Abstract
Proteins denature not only at high, but also at low temperature as well as high pressure. These denatured states are not easily accessible for experiment, because usually heat denaturation causes aggregation, whereas cold or pressure denaturation occurs at temperatures well below the freezing point of water or pressures above 5 kbar, respectively. Here we have obtained atomic details of the pressure-assisted, cold-denatured state of ubiquitin at 2,500 bar and 258 K by high-resolution NMR techniques. Under these conditions, a folded, native-like and a disordered state exist in slow exchange. Secondary chemical shifts show that the disordered state has structural propensities for a native-like N-terminal β-hairpin and α-helix and a nonnative C-terminal α-helix. These propensities are very similar to the previously described alcohol-denatured (A-)state. Similar to the A-state, 15N relaxation data indicate that the secondary structure elements move as independent segments. The close similarity of pressure-assisted, cold-denatured, and alcohol-denatured states with native and nonnative secondary elements supports a hierarchical mechanism of folding and supports the notion that similar to alcohol, pressure and cold reduce the hydrophobic effect. Indeed, at nondenaturing concentrations of methanol, a complete transition from the native to the A-state can be achieved at ambient temperature by varying the pressure from 1 to 2,500 bar. The methanol-assisted pressure transition is completely reversible and can also be induced in protein G. This method should allow highly detailed studies of protein-folding transitions in a continuous and reversible manner.
Keywords: protein unfolding, thermodynamics, protein dynamics, heteronuclear NMR
It has long been known that proteins unfold not only at high temperatures, but also at high pressures (1) as well as low temperatures (2). The so-called heat, pressure, and cold denaturations can be described in a unified way by Hawley’s theory (1, 3). This theory assumes a simplified two-state model of protein unfolding, where the free energy difference between folded and unfolded states is a general parabolic function of temperature and pressure:
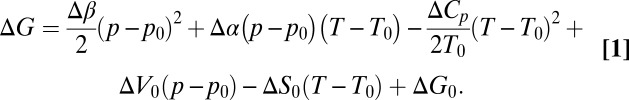 |
In Eq. 1 Δ indicates the difference of the respective value between the denatured and the native state; β is the compressibility factor 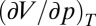 ; α is the thermal expansivity factor
; α is the thermal expansivity factor 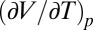 ; Cp is the heat capacity
; Cp is the heat capacity 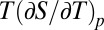 ; and p0, T0 is an arbitrarily chosen reference point. The phase boundary between denatured and native states is then given by the condition ΔG = 0, which corresponds to a tilted ellipse within the pT plane for commonly observed values of Δβ < 0, ΔCp > 0, and Δα > 0. This two-state model is clearly an oversimplification, because both folded and unfolded states can be heterogeneous, and due to the paucity of data it is also unclear whether the heat-, cold-, and pressure-denatured states are identical. Nevertheless the model presents a valuable general framework to pinpoint the main contributing thermodynamic entities, which in the case of ΔCp and ΔS0 are clearly linked to protein hydration and the hydrophobic effect (4–8).
; and p0, T0 is an arbitrarily chosen reference point. The phase boundary between denatured and native states is then given by the condition ΔG = 0, which corresponds to a tilted ellipse within the pT plane for commonly observed values of Δβ < 0, ΔCp > 0, and Δα > 0. This two-state model is clearly an oversimplification, because both folded and unfolded states can be heterogeneous, and due to the paucity of data it is also unclear whether the heat-, cold-, and pressure-denatured states are identical. Nevertheless the model presents a valuable general framework to pinpoint the main contributing thermodynamic entities, which in the case of ΔCp and ΔS0 are clearly linked to protein hydration and the hydrophobic effect (4–8).
