Abstract
Chemically induced dimerization is an important tool in chemical biology for the analysis of protein function in cells. Here we report the use of the natural product fusicoccin (FC) to induce dimerization of 14–3-3–fused target proteins with proteins tagged to the C terminus (CT) of the H+-ATPase PMA2. To prevent nonproductive or detrimental interactions of the 14-3-3 proteins and CT fusions with endogenous cell proteins, their interaction surface was engineered to facilitate FC-induced dimerization exclusively between the introduced protein constructs. Live-cell imaging documented the reversible FC-induced translocation of 14-3-3 and CT to different cell compartments depending on localization sequences fused to their dimerization partner protein. The functionality of this system was demonstrated by the FC-induced importation of the NF-κB-CT into the nucleus. In HeLa cells, FC-mediated dimerization of the NF-κB-CT with a constitutively nuclear-localized 14-3-3 protein led to an NF-κB–specific cellular response by inducing IL-8 secretion.
Keywords: protein–protein interaction, cell biology, protein engineering, fluorescence microscopy, surface plasmon resonance
Chemically induced dimerization (CID) mediated by cell-permeable, small molecules is a tool to induce the association of transfected proteins containing a ligand-binding domain fused to the protein under investigation. Since its initial introduction in 1993 (1), CID has become a valuable tool in chemical biology for elucidating biological phenomena at the protein level. In the ground-breaking work of Spencer et al. (1), a homodimeric derivative of the natural product FK506, FK1012, which binds to the immunophilin FKBP (FK506-binding protein), was used to trigger intracellular signaling by inducing dimerization of a fusion protein of FKBP and the ζ chain of the T-lymphocyte antigen receptor (TCR). In addition to homodimerizers, which combine two copies of their binding partners, heterodimerizers are used to combine two different proteins or protein domains. The classic example of a heterodimerizer is the bifunctional natural product rapamycin, which binds to FKBP and the FKBP rapamycin-binding domain (FRB) of the FKBP rapamycin-associated protein, FRAP (2, 3). Numerous biological processes in which rapamycin has been identified as the dimerizer have been studied (4). However, with rapamycin-induced dimerization of proteins fused to FKBP and FRB or alternative CID systems that are based on FKBP proteins, nonproductive interactions often occur because of the binding of the dimerizer to the abundant endogenous mammalian FKBP (1–3, 5–7). In addition, these applications are limited by the instability, toxicity, or expense of the dimerizers. Alternative cellular CID systems that are based on the bacterial proteins GyrB (8) and phytohormones such as abscisic acid (ABA) (9) and the gibberellin analog GA3-AM (10) have been reported. However, in most cases the mentioned CID systems are not reversible by mere medium exchange, most likely because of the binding of the molecules to the target proteins with affinities in the nanomolar range.
In the present study, our goal was to create a CID system that introduces a dimerizer that is reversible and whose components do not interfere with critical endogenous processes and do not cause nonproductive interactions. The natural product fusicoccin (FC), a diterpenoid glycoside with a dicyclopenta[a,d]cyclooctene skeleton (5-8-5 ring system; Fig. 1A) (11), was chosen as the dimerizing agent in this study. FC is known to stabilize the interaction of 14-3-3 proteins with the C-terminal regulatory domain of the plant plasma membrane H+-ATPase (PMA) by binding simultaneously to both proteins, thereby closing a gap in the otherwise weak interaction interface (12). This property and the commercial affordability of FC are important criteria for the development of this CID system. The C terminus of the proton pump is an autoinhibitory region that is bound to the rest of the protein in the inactive state. In the active state, a phosphorylation of its penultimate threonine residue triggers the binding of 14-3-3 proteins (13).
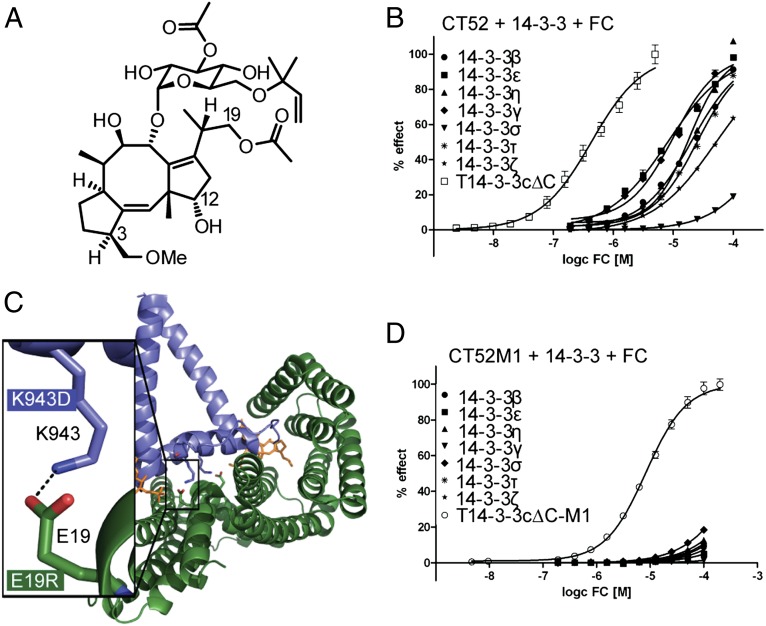
Fig. 1.
Engineering the T14-3-3cΔC/CT52 interface abolishes FC-dependent interaction of CT52 with human 14-3-3 proteins. (A) Structure of FC. (B) SPR measurements performed to calculate the EC50 values of FC on the CT52/14-3-3 interaction. FC was titrated in presence of 10 µM 14-3-3 in stepwise 1:1 dilutions onto immobilized CT52. EC50 values were calculated by plotting RU values of the eight SPR measurements against the corresponding FC concentration on a logarithmic scale and fitting the curves using the four parameter logistic nonlinear regression model (Table 1). For RU values plotted against time see SI Appendix, Fig. S2C. (C) Crystal structure of T14-3-3cΔC/CT52/FC (ref. 17, PDB ID 2O98) with a detailed view on the electrostatic interaction between E19 of T14-3-3cΔC (dark green) and K943 of CT52 (blue). The salt bridge was used for a charge reversal with the goal of preserving binding of CT52 and T14-3-3cΔC and simultaneously eliminating any other possible interactions. (D) FC titration in the presence of 10 µM 14-3-3cΔC onto immobilized CT52M1. EC50 values (Table 1) were calculated as described above. For RU values plotted against time, see SI Appendix, Fig. S4A.
The 14-3-3 proteins belong to a family of highly conserved acidic proteins that can be found in all eukaryotic cells. Many organisms contain multiple isoforms of 14-3-3 proteins; in mammals, seven isoforms (β, ɛ, η, γ, σ, τ, and ζ) have been identified. They regulate a multitude of biological processes in different eukaryotic organisms by binding to more than 200 different target proteins at a phosphorylated serine or threonine residue (14), resulting in modulation of their subcellular localization, enzymatic activity, or ability to interact with further proteins (15, 16). Biochemical studies showed that FC induces binding of tobacco 14-3-3 (T14-3-3c) with the last 52 C-terminal amino acids (CT52) of the tobacco H+-ATPase PMA2 even in its unphosphorylated state (17). Furthermore, three amino acid substitutions (S938A, T955D, and V956I) in the PMA2 peptide as well as a truncation of the C terminus of T14-3-3c (T14-3-3cΔC, residues 1–242), which is an inhibitor of the 14-3-3/ligand interaction (18), resulted in a higher binding affinity (17). The crystal structure of the T14-3-3cΔC/CT52/FC complex finally revealed that only the last 30 C-terminal amino acids of PMA2 are in direct contact with the T14-3-3cΔC protein and that a 22-amino acid helix functions as a spacer to position this interaction motif into the central binding channel of the T14-3-3cΔC protein (Fig. 1C) (17).
