Abstract
The human sensor of double-stranded RNA (dsRNA) oligoadenylate synthetase 1 (hOAS1) polymerizes ATP into 2′,5′-linked iso-RNA (2-5A) involved in innate immunity, cell cycle, and differentiation. We report the crystal structure of hOAS1 in complex with dsRNA and 2′-deoxy ATP at 2.7 Å resolution, which reveals the mechanism of cytoplasmic dsRNA recognition and activation of oligoadenylate synthetases. Human OAS1 recognizes dsRNA using a previously uncharacterized protein/RNA interface that forms via a conformational change induced by binding of dsRNA. The protein/RNA interface involves two minor grooves and has no sequence-specific contacts, with the exception of a single hydrogen bond between the -NH2 group of nucleobase G17 and the carbonyl oxygen of serine 56. Using a biochemical readout, we show that hOAS1 undergoes more than 20,000-fold activation upon dsRNA binding and that canonical or GU-wobble substitutions produce dsRNA mutants that retain either full or partial activity, in agreement with the crystal structure. Ultimately, the binding of dsRNA promotes an elaborate conformational rearrangement in the N-terminal lobe of hOAS1, which brings residues D75, D77, and D148 into proximity and creates coordination geometry for binding of two catalytic Mg2+ ions and ATP. The assembly of this critical active-site structure provides the gate that couples binding of dsRNA to the production and downstream functions of 2-5A.
Keywords: nucleotidyl transferase, cytokine, interferon response, PAP1, CCA-adding enzyme
Double-stranded RNA (dsRNA)-binding oligoadenylate synthetases OAS1, OAS2, OAS3, OASL, and their splicing isoforms comprise the cohort of 10 homologous proteins either known to or implicated in 2′,5′-linked iso-RNA (2-5A) synthesis in human cells (1, 2). For OASL, the 2-5A synthesis activity has not been demonstrated and presently it is classified as catalytically inactive. Cells respond to 2-5A by activating the transcription factors IRF-3 and NF-κB and by mounting the IFN response (3–5). The 2-5A pathway serves as a conserved mammalian signal of viral presence providing resistance to hepatitis C virus (6), West Nile virus (7), and other RNA and DNA viruses (1, 5, 7, 8). Broader roles of the 2-5A pathway in terminal differentiation of adipocytes (9), cell cycle (10), and BRCA1/IFN-γ–mediated apoptosis (11, 12) have emerged recently.
Members of the OAS family belong to the nucleotidyl transferase superfamily that also includes poly-A polymerase (PAP1) (13) and CCA-adding enzyme (14). OAS1/2/3, PAP1, and CCA-adding enzyme synthesize RNA without using an oligonucleotide template. However, OAS1/2/3 have important distinctions: OAS1/2/3 synthesize 2′,5′-linked instead of 3′,5′-linked RNA; OAS1/2/3 do not require a prebound RNA primer; and, in contrast to the constitutively active PAP1 and CCA-adding enzyme, OAS1/2/3 are normally repressed and require binding of dsRNA for activity. The requirement for dsRNA binding reflects the unique biology of OAS1/2/3 as sensors of double-stranded RNA in the cytosol. It is largely unknown how the OAS family members recognize dsRNA and recruit it for regulation of 2-5A synthesis. Here we cocrystallized human OAS1 (hOAS1) with dsRNA containing 18 bp and with a substrate analog, 2′-deoxy ATP (dATP), which establishes the molecular basis for dsRNA recognition by OAS1.
Results and Discussion
Overview of the hOAS1●dsRNA●dATP Ternary Complex.