The cold-denatured state of proteins is usually not easily accessible because cold denaturation temperatures are almost always below the freezing point of water. An exception has been reported for the protein L9, where the reduction of hydrophobic interactions by a destabilizing leucine to alanine point mutation led to a heterogeneous cold-denatured state above 273 K with native and nonnative structural elements (9). Supercooled solutions of native proteins have been investigated by NMR by Szyperski and Mills in small capillaries that prevent water freezing (10). However, only in the case of the cold-shock protein of Bacillus caldolyticus, cold denaturation could be observed, but no structural data were presented (11). For ubiquitin, cold denaturation under normal pressure has not been observed down to temperatures of ∼233 K in small capillaries (12) or in micrometer-scale water-in-oil emulsions (13). In contrast, in a nanoscale reverse micelle system progressive heterogeneous disappearance of native state ubiquitin NMR resonances was already observed at temperatures below 253 K, but no new resonances of a cold-denatured state could be detected (14). Apparently, the much reduced water entropy in the small micelle volume increases the cold denaturation temperature. However, the confinement of the protein in the micelle also limits conformational averaging such that heterogeneous populations disappear in the noise.
The application of high pressure presents a further strategy to move cold denaturation into an observable range, because it increases the cold denaturation temperature according to Eq. 1 and its associated parameters and at the same time reduces the freezing point of water. Thus, water can be kept liquid down to 251 K at a pressure of 2.07 kbar (3). Pressure-assisted cold denaturation has been followed for ubiquitin (15–17), using high-pressure quartz NMR tubes. The analysis of chemical shift data, resonance intensities, and 15N relaxation showed that the pressure-assisted cold unfolding of ubiquitin is not a simple two-state process, but that several intermediates exist at 3 kbar and pH 4.6. At 273 K one intermediate is locally unfolded in the segments of residues 33–42 and 70–76. At a pressure of 2 kbar and temperatures below 273 K (18), a further set of resonances is observed that was identified with a floppy unfolded state. However, due to the small sample volume of about 15 μL of the conventional pressure cells used, sensitivity was severely limited and no structural information on the pressure-assisted cold-denatured state was derived.
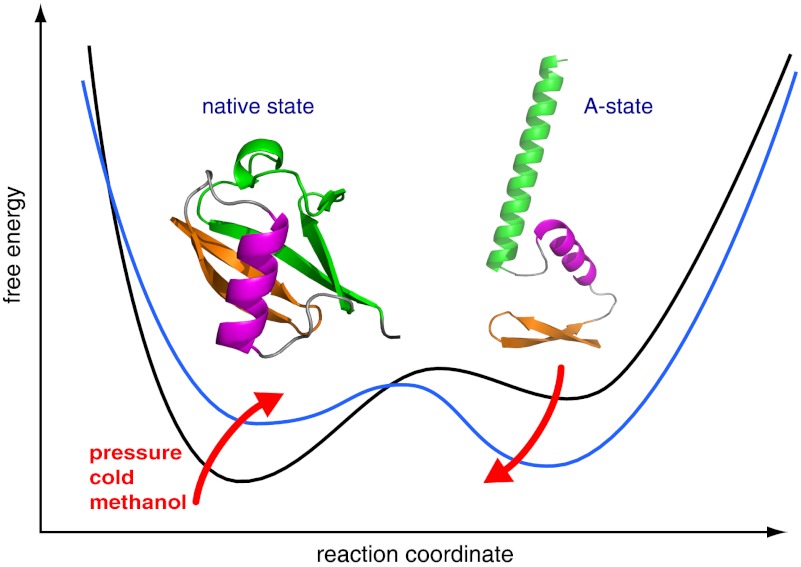
Here, we have characterized the pressure-assisted cold denaturation of ubiquitin, using a newly developed high-pressure NMR cell (Daedalus Innovations LLC) that provides much higher sensitivity by a sample volume of ∼120 μL. The obtained sequence-specific chemical shift assignments and 15N relaxation data at pressures from 1 to 2,500 bar show that the cold-denatured state is an unfolded ensemble with a high propensity for a first β-hairpin and α-helix that are similar to the native state. In contrast, the C terminus has high propensity for a nonnative second α-helix. These structural propensities are very similar to ubiquitin’s A-state (19, 20), which is induced by 60% methanol at pH 2 (21) at ambient temperature, as well as to ubiquitin’s urea-denatured state (22, 23). The close similarity between the pressure-assisted cold-denatured state and the A-state is corroborated by the fact that the transition from a native to an A-state–like NMR spectrum also can be induced at pH 4.5 and 45% methanol at 308 K by the application of pressure. All of the described forms of destabilization of ubiquitin’s native state seem to lead to a similar unfolded-state ensemble, suggesting a robust folding pathway via ubiquitin’s first β-hairpin and α-helix.