Here we show that FC can be used as a chemical tool to induce dimerization of T14-3-3cΔC and CT52 fusion proteins, e.g., those governing nuclear export, nuclear import, or plasma membrane recruitment. In principle, this system can be used to confer chemically induced proximity to any protein pair of choice. Engineering the interface of the T14-3-3cΔC/CT52 complex abrogated unproductive and potentially detrimental interactions of T14-3-3cΔC and CT52 with endogenous proteins in the transfected cells. To demonstrate the use of this system, we chose to modulate the subcellular localization of the transcriptional factor NF-κB to induce secretion of IL-8.
Results
T14-3-3cΔC/CT52 Interface Engineering Abrogates FC-Induced Interaction of CT52 with Human 14-3-3 Proteins.
The 14-3-3 proteins bind to and regulate several hundreds of proteins in eukaryotic cells (14). To develop a CID system involving FC, T14-3-3cΔC, and the PMA2 peptide CT52 in mammalian cells, we first focused on abrogating all unproductive or potentially harmful interactions of cotransfected T14-3-3cΔC and CT52 with endogenous proteins in mammalian cells. By mutating the interaction surface of this protein pair, we were able to prevent T14-3-3cΔC from binding to any endogenous 14-3-3 protein partners and the PMA2 construct from engaging cellular wild-type 14-3-3 proteins. To analyze the protein–protein interactions, far-Western blotting, surface-based fluorescence assay (SBFA), and surface plasmon resonance (SPR) were used. For far-Western blotting and SBFA experiments we used the PMA2 construct CT66, which contained the last 66 C-terminal amino acids of PMA2 fused to GST (GST-CT66) and which carried three amino acid substitutions, S938A, T955D, and V956I (SI Appendix, Fig. S1), which, as we previously reported, enhance the binding to T14-3–3cΔC (17). The 66-amino acid construct is more stable than its 52-amino acid counterpart when fused to GST and therefore is better suited to biochemical assays that necessitate a GST tag. With the exception of 14-3-3σ, FC-dependent binding of the tobacco and the other six human 14-3-3 proteins to the immobilized wild-type PMA2 peptide could be detected (SI Appendix, Fig. S2 A and B). These results were confirmed by SPR (Fig. 1B and SI Appendix, Fig. S2C). Here, the PMA2 construct CT52 with the same amino acid substitutions (SI Appendix, Fig. S1) was immobilized on a biosensor chip to measure FC-induced binding of all human and tobacco 14-3-3 proteins. The calculated EC50 values are listed in Table 1.
Table 1.
Steady-state kinetic parameters for the interaction of CT52 and CT52M1 with 14–3-3 proteins
| Protein | CT52 EC50 (µM) ± SEM | CT52M1EC50 (µM) ± SEM |
| T14-3–3cΔC | 0.58 ± 0.04 | n.d. |
| T14-3–3cΔC-M1 | n.d. | 8.30 ± 0.01 |
| 14–3-3ɛ | 10.70 ± 0.03 | n.m. |
| 14–3-3γ | 13.06 ± 0.05 | n.m. |
| 14–3-3η | 22.93 ± 0.03 | n.m. |
| 14–3-3β | 26.76 ± 0.02 | n.m. |
| 14–3-3τ | 33.50 ± 0.01 | n.m. |
| 14–3-3ζ | 68.59 ± 0.01 | n.m. |
| 14–3-3σ | 557.50 ± 0.03 | n.m. |
n.d., not determined; n.m., not measureable.
To prevent unwanted binding of CT52 and T14-3-3cΔC to endogenous partner proteins, a complementary charge exchange of an electrostatically engaged amino acid pair within the binding interface of the complex was carried out (E19 of T14-3-3cΔC and K943 of CT52; Fig. 1C). We exchanged E19 of T14-3-3cΔC for positively charged amino acids (E19R/K) and K943 of CT52 for negatively charged amino acids (K943D/E). Far-Western blotting of all four mutated variants confirmed binding of the 14-3-3 constructs to the immobilized GST-CT66 fusions; the strongest binding was that of T14-3-3cΔC-E19R (M1) to both CT66 variants (CT66-K943D/E) (SI Appendix, Fig. S3A). Quantification by means of SBFA confirmed this finding (SI Appendix, Fig. S3B). T14-3-3cΔC-M1 gains about 50–60% of the affinity of the T14-3-3cΔC/CT52 complex binding, whereas T14-3-3cΔC-E19K reaches just 10%. To enhance the binding of T14-3-3cΔC-M1 to CT66-K943D/E, we additionally introduced three further mutations in the PMA2 constructs that already had led to enhanced binding in the T14-3-3cΔC/CT52 complex as described above: CT66-K943D-S938A-T955D-V956I (CT66M1) and CT66-K943E-S938A-T955D-V956I (CT66M2), SI Appendix, Fig. S1) (17). The binding affinity of T14-3-3cΔC-M1 to CT66M1 is comparable to the binding affinity of T14-3-3cΔC to CT52 (SI Appendix, Fig. S3C). These results revealed that T14-3-3cΔC-M1/CT66M1 is the best-engineered protein pair, with an EC50 value of 8.30 ± 0.01 µM as measured by SPR (Fig. 1D, SI Appendix, Fig. S4A, and Table 1). Far-Western blotting of this interaction confirmed this finding (SI Appendix, Fig. S3D). Furthermore, we tested the binding of human 14-3-3 to CT66M1 by means of far-Western blotting, which showed that introduction of the charge-reversal mutation (K943D) into the corresponding PMA2 construct resulted in complete abrogation of FC-induced interaction with all seven human 14-3-3 isoforms (SI Appendix, Fig. S3D). The same result could be determined by SBFA (SI Appendix, Fig. S3E) and SPR (EC50 not measurable) (Fig. 1D, SI Appendix, Fig. S4A, and Table 1).
T14-3-3cΔC-M1/CT52M1 Interface Engineering Abrogates Interaction of T14-3-3cΔC-M1 with Human Partner Proteins.
