Abstract
Endocannabinoid, particularly 2-arachidonoyl glycerol (2-AG), signaling has recently emerged as a molecular determinant of neuronal migration and synapse formation during cortical development. However, the cell type specificity and molecular regulation of spatially and temporally confined morphogenic 2-AG signals remain unexplored. Here, we demonstrate that genetic and pharmacological manipulation of CB1 cannabinoid receptors permanently alters cholinergic projection neuron identity and hippocampal innervation. We show that nerve growth factor (NGF), implicated in the morphogenesis and survival of cholinergic projection neurons, dose-dependently and coordinately regulates the molecular machinery for 2-AG signaling via tropomyosine kinase A receptors in vitro. In doing so, NGF limits the sorting of monoacylglycerol lipase (MGL), rate limiting 2-AG bioavailability, to proximal neurites, allowing cell-autonomous 2-AG signaling at CB1 cannabinoid receptors to persist at atypical locations to induce superfluous neurite extension. We find that NGF controls MGL degradation in vitro and in vivo and identify the E3 ubiquitin ligase activity of breast cancer type 1 susceptibility protein (BRCA1) as a candidate facilitating MGL’s elimination from motile neurite segments, including growth cones. BRCA1 inactivation by cisplatin or genetically can rescue and reposition MGL, arresting NGF-induced growth responses. These data indicate that NGF can orchestrate endocannabinoid signaling to promote cholinergic differentiation and implicate BRCA1 in determining neuronal morphology.
Keywords: axon guidance, basal forebrain, neurotrophin, protein stability, choline acetyltransferase
CB1 cannabinoid receptors (CB1Rs) are the targets of marijuana (Cannabis spp.)-derived phytocannabinoids and endocannabinoids, including 2-arachidonoyl glycerol (2-AG) (1). 2-AG liberated from postsynaptic neurons serves as a retrograde messenger to limit neurotransmitter release from many mature presynapses (1). However, 2-AG signaling also emerges as a molecular determinant of cortical development (2–4). Despite recent advances, the contribution of endocannabinoids (specifically 2-AG) to neuronal diversification and the development of synaptic connectivity in subcortical territories remains unknown. Cholinergic projection neurons within a continuum of magnocellular nuclei in the basal forebrain (5) are appealing candidates for developmental 2-AG actions considering their CB1R expression (1) and the endocannabinoid sensitivity of acetylcholine release in adulthood (6).
Cholinergic afferents innervating the cerebral cortex are essential for learning and memory (5, 7). A favored approach to rescue cholinergic neurotransmission under disease conditions relies on neurotrophins to maintain the molecular identity, synaptic signaling, and survival of cholinergic neurons (8, 9). Nerve growth factor (NGF) appears particularly efficacious to promote the phenotypic differentiation and synaptic connectivity of postnatal cholinergic projection neurons (9, 10), as illustrated by experimental studies exploiting recombinant NGF-neutralizing antibodies (7), and the genetic manipulation of tropomyosine kinase (Trk)A receptors (9) to disrupt cholinergic development. Nevertheless, our knowledge of NGF’s mechanism of action on cholinergic neurons populating the primordial basal forebrain is fragmented. In particular, it is unknown whether NGF recruits “molecular effectors,” including 2-AG signals, to convert uncoordinated cholinergic sprouting (10, 11) into ordered axonal growth.
Here, we show that fetal cholinergic projection neurons coordinately express CB1Rs, sn-1-diacylglycerol lipases [DAGLα/β isoforms generate 2-AG from diacylglycerol (12, 13)] and monoacylglycerol lipase (MGL) [which degrades the bulk of 2-AG in the nervous system (14)], and use 2-AG signals to colonize the basal forebrain and to innervate cortical targets. We find that NGF can enhance 2-AG signaling by up-regulating breast cancer type 1 susceptibility protein (BRCA1), an E3 ubiquitin ligase (15), the ability of which to promote MGL degradation in motile neurite domains can convert neurotrophin action into cholinergic growth in vitro. By taking advantage of BRCA1’s cisplatin sensitivity (15), we rescued MGL in cholinergic growth cones, introducing a “stop” signal for NGF-induced growth responses.
Results and Discussion
CB1R Requirement of Cholinergic Projection Neuron Development.
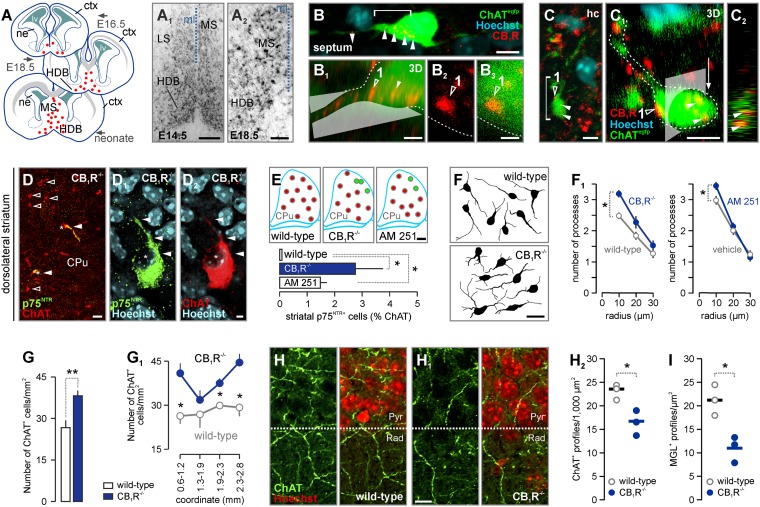
We have recently shown that endocannabinoid-mediated axonal growth and guidance requires the precisely ordered molecular assembly of 2-AG signaling networks during corticogenesis (2, 16). Here, we defined, by in situ hybridization, that CB1R mRNA was present in cholinergic territories, particularly the medial septum (MS) (Fig. 1 A–A2 and Fig. S1 A–A2) from embryonic day (E)14.5 until birth in mice. We found CB1Rs perisomatically in and distributed along the processes of bipolar genetically tagged choline acetyltransferase (ChAT)+ neurons (Fig. 1 B–B3), reminiscent of migrating CB1R+ γ-aminobutyric acid-containing interneurons (17). In addition, CB1Rs were localized to cholinergic axons and their putative growth cones traversing the hippocampus (Fig. 1 C–C2) and the corpus callosum (Fig. S1E2).