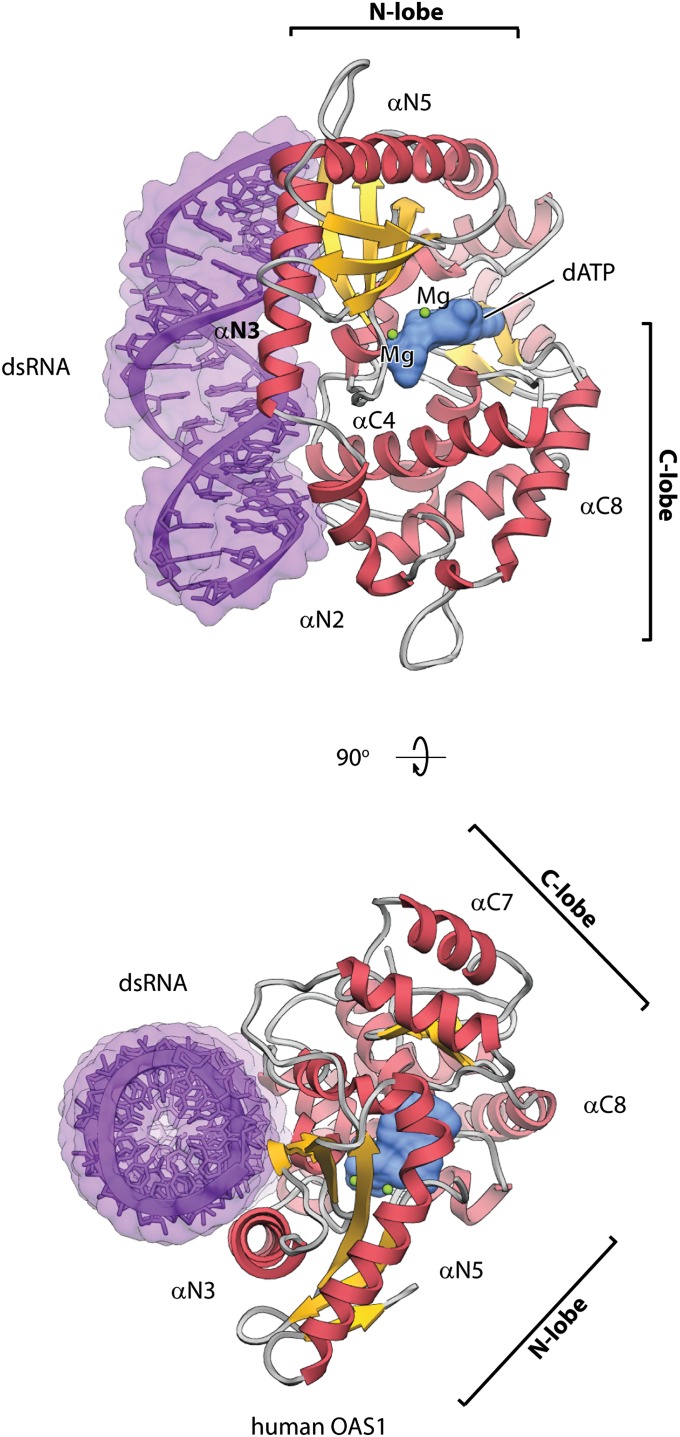
To understand how OAS1 recognizes dsRNA, we conducted cocrystallization screening with hOAS1 and dsRNA sequences derived from RNA constructs known to activate the sensor (2). Single cocrystals were obtained only with dsRNA having 18 bp and a serendipitously constructed sequence GGCUUUUGACCUUUAUGC. The structure of the ternary complex was determined at 2.7 Å resolution (Table S1). In the cocrystal structure, hOAS1 is bound to one face of the RNA double-helix (Fig. 1A and Figs. S1 and S2) and the dsRNA termini are unobstructed by the protein (Table S2). dsRNA is bound to the protein surface at the junction of the N- and C-terminal lobes of hOAS1 (Fig. 1A); however, the majority (22 of 30) of protein residues at the protein/RNA interface are contributed by the N lobe. The dATP molecule is bound to the pocket at the opposite face of hOAS1, as has been implicated with porcine OAS1 (pOAS1; 75% sequence identity with hOAS1); albeit, the latter was crystallized only in the inactive form, in the absence of dsRNA or dATP (13). The binding of dATP to hOAS1 involves two Mg2+ ions bridging the triphosphate moiety and the active site triad, D75, D77, and D148 (Fig. S3A). The crystal packing of the hOAS1 ternary complex occurs via end stacking of the dsRNA, suggesting a possible arrangement of hOAS1 on long dsRNA.
Fig. 1.
Structure of the hOAS1●dsRNA●dATP ternary complex. The complex is shown in two orientations related by 90° rotation. Protein is shown as ribbons; dsRNA (purple) and dATP (light blue) are shown as molecular surfaces. Two Mg2+ ions are shown as spheres (green). dsRNA is bound at the junction of the N lobe and the C lobe of hOAS1.
Recognition of dsRNA.
The cocrystal structure of hOAS1 with dsRNA reveals the RNA sequence requirements of hOAS1. Early in vivo and in vitro work with OAS1 suggested that hOAS1 may bind dsRNA without sequence specificity. This conclusion follows from the ability of poly-IC duplexes to activate both the IFN response in cells (5) and synthesis of 2-5A by purified OAS1 (13). However, the presence of RIG-I, MDA-5, TLR-3, and possibly other sensors of dsRNA complicates the interpretation of cell-based experiments, whereas activation of recombinant OAS1 by poly-IC does not dismiss a possibility that hOAS1 may have sequence preferences. Indeed, a preferred sequence motif, NNWWNNNNNNNNNWGN, has been discovered recently by in vitro selection (2). The cocrystal structure of hOAS1 with dsRNA provides the molecular framework for understanding of dsRNA recognition by hOAS1 and explains the surprising benefit from a single guanosine near one of the dsRNA termini.