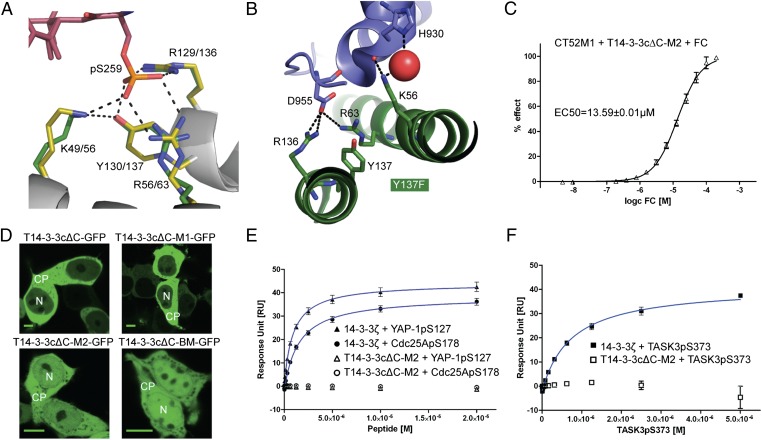
Because the CT52M1 construct lost the ability to bind human 14-3-3 proteins, we next focused on further engineering T14-3-3cΔC-M1 to minimize interference with the recipient cell physiology. Nearly all known 14-3-3–interacting proteins in humans and other eukaryotes bind to 14-3-3 proteins in a serine/threonine phosphorylation-dependent manner (19). The essential structural feature allowing 14-3-3 proteins to accommodate the corresponding phosphorylated binding partners is a strictly conserved, basic phospho-accepting site consisting of two arginines, a lysine, and a tyrosine which coordinate the phosphate moiety of the respective binding partners (Fig. 2A) (20). As we demonstrated previously for the interaction of the phosphopeptide C-RAF255–264pS259 and 14-3-3σΔC [Protein Data Bank (PDB) ID 3IQJ], polar contacts are established between the phosphorylated amino acid and all four aforementioned residues (21). To visualize that T14-3-3cΔC also is able to interact with the C-RAF peptide, we superimposed the 14-3-3σΔC/C-RAF255–264pS259 structure (yellow/magenta in Fig. 2A) with T14-3–3cΔC (PDB ID 1O9F) (green in Fig. 2A) (12). In comparison, the phospho-mimicking aspartate in the PMA2 peptide establishes a salt bridge solely with the two arginines of the basic pocket of T14-3-3cΔC, R136 and R63 (PDB ID 2O98) (17), with no polar contact to either K56 or Y137 (Fig. 2B). Instead, K56 of T14-3-3cΔC contacts the backbone of the PMA2 peptide and H930 via a water molecule, and Y137 shows no interaction. Therefore, we replaced Y137 of T14-3-3cΔC-M1 to phenylalanine (T14-3-3cΔC-M2) to abrogate the H-bond between its hydroxyl moiety and the phosphate moiety (Fig. 2A). First, we tested the effect of T14-3-3cΔC-M2 with respect to the FC-dependent binding to CT52M1 by means of far-Western blotting and detected a strong signal (SI Appendix, Fig. S4B). This result was confirmed by coupling CT52M1 to a biosensor chip and titrating FC in the presence of T14-3-3cΔC-M2, leading to the calculation of an EC50 of 13.59 ± 0.01 µM (Fig. 2C and SI Appendix, Fig. S4C). To investigate the localization of the T14-3-3cΔC variants in human cells, we transiently expressed the corresponding GFP constructs in HEK293T cells and monitored the green fluorescence. T14-3-3cΔC and T14-3-3cΔC-M1 were localized predominantly in the cytoplasm (Fig. 2D). However, the Y137F amino acid substitution abrogated this strong cytoplasmic localization, because we found T14-3-3cΔC-M2 evenly distributed in cytoplasm and nucleus (Fig. 2D). As a control and to gain an impression of the subcellular localization of a T14-3-3cΔC binding mutant, we replaced all amino acids that coordinate the phosphate moiety with the binding partners for alanine (T14-3-3cΔC-BM). When transiently expressed in HEK293T cells, this variant localized similarly to T14-3-3cΔC-M2, in the cytoplasm as well as in the nucleus (Fig. 2D), likewise suggesting a successful abrogation of the interaction of T14-3-3cΔC-M2 with endogenous proteins in human cells. To analyze possible interactions of T14-3-3cΔC-M2 with endogenous 14-3-3–binding proteins of human cells in vitro, we tested three well-known 14–3-3 partners, the cell-cycle phosphatase Cdc25A (22), the transcription coactivator YAP-1 (23, 24), and the K+ channel TASK3 (25), by means of SPR. For these studies, peptides of about 23–38 amino acids were chosen, each comprising the corresponding phosphorylated interaction site in each of these partners: Cdc25ApS178, YAP-1pS127, and TASK3pS373 (for peptide sequences see SI Appendix, Table S1). First 14-3-3ζ was immobilized onto a biosensor chip, and the phosphopeptides were titrated until saturation was reached (Fig. 2 E and F and SI Appendix, Fig. S5A). These data show interactions between 14-3-3ζ and the selected binding partners with Kd values ranging from 1.53–0.94 µM (Table 2). Furthermore, SPR experiments provided evidence that T14-3-3cΔC is able to interact with the three phosphopeptides with comparable Kd values (SI Appendix, Fig. S5B). Finally, we conducted SPR experiments to show that introduction of the Y137F substitution was sufficient to abrogate phosphorylation-dependent interactions with the 14–3-3 binding motifs of Cdc25ApS178, TASK3pS373, and YAP-1pS127 (Fig. 2 E and F and SI Appendix, Fig. S5C).
Fig. 2.
T14-3-3cΔC-M1/CT52M1 interface engineering abrogates the interaction of T14-3-3cΔC-M1 with 14–3-3 partner proteins. (A) Superimposition of 14-3-3σΔC/C-RAF255–264pS259 (yellow/magenta) and T14-3-3cΔC (green) demonstrates the coordination of a phosphopeptide in the 14-3-3–binding groove (PBD IDs 3IQJ and 1O9F) (12, 21). (B) Detailed view of the phospho-acceptor pocket of the T14-3-3cΔC/CT52/FC complex (PBD ID 2O98) (17). The phosphate-mimicking D955 of CT52 (blue) is coordinated by R63 and R136 of T14-3-3cΔC (dark green) but not by Y137 or K56. (C) SPR titration of FC in the presence of 10 µM T14-3-3cΔC-M2 onto immobilized CT52M1 and its corresponding fitting curve for EC50 calculation (13.59 ± 0.01 µM). (D) Confocal microscopy images of HEK293T cells transfected with plasmids encoding for T14-3-3cΔC-GFP variants. T14-3-3cΔC-GFP and T14-3-3cΔC-M1-GFP are located in the cytoplasm (CP), whereas T14-3–3cΔC-M2-GFP and T14-3-3cΔC-BM-GFP are evenly distributed between cytoplasm and nucleus (N). (Scale bars, 10 μm.) (E and F) SPR measurements of three phosphopeptides containing the 14–3-3–binding motifs of Cdc25C, TASK3, and YAP-1 to immobilized 14-3-3ζ (E) and T14-3-3cΔC-M2 (F). Binding affinity was measured by stepwise titration of the peptides in 1:1 dilutions and fitting the resulting curves using steady-state kinetics (see also Fig. 5A and C). Corresponding Kd values are listed in Table 2. No binding could be monitored for T14-3-3cΔC-M2.
Table 2.
Steady-state kinetic parameters for the interaction of 14–3-3ζ, T14-3–3cΔC, and T14-3–3cΔC-M2 with three phosphopeptides each containing the 14–3-3 binding motif
| Phosphopeptide | 14–3-3ζ Kd (µM) ± SEM | T14-3–3cΔC Kd (µM) ± SEM | T14-3–3cΔC-M2 Kd (µM) ± SEM |
| Cdc25ApS178 | 1.53 ± 0.12 | 1.95 ± 0.22 | n.m. |
| TASK3pS373 | 0.75 ± 0.06 | 0.46 ± 0.08 | n.m. |
| YAP-1pS127 | 0.94 ± 0.09 | 0.89 ± 0.05 | n.m. |
n.m., not measurable.
Time Course and Reversibility of FC-Induced Nuclear Exclusion of mCherry-CT52M1.
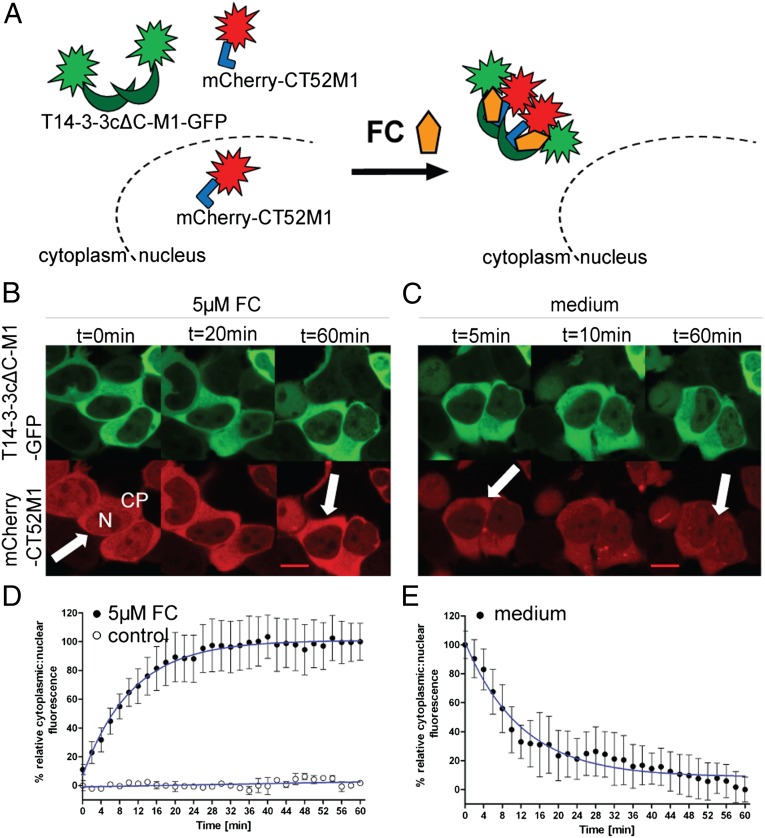
To use the principle ability of FC to induce dimerization of T14-3-3cΔC with the soluble C terminus of PMA2, we first transfected HEK293T cells with CT52M1 fused to mCherry and studied its localization by confocal microscopy. Because mCherry-CT52M1 was distributed evenly throughout the cell, we used the mainly cytoplasmic T14-3-3cΔC-M1 to show the FC-dependent binding of the mutated T14-3-3cΔC and CT52M1 in human cells (Fig. 3A). Addition of 5 µM FC to the medium did not influence T14-3-3cΔC-M1 but resulted in a strong decrease in the fluorescence of the nuclear population of mCherry-CT52M1 and a simultaneous increase of its cytoplasmic fluorescence within the first 20 min. Virtually all cellular fluorescence of mCherry-CT52M1 was located in the cytoplasm after 60 min, presumably because of FC-induced complexation of CT52M1 with the cytoplasmically localized T14-3-3cΔC-M1 (Fig. 3B and Movie S1). Quantitative analysis of the intensity of nuclear and cytoplasmic mCherry-CT52M1 fluorescence showed half-maximal effect, including translocation into the nucleus, after 7.4 min (Fig. 3D). In unstimulated cells, we could not detect any changes of the cellular fluorescence of CT52M1 by either visual or quantitative analysis (Fig. 3D and SI Appendix, Fig. S6A). Furthermore, we tested the possible reversibility of the chemically induced dimerization by washing the cells several times with DMEM after a complete nuclear exclusion of CT52M1 and found that this process was fully reversible within the same time-frame (Fig. 3 C and E and Movie S2).