Fig. 1.
Developmental organization and roles of 2-AG signaling in cholinergic neurons. (A) Schema of the fetal basal forebrain. Red dots indicate the general position of p75NTR+/ChAT+ cholinergic projection neurons. ctx, cortex; HDB, horizontal diagonal band of Broca; LS, lateral septum; ne, neuroepithelium. (A1–A2) CB1R mRNA localization in the MS and HDB at successive developmental stages. ml, midline. (B–B3) CB1Rs (arrowheads) are trafficked along the leading process of EGFP+ cholinergic neurons. A CB1R cluster viewed along the orthogonal axis is labeled as “1” in B2 and B3. Three-dimensional rendering is indicated by “3D.” (C–C2) CB1R+ cholinergic growth cone navigating in the neonatal hippocampus (hc). Putative cell surface-associated and intracellular CB1Rs are denoted by “1” and arrowheads, respectively. (D) A subset of ChAT+ neurons in the rostral dorsolateral striatum [caudate putamen (CPu)] unexpectedly coexpressed p75NTR (solid arrowhead) in CB1R−/− and AM 251-treated fetuses (Fig. S2C). Open arrowheads label ChAT+ prospective striatal interneurons. (E) Analysis of p75NTR+/ChAT+ striatal neurons at E18.5 (n = 3/group) revealed ectopic p75NTR+ cholinergic neurons (green) interspersed with cholinergic interneurons (red) in the dorsolateral striatum in CB1R−/− or AM 251-treated wild-type mouse fetuses. (F and F1) Sholl analysis of cholinergic projection neurons at E18.5 revealed an increase in neurite numbers emanating from cholinergic somata upon disrupted CB1R function. (G and G1) Quantitative densitometry of ChAT+ cholinergic neurons at select mediolateral coordinates in the striatum of adult CB1R−/− and wild-type mice. Surplus cholinergic neurons accumulated in the dorsal striatum. (H–H2) The density of ChAT+ profiles in stratum pyramidale of the CA1 subfield was significantly reduced in CB1R−/− animals relative to wild-type littermates. Pyr, stratum pyramidale; Rad, stratum radiatum. (I) Reduced density of MGL+ profiles in the stratum pyramidale of hippocampal CA1 in CB1R−/− animals relative to wild-type controls. Data were expressed as means ± SEM. **P < 0.01; *P < 0.05. (Scale bars: E, 200 µm; A1 and A2, 100 µm; B, D, and F, 10 µm; H1, 5 µm; B1, B3, C, C1, and D2, 3 µm.)
If endocannabinoid signaling at CB1Rs impacts cholinergic cell identity and differentiation, then their pharmacological or genetic disruption might impair the developmental organization of the cholinergic basal forebrain. In CB1R−/− fetuses, we observed ectopic localization of cholinergic neurons with a projection cell-like neurochemical makeup, coexpressing ChAT, the low-affinity neurotrophin receptor p75 (p75NTR), and the vesicular acetylcholine transporter (VAChT) (18), in the fetal dorsolateral striatum, which otherwise lacked p75NTR+ cells in wild-type littermates (5) (Fig. 1 B–E and Fig. S2 A–B1). CB1R antagonism by AM 251 during pregnancy recapitulated genetic loss of function (Fig. 1E and Fig. S2 B and C). Ectopic p75NTR+/ChAT+ neuron density is likely an underestimate because our analysis coincided with the expressional onset of both markers.
Loss of CB1R function disrupts neuronal morphogenesis (2, 16). Therefore, we asked whether neurite complexity of cholinergic neurons routing normally to the fetal basal forebrain is impaired upon manipulating CB1Rs. Sholl analysis of septal ChAT+ neurons (E18.5) demonstrated significantly increased numbers of neurites emanating from cholinergic somata in CB1R−/− and AM 251-exposed fetuses (Fig. 1 F and F1).
Next, we tested whether disrupted neurodevelopmental 2-AG signaling imposes permanent modifications to the cholinergic component of the basal forebrain. We found a significantly increased density of ChAT+ neurons in the striatum of adult CB1R−/− mice relative to their littermate controls (Fig. 1 G and G1 and Fig. S2 D–D2), suggesting that misrouted cholinergic neurons can survive and integrate in the postnatal striatum.
The altered morphology of fetal cholinergic projection neurons upon CB1R manipulation, together with prominent CB1R localization along cholinergic axons in the fetal hippocampus (Fig. 1C), prompted us to test the integrity of the septohippocampal pathway in adult CB1R−/− mice. We found the loss of cholinergic processes in the pyramidal layer of the cornu Ammonis hippocampal subfield (CA)1 (Fig. 1 H–H2 and Fig. S2E) coincident with significantly reduced MGL immunoreactivity in putative presynapses of the same layer in CB1R−/− mice (Fig. 1I and Fig. S2 F–G1). Thus, our data imply that 2-AG signaling at CB1Rs contributes to defining the neurochemical specificity, final positions, morphology, and connectivity of basal forebrain cholinergic neurons.
Cell-Autonomous 2-AG Signaling in Fetal Cholinergic Neurons.