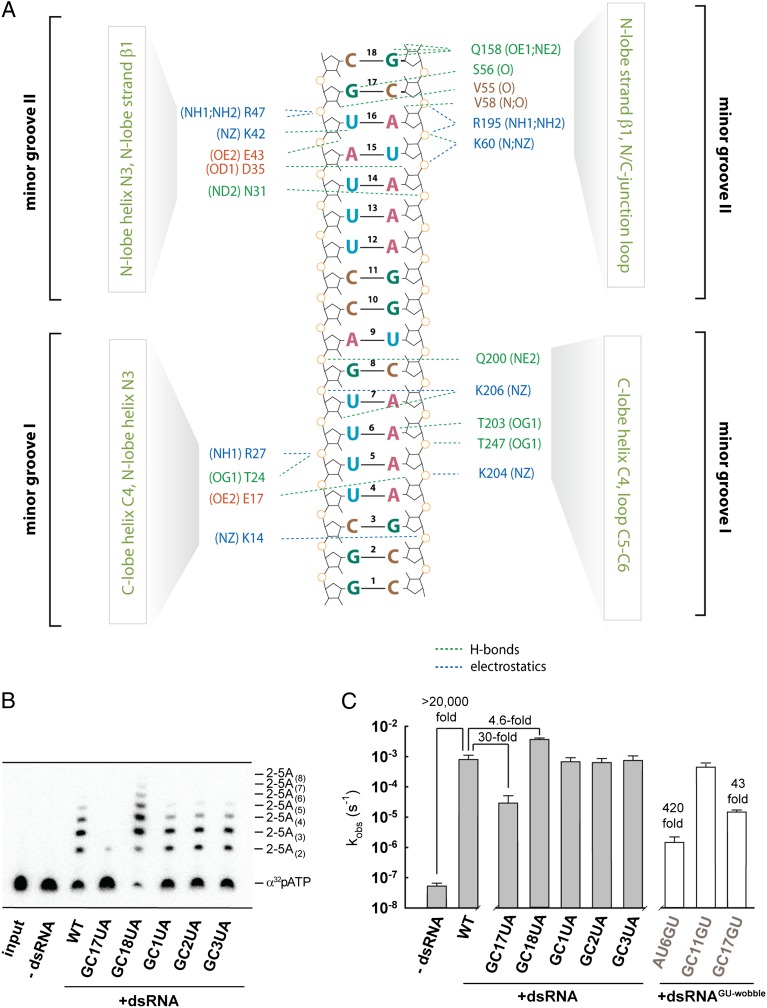
Human OAS1 recognizes dsRNA using a unique protein surface, distinct from the surface committed by the structurally related CCA-adding enzyme to recognition of the double-stranded tRNA acceptor stem (Fig. S4A). The surface in hOAS1 contains two dsRNA-binding sites located ∼30 Å apart (Fig. 2A). This arrangement creates a structurally uncharacterized (15) (Dataset S1) dsRNA-binding motif induced by binding of dsRNA, which recognizes two adjacent minor grooves. Recognition of two minor grooves sets a minimum length requirement for dsRNA at ∼17 bp, distinguishing OAS1 from two dsRNA sensors in the IFN response cocrystallized with dsRNA previously, which bind either long extracellular dsRNAs with 44 bp (TLR-3) (16) or short, blunt-ended cytosolic dsRNA with 8 bp (RIG-I) (17) (Table S2). Human OAS1 distorts dsRNA considerably from an ideal A-form helix (Table S2) by bending the ribophosphate backbone. The protein/RNA interface buries 3,042 Å2 of surface area (Table S2) and is stabilized by hydrogen bonds and by electrostatic interactions conserved in the OAS family (Fig. S5). Mutations K42E, R195E, K199E, K199M, K204E, and R210A, which map to the protein/RNA interface identified herein, have been reported to impair OAS1 activity by 60- to 2,500-fold (13, 14, 18) (Fig. S6).
Fig. 2.
Protein/RNA interface and the 2-5A synthesis activity of hOAS1. (A) Overview of the protein/RNA interface formed upon recognition of two dsRNA minor grooves by hOAS1. The minor groove with base pairs 13–18 is recognized by the protein residues located predominantly in the N lobe of hOAS1. The minor groove with base pairs 2–8 is recognized by the protein residues located predominantly in the C lobe of hOAS1. (B) Ten-minute end point assay for hOAS1 activation by dsRNA. Synthesis of a range of 2-5A species was analyzed by gel electrophoresis using α32p-ATP for visualization (Methods). (C) Rate constants (kobs) for 2-5A synthesis obtained with wild-type and mutant dsRNA using single-exponential fitting of time courses. Error bars show SEs from two measurements of kobs.
