Abstract
The main goal of brain tumor surgery is to maximize tumor resection while preserving brain function. However, existing imaging and surgical techniques do not offer the molecular information needed to delineate tumor boundaries. We have developed a system to rapidly analyze and classify brain tumors based on lipid information acquired by desorption electrospray ionization mass spectrometry (DESI-MS). In this study, a classifier was built to discriminate gliomas and meningiomas based on 36 glioma and 19 meningioma samples. The classifier was tested and results were validated for intraoperative use by analyzing and diagnosing tissue sections from 32 surgical specimens obtained from five research subjects who underwent brain tumor resection. The samples analyzed included oligodendroglioma, astrocytoma, and meningioma tumors of different histological grades and tumor cell concentrations. The molecular diagnosis derived from mass-spectrometry imaging corresponded to histopathology diagnosis with very few exceptions. Our work demonstrates that DESI-MS technology has the potential to identify the histology type of brain tumors. It provides information on glioma grade and, most importantly, may help define tumor margins by measuring the tumor cell concentration in a specimen. Results for stereotactically registered samples were correlated to preoperative MRI through neuronavigation, and visualized over segmented 3D MRI tumor volume reconstruction. Our findings demonstrate the potential of ambient mass spectrometry to guide brain tumor surgery by providing rapid diagnosis, and tumor margin assessment in near–real time.
Surgery is a key component in the treatment of brain tumors. Substantial evidence suggests that a smaller volume of postoperative residual tumor is associated with an improved prognosis (1–4). One of the greatest barriers to achieving optimal surgical results in infiltrative tumors is the difficulty in distinguishing tumor from normal brain (5–7). Failure to discern where the border of the lesion ends and viable, minimally infiltrated brain begins increases the likelihood of inadequate resection and neurological damage. Although maximal surgical resection with the goal of gross total tumor resection is desirable, in practice, delineation of resection margins is difficult because tumors can closely resemble healthy brain, often infiltrate brain tissue, and may be immediately adjacent to critical functional brain tissue. Aggressive resection can increase the risk for postoperative neurological deficits (7). For example, brain tumors located near critical brain areas subserving primary motor, sensory, or language functions are difficult to resect maximally while avoiding postoperative neurological deficits (8). Therefore, the principal challenge and objective of neurosurgical intervention is to maximize the resection of tumor, and minimize the potential for neurological deficit by preserving critical tissue. Intraoperatively acquired images can provide the neurosurgeon with the information needed to perform real-time, image-guided surgery.
A variety of mapping techniques have been developed to provide the surgeon with an understanding of the relationship of the tumor to surrounding key cortical areas, but none offers the molecular precision required to delineate tumor boundaries and evaluate the extent of infiltration. Intraoperative MRI, developed at Brigham and Women’s Hospital (BWH), created an opportunity for collecting information about the extent of tumor resection during surgery (5, 6). Although brain tumor resection typically requires multiple hours, intraoperative MRI can be completed and information evaluated within an hour. However, MRI has limited ability to distinguish residual tumor from surrounding normal brain (9). In consequence, there is a need for more detailed molecular information to be acquired on a timescale closer to real time than can be supplied by MRI.
Standard histopathology methods are the gold standard for detecting the presence of tumor and can provide diagnostic information during surgery within an hour, but are typically limited to the evaluation of one or very few samples. The most useful ancillary tests in diagnostic pathology exploit changes in nucleic acids that are at the foundation of tumors: DNA point mutations, chromosomal translocations, changes in copy number, and most recently, the changes in mRNA levels (10). Beyond nucleic acids, data on proteins and lipids in tissue can reveal important information about physiology and disease. Proteomic information is used regularly in pathology, albeit in a low-throughput manner using immunohistochemistry. The lipid constituents of tissues, however, remain entirely unused in diagnostic practice as lipidomic information has until recently been impractical to acquire. Now, desorption electrospray ionization mass spectrometry (DESI-MS) (11), as well as related MS-imaging methods (12), offer the possibility of incorporating lipid information into tumor diagnostics. DESI is one of a recently developed group of ionization techniques in MS in which samples are examined in the ambient environment with minimal pretreatment (13–17). Using DESI, direct lipid analysis from biological samples such as tissue sections can be performed rapidly and routinely. This microextraction experiment takes less than a second and gives a large amount of information on an individual small spot (pixel) on the unmodified tissue. Samples can be examined also by scanning them in 2D space while a spray of charged droplets impinges on the tissue, extracting lipids into secondary microdroplets that are continuously transferred to a mass spectrometer. Chemical information obtained from such scans is then represented in the form of 2D images recording relative abundances and spatial distributions of specific ions. In a single experiment, DESI-MS allows the characterization and imaging of many lipids including fatty acyls, glycerophospholipids, glycerolipids, and sphingolipids (18). DESI-MS imaging of tissue sections from banked tissue has been shown to enable disease diagnosis based on lipid profiles of different human diseases, such as bladder (19), kidney (20), prostate (21), and brain cancer (22, 23). These studies demonstrate the potential value of DESI in providing tissue diagnosis in a few seconds for intraoperative use, a task also being investigated by related mass-spectrometry methods (24).
The World Health Organisation (WHO) recognizes over 125 types of brain tumors according to histopathological evaluation. Tissue is differentiated according to cellular composition, including characteristics of astrocytic, ependymal, meningeal, neuronal, oligodendrocytic cells, and according to tumor origin. The WHO classification system also grades malignancy according to such characteristics as proliferation, cellular and nuclear morphology, necrosis, and the presence of abnormal vasculature (25). Glial tumors, gliomas, account for 30% of all intracranial tumors (26). The main subtypes are astrocytoma and oligodendroglioma, of which about 70% are grade III or higher. Grades II–IV are characterized by the absence of a discrete tumor boundary as tumor cells infiltrate into surrounding brain tissue. The most malignant form, glioblastoma (astrocytoma WHO grade IV), is refractory to treatment strategies and is associated with a median survival of 12–15 mo (27). Meningiomas are brain tumors that develop from the meninges, the tissue covering the brain and spinal cord, and are the most common type of brain tumor (34.7%) (26). Most meningiomas are slow growing, but many of these must still be removed due to resulting neurological complications. A small subset of meningiomas is more aggressive, and also requires surgical intervention. In contrast to gliomas, meningiomas usually feature a very clear border between tumor and normal brain.