Fig. 3.
FC-induced nuclear exclusion of mCherry-CT52M1. (A) Schematic representation of FC-dependent nuclear export of mCherry-CT52M1. (B and C) Time-course images of HEK293T cells transfected with plasmids encoding for T14-3-3cΔC-M1-GFP and mCherry-CT52M1 at stated times, monitoring nuclear exclusion of mCherry-CT52M1 fluorescence after the addition of 5 µM FC (B) and its reverse translocation after the cells were washed with medium (C). See SI Appendix, Fig. S6A for control cells treated with solvent (3.3% ethanol). (Scale bars, 10 μm.) CP: cytoplasm; N, nucleus. (D and E) Quantification of mCherry-CT52M1 translocation in response to 5 µM FC and the analysis of untreated cells (control) (D) and of the reversibility of mCherry-CT52M1 translocation (E). Mean cytoplasmic and nuclear fluorescent intensities were calculated every 2 min and plotted as a ratio against time. The data represent mean values (± SEM) from three experiments with at least two cells per experiment; n = 6–9. To obtain the EC50 curve, fitting was performed using nonlinear regression and one-phase exponential association.
Nuclear Accumulation and Plasma Membrane Recruitment of an Engineered FC Target Protein.
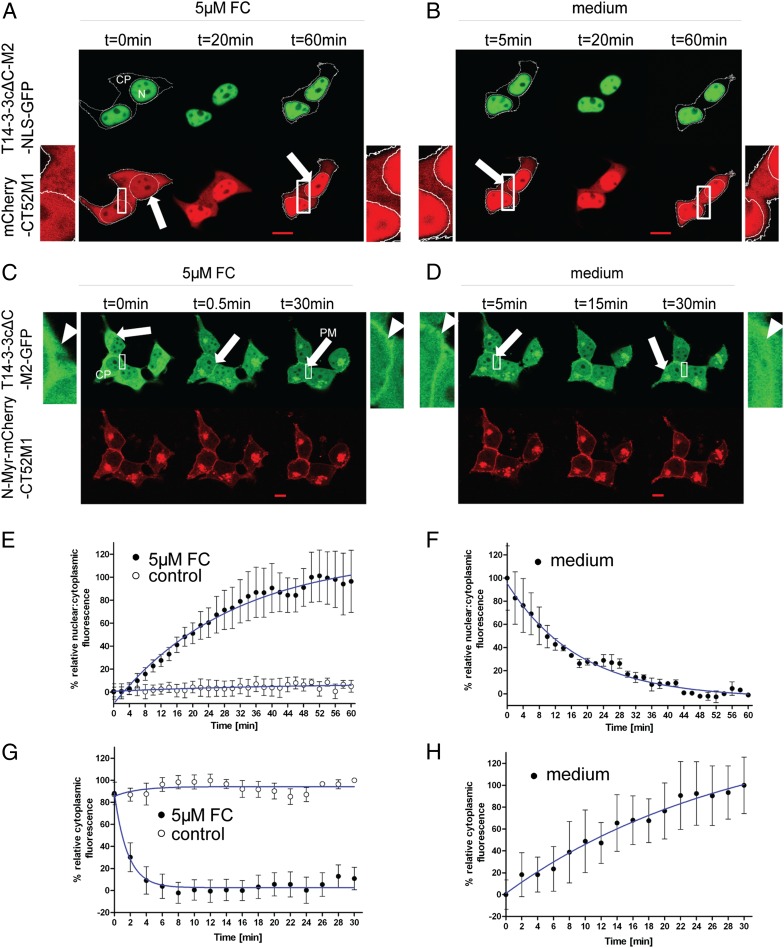
To demonstrate the expandability of the CID system to relocate proteins to different compartments, we used a nuclear localization sequence (NLS) from the cell-cycle phosphatase Cdc25A (268STRSVLKRPERSQEESPPGSTKRRKSMSGA297) (26), which was expressed as a fusion with T14-3-3cΔC-M2-GFP (T14-3-3cΔC-M2-NLS-GFP). When cotransfected with mCherry-CT52M1 in HEK293T cells, this construct was observed exclusively in the nucleus, whereas mCherry-CT52M1 was localized in the nucleus and cytoplasm as described above (Fig. 4A). After the addition of 5 µM FC to the medium of the transfected cells, nuclear fluorescence of mCherry-CT52M1 began to increase within minutes, concomitant with a decrease in its cytoplasmic fluorescence (Fig. 4A and Movie S3). Quantification of the intensity of nuclear and cytoplasmic mCherry-CT52M1 fluorescence showed an increase in the nuclear/cytoplasmic fluorescence, including translocation through the nuclear membrane, reaching the half-maximal effect after 20.7 min (Fig. 4E). This translocation occurred dose dependently with an EC50 of 1.22 ± 0.04 µM FC (SI Appendix, Fig. S8). Untreated transfected cells showed no translocation of mCherry-CT52M1 into the nucleus (Fig. 4E and SI Appendix, Fig. S6B). Furthermore, the nuclear fraction of HEK293T cells transfected with the same plasmids was analyzed by Western blotting before and after FC treatment using an anti-mCherry antibody (SI Appendix, Fig. S9); mCherry accumulation in the nuclear fraction was detected only in the FC-treated cells. We also analyzed the reversibility of the CT52M1 nuclear accumulation as described above and found that this process also is fully reversible after sufficient rinsing with DMEM (Fig. 4 B and F and Movie S4).
Fig. 4.