The recognition of dsRNA involves several 2′-OH groups, as observed commonly with dsRNA binding proteins, and several nucleobases (Fig. 2A and Table S2). Modeling of different nucleotides in the dsRNA suggests that nearly all substitutions would be tolerated without changing the number of hydrogen bonds to the protein. The only exceptions occur for the base pairs GC17 and GC18, where GC/UA mutations are predicted to disrupt one hydrogen bond to hOAS1. To test the importance of these contacts, we measured activation of hOAS1 by GC/UA-substituted dsRNA, using ATP with trace α32p-ATP as the substrate (Fig. 2 B and C). The GC17UA substitution impaired the dsRNA response of hOAS1 by 30-fold, whereas the GC18UA substitution had a small stimulatory effect (Fig. 2C). The absence of sequence recognition at position 18 suggests that only two hydrogen bonds to Q158 that are preserved upon GC18AU mutation may be important for binding (Fig. S7). Alternatively, Q158 may contribute little to dsRNA recognition because of its location in a surface loop that could disfavor Q158 binding entropically. In contrast, position 17 has a clear functional preference for GC over UA, revealing the importance of the G17/β1 hydrogen bond (Fig. S7). As expected, mutations at the opposite terminus of the dsRNA had no effect on activation of hOAS1 (Fig. 2C) because of the absence of nucleobase/protein contacts.
The relaxed overall sequence specificity of hOAS1 indicated by the crystal structure agrees with the biochemical evidence (2) and with the expectation that hOAS1 should detect diverse pathogenic dsRNA sequences. TLR-3 and RIG-I also recognize dsRNA without sequence specificity (16, 17); however, TLR-3 can discriminate between pathogen and host dsRNA by facing the extracellular environment or endosomal compartments, whereas RIG-I prefers blunt-ended dsRNA with 5′-triphosphates (19), generally indicative of viral infections. In contrast to these sensors, hOAS1 lacks exclusive compartmentalization or structural features to discriminate between sufficiently long host and pathogen dsRNA.
Perhaps, binding to dsRNA with 17 base pairs provides a strategy for avoiding short dsRNA elements in host RNA. Longer dsRNA elements might activate hOAS1, at least partially, if they become accessible to this sensor, and may have roles in connecting the 2-5A pathway to cell cycle, differentiation, and apoptosis (9–12). Using bioinformatics we identified a plethora of dsRNA with 17 or more continuous base pairs in human mRNA, microRNA and long intergenic noncoding RNA (lincRNA) (Fig. S8 and SI Text). Inclusion of GU-wobble base pairs in the search algorithm increased the number of predicted long dsRNA segments in mRNAs and in lincRNAs by 102- and 10-fold, respectively (Fig. S8). To determine whether the pool of dsRNA with GU wobbles could contain potential activators of hOAS1, we tested the effects of GU-wobble substitutions on recognition by hOAS1 (Fig. 2C, open bars). GU wobbles at the protein/RNA interface inhibited the 2-5A synthesis by 43- to 420-fold, indicating the importance of Watson–Crick base pairs at these positions. In contrast, a GU-wobble mutation in the middle of the dsRNA was readily tolerated (Fig. 2 A and C, base pair 11), which could be explained by the absence of protein–RNA interactions in this RNA region. Thus, optimal dsRNA activators of hOAS1 may contain GU wobbles in certain positions, expanding the pool of possible targets of hOAS1.
Mechanism of OAS1 Activation.
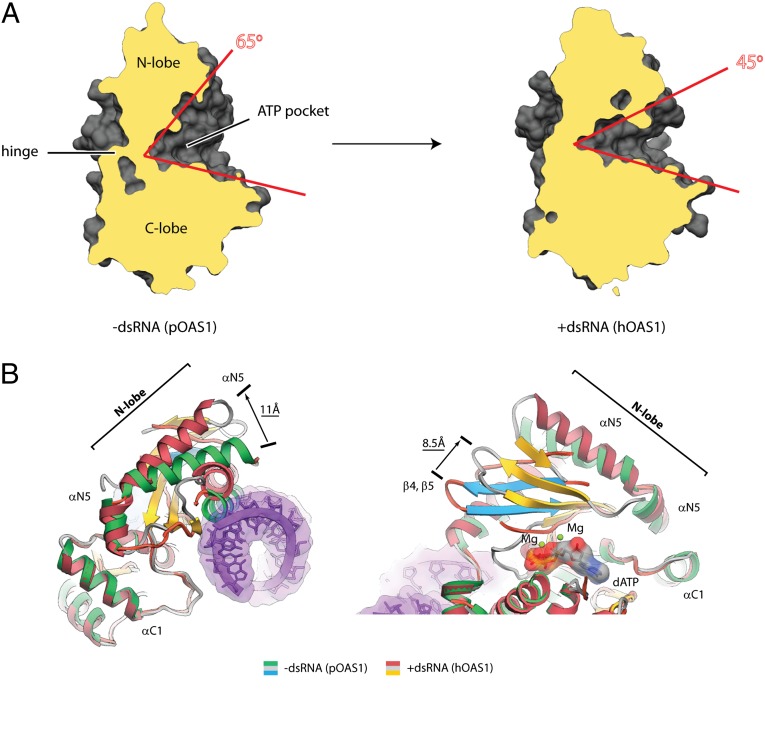
Repression and activation of the polymerase activity distinguishes OAS1 from the constitutively active nucleotidyl transferases PAP1 (13) and CCA-adding enzyme (14). Human OAS1 used here exhibits no detectable activity in the absence of dsRNA and undergoes more than 20,000-fold activation upon dsRNA binding (Fig. 2 B and C). Based on the crystal structure of the inactive apo-state of porcine OAS1 (pOAS1), it has been proposed that binding of dsRNA could activate OAS1 by a conformational change that moves its N and C lobes via the interlobe hinge to widen the active site cleft between the lobes (13).