Using a set of banked tumor samples, we have previously demonstrated that gliomas of different subtypes (oligodendrogliomas, astrocytomas, and oligoastrocytomas), grades and cell concentrations (a marker of tumor infiltration into normal tissue) could be rapidly classified by DESI-MS imaging using multivariate statistical analysis and machine learning (23). In the present study, we expanded our classification strategy for brain cancers by incorporating meningioma as a tumor type. The more comprehensive classification system is tested and validated through application to the diagnosis and tumor margin assessment of stereotactically registered surgical specimens from a cohort of five patients receiving neurosurgical care at BWH. DESI-MS imaging data and statistical classification results are correlated to histopathology for validation, and results for stereotactically registered samples are correlated to preoperative MRI through neuronavigation and visualized over segmented 3D MRI tumor volume reconstruction. Tumor margins are defined by evaluating the results for tumor grade and tumor cell concentration in the samples removed from peripheral regions of the tumor. Our results demonstrate an extensive clinical and interdisciplinary workflow that uses DESI-MS imaging as an analytical platform that can contribute molecular information in near–real time to support surgical decision making.
Results and Discussion
Classification Models for Brain Tumors.
First, we developed a classification model that would distinguish the two main types of primary intracranial tumors characterized by different invasive phenotypes: gliomas and meningiomas. DESI-MS imaging data from 21 glioma samples, of different subtypes and histological grades, and 11 meningioma samples were used as a training set. The classifier generated using support vector machine (SVM) supervised learning model had an overall recognition capability of 100% with 99.7% cross-validation. We then applied this classification model of brain tumor type to a validation set of 8 meningiomas and 15 gliomas. The results are shown in Table S1. It was remarkable that no discrepancies were observed for any of the specimens tested, with 100% agreement between results for brain tumor type and histopathological examination. The mass spectra showed distinctive lipid profiles between gliomas and meningiomas, as discussed next.
Molecular Diagnosis of Meningiomas by DESI-MS Imaging.
Banked meningioma tissue samples had a characteristic lipid profile in the negative-ion mode mass spectra with prominent peaks observed for plasmenylethanolamines (pls-PE), phosphatidylinositols (PI), and phosphatidylserines (PS). For example, for meningioma sample MG9, we observed high relative abundances for the ions at m/z 788.3, PS(36:1), m/z 885.4, PI(38:4), and m/z 810.2, PS(38:4); and lower relative abundances of ions m/z 700.4, pls-PE(34:1), m/z 722.3, pls-PE(36:4), m/z 750.3, pls-PE(38:4), m/z 760.3, PS(34:1), m/z 766.3, PE(38:4), m/z 812.3, PS(38:3), m/z 834.3, PS(40:6), m/z 835.5, PI(34:1), m/z 836.3, PS(40:5), m/z 838.3, PS(40:4), m/z 887.4, PI(38:3), and m/z 913.4, PI(40:4). Lipid species were tentatively identified based on collision-induced dissociation (CID) tandem MS experiments and comparison with CID data obtained previously on lipid species from human gliomas (22, 23). Meningioma lipid profiles (Fig. S1) were characteristically different from those previously reported for human gliomas by DESI-MS, with complete absence of the sulfatide ions commonly observed in our studies of low-grade gliomas (22). DESI-MS ion images showed relatively homogeneous distributions of the main lipid ions throughout the tissue samples. It is interesting to note that for meningioma sample MG12, a small region within the DESI-MS ion images showed a distinctive lipid profile characteristic of gray matter (Fig. S2). Histopathological evaluation revealed that the region consisted of normal brain gray matter abutting the meningioma tissue. This example highlights the ability to determine margins between meningioma and nonneoplastic brain tissue based on DESI-MS lipid profiles.
Diagnosis of Tissue Specimens from Five Stereotactic Surgical Cases.
Next, we further validated the classification system using samples obtained from five surgeries that were performed in the Advanced Multi-Modality Image Guided Operating suite and in standard operating rooms at BWH, Harvard Medical School. During the course of tumor resection, the neurosurgeon used a stereotactic neuronavigation pointer (Brainlab) to identify the sampling position on preoperative MRI and register 3D coordinates for most of the samples obtained. The samples obtained intraoperatively were sectioned and analyzed ex situ using DESI-MS imaging. Among these surgical cases, four were diagnosed as glioma by histopathology (three oligodendroglioma and one astrocytoma), and one as a meningioma. We first classified all specimens by brain tumor type discrimination as either meningioma or glioma. Gliomas were further subclassified to determine glioma subtype, grade, and tumor cell concentration (23). Results for these cases are presented in Table 1 and Table S2 and supported by a detailed discussion of mass spectral and statistical results. We correlate our findings with histopathology, and discuss the implications of our results for potential clinical application.
Table 1.