FC-induced nuclear accumulation of mCherry-CT52M1 and plasma membrane recruitment of T14-3-3cΔC-M2-GFP. (A and B) Time-series images of HEK293T cells transfected with plasmids encoding for T14-3-3cΔC-M2-NLS-GFP and mCherry-CT52M1 at stated times, showing nuclear accumulation of mCherry-CT52M1 fluorescence after the addition of 5 µM FC (A) and its reverse translocation after the cells were rinsed with FC-free medium (B). The additional images on the right and left of the center panel are enlarged from the respective boxed areas. Arrows indicate the relative increase in fluorescence intensity in the nucleus concomitant with the relative decrease in fluorescence intensity in the cytoplasm. (C and D) Corresponding time courses of cotransfected T14-3-3cΔC-M2-GFP and N-Myr-mCherry-CT52M1, showing partial colocalization of both proteins at the plasma membrane (arrows) after the addition of 5 µM FC. Enlarged images of the boxed areas in the center panels show the relative increase in fluorescence intensity at the plasma membrane concomitant with relative decrease in fluorescence intensity in the cytoplasm (arrowheads) (C) and reverse translocation of T14-3-3cΔC-M2-GFP into the cytoplasm after the cells were washed with medium (D). See SI Appendix, Fig. S6 B and C for control cells treated with solvent (3.3% ethanol). (Scale bars, 10 μm.) (E and F) Quantification of mCherry-CT52M1 translocation efficiency in response to 5 µM FC and the analysis of untreated cells (E) and of the corresponding reversibility of mCherry-CT52M1 translocation (F). Mean nuclear and cytoplasmic fluorescent intensities of mCherry-CT52M1 were calculated every 2 min and plotted as a ratio against time. (G and H) Quantification of T14-3-3cΔC-M2-GFP translocation in response to 5 µM FC and the analysis of untreated cells (G) and of the corresponding reversibility T14-3-3cΔC-M2-GFP translocation (H). Here the mean cytoplasmic fluorescent intensities of T14-3-3cΔC-M2-GFP were determined and plotted against time. The data represent mean values (± SEM) from three experiments with at least two cells per experiment; n = 6–14.
We also analyzed whether the CID system can be used to recruit a protein to the plasma membrane. To do so, an N-terminal plasma membrane-targeting sequence (N-Myr) from c-Src kinase (27) (MGSSKSKPKDPSQR) was coupled to mCherry-CT52M1 (N-Myr-mCherry-CT52M1). This construct localized at the plasma membrane and presumably the Golgi apparatus, which is in line with the reported subcellular distribution of c-Src (28). T14-3-3cΔC-M2-GFP was distributed evenly throughout the cell (Fig. 4C). After application of 5 µM FC, GFP fluorescence at the plasma membrane started to increase within a few seconds, and GFP fluorescence in the cytoplasm decreased. Monitoring the time course for 30 min showed colocalization of both proteins at the plasma membrane, suggesting FC-induced dimerization of T14-3-3cΔC-M2-GFP with the plasma membrane-localized N-Myr-mCherry-CT52M1. Hence, plasma membrane recruitment of T14-3-3cΔC-M2-GFP was induced at the addition of FC (Fig. 4C and Movie S5). Quantitative analysis of the intensity of cytoplasmic T14-3-3cΔC-M2-GFP fluorescence showed that the fluorescence intensity of the cytoplasm decreased, reaching the half-maximal effect after 72 s (Fig. 4G). In contrast, untreated cells did not show any changes in the cellular fluorescence of T14-3-3cΔC-M2-GFP at the plasma membrane (Fig. 4G and SI Appendix, Fig. S6C). Recruitment of T14-3-3cΔC-M2-GFP to the plasma membrane was reversed by rinsing the cells with DMEM as described in the previous experiments (Fig. 4 D and H and Movie S6). For easy and individual use of our CID system, we created mammalian eukaryotic expression vectors containing genes for the FC-binding proteins (for plasmid maps, see SI Appendix, Fig. S10).
Nuclear Accumulation of NF-κB (p65) in Response to FC Treatment.
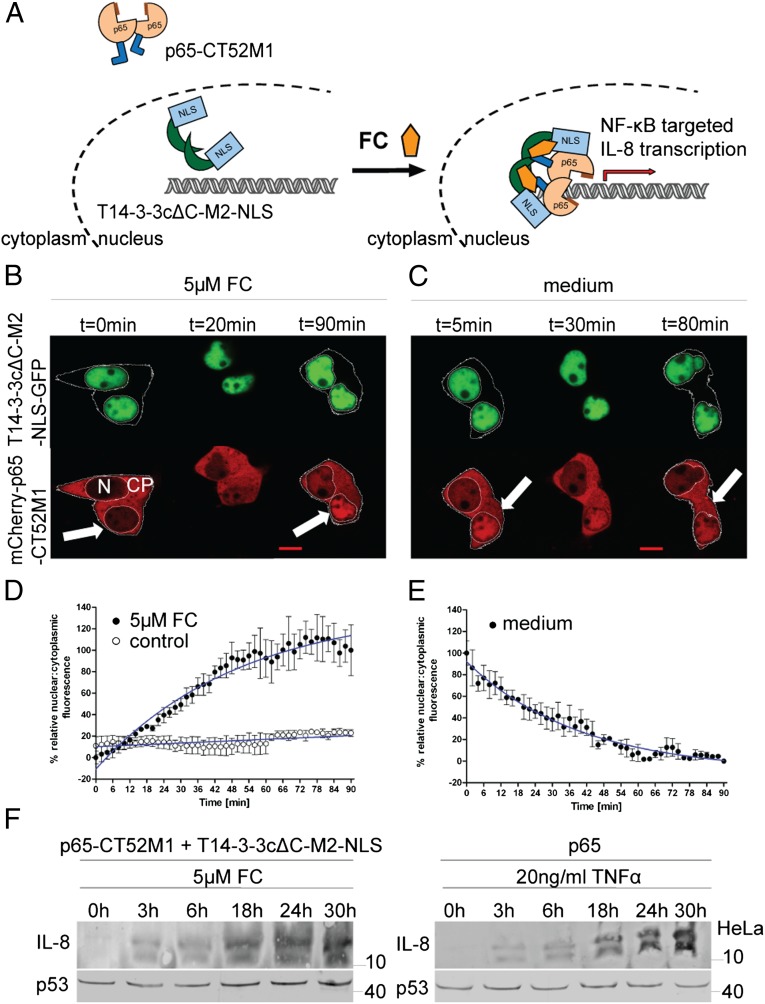
We further tested the applicability of the described CID system by modulating the subcellular localization of the transcription factor NF-κB. NF-κB is assembled as a homo- or heterodimer from a set of different possible subunits, including p65 (RelA), c-Rel, RelB, p50, and p52 (29). In the inactive state, NF-κB is bound to IκB in the cytoplasm (29). Stimulation of the cells by TNF-α leads to phosphorylation of IκB by IκB kinase followed by ubiquitination and then degradation of IκB. This process renders the NLS of NF-κB accessible to the nuclear import machinery (30). After entering the nucleus, NF-κB induces the transcription of subunit-specific target genes that are involved in cellular responses, including immune and inflammatory regulation (SI Appendix, Fig. S11) (30). We fused the NF-κB subunit p65 to mCherry-CT52M1 (mCherry-p65-CT52M1) to translocate it into the nucleus by FC-mediated binding to coexpressed T14-3-3cΔC-NLS-M2-GFP (Fig. 5A). The latter was localized in HEK293T cells in the nucleus as described before, whereas mCherry-p65-CT52M1 was found predominantly in the cytoplasm (Fig. 5B). Addition of 5 µM FC to the medium of the cells did not change the nuclear population of T14-3-3cΔC-NLS-M2-GFP but increased the intensity of nuclear mCherry-p65-CT52M1 fluorescence concomitant with a decrease of its cytoplasmic fluorescence ∼20 min later (Fig. 5B and Movie S7). Differences among cells in the magnitude of this effect may be caused by disparities in transfection and translation efficiencies of the two proteins. The time course of 90 min shows a shift of mCherry-p65-CT52M1 cellular fluorescence, suggesting the formation of an FC-controlled complex of mCherry-p65-CT52M1 and T14-3-3cΔC-NLS-M2-GFP. Subsequent quantification analysis of the intensities of nuclear and cytoplasmic mCherry-p65-CT52M1 fluorescence allowed a deeper insight into this translocation process. The nuclear/cytoplasmic fluorescence ratio increased, reaching the half-maximal effect, including translocation through the nuclear membrane, after 36.5 min (Fig. 5D). As a control, the untreated transfected cells did not show any shifts in the fluorescence intensity of mCherry-p65-CT52M1 (Fig. 5D and SI Appendix, Fig. S6D). Furthermore cells transfected with NLS-GFP or 14-3-3ɛ-NLS-GFP and mCherry-p65-CT52M1 did not show any shifts in the fluorescence intensity of mCherry-p65-CT52M1 with FC treatment (SI Appendix, Fig. S7D). We tested the reversibility of mCherry-p65-CT52M1 nuclear accumulation and demonstrated that this process is reversible, because rinsing with medium causes the mCherry-p65-CT52M1 fluorescence to shift back into the cytoplasm within 90 min (Fig. 5 C and E and Movie S8). Finally, we tested whether FC-induced dimerization and nuclear translocation also would influence the physiological functionality of p65 by monitoring protein secretion of its target gene, IL-8. We cotransfected HeLa cells with p65-CT52M1 and T14-3-3cΔC-M2-NLS and analyzed IL-8 secretion during a time course of 30 h. Increasing IL-8 accumulation after incubation with FC was observed (Fig. 5F), suggesting FC-dependent dimerization of p65-CT52M1 with T14-3-3cΔC-M2-NLS, which in turn drives p65-CT52M1 into the nucleus where p65 induces IL-8 expression (Fig. 5A). Untransfected HeLa cells treated with same amount of FC did not show IL-8 secretion during the same time course (SI Appendix, Fig. S12). The addition of TNF-α to p65-transfected HeLa cells served as a positive control for IL-8 secretion (Fig. 5F), displaying the same time course as the FC-induced effect.