The crystal structure of the active complex with dsRNA bound now shows that both lobes remain in the same position relative to the hinge. Contrary to the predictions, the active site becomes narrower (Fig. 3A), as the N lobe undergoes a major conformational rearrangement with displacements of secondary structure elements exceeding 10 Å. The conformational change moves helix N3 from the position that blocks the dsRNA binding site to the position that forms a part of the protein/dsRNA interface (Fig. S9). The C terminus of helix N5 moves by 11 Å outward from dsRNA, whereas the position of the N terminus remains unchanged (Fig. 3B). Finally, the β-sheet floor slides by as much as 8.5 Å toward helix N5 (Fig. 3B).
Fig. 3.
Global conformational changes in OAS1 upon dsRNA binding. (A) Side view of the interlobe groove in pOAS1 and in hOAS1. Clipping planes cut the structures at the same position. (B) Superposition of pOAS1 in the inactive conformation (PDB code 1PX5) with hOAS1 in the active conformation (present work). Movement of helix N5 away from the protein/RNA interface and sliding of the β-strand floor that contains the active site residues D75, D77, and D148 are shown.
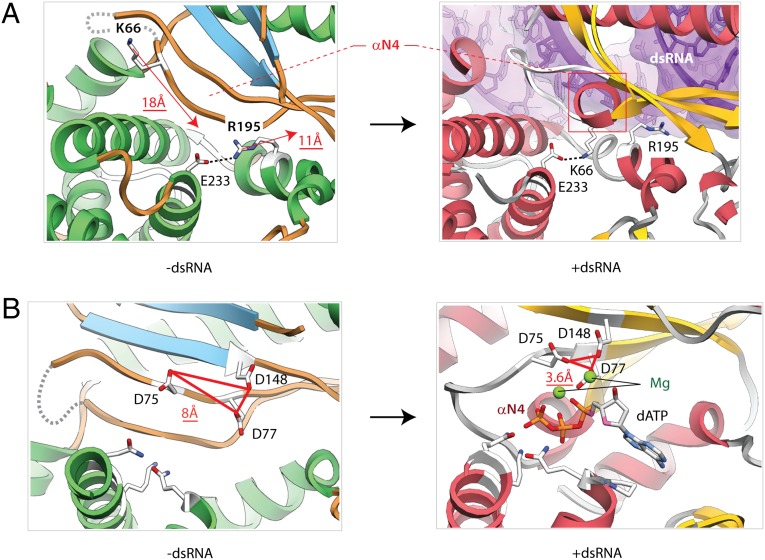
The rearrangements propagate from the protein/RNA interface toward the active site. In notable 18 Å and 11 Å movements, two basic side chains, K66 and R195, exchange their positions next to glutamate E233 (Fig. 4A and Fig. S5B). The arginine-lysine exchange places residue R195 at the protein/RNA interface and results in the formation of a new helix N4 from a previously existing strand (Fig. 4A, and Figs. S10 and S11). This conserved element of the structure is remarkable because it begins with strand β1 hosting the highest density of protein/RNA contacts (Fig. S10), including the functionally important interaction with the nucleobase G17 (Fig. S7), continues with the dsRNA-induced helix N4, and ends with strand β2 that harbors the active site residues D75 and D77. We refer to this element (residues 54–77) as RNA-induced catalytic structure (RICS).
Fig. 4.
RNA-induced conformational changes in the active site. (A) Binding of dsRNA extracts arginine R195 from the core of the protein and positions it at the protein/RNA interface. The exchange between residues K66 and R195 is coupled to the formation of the new helix, N4. The movement of the amino acids is shown by red arrows. (B) Conformational rearrangement positions the active site residues D75, D77, and D148 compactly for coordination of two Mg2+ ions (green spheres) and for binding of dATP. Red triangles show the inactive and active arrangements of the acidic triad.
RICS activates OAS1 by placing three acidic residues D75, D77, and D148 compactly, thereby providing the coordination geometry for chelation of two Mg2+ ions, binding of ATP and catalysis (Fig. 4B). Mutations D75A and D77A have been shown to inactivate OAS1 (14). The role of the D148 residue has not been tested previously. To confirm that D148 is also a part of the active site, we purified hOAS1 with a D148A mutation and tested its activation by dsRNA. The D148A mutation inhibited hOAS1 by ∼145-fold (Fig. S6C), confirming the importance of D148 for 2-5A synthesis and completing the view of the activation mechanism.