Classification results for surgical specimens from surgical cases 2, 3, 4, and 8
| Tumor type (%) |
Glioma subtype (%) |
Glioma grade (%) |
Glioma concentration (%) |
|||||||||
| Name | Histological diagnosis | GL | MG | A | OA | O | II | III | IV | Low | Med | High |
| Case 2 | ||||||||||||
| S7 | O, II, med | 95 | 5 | 3 | 0 | 97 | 97 | 3 | 0 | 1 | 99 | 0 |
| S8 | O, II, low | 80 | 20 | 13 | 1 | 86 | 58 | 32 | 11 | 48 | 44 | 7 |
| S10 (B) | O, II, low | 100 | 0 | 0 | 0 | 100 | 15 | 59 | 26 | 43 | 57 | 0 |
| S11 (A) | O, II, low | 100 | 0 | 0 | 0 | 100 | 27 | 52 | 21 | 41 | 59 | 0 |
| S12 (C) | O, II, med | 92 | 8 | 45 | 0 | 55 | 100 | 0 | 0 | 8 | 92 | 0 |
| S13 (D) | O, II, med | 95 | 5 | 2 | 0 | 98 | 100 | 0 | 0 | 7 | 88 | 5 |
| S15 | O, II, low | 73 | 27 | 40 | 0 | 60 | 53 | 27 | 20 | 0 | 60 | 40 |
| Total | 90 | 10 | 10 | 1 | 89 | 67 | 24 | 9 | 27 | 69 | 4 | |
| Case 3 | ||||||||||||
| S16 | O–III, high | 22 | 78 | 20 | 4 | 76 | 4 | 76 | 21 | 1 | 10 | 89 |
| S17 (B) | O–III, high | 10 | 90 | 37 | 18 | 46 | 3 | 85 | 11 | 1 | 29 | 70 |
| S18 | O–III, high | 11 | 89 | 23 | 12 | 64 | 1 | 88 | 12 | 7 | 27 | 67 |
| S19 | O–III, high | 86 | 14 | 7 | 26 | 67 | 0 | 76 | 24 | 4 | 79 | 17 |
| S20 (C) | O–III, high | 83 | 17 | 21 | 7 | 71 | 1 | 93 | 6 | 11 | 35 | 54 |
| S21 (D) | O–III, med | 97 | 3 | 43 | 17 | 40 | 14 | 43 | 43 | 8 | 46 | 46 |
| S22 (A) | O–III, high | 51 | 49 | 0 | 0 | 100 | 5 | 94 | 1 | 1 | 67 | 32 |
| S27 | O–III, med | 69 | 31 | 23 | 0 | 76 | 9 | 53 | 38 | 4 | 22 | 74 |
| Total | 50.2 | 49.8 | 20 | 12 | 68 | 4 | 76 | 20 | 4 | 40 | 56 | |
| Case 4 | ||||||||||||
| S23 | meningioma | 0 | 100 | |||||||||
| S24 | meningioma | 0 | 100 | |||||||||
| Total | 0 | 100 | ||||||||||
| Case 8 | ||||||||||||
| D12 (BL02) | A–IV, high | 100 | 0 | 21 | 8 | 71 | 38 | 0 | 63 | 58 | 0 | 42 |
| D13 (BL04) | A–IV, low | 100 | 0 | 26 | 73 | 2 | 4 | 0 | 96 | 66 | 32 | 1 |
| D14 (BL01) | A–IV, high | 100 | 0 | 58 | 42 | 0 | 4 | 0 | 96 | 66 | 13 | 21 |
| D15 (BL05) | A–IV, low | 100 | 0 | 16 | 84 | 0 | 25 | 0 | 75 | 65 | 33 | 2 |
| D16 (BL07) | A–IV, low | 100 | 0 | 31 | 69 | 0 | 44 | 0 | 56 | 69 | 24 | 7 |
| D17 (BL08) | A–IV, low | 100 | 0 | 19 | 81 | 0 | 25 | 0 | 75 | 44 | 56 | 0 |
| D18 (BL10) | A–IV, med | 100 | 0 | 5 | 2 | 93 | 20 | 29 | 52 | 7 | 0 | 93 |
| D19 (BL11) | A–IV, med | 100 | 0 | 56 | 30 | 14 | 8 | 1 | 91 | 12 | 51 | 37 |
| D20 (BL12) | A–IV, med | 100 | 0 | 14 | 86 | 0 | 11 | 0 | 89 | 17 | 80 | 3 |
| Total | 100 | 0 | 34 | 56 | 10 | 12 | 2 | 86 | 48 | 34 | 18 | |
Results indicate the percent of pixels within each image that were assigned to a given class. For each case, samples for which stereostactic information was available are marked by their position in parenthesis using alpha and alphanumeric labeling systems. Italicized results are in apparent disagreement with diagnosis and further discussed in the text. A, astrocytoma; GL, glioma; Med, medium; MG, meningioma; OA, oligoastrocytoma; O, oligodendroglioma.
Surgical case 2.
Surgical case 2 was clinically diagnosed as a glioma, subtype oligodendroglioma, WHO grade II, which recurred from a previous oligodendroglioma grade II. We evaluated seven surgical samples obtained from this case (S7, S8, S10–S13, and S15) by DESI-MS: stereotactic information was available for four of them (S11, position A; S10, position B; S12, position C; and S13, position D). DESI mass spectra revealed lipid profiles that were characteristic of a low-grade glioma. For example, sample S10 showed a lipid profile characteristic of grade II oligodendroglioma at low-tumor-cell concentration (23). Oligodendrogliomas are characterized by having high relative abundances of the ions m/z 750.4 assigned to lipid species pls-PE(38:4); m/z 788.2, PS(36:1); m/z 810.3, PS(38:4); m/z 834.3, PS(40:6); m/z 838.3, PS(40:4); and m/z 885.5, PI(38:4), as well as ions corresponding to free fatty acids such as palmitic acid FA(16:0), m/z 255.3, stearic acid FA(18:0), m/z 283.3, and arachidonic acid FA(20:4), m/z 303.2, and fatty acid dimers in the region of m/z 500 to m/z 650. Changes in the relative abundances of ions due to certain lipid species are observed as a function of grade for oligodendrogliomas. For example, the ratio between m/z 838.3, PS(40:4) and m/z 834.3, PS(40:6), as well as that between m/z 838.3, PS(40:4), and m/z 885.5, PI(38:4), increases as the grade of the oligodendroglioma increases.
