1985. Microliths in Indonesian flaked stone industries. Recent Advances in Indo-Pacific Prehistory, eds Misra V, Bellwood P (Oxford & IBH, New Dehli, India)
- 44.Beaton J. 1977. Dangerous harvest: Investigations in the late prehistoric occupation of upland south-east central Queensland. PhD thesis (Australian National University, Canberra, Australia)
- 45. Hiscock P (2008) Archaeology of Ancient Australia (Routledge, London)
- 46.Gollan K. 1985. Prehistoric dogs in Australia: An Indian origin? Recent Advances in Indo-Pacific Prehistory, eds Misra V, Bellwood P (Oxford & IBH, New Dehli, India)
- 47.Savolainen P, Leitner T, Wilton AN, Matisoo-Smith E, Lundeberg J. A detailed picture of the origin of the Australian dingo, obtained from the study of mitochondrial DNA. Proc Natl Acad Sci USA. 2004;101(33):12387–12390. doi: 10.1073/pnas.0401814101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eriksson A, Manica A. Effect of ancient population structure on the degree of polymorphism shared between modern human populations and ancient hominins. Proc Natl Acad Sci USA. 2012;109(35):13956–13960. doi: 10.1073/pnas.1200567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sankararaman S, Patterson N, Li H, Pääbo S, Reich D. The date of interbreeding between Neandertals and modern humans. PLoS Genet. 2012;8(10):e1002947. doi: 10.1371/journal.pgen.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.