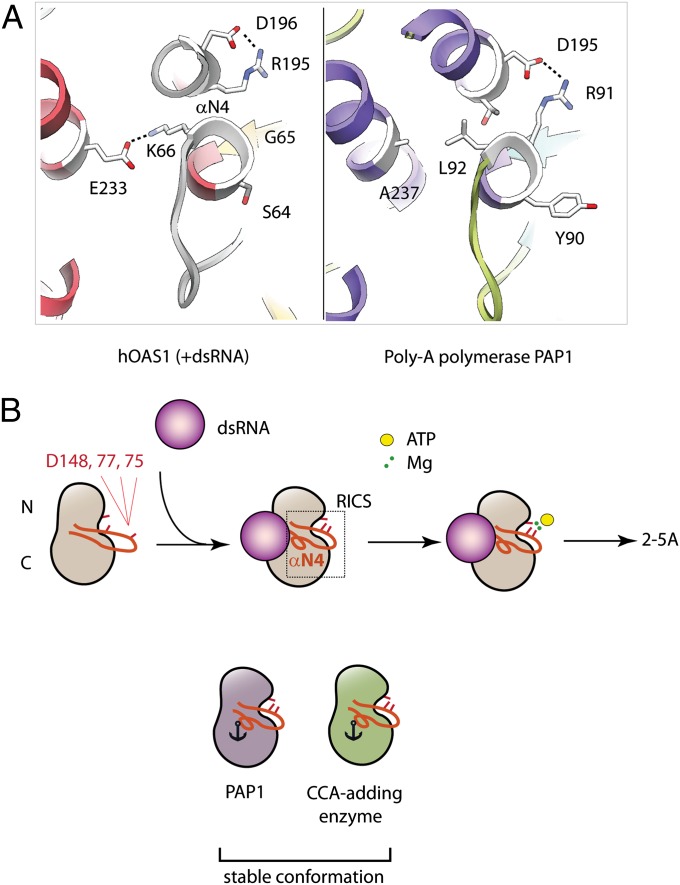
The conformational flexibility of RICS resembles that of the activation loop in protein kinases (20) (although RICS is not known to have phosphorylation sites). The analogy between OAS1 and protein kinases is reinforced by the observation that both RICS and the activation loop terminate with conserved aspartates [D75 in RICS and D from the DFG triad (21) in protein kinases], which are essential for coordination of the magnesium-ATP moiety in both protein families. RICS is absent in the constitutively active PAP1 and CCA-adding enzyme that contain hydrophobic and bulky residues in the helix equivalent to the helix N4 of OAS1, and lack the K66/R195 pair (Fig. 5A and Fig. S4B). Thus, RICS represents a prominent feature of the OAS family acquired for signal transduction.
Fig. 5.
Model for OAS1 regulation by dsRNA. (A) Comparison of the helix N4 in OAS1 (Left) with the structurally equivalent helix in poly-A polymerase PAP1 (Right; PDB ID 2Q66). (B) Active site residues of OAS1 D75, D77, and D148 face divergent directions in the ground state and are not compatible with coordination of Mg2+ ions and binding of ATP. dsRNA induces RICS and brings the residues D75, D77, and D148 together to enable binding of ATP●Mg2+. It is possible that ATP stabilizes RICS further upon binding to the OAS1●dsRNA complex, finalizing the conformational change. The dsRNA-independent CCA-adding enzyme and PAP1 are locked in the constitutively active state because their RICS-equivalent structures contain bulky hydrophobic residues anchored in the protein core.
The structure of hOAS1 provides a glimpse into the 2′,5′-specificity of hOAS1. Superposition of this structure with the structures of PAP1 and CCA-adding enzyme shows a unique orientation of the sugar and the base in dATP bound to hOAS1 (Figs. S4C and S12). This difference could provide a part of the specificity gate that favors 2′,5′-linkages. A comprehensive understanding of this mechanism must await a structure of hOAS1 with a bound donor of the 2′-OH group.
Conclusions
We show that hOAS1 has no detectable unstimulated activity and undergoes at least 104-fold activation by dsRNA. Thus, hOAS1 is regulated exquisitely to protect cells from rogue 2-5A signaling. Our studies establish the precise molecular basis for this regulation, which should apply to the entire OAS family and provide a common strategy for recognition of intracellular dsRNAs by these sensors. The cocrystal structure of hOAS1 captured with dsRNA reveals an unusual example of evolution in the nucleotidyl transferase family, wherein one of the constitutively active ancient nucleotidyl transferases involved in mRNA or tRNA maturation must have acquired a flexible loop in the core of the protein globule and used this flexibility to couple dsRNA binding to the assembly of the active site (Fig. 5B). The structural evidence suggests that hOAS1 lacks features to discern between host and pathogen dsRNA and raises a possibility that hOAS1 could recognize self-RNA with sufficiently long dsRNA segments. We show that hOAS1 could be activated not only by dsRNA with Watson–Crick base pairs, but also by dsRNA with GU wobbles, abundant in the human mRNA and lincRNA transcriptomes. It remains to be seen whether interactions with some of these RNAs may have biological significance, perhaps adding unexplored dimensions to hOAS1 signaling.