By histopathological evaluation of an hematoxylin and eosin (H&E)-stained serial tissue section, this sample (S10) was diagnosed as an oligodendroglioma, WHO grade II–III, with a low to moderate concentration of tumor cells. Most of the samples analyzed for this case, such as samples S7 and S12, were diagnosed as low-grade oligodendroglioma with moderate tumor cell concentration, which agreed with the lipid information observed in the DESI mass spectra. Some other samples, such as S8, presented very low tumor cell concentration infiltrating into adjacent normal tissue. With DESI-MS analysis, we were able to appropriately classify all of these samples as gliomas (Table 1). When tested for glioma subtype, grade, and tumor cell concentration, agreement was achieved for all samples within all classes (Table 1). For example, sample S7 was tested and correctly classified as 97% oligodendroglioma, 97% grade II, and 99% medium tumor cell concentration. Differences in classification results for the samples were observed mostly for tumor grade (varying between grades II and III) and tumor cell concentration (varying between low and medium concentration). Sample S10 was classified as 100% oligodendroglioma, but 59% as grade III and 57% as medium tumor cell concentration. These differences in grade and concentration between samples from the same surgical case are highly anticipated and reflect the heterogeneity in cell concentration often observed toward the margins of tumors, where the neoplastic cells infiltrate into neighboring nonneoplastic tissue, as confirmed by histopathological evaluation.
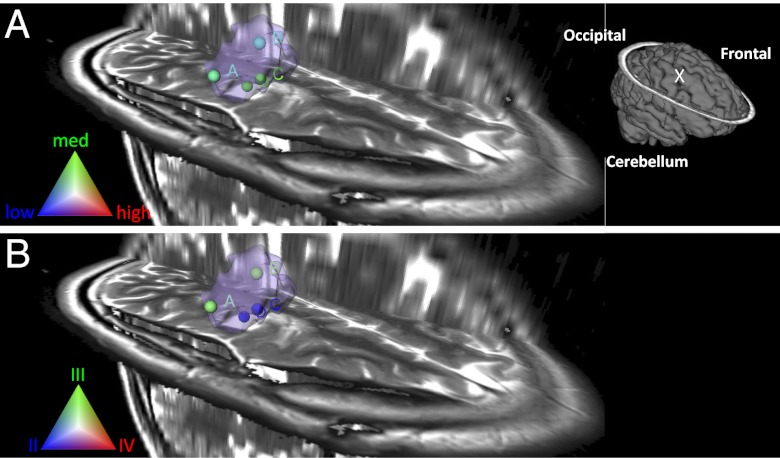
Combining DESI-MS information obtained for all samples analyzed, this case was accurately classified as 90% glioma, 89% oligodendroglioma subtype, 67% grade II, and 69% medium tumor cell concentration. Although determination of tumor type/subtype is important for diagnosis, tumor grade and cell concentration are more valuable to guide surgical resection. Visualization of the classification results for grade and tumor cell concentrations is shown in Fig. 1 for each stereotactically registered sample in the segmented 3D MRI tumor volume reconstruction. The approach allows visualization of diagnostic information as it relates to tumor growth and allows the definition of margins as the tumor infiltrates into surrounding normal brain. As presented in Fig. 1, specimens of lower tumor cell concentration were from the margins of the tumor, as reconstructed by 3D MRI. It is interesting to note that samples S10 (position B) and S11 (position A) were principally classified as WHO grade III and are from the margins of the tumor, and samples S12 (position C) and S13 (position D), classified as 100% grade II, are from the tumor mass center. The information on grade and tumor cell concentration in different positions of the tumor volume could be highly valuable to guide tumor resection and may shed light on the growth properties of gliomas.
Fig. 1.
Glioma tumor cell concentration (A) and grade classification (B) results for surgical case 2 visualized in segmented preoperative 3D MRI volume reconstruction of the tumor and surrounding regions. Tumor volume is represented in light purple. Schematic of the full brain showing tumor position (“X”) is shown in (A).
Surgical case 3.
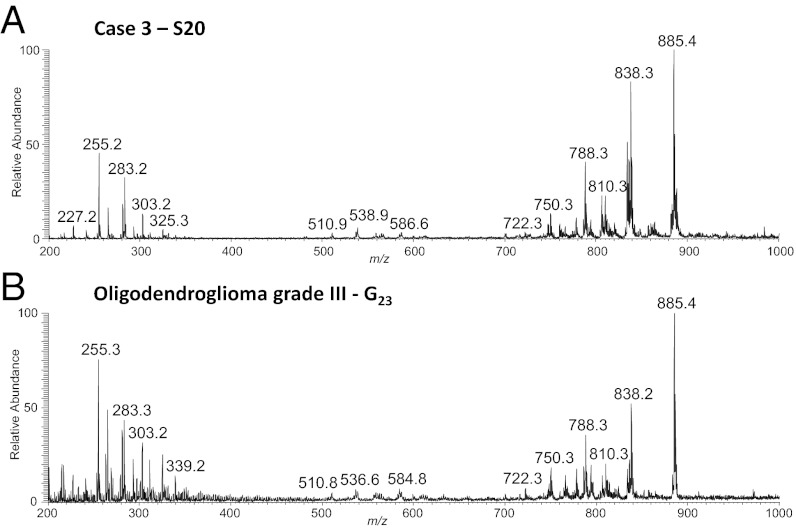
Surgical case 3 consisted of an anaplastic oligodendroglioma, WHO grade III. Eight surgical specimens (S16–S22, S27) obtained from this case were analyzed by DESI. Four of these were stereotactically registered (S17, S20, S21, and S22, displayed as positions B, C, D, and A, respectively). Negative-ion-mode mass spectra showed characteristic lipid profiles for oligodendroglioma grade III comprised of mainly pls-PE, PS, and PI lipid species with PS(40:4) m/z 838.3, and PI(38:4) m/z 885.5 present in much higher relative abundances than commonly observed in other glioma subtypes (23). DESI-MS ion images for the most abundant lipid species for sample S20 are shown in Fig. S3. Note that the phospholipid profile for this same sample is remarkably similar to that previously reported by us from an unrelated patient with an oligodendroglioma grade III (Fig. 2) (23). Histopathological evaluation of an H&E-stained serial section confirmed that the specimen consisted of a grade III oligodendroglioma, of medium-high tumor cell concentration. Similar data were obtained for the other specimens analyzed from this case.