Fig. 5.
FC-induced nuclear accumulation of NF-κB (p65). (A) Schematic representation of FC-induced nuclear accumulation of p65-CT52M1. (B and C) Time-course images of HEK293T cells transfected with plasmids encoding for T14-3-3cΔC-M2-NLS-GFP and mCherry-p65-CT52M1 at stated times, monitoring nuclear accumulation of mCherry-p65-CT52M1 fluorescence after the addition of 5 µM FC (B) and its reverse translocation after the cells were washed with medium (C). See SI Appendix, Fig. S6D for control cells treated with solvent (3.3% ethanol). (Scale bars, 10 μm.) CP, cytoplasm; N, nucleus. (D and E) Quantification of mCherry-p65-CT52M1 translocation in response to 5 µM FC and the analysis of untreated cells (control) (D) and of the reversibility of mCherry-p65-CT52M1 translocation (E). Mean nuclear and cytoplasmic fluorescent intensities were analyzed as described in Fig. 4 E and F. (F) Induction of IL-8 expression by FC in HeLa cells cotransfected with plasmids encoding for T14-3-3cΔC-M2-NLS and p65-CT52M1 demonstrated by Western blotting. Secreted IL-8 was detected upon treatment with 5 µM FC during a time course of 30 h (Left). For a positive control, p65-transfected HeLa cells were treated with 20 ng/mL TNF-α for the same time course (Right). Endogenous p53 was used as a loading control.
Discussion
In this study we developed a CID system based on FC as a dimerizer molecule. Because FC can bind simultaneously to two different proteins, it can be added to the still limited number of known heterodimerizers (for an overview of CID systems, see SI Appendix, Table S2). One of the advantages of using FC as a chemical tool is that it binds with significant affinity only to the binary complex of T14-3-3cΔC and CT52 but is unable to bind to each single component independently (12, 17), thus avoiding the likelihood of nonproductive interactions that often are observed in other CID systems (1–3, 6, 8–10, 31). A further advantage of this FC-based CID system is that CT52M1 is a short peptide of 6.5 kDa, minimizing possible steric interferences during complex formation. Hence, CT52M1 is a tag that can be fused in a single copy to a target protein of choice without interfering with biological functions. Among the components of the current CID systems, CT52M1 shows the smallest molecular weight. In fact, our example of the FC-induced nuclear import of a p65-CT52M1 fusion followed by IL-8 secretion clearly demonstrates the preservation of physiological functionality of the fused target protein. In the FC-dependent system we have shown that the exogenously applied effective concentration of 5 µM FC can be reduced to 0.625 µM (EC50 = 1.22 µM FC) (SI Appendix, Fig. S8). The effect can repeatedly be fully reversed simply by rinsing the cells with FC-free medium, thereby allowing the target protein to return to its original localization in the cell (Movies S9–S12). To the best of our knowledge, such a gentle reversibility has not been reported with current CID systems. The FK1012-induced homodimerization of three molecules of FKBP that are coupled to the intracellular domain of the TCR could be reversed by addition of FK506 (FK1012 monomer) (1). Excess FK506 competes with FK1012 for the FKBP-binding groove and replaces FK1012. Ho et al. (2) showed that the FK506-induced heterodimerization of FKBP with calcineurin can be prevented by rapamycin. Rapamycin binds FKBP but not calcineurin. As described above, both ligands, FK506 and rapamycin, strongly influence cell physiology by binding to endogenous FKBP. The use of these molecules as inverters of the dimerization therefore is critical.
To minimize potentially harmful interactions of the CT52 construct with endogenous 14-3-3 proteins of the target cells, we conducted a complementary charge exchange of an electrostatically engaged amino acid pair in the T14-3-3cΔC/CT52–binding interface. The K943D substitution in CT52 was sufficient to abrogate completely the binding to human wild-type 14-3-3 proteins in the presence of FC. However, an EC50 of 8.30 ± 0.01 µM was measured for the pair of engineered proteins T14-3-3cΔC-E19R and CT52-K943D, demonstrating that the essential salt bridge contact between T14-3-3cΔC and CT52 is conserved in the presence of these complementary amino acid substitutions and still allows effective complex formation. The ability to bind to endogenous, phosphorylated 14-3-3 partner proteins could cause unwanted side effects in experiments with mammalian cells. Therefore, additional engineering efforts were necessary to abrogate this phosphorylation-dependent binding. The replacement of Y137 by a phenylalanine in T14-3-3cΔC-M1 was sufficient to impede the interaction of T14-3-3cΔC with phosphorylated native binding partners, presumably because of the loss of an important H-bond to the phosphorylated moiety of the respective native binding partners (Fig. 2A). Because the hydroxyl moiety of Y137 also interacts with K56 in the basic binding pocket of T14-3-3c, the Y137F exchange might further affect the complex stabilizing influence of this rather flexible lysine residue and thus cause the loss of two amino acid contacts to the phosphorylated moiety of the native binding partners. The results further show that a coordination of the phosphorylated moiety by the remaining R63 and R136 residues of the T14-3-3cΔC-M1–binding pocket is not possible and reveals that Y137 within the 100%-conserved basic pocket of 14-3-3 proteins is essential for the coordination of the phosphorylated binding partners. Furthermore, one effect of the Y137F exchange is a change in the strictly cytoplasmic subcellular localization of T14-3-3cΔC-M1 to an even distribution of T14-3-3cΔC-M2 throughout the cell. One explanation for this effect could be that the interaction of T14-3-3cΔC-M2 with cytosolic partner proteins such as cofilin (32) and vimentin (33) is abolished. This effect is similar that reported by Paul et al. (34), who demonstrated that the subcellular localization of different 14-3-3 proteins is dependent on their ability to interact with their partner proteins. Abrogation of this interaction with a chemical compound (AICAR) or a high-affinity 14-3-3–binding peptide (R18) led to a more even distribution of GFP-14-3-3 isoforms in different plant cell types. Taken together, these results show that the interface of the FC-binding proteins CT52 and T14-3-3cΔC was engineered successfully so that interactions of human 14-3-3 proteins with CT52M1 and the interactions of endogenous proteins with T14-3-3cΔC-M2 are abolished in the targeted cell. However, we cannot fully rule out interactions of T14-3-3cΔC-M1, which is needed for nuclear exclusion of a target protein, with endogenous partners in mammalian cells.