Methods
Statement of Errors.
All observed rate constants (kobs) were measured from single-exponential fitting using multiple data points. The measurements of kobs were repeated twice on two different days. The observed uncertainties were small compared with the effects we describe. Experimental errors are shown by error bars, when appropriate.
Protein Preparation.
The coding region for the p46 isoform of hOAS1 was cloned into pGEX-6P (GE Healthcare Life Sciences) and a stop codon was introduced at position 347. Human OAS1 was expressed as a GST fusion protein in Escherichia coli BL21 (DE3)-CodonPlus RIPL (Agilent Technologies). Cells were lysed on an EmulsiFlex C3 (Avestin) in buffer containing 20 mM Hepes (pH 7.4), 300 mM NaCl, 5 mM MgCl2, 0.1 mM EDTA, 10% (vol/vol) glycerol, 2 mM DTT, and 1% Triton X-100. Crude lysates were clarified by centrifugation at 35,000 × g for 30 min, at 4 °C. Clarified lysates were affinity-purified using glutathione Sepharose (GE Healthcare Life Sciences) and the GST tag was removed with Prescission protease (GE Healthcare Life Sciences). Human OAS1 was purified to more than 95% purity by Superdex 200 size-exclusion chromatography.
OAS1 Activity Assay.
Starting reaction mixtures containing hOAS1 and dsRNA in 9 μL of reaction buffer were preincubated for 10 min at 20 °C. Following preincubation, 1 μL of 10 mM ATP, trace-labeled with α32p-ATP, was added to initiate the reactions. The final reactions contained 3 μM hOAS1, 10 μM dsRNA, 20 mM Hepes (pH 7.4), 70 mM NaCl, 2 mM Mg(OAc)2, 4 mM DTT, 1 mM ATP, < 1 nM α32p-ATP, and 10% (vol/vol) glycerol. Aliquots (1 μL) were withdrawn at different time intervals and added to a stop solution (10 M urea, 0.002% bromophenol blue, 0.002% xylene cyanol). Samples were analyzed by denaturing 20% (wt/vol) PAGE and visualized by phosphorimaging on Typhoon FLA-7000 (GE Healthcare). Images were quantified using GelQuant.NET software (http://biochemlabsolutions.com/GelQuantNET.html). The observed rate constants were obtained by single-exponential fitting in SigmaPlot (www.sigmaplot.com). As no activity could be detected with hOAS1 in the absence of dsRNA over a 24-h incubation period, the kobs for the “-dsRNA” reactions in Fig. 2C represents the upper limit obtained from single-exponential fitting to the fastest possible rate consistent with the data, plus two SEs of the fit.
Crystallization of the hOAS1●dsRNA●dATP Ternary Complex.
dsRNA was reconstituted from cRNA strands purchased from Dharmacon. The crystallization mixture contained 15 mg/mL hOAS1, 550 μM dsRNA, 2 mM dATP (Sigma), 10 mM Hepes (pH 7.4), 350 mM NaCl, and 5 mM MgCl2. The reservoir solution contained 100 mM Hepes (pH 7.4) and 20% (vol/vol) PEG 200. Crystals were grown using the hanging drop vapor diffusion method and were cryo-protected in reservoir solution with 25% (vol/vol) ethylene glycol.
Diffraction Data Collection and Analysis.