Fig. 2.
(A) Negative-ion mode DESI-MS mass spectra obtained from surgical sample S20. The lipid profile observed is highly characteristic of oligodendroglioma WHO grade III. (B) Representative negative-ion mode DESI-MS mass of anaplastic oligodendroglioma grade III, obtained for an unrelated sample, G23.
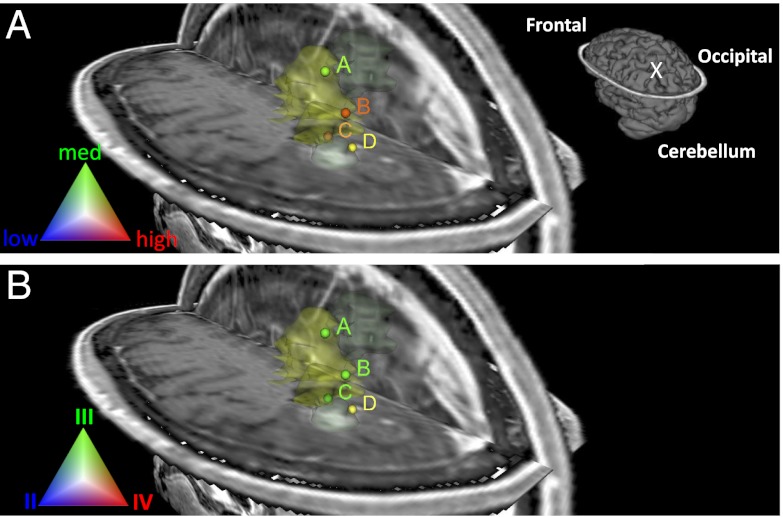
DESI-MS spectra obtained for the five samples of case 3 were initially tested for brain tumor type, followed by glioma subtype, grade, and tumor cell concentration (Table 1). Three of the eight specimens analyzed (samples S16, S17, and S18) were classified as predominantly meningioma. It is interesting to note that histopathological evaluation of these samples revealed significant heterogeneity with regions of calcification (S16) and blood vessels with prominent lymphocytic infiltration (S17). These morphological features are not distinguished by the classifier and could account for the misclassification of the pixels within those regions as meningioma. Overall, combining the mass-spectral information for all samples, the case was classified as an oligodendroglioma WHO grade III with medium-to-high tumor cell concentration, matching the histopathology assessment (for details, see Supporting Information). Visualization of the tumor cell concentration and grade results obtained over 3D MRI is shown in Fig. 3. Samples of high tumor cell concentration (positions B, C, and D) were obtained from the tumor margins raising the concern that residual tumor remained at the resection margin. Although this might be in part explained by a location registration error induced by a brain shift, it emphasizes the need for real-time tissue characterization modalities. This information has the potential to be used by surgeons during surgical interventions to enable tumor resection beyond the radiographic margins, with the potential to minimize recurrence.
Fig. 3.
Glioma tumor cell concentration (A) and grade classification (B) results for surgical case 3 visualized in segmented preoperative 3D MRI volume reconstruction of the tumor and surrounding regions. Tumor volume is represented in light yellow. The samples marked B, C, and D in each image (yellow to red dots) represent medium-high tumor cell concentrations near the tumor margins; this raises concern that the margin is inaccurately determined by MRI and hence not completely resected. Schematic of the full brain showing tumor position (“X”) is shown in (A).
Surgical case 4.
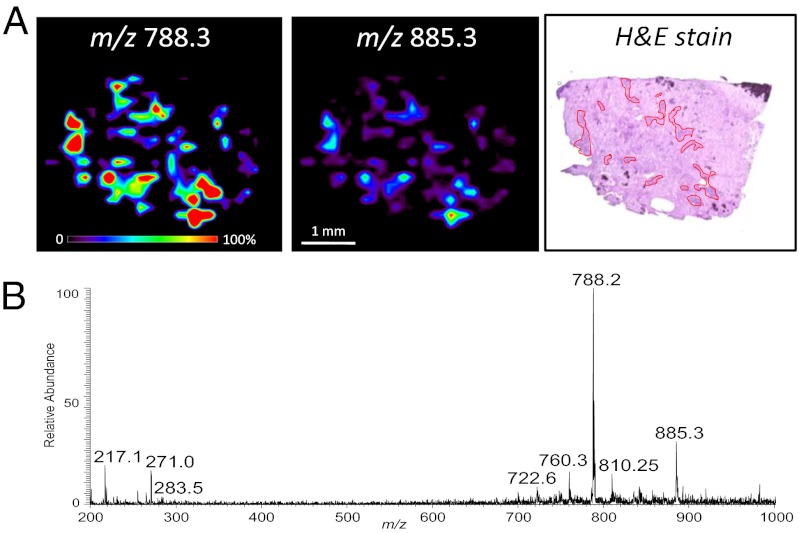
Surgical case 4, a meningioma WHO grade I, had some atypical features. The meningioma focally invaded surrounding bone. Four surgical samples obtained for this case (S23–S26) were imaged by DESI-MS in the negative-ion mode. The lipid profile observed for sample S23 was very similar to those previously obtained from banked meningioma samples, with high abundances of the ions at m/z 788.3, PS(36:1), m/z 885.3, PI(38:4), and m/z 810.2, PS(38:4), and lower relative abundances of ions such as m/z 700.4, pls-PE(34:1), m/z 722.3, pls-PE(36:4), and m/z 760.3, PS(34:1) (Fig. 4). It is interesting to note that DESI-MS ion images of these lipid species clearly showed that meningioma signal was obtained only from focal regions scattered within the tissue section. An H&E-stained serial section revealed that the specimen was mainly normal dura mater with clusters of meningioma cells spatially correlating with the DESI-MS signals of meningioma. A very low total abundance of ions was observed in the regions of dense connective tissue, with almost complete absence of lipid signals and higher relative abundances of the ions m/z 217.0 and 271.1 and other background signals (Fig. S4). Similar results were observed for tissue sections obtained from sample S24, and almost no signal was obtained for samples S25 and S26, which consisted mainly of connective tissue.