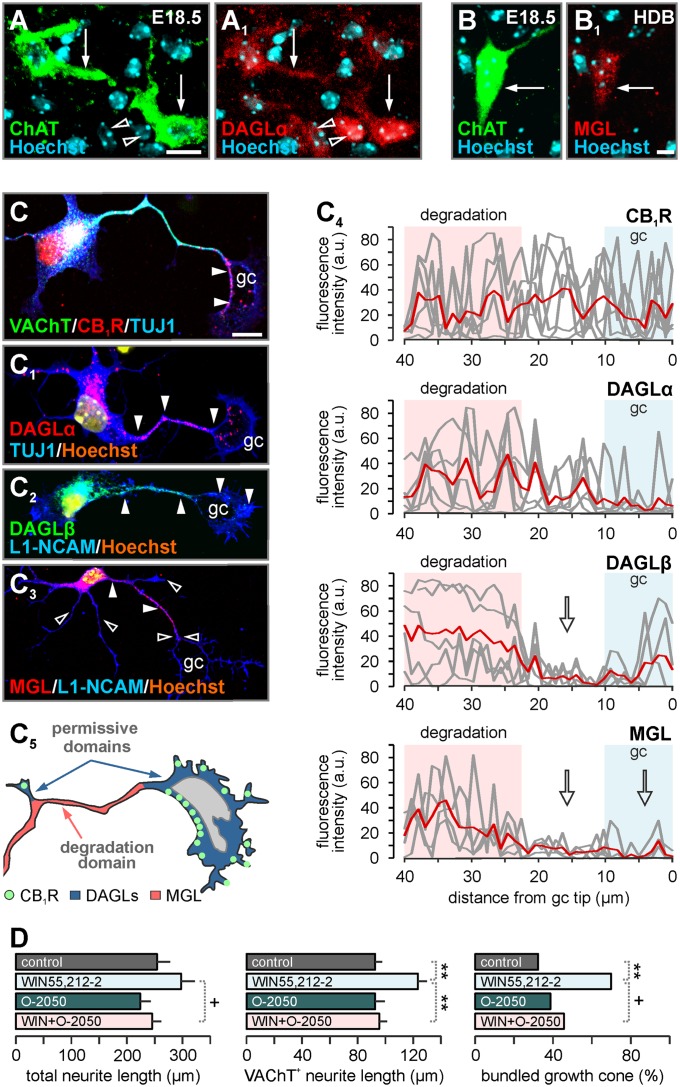
CB1R expression is spatially and temporally coordinated with 2-AG synthesis and degradation in the developing cerebrum (2). DAGLα, producing 2-AG (12) (Fig. 2 A and A1 and Fig. S1I), and MGL (Fig. 2 B and B1) were localized to the perikarya and processes of cholinergic projection neurons by E18.5. Cholinergic neurons isolated from the fetal basal forebrain retained the expression of VAChT, CB1Rs, and 2-AG metabolic enzymes in vitro (Fig. 2 C–C3). DAGLα/β and MGL were differentially targeted along the developing VAChT+ primary neurite (Fig. 2C4), the prospective axon (18). MGL was restricted to the proximal neurite stem. In contrast, CB1R and DAGLα/β partitioned to the distal neurite, suggesting the spatial confinement of 2-AG signaling to the motile neurite segment including the growth cone (Fig. 2C5). Accordingly, agonist-induced CB1R activity facilitated neurite outgrowth and inhibited growth cone differentiation (16) (Fig. 2D). These data suggest the dominance of cell-autonomous 2-AG signaling during cholinergic development.
Fig. 2.
Cholinergic neurons possess cell-autonomous 2-AG signaling that regulates neurite outgrowth. (A and B) Cholinergic neurons (arrows) harbor DAGLα (A1) and MGL (B1) to produce and degrade 2-AG, respectively. Open arrowheads indicate lack of colocalization. (C–C3) Cholinergic neurons retain phenotypic identity markers (VAChT+/TrkA+) and the ability for 2-AG signaling in vitro. Solid arrowheads denote the sites of subcellular CB1R, DAGLα/β, and MGL recruitment along the primary neurite and within growth cones (gc). CB1Rs accumulate at the collapsing surface of the growth cone. Open arrowheads point to DAGLβ or MGL-sparse domains. L1-NCAM, L1 neural cell adhesion molecule; TUJ1, β-III-tubulin. (C4) Fluorescence-intensity plots measured in individual growth cones and adjoining distal neurite segments (n = 6–8/group) revealed the segregated distribution of 2-AG signaling components. Arrows indicate MGL and DAGLβ sparse domains. a.u., arbitrary units. (C5) Molecular organization of 2-AG signaling networks in cholinergic axons and growth cones, suggesting prevailing focal and protrusive 2-AG signaling in growth domains. (D) CB1R agonist treatment induces neurite outgrowth and retains undifferentiated growth cone morphology in vitro. O-2050 was used to verify CB1R involvement (n ≥ 25 per treatment). Data were expressed as means ± SEM. **P < 0.01; *P < 0.05; +P < 0.1. (Scale bars: A, 20 µm; B1 and C, 10 µm.)
NGF Regulates 2-AG Signaling.