Because the natural target of FC is the complex of a plant 14-3-3 protein and the C-terminal regulatory domain of PMA, which also is specific for higher plants, unwanted effects of the chemical dimerizer itself in mammalian cell physiology are not likely. The 14-3-3–binding motif of PMA compared with conventional motifs, defined as mode I (RSX-pS/T-XP) and mode II (RXXX-pS/T-XP) (35), is special, because it ends after the +1 amino acid (YpTV-COOH) and is referred to as “mode III” (12). In human cells, of the more than 200 known 14-3-3–interacting proteins, only five are reported to carry mode III-recognition motifs (19): the IL-9 receptor (IL9R), the K+ channel TASK3, platelet glycoprotein Ibα (GpIbα), the G protein-coupled receptor GBR15, and the SLIT- and TRK-like family member 1 (SLITRK1). Of these, SLITRK1 and IL9R display an aspartate or phenylalanine, respectively, at the +1 position, and neither is compatible with FC, probably because their rather bulky side chains impede binding of FC to the binary complex (14-3-3/IL9R, 14-3-3/SLITRK1). The other three proteins contain either a valine or a leucine; these amino acids also are found at this position in different PMA isoforms and therefore probably are addressable by FC when complexed with 14-3-3. Compared with other chemical dimerizers, the number of possible interfering interactions is fairly low. We certainly cannot rule out unknown off-target effects of FC, but we have not experienced any deleterious effects of FC in HEK293T or in HeLa cells up to a concentration of 60 µM. Because much lower concentrations of FC (0.625–5 µM) are sufficient for induction of dimerization, we do not expect significant adverse effects that would call the observations into question. We demonstrated the usability of this CID system to induce a physiologically important cellular response by translocating the transcription factor NF-κB (p65) into the nucleus and thereby activating NF-κB signaling, resulting in target gene expression examined by the example of IL-8 biosynthesis. The continuous increase in IL-8 expression was in line with the time course of IL-8 expression induced by TNF-α, the physiological activator of the NF-κB pathway. The CID system is especially interesting for the analysis of signaling pathways that are dependent on the nuclear import of one component (NF-κB, nuclear receptors, or β-catenin), because it potentially enables the experimental distinction between the mere fact of nuclear localization and the liberation of regulatory proteins such as IκB, HSP90, or axin. Furthermore, because this analysis is put under the chemical control of an exogenous applicable dimerizer, the role of the different subcellular localizations can be studied in every developmental stage or physiological context of a given biological system. FC-induced secretion of protein factors also could be used in a clinical setting, for example to control chemically the release of insulin or human growth hormone.
Here we have reported a CID system that complements available CID systems with dimerizers such as rapamycin, ABA, or GA3-AM potentially to address several different signaling pathway components in parallel, because our CID system does not overlap mechanistically with any of the already available ones. Taken together, the FC-dependent CID system and its engineered binding proteins CT52M1/T14-3-3cΔC-M2 described in this study represent a widely applicable and reversible chemical dimerization tool that does not cause apparent side effects in mammalian physiology.
Methods
Peptide Synthesis.
The phosphopeptides were synthesized by GenScript (with the exception of Cdc25ApS178, which was purchased from Caslo), each with purity >95% (wt/wt). All three peptides were resuspended in distilled water to a final concentration of 2 mM.
Far-Western Blot Analysis.
Ten micrograms of purified GST-fused CT66 variants were separated by 15% SDS/PAGE after boiling before the gel was loaded and were transferred to nitrocellulose membrane. Nonspecific sites were blocked by incubation for 1 h at room temperature with 5% (wt/vol) dry milk in 50 mM Tris⋅HCl (pH 7.8), 150 mM NaCl, and 1 mM MgCl2 (Tris-buffered saline, TBS). The membrane was incubated overnight at 4 °C with purified His6-tagged 14-3-3 proteins diluted to 500 µg/mL in 20 mM Hepes/NaOH (pH 7.5), 20% (wt/vol) glycerol, 2 mM MgCl2, and 0.5 mM Tris (2-carboxyethyl) phosphine (TCEP) in the presence of 5 µM FC or solvent (3.3% ethanol, vol/vol) under gentle agitation. After washing with TBS, bound His6-tagged 14-3-3 was visualized by immunodetection with mouse monoclonal anti–RGS-His6 antibody (Qiagen) in combination with goat anti-mouse antibody conjugated with alkaline phosphatase (Cell Signaling Technology) and the enzyme substrates 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium (BCIP/NBT). The antibodies were used at a dilution of 1:1,000 in 2% BSA in TBS.
Western Blot Analysis.
IL-8 secretion was analyzed for 38 h after transient expression of HeLa cells cotransfected with T14-3-3cΔC-M2-NLS and p65-CT52M1 that were treated with 5 µM FC. Control cells cotransfected with p65 were treated with 20 ng/mL TNF-α. After 0, 3, 6, 18, 24, and 30 h PMSF was added to the medium to a final concentration of 0.5 mM; then the medium was collected and evaporated to a final volume of 1 mL. Medium samples (150 µg), boiled prior loading, were run on 15% (wt/vol) SDS/PAGE gels and transferred to PVDF membrane. Nonspecific sites were blocked as described above. IL-8 was detected by mouse monoclonal anti–IL-8 antibody (Abcam) in combination with the secondary antibody and detection method described previously. The antibodies were used at a dilution of 1:1,000 in 2% (wt/vol) BSA in TBS.
SPR Analysis.
The measurements were carried out at 25 °C in a BiaCore T100 system (GE Healthcare Life Sciences). To measure the EC50 of FC on each CT52/14-3-3 complex, CT52 variants were covalently immobilized onto a CM5 biosensor chip according to the manufacturer’s instructions. FC was titrated at the indicated concentrations stepwise in 1:1 dilutions in the presence of 10 µM purified 14-3-3 proteins and was injected over a time course of 180 s (contact time) with a flow rate of 30 µL/min. Subsequently the FC/14-3-3 solution was washed for 180 s (dissociation time) with 10 mM Hepes (pH 7.4), 150 mM NaCl, 2 mM MgCl2, and 0.05% Tween20. The sensor chip was regenerated after each injection with 2 M MgCl2. Nonspecific binding was eliminated by subtracting the background signal of the reference cell from the immobilized cell signals at each injection. To calculate EC50, the response units (RU) were plotted against the corresponding FC concentration on a logarithmic scale, and the resulting curves were fitted using steady-state kinetics. To measure the binding affinity of phosphopeptides to 14-3-3 proteins, 14-3-3 variants were immobilized onto the CM5 chip, and the phosphopeptides were titrated at the indicated concentrations stepwise in 1:1 dilutions. To determine the Kd values, the RUs were plotted against the corresponding peptide concentration, and the resulting curves were fitted using steady-state kinetics. For statistical analysis and curve fitting, GraphPad Prism was used. Each point represents the mean of three measurements; error bars represent the SEM.
Cell Lines, Culture Conditions, and Transfection.
HEK293Tand HeLa cells were obtained from the American Type Culture Collection. HEK293T cells were cultured in DMEM supplemented with 10% (vol/vol) FBS and 0.3% antibiotics at 37 °C in 10% CO2 atmosphere. HeLa cells were grown in Minimum Essential Medium with Earle’s salts, plus 10% (vol/vol) FBS, 0.3% antibiotics, and 1% (wt/vol) nonessential amino acids at the same culture conditions. For confocal microscopy HEK293T cells were plated on 10-mm glass coverslips in 24-well plates at 6 × 104 cells per well in 1 mL medium. After 24 h, cells were transiently transfected with appropriate plasmids using Fugene 6 (Roche) according to the manufacturer’s instruction. The optimized ratio of DNA:Fugene 6 used for such transfections was 200 ng DNA with 1 µL Fugene 6. For Western blot analysis HeLa cells were plated in six-well plates at 2.5 × 105 cells per well in 2 mL medium and were grown for 24 h before transfection. Cells were transfected using Fugene 6, at an optimized ratio of 1 µg DNA to 5 µL Fugene 6 per well. The DNA concentration used for single transfections also was used for dual transfections.
Live-Cell Imaging, Microscopy, and Image Analysis.