X-ray diffraction data were recorded on beam line X29 at Brookhaven National Laboratory in Upton, NY. The datasets were collected using X-ray wavelength 1.075 Å and oscillation angle of 1°. The data were indexed, integrated and scaled in XDS (22). Five percent of the reflections were marked as a test-set. Crystals of the ternary complex hOAS1●dsRNA●dATP belong to space group P212121 (Table S1) and contain one copy of the complex in the asymmetric unit. Molecular replacement solution was found using PHASER (23) after anisotropy correction of the data (24). Initially, PDB entry 1PX5 was used as a search model, but produced no solution despite the 80% sequence homology with hOAS1. This outcome was later explained by the vast conformational change in OAS1 upon dsRNA binding. The correct solution was found with the C lobe of PDB entry 1PX5. The electron density for the N lobe, the dsRNA containing the expected number of base pairs, and the triphosphate moiety of dATP were observed at this stage and allowed building the complete structure. Following initial rigid-body refinement in PHENIX (25), the structure was built iteratively in COOT (26) and refined using simulated annealing (2,000 K) and TLS (one TLS group contained all PDB chains) refinement in PHENIX. Fourier Fobs − Fcalc difference maps were used for interpretation of new electron density. The orientation of the dsRNA duplex was determined from an unbiased simulated-annealing map and a biased map calculated with reverse dsRNA (Fig. S1). The molecule of dATP was modeled during the final stages of structure building. Two additional spherical densities were observed near the triphosphate group and the D75, D77, and D148 residues of hOAS1, indicating the presence of two cations. Although diffraction data alone do not distinguish between the two cations present in the crystallization buffer, Na+ and Mg2+, magnesium was modeled based on high-resolution structures of other nucleotidyl transferases, ion-ligand geometry, and cooperative activation of hOAS1 by Mg2+ (Fig. S3). Data integration statistics suggests that diffraction extends at least to 2.5 Å [I/σI = 1.26 and CC (1/2) = 75.8% in the 2.5–2.6 Å shell]. However, a model-vs.-data test shows that only data that extend to 2.7 Å improve the overall model quality (Fig. S13). Final structures were visualized with University of California at San Francisco Chimera (27) and PyMol 1.2 (DeLano Scientific). The resulting model has excellent stereochemical parameters (bonds = 0.003 Å, angles = 0.776°), no Ramachandran outliers, and crystallographic R/Rfree = 0.23/0.28 (Table S1).
Supplementary Material
Acknowledgments
We thank David Botstein (Princeton University), Andrei Korostelev (University of Massachusetts), Carl Correll (Rosalind Franklin University), and members of the A.K. laboratory for reading the manuscript and making valuable suggestions, and Gena Whitney for technical assistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4IG8).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218528110/-/DCSupplemental.
References
- 1.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8(7):559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kodym R, Kodym E, Story MD. 2′-5′-Oligoadenylate synthetase is activated by a specific RNA sequence motif. Biochem Biophys Res Commun. 2009;388(2):317–322. doi: 10.1016/j.bbrc.2009.07.167. [DOI] [PubMed] [Google Scholar]
- 3.Chakrabarti A, Jha BK, Silverman RH. New insights into the role of RNase L in innate immunity. J Interferon Cytokine Res. 2011;31(1):49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silverman RH. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol. 2007;81(23):12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448(7155):816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, Kang H, Ji Y, Chen X. Evaluate the relationship between polymorphisms of OAS1 gene and susceptibility to chronic hepatitis C with high resolution melting analysis. Clin Exp Med. 2012 doi: 10.1007/s10238-012-0193-6. 10.1007/s10238-012-0193-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kajaste-Rudnitski A, et al. The 2′,5′-oligoadenylate synthetase 1b is a potent inhibitor of West Nile virus replication inside infected cells. J Biol Chem. 2006;281(8):4624–4637. doi: 10.1074/jbc.M508649200. [DOI] [PubMed] [Google Scholar]
- 8.Lim JK, et al. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog. 2009;5(2):e1000321. doi: 10.1371/journal.ppat.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabre O, et al. RNase L controls terminal adipocyte differentiation, lipids storage and insulin sensitivity via CHOP10 mRNA regulation. Cell Death Differ. 2012;19(9):1470–1481. doi: 10.1038/cdd.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Haj L, Blackshear PJ, Khabar KS. Regulation of p21/CIP1/WAF-1 mediated cell-cycle arrest by RNase L and tristetraprolin, and involvement of AU-rich elements. Nucleic Acids Res. 2012;40(16):7739–7752. doi: 10.1093/nar/gks545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ireland DD, et al. RNase L mediated protection from virus induced demyelination. PLoS Pathog. 2009;5(10):e1000602. doi: 10.1371/journal.ppat.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullan PB, et al. The 2,5 oligoadenylate synthetase/RNaseL pathway is a novel effector of BRCA1- and interferon-gamma-mediated apoptosis. Oncogene. 2005;24(35):5492–5501. doi: 10.1038/sj.onc.1208698. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol Cell. 2003;12(5):1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- 14.Torralba S, Sojat J, Hartmann R. 2′-5′ oligoadenylate synthetase shares active site architecture with the archaeal CCA-adding enzyme. Cell Mol Life Sci. 2008;65(16):2613–2620. doi: 10.1007/s00018-008-8164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320(5874):379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang F, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479(7373):423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar SN, et al. Natural mutations in a 2′-5′ oligoadenylate synthetase transgene revealed residues essential for enzyme activity. Biochemistry. 2005;44(18):6837–6843. doi: 10.1021/bi0502893. [DOI] [PubMed] [Google Scholar]
- 19.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125(6):1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 21.Dar AC, Lopez MS, Shokat KM. Small molecule recognition of c-Src via the Imatinib-binding conformation. Chem Biol. 2008;15(10):1015–1022. doi: 10.1016/j.chembiol.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 23.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strong M, et al. Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103(21):8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.