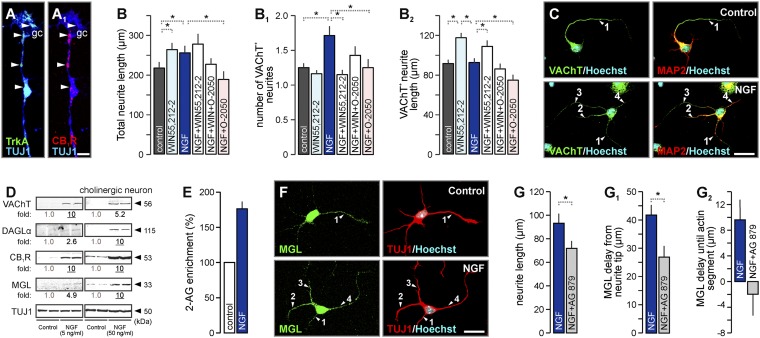
Endocannabinoids are unlikely to function as a solitary signaling system to define cholinergic morphology and connectivity, particularly because CB1R deletion does not arrest cholinergic differentiation or survival. In contrast, NGF is required for cholinergic projection neurons to survive, reach morphological and neurochemical maturity, and establish and maintain axonal projections (9). We hypothesized that NGF signaling might use endocannabinoids and that the coordinated action of neurotrophin and 2-AG signals could determine cholinergic morphology, particularly axonal complexity. Coincident targeting of TrkA and CB1Rs to cholinergic growth cones identified a subcellular platform for molecular interactions (Fig. 3 A and A1). NGF [2–4 d in vitro (DIV)] (10) increased neurite outgrowth (Fig. 3B). In doing so, NGF triggered the formation of multiple VAChT/Tau-2/collapsin response mediator protein 2 (CRMP-2)+ processes (Fig. 3 B1 and C and Fig. S3 A and B), suggesting that NGF alters neuronal polarity (and probably induces a multiaxonal phenotype) on the expense of the elongation of the primary VAChT+ neurite (Fig. 3B2) but not cholinergic commitment or survival (Fig. S3C).
Fig. 3.
NGF regulates 2-AG signaling in cholinergic neurons. (A and A1) TrkA and CB1Rs in cholinergic neurites and growth cones. (B and C) Agonist (WIN 55,212-2)-induced signaling at CB1Rs reinstated neuronal polarity (B1) and molecular identity (B2) in NGF-treated cholinergic neurons. In contrast, O-2050 blunted NGF effects. VAChT+ neurites were numbered (C). (D) NGF dose-dependently induced VAChT expression, reflecting cholinergic differentiation, and up-regulated 2-AG metabolism (DAGLα, MGL) and CB1R levels. (E) NGF enhanced 2-AG levels in basal forebrain cultures (4 DIV). (F) MGL ventured into multiple neurites in NGF-treated neurons. AG 879 decreased neurite length (G) and stabilized MGL in distal actin-rich neurite segments (G1 and G2). Experimental details are given in Table S2. Data were expressed as means ± SEM. *P < 0.05; (n = 22–28 observations per group). (Scale bars: C and F, 20 µm; A1, 5 µm.)
If CB1Rs escape desensitization and 2-AG signaling remains nonsaturated and dynamic upon NGF treatment, then pharmacological manipulation of CB1Rs might modify the ensuing cholinergic phenotype. Accordingly, WIN55,212-2, a CB1R agonist (16), induced neurite outgrowth (Fig. 3B), reinstated VAChT+ neurite identity (Fig. 3B1), and extended the VAChT+ neurite (the quiescent axon) in NGF-treated cholinergic neurons (Fig. 3B1 and Fig. S3E) while maintaining cholinergic growth cones in undifferentiated, motile states (Fig. S3D). Next, we tested whether NGF-induced neurite outgrowth requires CB1R activation. O-2050, a silent CB1R antagonist (2), occluded NGF-induced morphogenesis, being particularly potent to reduce the outgrowth and number of VAChT+ neurites (Fig. 3 B and C and Fig. S3E). Similarly, DAGL inhibition by O-3841 (SI Text) arrested cholinergic neuritogenesis (Fig. S3 F and G).
Cholinergic neurons responded to NGF by up-regulating CB1R, DAGLα, and MGL protein levels in a time-dependent (Fig. S4 A and B) and dose-dependent (Fig. 3D) fashion, resulting in increased intracellular 2-AG concentrations (Fig. 3E). NGF induced MGL (Fig. 3F) and CB1R (Fig. S4C) accumulation at atypical locations in the proximal stem of multiple neurites. Tyrphostin (AG 879), an inhibitor of TrkA signaling (19), acutely reduced neurite outgrowth (Fig. 3G) and coincidently allowed MGL to venture into the actin-rich motile neurite tip (Fig. 3 G1 and G2 and Fig. S4E). This suggests that TrkA activation can adjust the length of 2-AG–responsive neurite segments by regulating MGL availability. Cumulatively, we interpret these findings that NGF alters the morphological complexity of cholinergic neurons by compartmentalizing 2-AG degradation, allowing 2-AG to facilitate the simultaneous outgrowth of multiple CB1R-laden neurites.
NGF Affects MGL Stability in Cholinergic Neurons.