The high similarity of methanol- and pressure-destabilized states may point to a link between pressure unfolding and the hydrophobic effect. A similar transition from the native to a partially unfolded state can be induced for protein G in the presence of methanol at a pressure of 2,500 bar and ambient temperatures.
Results
Resonance Assignments.
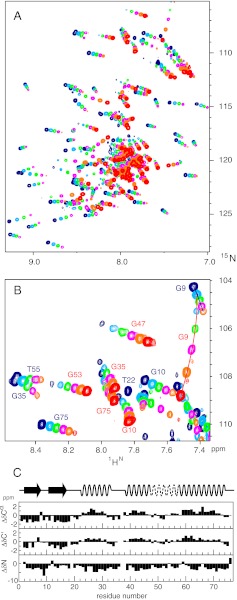
When ubiquitin is cooled from ambient to low temperatures at 2,500 bar (pH 6.5), the onset of cold denaturation becomes visible in 1H-15N heteronuclear single quantum coherence (HSQC) spectra at about 278 K by the appearance of a new set of resonances in a narrow spectral region typical for the expected disordered polypeptide of the cold-denatured state. At 258 K both the native and the cold-denatured state show a maximum number of cross peaks at about equal intensities (Fig. 1). The spectral changes induced by pressure and cold temperature are completely reversible. Apparently at 258 K and 2,500 bar the cold-denatured and the native state coexist in an approximate 1:1 equilibrium in slow exchange on the chemical shift scale. No cross peaks could be detected in an exchange spectroscopy (EXSY) experiment with a mixing time of 3.2 s, which sets a lower limit to the exchange time of about 20 s.
Fig. 1.
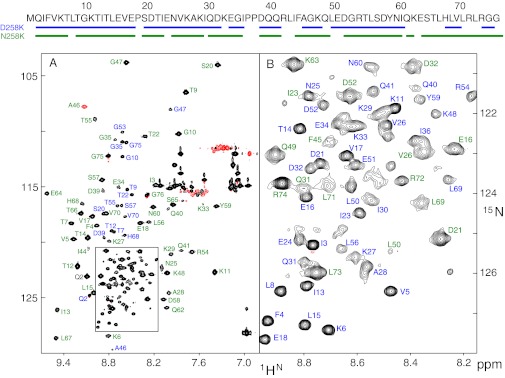
(A) 1H-15N HSQC spectrum of uniformly 15N/13C-labeled ubiquitin at 258 K and 2,500 bar (pH 6.5). Peaks are labeled with assignment information for the folded state (green) and the cold-denatured state (blue). Negative contour lines are shown in red. The amino acid sequence of ubiquitin is indicated at the top with assigned residues underlined for both states. The spectrum was acquired for 1.5 h at a sample concentration of 1 mM on an 800-MHz spectrometer. (B) Enlarged region of the box shown in A.
Assignments of the folded state at 258 K and 2,500 bar were obtained by following the 1H-15N-HSQC spectra from ambient conditions through a combined pressure and temperature path (298 K/1 bar → 298 K/2,500 bar → 258 K/2,500 bar). As the temperature is decreased, the folded-state cross peaks broaden to some extent in a heterogeneous way with disappearance of a few resonances, indicating varied disorder of these residues. In total 61 backbone resonances could be assigned (Fig. 1). Differential broadening of the folded state by pressure has been observed before at room temperature (pH 4.6, 3,000 bar), using 15N transverse relaxation measurements (17). At ambient and at low temperature, the affected residues are located mainly in the region D21–I44, indicating the destabilization of ubiquitin’s α-helix and of the irregular region connecting the α-helix to strand β3 by high pressure.
Resonances of the cold-denatured state were assigned from a combination of triple-resonance 3D experiments. The quality of these spectra at 258 K and 2,500 bar is very good and allowed the unambiguous backbone resonance assignment for 57 residues in the cold-denatured state and also confirmed the assignment of the folded state. These data cover 80% of all nonproline residues. Unassigned residues in the denatured state include the N-terminal methionine and several residues in the C-terminal tail (Fig. 1).
Secondary Structure Information from 13Cα, 13C′, and 15N Chemical Shifts.