Confocal microscopy was carried out 38 h after transient expression in a humidified CO2 incubator (37 °C, 10% CO2) using a Leica TCS SP2. Excitation of GFP was performed using an argon ion laser at 488 nm, and mCherry fluorescence was excited using a green GreNe ion laser at 543 nm. HEK293T cells were treated with FC just before detection by replacing 1/10th of the medium volume in the dish with an appropriate concentration of FC. Data analysis was carried out with ImageJ version 1.46a and involved determination of mean fluorescence intensities for nucleus, cytoplasm, and plasma membrane every 2 min.
Supplementary Material
Acknowledgments
We thank Prof. Michael R. H. White for the p65 vector, Prof. Alfred Wittinghofer for helpful discussions, and Dr. Martin Winkler and Dr. Jeffrey Simard for critical reading of the manuscript. We appreciate the work done by the Dortmund Protein Facility (Max Planck Institute of Molecular Physiology). This work was supported by a Ph.D. scholarship from the Max Planck Institute of Molecular Physiology (to M.S.). C.O. received support from AstraZeneca, Bayer CropScience, Bayer Healthcare, Boehringer Ingelheim, and Merck KGaA.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.J.L. is a guest editor invited by the Editorial Board.
See Author Summary on page 1579 (volume 110, number 5).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212990110/-/DCSupplemental.
References
- 1.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262(5136):1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 2.Ho SN, Biggar SR, Spencer DM, Schreiber SL, Crabtree GR. Dimeric ligands define a role for transcriptional activation domains in reinitiation. Nature. 1996;382(6594):822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 3.Rivera VM, et al. A humanized system for pharmacologic control of gene expression. Nat Med. 1996;2(9):1028–1032. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- 4.Putyrski M, Schultz C. Protein translocation as a tool: The current rapamycin story. FEBS Lett. 2012;586(15):2097–2105. doi: 10.1016/j.febslet.2012.04.061. [DOI] [PubMed] [Google Scholar]
- 5.Liberles SD, Diver ST, Austin DJ, Schreiber SL. Inducible gene expression and protein translocation using nontoxic ligands identified by a mammalian three-hybrid screen. Proc Natl Acad Sci USA. 1997;94(15):7825–7830. doi: 10.1073/pnas.94.15.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belshaw PJ, Ho SN, Crabtree GR, Schreiber SL. Controlling protein association and subcellular localization with a synthetic ligand that induces heterodimerization of proteins. Proc Natl Acad Sci USA. 1996;93(10):4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clemons PA et al. (2002) Synthesis of calcineurin-resistant derivatives of FK506 and selection of compensatory receptors. Chem Biol 9(1):49–61. [DOI] [PubMed]
- 8.Farrar MA, Alberol-Ila J, Perlmutter RM. Activation of the Raf-1 kinase cascade by coumermycin-induced dimerization. Nature. 1996;383(6596):178–181. doi: 10.1038/383178a0. [DOI] [PubMed] [Google Scholar]
- 9.Liang F-S, Ho WQ, Crabtree GR. Engineering the ABA plant stress pathway for regulation of induced proximity. Sci Signal. 2011;4(164):rs2. doi: 10.1126/scisignal.2001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyamoto T, et al. Rapid and orthogonal logic gating with a gibberellin-induced dimerization system. Nat Chem Biol. 2012;8(5):465–470. doi: 10.1038/nchembio.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ballio A, et al. Fusicoccin: A new wilting toxin produced by Fusicoccum amygdali del. Nature. 1964;203:297–297. [Google Scholar]
- 12.Würtele M, Jelich-Ottmann C, Wittinghofer A, Oecking C. Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J. 2003;22(5):987–994. doi: 10.1093/emboj/cdg104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svennelid F, et al. Phosphorylation of Thr-948 at the C terminus of the plasma membrane H(+)-ATPase creates a binding site for the regulatory 14-3-3 protein. Plant Cell. 1999;11(12):2379–2391. doi: 10.1105/tpc.11.12.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermeking H, Benzinger A. 14-3-3 proteins in cell cycle regulation. Semin Cancer Biol. 2006;16(3):183–192. doi: 10.1016/j.semcancer.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Bridges D, Moorhead GBG. 14-3-3 proteins: A number of functions for a numbered protein. Sci STKE. 2005;2005(296):re10. doi: 10.1126/stke.2962005re10. [DOI] [PubMed] [Google Scholar]
- 16.Morrison DK. The 14-3-3 proteins: Integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009;19(1):16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ottmann C, et al. Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+ -ATPase by combining X-ray crystallography and electron cryomicroscopy. Mol Cell. 2007;25(3):427–440. doi: 10.1016/j.molcel.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 18.Truong AB, Masters SC, Yang H, Fu H. Role of the 14-3-3 C-terminal loop in ligand interaction. Proteins. 2002;49(3):321–325. doi: 10.1002/prot.10210. [DOI] [PubMed] [Google Scholar]
- 19.Johnson C, et al. Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem J. 2010;427(1):69–78. doi: 10.1042/BJ20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rittinger K, et al. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol Cell. 1999;4(2):153–166. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- 21.Molzan M, et al. Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling. Mol Cell Biol. 2010;30(19):4698–4711. doi: 10.1128/MCB.01636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conklin DS, Galaktionov K, Beach D. 14-3-3 proteins associate with cdc25 phosphatases. Proc Natl Acad Sci USA. 1995;92(17):7892–7896. doi: 10.1073/pnas.92.17.7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15(10):1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schumacher B, Skwarczynska M, Rose R, Ottmann C. Structure of a 14-3-3σ-YAP phosphopeptide complex at 1.15 A resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66(Pt 9):978–984. doi: 10.1107/S1744309110025479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Kelly I, Butler MH, Zilberberg N, Goldstein SAN. Forward transport. 14-3-3 binding overcomes retention in endoplasmic reticulum by dibasic signals. Cell. 2002;111(4):577–588. doi: 10.1016/s0092-8674(02)01040-1. [DOI] [PubMed] [Google Scholar]
- 26.Källström H, Lindqvist A, Pospisil V, Lundgren A, Rosenthal CK. Cdc25A localisation and shuttling: Characterisation of sequences mediating nuclear export and import. Exp Cell Res. 2005;303(1):89–100. doi: 10.1016/j.yexcr.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Cross FR, Garber EA, Pellman D, Hanafusa H. A short sequence in the p60src N terminus is required for p60src myristylation and membrane association and for cell transformation. Mol Cell Biol. 1984;4(9):1834–1842. doi: 10.1128/mcb.4.9.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiu VK, et al. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4(5):343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 29.Beg AA, Baldwin AS., Jr The I κ B proteins: Multifunctional regulators of Rel/NF-κ B transcription factors. Genes Dev. 1993;7(11):2064–2070. doi: 10.1101/gad.7.11.2064. [DOI] [PubMed] [Google Scholar]
- 30.Wu JT, Kral JG. The NF-kappaB/IkappaB signaling system: A molecular target in breast cancer therapy. J Surg Res. 2005;123(1):158–169. doi: 10.1016/j.jss.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Czlapinski JL, et al. Conditional glycosylation in eukaryotic cells using a biocompatible chemical inducer of dimerization. J Am Chem Soc. 2008;130(40):13186–13187. doi: 10.1021/ja8037728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gohla A, Bokoch GM. 14-3-3 regulates actin dynamics by stabilizing phosphorylated cofilin. Curr Biol. 2002;12(19):1704–1710. doi: 10.1016/s0960-9822(02)01184-3. [DOI] [PubMed] [Google Scholar]
- 33.Satoh J-I, Yamamura T, Arima K. The 14-3-3 protein epsilon isoform expressed in reactive astrocytes in demyelinating lesions of multiple sclerosis binds to vimentin and glial fibrillary acidic protein in cultured human astrocytes. Am J Pathol. 2004;165(2):577–592. doi: 10.1016/s0002-9440(10)63322-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paul A-L, Sehnke PC, Ferl RJ. Isoform-specific subcellular localization among 14-3-3 proteins in Arabidopsis seems to be driven by client interactions. Mol Biol Cell. 2005;16(4):1735–1743. doi: 10.1091/mbc.E04-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yaffe MB, et al. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91(7):961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]