Fig. 4.
DESI-MS imaging of surgical sample S23 from case 4, meningioma. (A) Negative mode DESI-MS ion images showing the distribution of m/z 788.3 and m/z 885.3. Optical image of H&E-stained serial section is shown with the main regions containing meningioma cells delineated with red lines within connective tissue. The distribution of meningioma cells observed by microscopy correlates with the distribution of ion signal characteristic of meningiomas from the DESI-MS images. (B) Negative-ion mode mass spectrum of meningioma region of tissue section, showing characteristic meningioma lipid profile.
DESI-MS imaging results for samples S23 and S24 were further evaluated with the classification model for brain cancer type. As shown in Table 1, these samples were classified as meningioma with 100% agreement. Although no stereotactic information is available for these samples, case 4 exemplifies the fact that our methodology provides a powerful tool for identifying and diagnosing different types of brain tumors. In addition, it demonstrates that small foci of tumor can be identified, even when embedded within large areas of nonneoplastic tissue.
Surgical case 8.
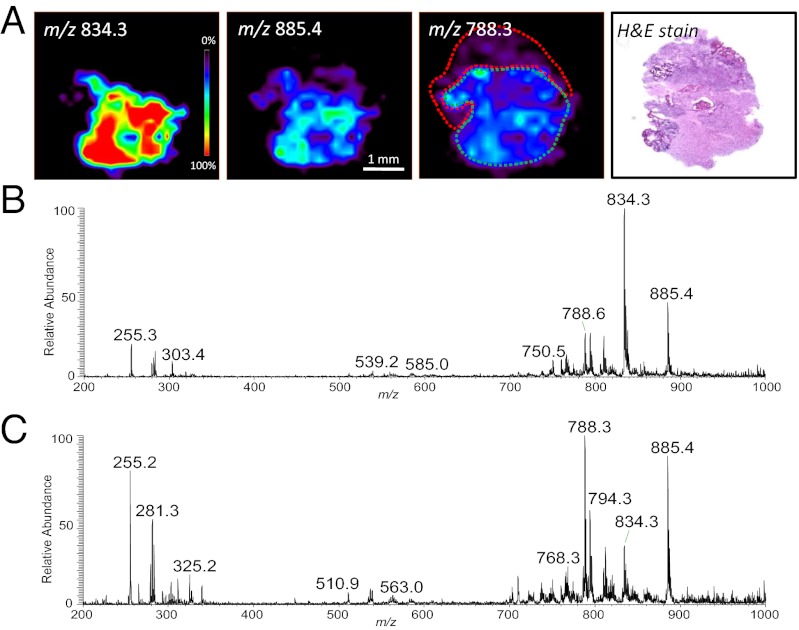
Case 8 was clinically diagnosed as an astrocytoma WHO grade IV (glioblastoma). The tumor was recurrent/residual, of moderate size, and was infiltrating adjacent brain parenchyma. Nine specimens, D12–D20, were obtained and analyzed by DESI-MS. The lipid profiles observed for the majority of the specimens indicated that most had low tumor cell concentrations, and were infiltrating normal brain tissue. In addition, DESI-MS ion images revealed high heterogeneity within some of the specimens. For example, two distinct regions were observed in the DESI-MS ion images obtained from sample D14 (Fig. 5). One region showed a lipid profile characteristic of astrocytoma grade IV, and the lipid profile of the remaining part of the tissue indicated lower tumor cell concentration infiltrating into gray matter. Histopathological evaluation of an H&E-stained serial tissue section confirmed the presence of a dense region with high tumor cell concentration adjacent to an area of low tumor cell concentration, spatially correlating to the regions detected in the DESI-MS images. Similar results to those obtained for sample D14 were observed for sample D12. A few other specimens (D18, D19, D20, and D21) showed overall medium tumor cell concentrations, and specimens D13, D15, D16, and D17 presented low tumor cell concentrations, information that was clearly reflected in the lipid profiles observed. Regions of normal cortex and white matter were observed in some samples such as D20 and D21. All samples were tested for tumor type and classified as 100% glioma (Table 1). However, when tested for subtype, the majority of the samples were classified as predominantly oligoastrocytoma, likely reflecting heterogeneity observed by histopathological evaluation. Overall the case was classified as 34% astrocytoma, 56% oligoastrocytoma, and 10% oligodendroglioma. Nevertheless, when tested for grade, all samples were classified as WHO grade IV, supporting the glioblastoma diagnosis (astrocytoma grade IV). Overall, the case was classified as 86% grade IV, 12% grade II, and 2% grade III. This heterogeneity is also observed in the tumor cell concentration classifier results. Different from the other cases discussed here, the results achieved for concentration were much more evenly distributed between the three classes (low, medium, or high) for these specimens. Sample D14 was classified as being 66% low tumor cell concentration, 13% medium, and 21% high tumor cell concentration, which correlates with histopathology observations. Samples D13, D15, D16, and D17, diagnosed by histopathology as having low tumor cell concentrations, were also correctly classified by the concentration classifier as mostly low tumor cell concentration; and samples D19 and D20 were also correctly classified as medium tumor cell concentration. Visualization of the classification results obtained for grade and tumor cell concentration over the segmented 3D MRI volume reconstruction of the tumor and surrounding regions is presented in Fig. S5. Although WHO grade IV was detected throughout the tumor volume, the tumor cell concentration varied in a way that is consistent with the successful detection of the tumor margin.