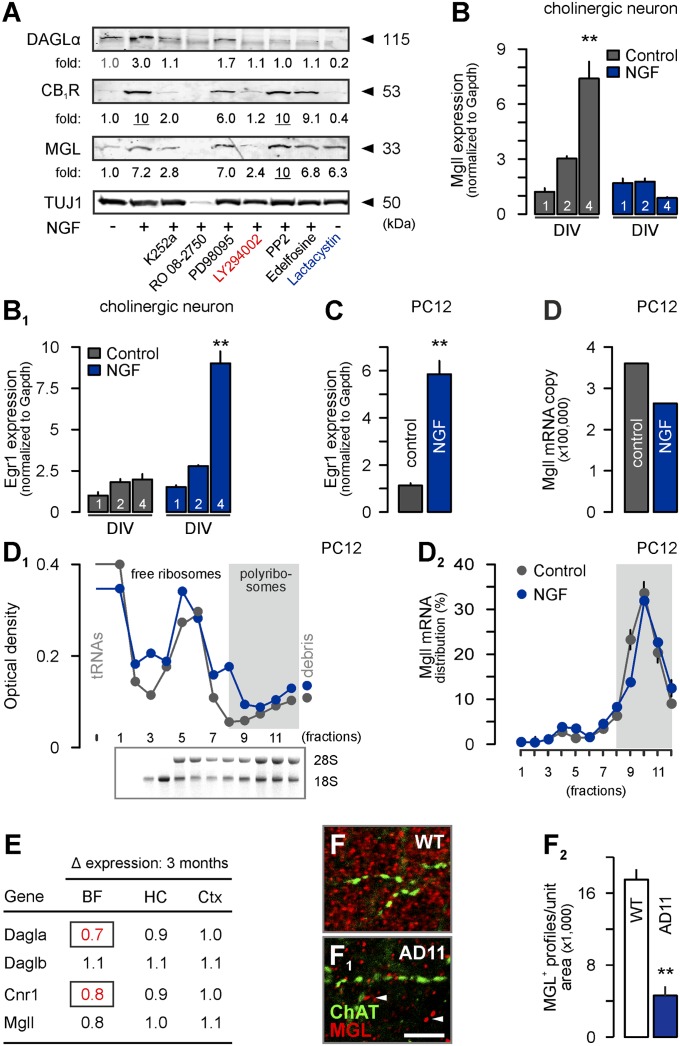
NGF affects the differentiation and connectivity of postnatal cholinergic neurons via TrkA/extracellular signal-regulated kinase (Erk) signaling (9). Here, we used a pharmacological approach to dissect NGF’s receptor requirements and downstream signaling inducing 2-AG signaling in fetal cholinergic neurons in vitro. Inhibition of TrkA phosphorylation by K252a eliminated the NGF-induced coordinated increase of DAGLα, CB1R, and MGL expression, verifying TrkA involvement (Fig. 4A). In contrast, p75NTR blockade limited cholinergic survival, confirming that p75NTR is a “dependence” (or survival) receptor in this cell type (20). We found that Erk inhibition (PD98095) reduced NGF-induced DAGLα and CB1R but not MGL protein levels, confirming neurotrophin-induced Erk signaling in cholinergic neurons (9). However, inhibition of the phosphatidylinositol-3-kinase (PI3K)/protein kinase B pathway associated with endocannabinoid-induced neurite outgrowth (21), but not the Src or phospholipase C pathways, eliminated NGF-dependent protein, particularly MGL expression (Fig. 4A). This finding is significant because MGL activity in growth domains can limit neurite outgrowth (2).
Fig. 4.
Molecular regulation of 2-AG signaling by NGF. (A) Pharmacological inhibition of TrkA signaling identified PI3K-dependent (LY294002) control of 2-AG signaling. Note that proteasome inhibition by lactacystin recapitulated NGF effects on retained MGL protein. NGF exposure failed to induce Mgll mRNA transcription (B) in cholinergic neurons. Egr1 (22) (B1) was used as positive control. Similarly, NGF induced Egr1 (C) but not Mgll mRNA transcription (D) in PC12 cells (48 h). (D1 and D2) Polysomal distribution of mRNAs (D1) and sedimentation profile of Mgll mRNAs (D2) in control vs. NGF-treated PC12 cells. (E) Whole-genome microarray analysis combined with comparative mRNA profiling showed retained Mgll mRNA expression in the cholinergic basal forebrain (BF) and its projection targets in AD11 mice. Statistically significant differential expression values were highlighted in red (P < 0.05). (F–F2) NGF deprivation reduced MGL protein levels and synaptic recruitment in the cerebral cortex of AD11 mice relative to wild-type (WT) controls. Data were expressed as means ± SEM; **P < 0.01. (Scale bar: F1, 3 µm).
Next, we explored the molecular mechanism by which NGF regulates MGL protein levels. We excluded NGF-dependent induction of MGL transcription by quantitative PCR (Fig. 4B), using early growth response protein (Egr)1 as positive control (22) (Fig. 4B1; see Fig. S5 A–A2 for CB1R and DAGLα/β). These findings raise the possibility of an NGF-induced increase in the translation efficacy of MGL (Mgll) mRNAs. We tested this hypothesis in pheochromocytoma cell line 12 (PC12) cells, which respond to NGF by increased Egr1 mRNA (Fig. 4C) and MGL protein expression (Fig. S5B), recapitulating cholinergic responsiveness. By using absolute PCR combined with sucrose gradient fractionation of mRNAs bound to polysomes, we find that NGF affected neither the copy number nor the recruitment of MGL (Mgll) mRNAs to free ribosomes or polyribosomes (Fig. 4 D–D2).
Our in vitro data support that MGL’s proteasomal degradation (2) may be a candidate mechanism to facilitate focal 2-AG signaling in CB1R+ cholinergic growth cones. Therefore, we sought to determine whether NGF posttranslationally controls MGL in vivo. Because available genetic tools favor the molecular dissection of NGF effects on cholinergic neurons during postnatal life (9), we used presymptomatic AD11 mice (≤3 mo of age) that express an NGF-neutralizing antibody (7) to assess NGF’s effects on MGL mRNA and protein when NGF withdrawal is yet to significantly disrupt cholinergic neurotransmission (Fig. 5C). By combining genome-wide microarray analysis and comparative mRNA profiling, we found that NGF deprivation significantly reduced CB1R and DAGLα but not MGL mRNA levels in the basal forebrain (Fig. 4E). In contrast, Western blotting demonstrated the robust loss of MGL protein (Fig. S5C). Quantitative morphometry revealed the loss of MGL+ presynapses in the cerebral cortex (Fig. 4 F–F2) and basal forebrain (Fig. S5 D and D1) in AD11 mice relative to wild-type controls, confirming that NGF regulates MGL protein but not mRNA levels in vivo.
Fig. 5.