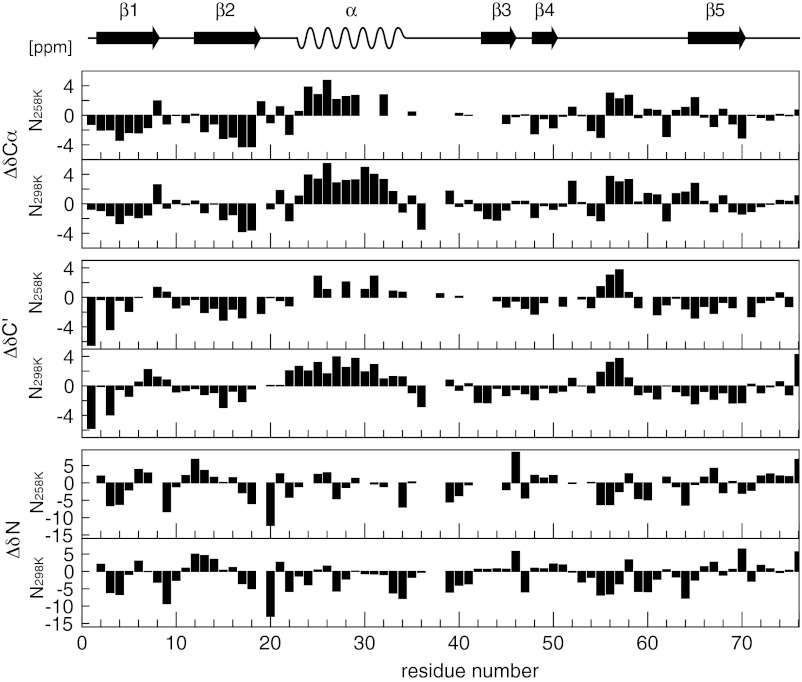
The deviations of observed chemical shifts for backbone nuclei from their random coil values, known as secondary chemical shifts (Δδ = δobs − δrc), can be used as a fast and accurate measure of polypeptide secondary structure (24, 25). Not surprisingly, the secondary shifts ΔδCα, ΔδC′, and ΔδN for the folded state at 258 K, 2,500 bar and at 298 K, 1 bar are almost identical (Fig. 2), indicating that the native state backbone conformation is almost completely preserved even at low temperature and high pressure. It is noted that referencing of the chemical shifts was achieved indirectly via the resonance frequency of water (26), which was calibrated to trimethylsilyl propionate in a separate experiment yielding a temperature and pressure dependence of the form δH2O(p, T)/ppm = 7.866–1.612·10−4 bar−1·p –1.025·10−2 K−1·T + 5.945·10−7 bar-1K−1·p·T (Fig. S1 and Table S1). The good agreement of the native state secondary shifts under both conditions indicates that this method is also adequate for obtaining secondary structure information at 258 K and 2,500 bar.
Fig. 2.
Secondary chemical shifts analysis (13Cα, 13C′, 15N) of the native state of ubiquitin at low temperature and high pressure (258 K, 2,500 bar) and at ambient conditions (293 K, 1 bar, pH 6.6). Secondary structure elements of the native state are drawn at the top.