Fig. 5.
DESI-MS imaging of surgical sample D14 (Case 8), astrocytoma grade IV, showing regions of heterogeneity as recognized by histopathological evaluation. (A) DESI-MS ion images showing the distribution of m/z 834.3, m/z 885.4, and m/z 788.3, and optical image of H&E-stained tissue section. (B) Low tumor cell concentration region infiltrating into gray matter (delineated in green in the same ion image). (C) DESI-MS mass spectrum of dense tumor region (delineated in red in the m/z 788.3 ion image), with high tumor cell concentration.
Surgical case 5.
Conclusion
We have developed a system based on lipid patterns for intraoperative molecular characterization of brain tumors, which could be applied to the molecular detection of the boundaries between healthy and neoplastic tissue. The results obtained in this exploratory study demonstrate the potential value of the chemical information obtained directly from tissue samples by DESI-MS analysis and the reliability of the classification system based on SVM in providing diagnostic information. We validated the analytical performance of the methodology by analyzing surgical samples ex situ, and demonstrated its potential in providing diagnosis and information on tumor margins. The information obtained on tumor cell concentration and grade from the specimens removed from tumor margins could provide surgeons with critical and previously unavailable information, which could rapidly guide surgical resections and improve management of patients with malignant brain tumors. Although the present results show that tumor margin assessment is possible using tumor cell concentration and grade classifiers, we expect inclusion of normal brain tissue into the classification models to facilitate visualization of tumor margins. This classification system will be integrated into the stereotactic MS-imaging platform currently under development at BWH for large-scale intrasurgical experiments aimed at fully validating the methodology. Efforts are underway at Purdue to create a small mass spectrometer capable of providing an intraoperative assessment of surgical specimens within an appropriate timeframe to guide decision making during brain tumor surgery. Furthermore, we anticipate that mass spectrometry may have important applications to other cancer surgeries where tumor margins are unclear.
Materials and Methods
Sample Collection.
Research subjects were recruited from surgical candidates at the neurosurgery clinic of the BWH, and gave written informed consent to the Partners Healthcare Institutional Review Board (IRB) protocols. Meningioma samples were obtained from the BWH Neurooncology Program Biorepository collection, and analyzed under approved IRB protocol.
Image-Guided Neurosurgery.
All surgeries were performed with auxiliary image guidance of the BrainLab Cranial 2.1 neuronavigation system (BrainLab). Preoperative MRI-imaging sequences included full T2 (1 × 1 × 2 mm, 100 × 100 slice matrix) and postcontrast T1 (1 × 1 × 1 mm, 256 × 256 slice matrix, 176 slices), and processed in the BrainLab iPlanNet 3.0 software.
DESI-MS Imaging.
The DESI ion source was a laboratory-built prototype, similar to a commercial source from Prosolia, Inc. The mass spectrometer used was an LTQ linear ion trap controlled by XCalibur 2.0 software (Thermo Fisher Scientific). DESI-MS experiments were carried out in the negative-ion mode using ion source parameters similar to those previously used (22, 23). The tissues were scanned using a 2D moving stage in horizontal rows separated by a 200-µm vertical step.
Histopathology Analysis.
H&E-stained serial tissue sections were optically scanned using the Mirax Micro 4SL telepathology system from Zeiss and tumor content was evaluated by a neuropathologist.
Statistical Analysis.
Classification models for glioma subtype, grade, and tumor cell concentration of gliomas had been previously developed using SVM. For this study, a classification model for brain tumor type (meningioma or glioma) was developed. The calculated models were used to classify spectra from selected subregions for each surgical sample.
3D Visualization of MRI and MS Data.
MRI data obtained were plotted in 3D Slicer (www.Slicer.org) (version 4.1). Mass-spectrometry data subjected to the described classification system were overlaid as stereotactic points rendered in color in relation to different grade or tumor cell concentration classification determinations.
Supplementary Material
Acknowledgments
The work received support from the James S. McDonnell Foundation; the Brain Science Foundation and the Daniel E. Ponton fund for the Neurosciences; the National Institutes of Health (NIH) Director's New Innovator Award (Grant 1DP2OD007383-01 to N.Y.R.A.); and the Klarman Family Foundation (A.J.G.). S.S. is supported by NIH Grant K08NS064168. L.S.E., A.K.J., and R.G.C. also thank the NIH (Grant 1R21EB009459). This project was supported by the National Center for Research Resources and the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health through Grants P41EB015898 and P41RR019703.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215687110/-/DCSupplemental.