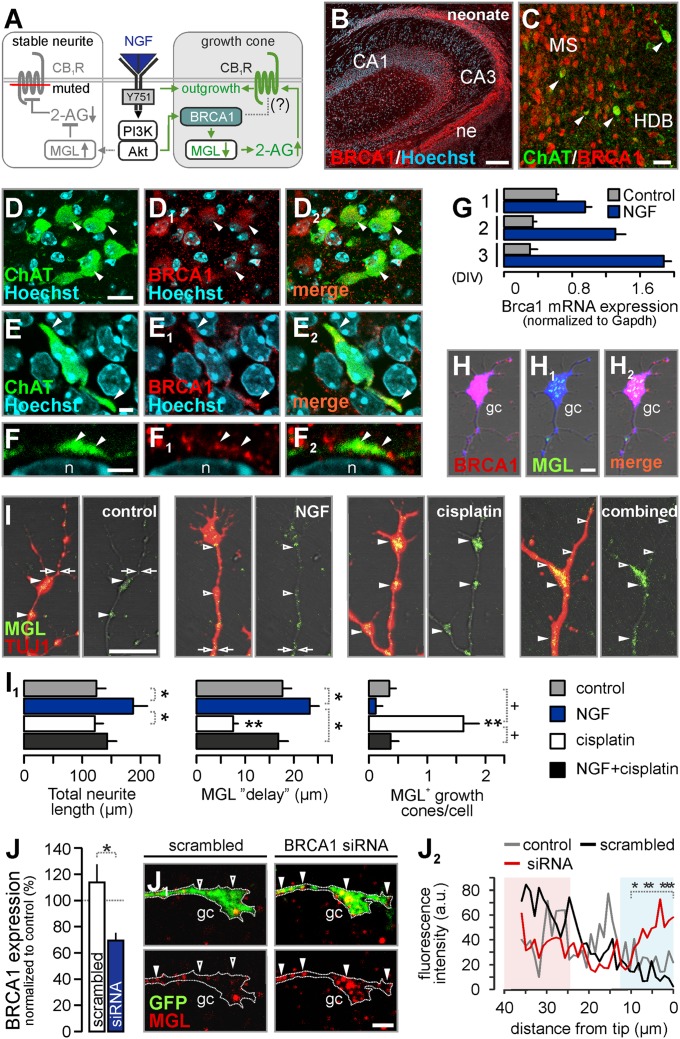
NGF recruits BRCA1 and controls MGL availability and distribution. (A) Proposed schema of NGF’s domain-specific molecular control of endocannabinoid signaling. (B and C) BRCA1 distribution in proliferating zones of the fetal hippocampus (B) and postmitotic neurons of the MS (C), including ChAT+ neurons. (D–D2) Noncholinergic septal neurons (arrowheads) also expressed BRCA1. (E–E2) Cholinergic neuron with migratory morphology and BRCA1 in the tip of the leading processes. (F–F2) ChAT+ process contained BRCA1. Arrowheads point to colocalizations; “n” identifies nucleus. (G) NGF increased Brca1 mRNA expression. (H–H2) BRCA1 and MGL immunoreactivities in growth cones in vitro. (I–I1) Cisplatin rescued NGF-induced degradation of MGL and reduced MGL’s exclusion (“MGL delay”) from NGF-exposed motile neurite tips. +P < 0.1; *P < 0.05; **P < 0.01 (n = 20–22 growth cones per group). (J) siRNA-mediated Brca1 silencing decreased BRCA1 immunoreactivity in cultured basal forebrain neurons. (J1 and J2) Reducing BRCA1 availability stabilized MGL in growth cones (*P < 0.05; n = 7 neuron per group). Arrowheads point to MGL immunoreactivity, whereas open arrowheads denote the lack thereof. Data were expressed as means ± SEM, except for J2, where mean values were plotted. (Scale bars: B, 150 µm; C and D, 10 µm; E, F, H1, I, and J1, 4 μm.)
BRCA1 Is Expressed in Cholinergic Neurons: Implications for MGL Turnover.
Inhibiting the proteasome by lactacystin stabilized MGL in cholinergic neurons (Fig. 4A). Therefore, we hypothesized that NGF could alter MGL protein turnover by a mechanism operating focally in cholinergic growth cones (Fig. 5A). BRCA1 possesses E3 ubiquitin ligase activity (15), accumulates at leading edges in migrating cells (23), and is expressed during brain development (24), particularly in proliferative zones (Fig. 5B). Therefore, we tested whether BRCA1 is a candidate ubiquitin ligase to destine MGL toward proteasomal degradation in cholinergic neurites. We detected BRCA1 in the basal forebrain (Fig. 5 C–D2), with BRCA1 being particularly noticeable in leading processes of cholinergic neurons (Fig. 5 E–F2).
Next, we addressed whether NGF regulates BRCA1 expression and whether altered BRCA1 levels correlate with those of MGL. NGF progressively induced BRCA1 mRNA expression in cultured cholinergic neurons (Fig. 5G) and PC12 cells (Fig. S6A). In AD11 mice, BRCA1 levels were slightly diminished (Fig. S5C), suggesting that reduced BRCA1 expression is sufficient to underpin physiological BRCA1 functions.
BRCA1’s subcellular distribution in cholinergic neurites, particularly in motile filopodia (Fig. 5 H–H2 and Fig. S6 B and B1), is mutually exclusive with MGL, suggesting that BRCA1 may contribute to regulating MGL turnover. We tested this possibility by exposing basal forebrain neurons to cisplatin (15). We show that inhibition of BRCA1’s ubiquitin ligase activity by the platinum-based anticancer drug (15) stabilized MGL in growth cones under control conditions (Fig. 5 I and I1). Moreover, cisplatin limited neurite outgrowth from NGF-treated cholinergic neurons (Fig. 5 I and I1) and PC12 cells (Fig. S6 D and D1) by reinstating MGL’s subcellular distribution (Fig. 5I). We validated these observations by showing MGL stabilization upon siRNA-mediated BRCA1 silencing in growth cones of basal forebrain neurons (Fig. 5 J–J2 and Fig. S6 E and E1), as well as the SH-SY5Y neuroblastoma cell line (Fig. S6F). Collectively, our findings highlight a mechanism coupling NGF signaling at TrkA receptors to neurite outgrowth via sequential regulation of BRCA1 and MGL in cholinergic neurons.