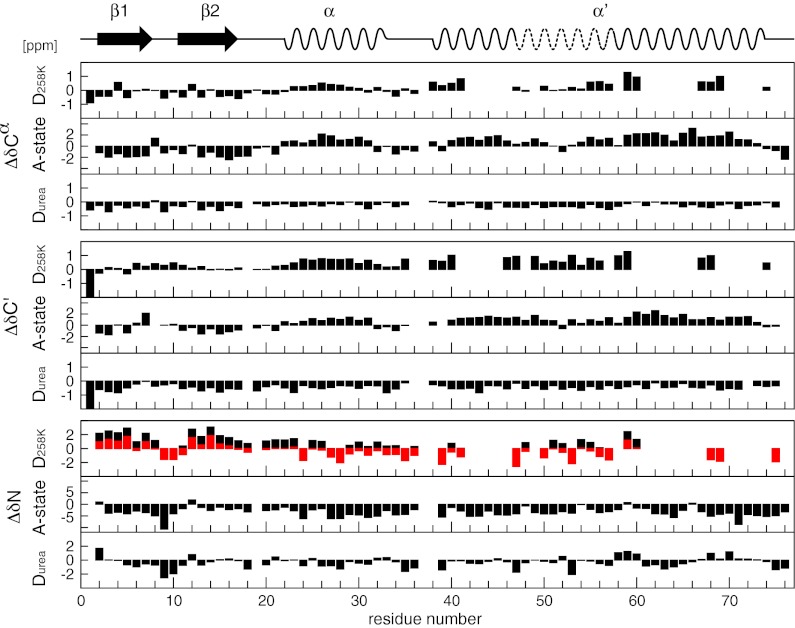
Fig. 3 shows the secondary shifts ΔδCα, ΔδC′, and ΔδN for the pressure-assisted cold-denatured state at 258 K, 2,500 bar. The negative secondary shifts for 13Cα and 13C′ in the region of residues 1–18 indicate a propensity for an extended β-type structure reminiscent of the native first β-hairpin. This is also supported by the 15N secondary shifts, which are positive in this region and show a dip for residues 9–11, indicative of a β-turn. It is revealing to compare these secondary shifts to respective secondary shifts of the A-state (19, 20) and the urea-denatured state (22) (Fig. 3). Both the A-state and the urea-denatured state have a very similar pattern to that of the cold-denatured state in this part of the sequence, albeit the size of ΔδCα and ΔδC′ values of the cold-denatured state and the urea-denatured state is about 25–50% of that of the A-state values. Because A-state and native-state secondary shifts are of very similar size in this region, it may be concluded that the first β-hairpin is populated to about 25–50% in the cold-denatured state.
Fig. 3.
Secondary chemical shift analysis (13Cα, 13C′, 15N) of the cold-denatured state (258 K, 2,500 bar), the urea-denatured state (298 K, 1 bar), and the A-state (298 K, 1 bar) of ubiquitin. Secondary structure elements of the A-state are drawn at the top. The red bars for the 15N secondary shift of the cold-denatured state (D258K) are shifted relative to standard referencing (black bars) by −1.41 ppm to account for the uniform pressure effect on solvent-exposed amide groups (main text).
Compared with the A-state and the urea-denatured state, 15N secondary shifts of the cold-denatured state show a distinct positive offset (Fig. 3), which was not noticed for the folded state at 258 K (Fig. 2, see above). To characterize this effect we have also determined the chemical shifts of a 15N-labeled nonapeptide as a function of pressure in the range from 1 to 2,500 bar and temperature from 258 to 298 K (Fig. S2). The 15N chemical shifts have an approximate linear dependence on pressure and temperature, which on average corresponds to coefficients of (2.93 ± 0.53)·10−4 ppm/bar and (−17.0 ± 9.3)·10−3 ppm/K. This indicates that the 15N chemical shift of an exposed amide is much more influenced by pressure and temperature than that of a buried amide or the chemical shift of 13Cα and 13C′ nuclei. Presumably, the compression of the hydrogen bonds between the exposed amide and water is responsible for this behavior. To correct for this effect relative to ambient conditions (298 K, 1 bar), the 15N secondary shift of the cold-denatured state at 258 K, 2,500 bar was adjusted by −1.41 ppm (Fig. 3, red bars), which brings the secondary shift pattern in closer agreement to that of the A-state and the urea-denatured state.
The similarity between the cold-denatured and the A-state continues beyond the first β-hairpin. Residues 23–32 show similar positive ΔδCα and ΔδC′ shifts for cold-denatured and A-states, indicative of an α-helix, which is located at the same position as the native α-helix. Their absolute size is about 20% (cold-denatured) or 50% (A-state) of the native secondary shifts, which indicates that both states contain respective fractional populations of native state in this region. Also the ΔδN shifts of the cold-denatured and the A-state have a similar sequence pattern. However, the rather small ΔδN values of the cold-denatured state may be affected by an overall offset resulting from inaccurate compensation of the nonspecific pressure effect on 15N shifts of exposed amides (see above). It is also noted that the urea-denatured state has close to zero or even negative ΔδCα and ΔδC′ shifts in the region of residues 23–32, which are not indicative of an α-helical conformation. In contrast, α-helical Cα-Cα contacts on the order of 10–20% of the native state have been found in structural ensembles calculated from extensive residual dipolar coupling (RDC), paramagnetic relaxation enhancement (PRE), and small angle X-ray scattering (SAXS) data (23). This may not necessarily be a contradiction because binding of urea apparently leads to backbone conformations, which are more extended than the typical random coil (27). This in turn would bias the observed chemical shift, which is averaged over the small fraction of α-helical and the large fraction of unfolded conformations, toward the extended values, i.e., to negative ΔδCα and ΔδC′ shifts.