References
- 1.Lacroix M, et al. A multivariate analysis of 416 patients with glioblastoma multiforme: Prognosis, extent of resection, and survival. J Neurosurg. 2001;95(2):190–198. doi: 10.3171/jns.2001.95.2.0190. [DOI] [PubMed] [Google Scholar]
- 2.Sanai N, Polley M-Y, McDermott MW, Parsa AT, Berger MS. An extent of resection threshold for newly diagnosed glioblastomas. J Neurosurg. 2011;115(1):3–8. doi: 10.3171/2011.2.jns10998. [DOI] [PubMed] [Google Scholar]
- 3.Smith JS, et al. Role of extent of resection in the long-term outcome of low-grade hemispheric gliomas. J Clin Oncol. 2008;26(8):1338–1345. doi: 10.1200/JCO.2007.13.9337. [DOI] [PubMed] [Google Scholar]
- 4.Stummer W, et al. ALA-Glioma Study Group Extent of resection and survival in glioblastoma multiforme: Identification of and adjustment for bias. Neurosurgery. 2008;62(3):564–576, discussion 564–576. doi: 10.1227/01.neu.0000317304.31579.17. [DOI] [PubMed] [Google Scholar]
- 5. Black PM, et al. (1999) Craniotomy for tumor treatment in an intraoperative magnetic resonance imaging unit. Neurosurgery 45(3):423–431, discussion 431–423. [DOI] [PubMed]
- 6. Black PM, et al. (1997) Development and implementation of intraoperative magnetic resonance imaging and its neurosurgical applications. Neurosurgery 41(4):831–842, discussion 842-835. [DOI] [PubMed]
- 7.Stummer W, et al. ALA-Glioma Study Group Counterbalancing risks and gains from extended resections in malignant glioma surgery: A supplemental analysis from the randomized 5-aminolevulinic acid glioma resection study. J Neurosurg. 2011;114(3):613–623. doi: 10.3171/2010.3.JNS097. [DOI] [PubMed] [Google Scholar]
- 8.Talos IF, et al. Supratentorial low-grade glioma resectability: Statistical predictive analysis based on anatomic MR features and tumor characteristics. Radiology. 2006;239(2):506–513. doi: 10.1148/radiol.2392050661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albert FK, Forsting M, Sartor K, Adams HP, Kunze S. Early postoperative magnetic resonance imaging after resection of malignant glioma: Objective evaluation of residual tumor and its influence on regrowth and prognosis. Neurosurgery. 1994;34(1):45–60, discussion 60–61. doi: 10.1097/00006123-199401000-00008. [DOI] [PubMed] [Google Scholar]
- 10.Ross JS, Hatzis C, Symmans WF, Pusztai L, Hortobagyi GN. Commercialized multigene predictors of clinical outcome for breast cancer. Oncologist. 2008;13(8):921–921. doi: 10.1634/theoncologist.2007-0248. [DOI] [PubMed] [Google Scholar]
- 11.Dill AL, Eberlin LS, Ifa DR, Cooks RG. Perspectives in imaging using mass spectrometry. Chem Commun (Cambridge) 2011;47(10):2741–2746. doi: 10.1039/c0cc03518a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seeley EH, Schwamborn K, Caprioli RM. Imaging of intact tissue sections: Moving beyond the microscope. J Biol Chem. 2011;286(29):25459–25466. doi: 10.1074/jbc.R111.225854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takáts Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306(5695):471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 14.Harris GA, Galhena AS, Fernández FM. Ambient sampling/ionization mass spectrometry: Applications and current trends. Anal Chem. 2011;83(12):4508–4538. doi: 10.1021/ac200918u. [DOI] [PubMed] [Google Scholar]
- 15.Laskin J, Heath BS, Roach PJ, Cazares L, Semmes OJ. Tissue imaging using nanospray desorption electrospray ionization mass spectrometry. Anal Chem. 2012;84(1):141–148. doi: 10.1021/ac2021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nemes P, Vertes A. Ambient mass spectrometry for in vivo local analysis and in situ molecular tissue imaging. Trac, Trends Anal Chem. 2012;34:22–34. [Google Scholar]
- 17.Watrous J, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA. 2012;109(26):E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eberlin LS, Ferreira CR, Dill AL, Ifa DR, Cooks RG. Desorption electrospray ionization mass spectrometry for lipid characterization and biological tissue imaging. Biophis Biochem Acta, Mol Cell Biol Lipids. 2011;1811(11):946–960. doi: 10.1016/j.bbalip.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dill AL, et al. Multivariate statistical identification of human bladder carcinomas using ambient ionization imaging mass spectrometry. Chemistry. 2011;17(10):2897–2902. doi: 10.1002/chem.201001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dill AL, et al. Multivariate statistical differentiation of renal cell carcinomas based on lipidomic analysis by ambient ionization imaging mass spectrometry. Anal Bioanal Chem. 2010;398(7–8):2969–2978. doi: 10.1007/s00216-010-4259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eberlin LS, et al. Cholesterol sulfate imaging in human prostate cancer tissue by desorption electrospray ionization mass spectrometry. Anal Chem. 2010;82(9):3430–3434. doi: 10.1021/ac9029482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eberlin LS, et al. Discrimination of human astrocytoma subtypes by lipid analysis using desorption electrospray ionization imaging mass spectrometry. Angew Chem Int Ed Engl. 2010;49(34):5953–5956. doi: 10.1002/anie.201001452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eberlin LS, et al. Classifying human brain tumors by lipid imaging with mass spectrometry. Cancer Res. 2012;72(3):645–654. doi: 10.1158/0008-5472.CAN-11-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schäfer KC, et al. Real time analysis of brain tissue by direct combination of ultrasonic surgical aspiration and sonic spray mass spectrometry. Anal Chem. 2011;83(20):7729–7735. doi: 10.1021/ac201251s. [DOI] [PubMed] [Google Scholar]
- 25.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. WHO Classification of Tumors of the Central Nervous System. Lyon, France: WHO Press; 2007. [Google Scholar]
- 26.CBTRUS 2012 CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2004-2008 (March 23, 2012 Revision) (Central Brain Tumor Registry of the United States, Hinsdale, IL). Available at http://www.cbtrus.org/2012-NPCR-SEER/CBTRUS_Report_2004-2008_3-23-2012.pdf. Accessed December 27, 2012. [Google Scholar]
- 27.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.