Conclusions
The contribution of endocannabinoid signaling to the formation of neuronal networks in the developing forebrain is increasingly appreciated (2, 4, 16). Available models implicate neurotrophin-induced Ca2+ increase as an upstream signal activating 2-AG generation (25) and emphasize DAGLα requirements of the initiation of neurite outgrowth (13). Our study provides an alternative to this “DAGL centric” view, favoring positional enzymatic ligand inactivation as a means of rate-limiting temporal and spatial endocannabinoid availability during developmental processes.
Although endocannabinoids are increasingly recognized for their impact on synapse structure and function (1, 16), the upstream regulation of endocannabinoid metabolism and signaling at CB1Rs remains poorly understood. Our observations, together with the proposed regulation of BRCA1 by CB1R activity (21, 26), highlight BRCA1 as a molecular “hinge” to integrate coexistent permissive signals and outline a bidirectional signaling loop that emerges as a key component of neuronal differentiation and synaptic coupling (21). Identifying a regulatory pathway involving TrkA and CB1R coincidently modulating BRCA1 acting as a “feedback amplifier” is broadly relevant to the orchestration of neurotrophin/endocannabinoid interplay in cancer (19, 26), pain (27), or neurodegeneration (1, 7). Understanding developmental mechanisms that allow the integration of specific neuronal subtypes into large-scale neuronal networks may be rewarding to device translational restorative strategies aimed to prevent disease-related modifications of synaptic connectivity. Our evidence that CB1R-mediated 2-AG signals convert NGF-induced axonal sprouting into a regulated growth process in cholinergic neurons reconciles the decade-long dilemma on NGF’s ineffectiveness in promoting the growth of cholinergic grafts, even when exogenous sources of NGF are present. Recognizing the reliance of NGF on secondary signaling systems to exert coordinated and directional growth presents possibilities for maintaining effective cholinergic neurotransmission under disease conditions.
Materials and Methods
Mice and Drug Treatment.
Tissues from wild-type and transgenic mice were characterized and processed as described (2). AM 251 was administered at a dose of 3 mg/kg, with male embryos harvested on E18.5 (SI Materials and Methods).
Histo- and Cytochemistry.
Multiple immunofluorescence labeling of fetal mouse brains, cultured neurons, and PC12 cells was performed by applying select mixtures of affinity-purified antibodies (Fig. S1 and Table S1). In situ hybridization was carried out by using digoxigenin-labeled riboprobes (16). Images were acquired on a Zeiss 710LSM confocal laser-scanning microscope (2) (SI Materials and Methods).
mRNA Detection and Protein Biochemistry.
Gene expression profiling was done using the two-color protocol by Agilent with reference experimental design. Quantitative PCRs were performed on a Bio-Rad MyIQ thermal cycler (2) using primer sets listed in Table S2. Polyribosome profiling in PC12 cells was on linear sucrose gradients (15–50%). Basal forebrain neurons or SH-SY5Y human neuroblastoma cells were transfected with either nontargeting (scrambled) siRNA or a pool of BRCA1-specific siRNAs (21). Protein samples from basal forebrains, cultured neurons after treatment (Table S3), and immortalized cells were analyzed under denaturing conditions. Antibodies used for Western blotting are listed in Table S1.
Supplementary Material
Acknowledgments
We thank P. Ernfors, T. Hökfelt, B. Lutz, M. Matteoli, M.-M. Poo, and R. A. Ross for valuable discussions and comments; C. Ledent and A. Zimmer for colony founders of CB1R−/− mice; M. I. Kotlikoff and B. Shui for ChAT(BAC)-EGFP mice; R. Razdan and V. di Marzo for O-3841; H. Martens for anti-VAChT antibodies; and G. A. Cameron for mass spectrometry. This study was supported by the Scottish Universities Life Science Alliance, Vetenskapsrådet, Hjärnfonden, the Novo Nordisk Foundation, European Commission Grant HEALTH-F2-2007-201159, Karolinska Institutet, and National Institutes of Health Grants DA023214, DA011322, and DA021696.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212563110/-/DCSupplemental.
References
- 1.Kano M, Ohno-Shosaku T, Hashimotodani Y, Uchigashima M, Watanabe M. Endocannabinoid-mediated control of synaptic transmission. Physiol Rev. 2009;89(1):309–380. doi: 10.1152/physrev.00019.2008. [DOI] [PubMed] [Google Scholar]
- 2.Keimpema E, et al. Differential subcellular recruitment of monoacylglycerol lipase generates spatial specificity of 2-arachidonoyl glycerol signaling during axonal pathfinding. J Neurosci. 2010;30(42):13992–14007. doi: 10.1523/JNEUROSCI.2126-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argaw A, et al. Concerted action of CB1 cannabinoid receptor and deleted in colorectal cancer in axon guidance. J Neurosci. 2011;31(4):1489–1499. doi: 10.1523/JNEUROSCI.4134-09.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu CS, et al. Requirement of cannabinoid CB(1) receptors in cortical pyramidal neurons for appropriate development of corticothalamic and thalamocortical projections. Eur J Neurosci. 2010;32(5):693–706. doi: 10.1111/j.1460-9568.2010.07337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mesulam MM, Mufson EJ, Wainer BH, Levey AI. Central cholinergic pathways in the rat: An overview based on an alternative nomenclature (Ch1-Ch6) Neuroscience. 1983;10(4):1185–1201. doi: 10.1016/0306-4522(83)90108-2. [DOI] [PubMed] [Google Scholar]
- 6.Gifford AN, Ashby CR., Jr Electrically evoked acetylcholine release from hippocampal slices is inhibited by the cannabinoid receptor agonist, WIN 55212-2, and is potentiated by the cannabinoid antagonist, SR 141716A. J Pharmacol Exp Ther. 1996;277(3):1431–1436. [PubMed] [Google Scholar]
- 7.Ruberti F, et al. Phenotypic knockout of nerve growth factor in adult transgenic mice reveals severe deficits in basal forebrain cholinergic neurons, cell death in the spleen, and skeletal muscle dystrophy. J Neurosci. 2000;20(7):2589–2601. doi: 10.1523/JNEUROSCI.20-07-02589.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibel M, Barde YA. Neurotrophins: Key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000;14(23):2919–2937. doi: 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez-Ortiz E, et al. TrkA gene ablation in basal forebrain results in dysfunction of the cholinergic circuitry. J Neurosci. 2012;32(12):4065–4079. doi: 10.1523/JNEUROSCI.6314-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hefti F, Will B. Nerve growth factor is a neurotrophic factor for forebrain cholinergic neurons; implications for Alzheimer’s disease. J Neural Transm Suppl. 1987;24:309–315. [PubMed] [Google Scholar]
- 11.Knusel B, Hefti F. Development of cholinergic pedunculopontine neurons in vitro: Comparison with cholinergic septal cells and response to nerve growth factor, ciliary neuronotrophic factor, and retinoic acid. J Neurosci Res. 1988;21(2-4):365–375. doi: 10.1002/jnr.490210228. [DOI] [PubMed] [Google Scholar]
- 12.Bisogno T, et al. Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J Cell Biol. 2003;163(3):463–468. doi: 10.1083/jcb.200305129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung KM, Astarita G, Thongkham D, Piomelli D. Diacylglycerol lipase-alpha and -beta control neurite outgrowth in neuro-2a cells through distinct molecular mechanisms. Mol Pharmacol. 2011;80(1):60–67. doi: 10.1124/mol.110.070458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinh TP, et al. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc Natl Acad Sci USA. 2002;99(16):10819–10824. doi: 10.1073/pnas.152334899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atipairin A, Canyuk B, Ratanaphan A. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by the platinum-based anticancer drugs. Breast Cancer Res Treat. 2011;126(1):203–209. doi: 10.1007/s10549-010-1182-7. [DOI] [PubMed] [Google Scholar]
- 16.Berghuis P, et al. Hardwiring the brain: Endocannabinoids shape neuronal connectivity. Science. 2007;316(5828):1212–1216. doi: 10.1126/science.1137406. [DOI] [PubMed] [Google Scholar]
- 17.Morozov YM, Torii M, Rakic P. Origin, early commitment, migratory routes, and destination of cannabinoid type 1 receptor-containing interneurons. Cereb Cortex. 2009;19(Suppl 1):i78–i89. doi: 10.1093/cercor/bhp028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felder E, Dechant G. Neurotrophic factors acutely alter the sorting of the vesicular acetyl choline transporter and the vesicular monoamine transporter 2 in bimodal sympathetic neurons. Mol Cell Neurosci. 2007;34(1):1–9. doi: 10.1016/j.mcn.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Rende M, et al. Role of nerve growth factor and its receptors in non-nervous cancer growth: Efficacy of a tyrosine kinase inhibitor (AG879) and neutralizing antibodies antityrosine kinase receptor A and antinerve growth factor: An in-vitro and in-vivo study. Anticancer Drugs. 2006;17(8):929–941. doi: 10.1097/01.cad.0000224459.13651.fd. [DOI] [PubMed] [Google Scholar]
- 20.Dechant G, Barde YA. Signalling through the neurotrophin receptor p75NTR. Curr Opin Neurobiol. 1997;7(3):413–418. doi: 10.1016/s0959-4388(97)80071-2. [DOI] [PubMed] [Google Scholar]
- 21.Bromberg KD, Ma’ayan A, Neves SR, Iyengar R. Design logic of a cannabinoid receptor signaling network that triggers neurite outgrowth. Science. 2008;320(5878):903–909. doi: 10.1126/science.1152662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin WF, et al. SH2B1beta enhances fibroblast growth factor 1 (FGF1)-induced neurite outgrowth through MEK-ERK1/2-STAT3-Egr1 pathway. Cell Signal. 2009;21(7):1060–1072. doi: 10.1016/j.cellsig.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Coene ED, et al. A novel role for BRCA1 in regulating breast cancer cell spreading and motility. J Cell Biol. 2011;192(3):497–512. doi: 10.1083/jcb.201004136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pulvers JN, Huttner WB. Brca1 is required for embryonic development of the mouse cerebral cortex to normal size by preventing apoptosis of early neural progenitors. Development. 2009;136(11):1859–1868. doi: 10.1242/dev.033498. [DOI] [PubMed] [Google Scholar]
- 25.Oudin MJ, Hobbs C, Doherty P. DAGL-dependent endocannabinoid signalling: Roles in axonal pathfinding, synaptic plasticity and adult neurogenesis. Eur J Neurosci. 2011;34(10):1634–1646. doi: 10.1111/j.1460-9568.2011.07831.x. [DOI] [PubMed] [Google Scholar]
- 26.De Petrocellis L, et al. The endogenous cannabinoid anandamide inhibits human breast cancer cell proliferation. Proc Natl Acad Sci USA. 1998;95(14):8375–8380. doi: 10.1073/pnas.95.14.8375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long JZ, et al. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol. 2009;5(1):37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.