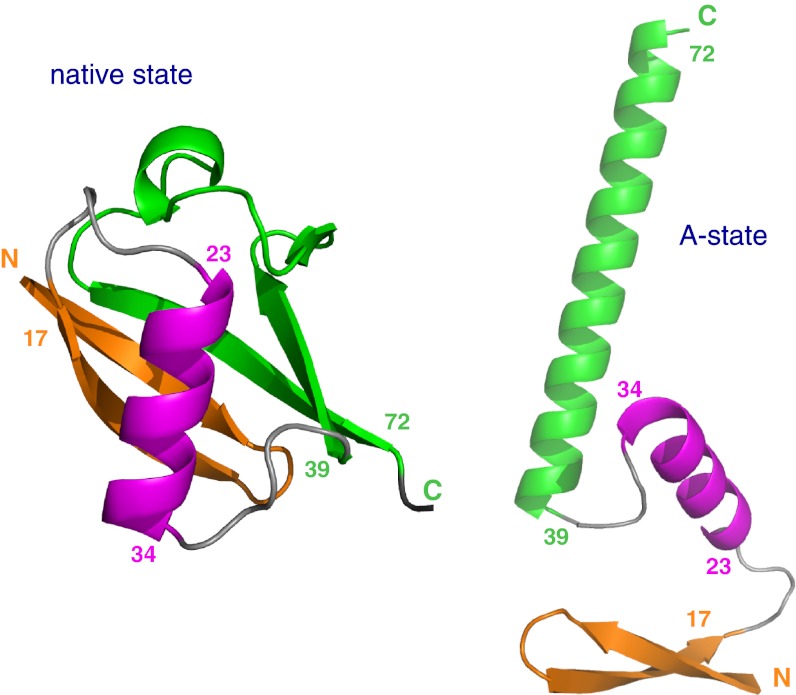
A high similarity between the cold-denatured and the A-state is also found in the second half of the protein for residues 38–76. In native state ubiquitin, this part largely consists of the antiparallel β-sheet β4/β3/β5 that closes the structure by its connection to strand β1. However, in the A-state, this part is switched to the long α-helix α′ (19, 20) (Fig. 4). The pattern of positive ΔδCα and ΔδC′ as well as mixed positive/negative ΔδN secondary shifts of the cold-denatured state are highly similar to those of the A-state (Fig. 3). It is noted that the center part of helix α′ has somewhat reduced secondary shifts both in the cold-denatured and in the A-state, indicative of a weakening of helix propensity—a feature that had been overlooked previously (19, 20). Again the absolute size of the secondary shifts is about 50% of the A-state values in this region. Thus, along the entire sequence the cold-denatured state has deviations from “pure random coil” behavior that correspond to ∼50% of the secondary structure propensities of the A-state.
Fig. 4.
Structural switch from native state (Left) to A-state (Right). Corresponding structural elements are coded by identical colors: N-terminal β-hairpin (orange), central α-helix (magenta), and C-terminal half (green). The C-terminal half switches from a β-sheet structure in the native state to the long α-helix α′ in the A-state. The structural scheme of the A-state is adapted from Brutscher et al. (19).
Backbone Dynamics.
The backbone dynamics of the two states at 258 K and 2,500 bar have been characterized by 15N T1, T2, and {1H}-15N NOE relaxation experiments. With the exception of the flexible C terminus beyond residue V70, rather uniform T1, T2, and {1H}-15N NOE values are observed for the folded-state resonances (Fig. S3A), indicative of a mostly homogeneous, well-folded structure. Excluding residues with obvious exchange contributions to T2 (28) or fast backbone motions (NOE < 0.6), the average T2/T1 ratio (0.035) corresponds to a rotational correlation time τc of 16.9 ns. Thus, τc is increased 4.1-fold compared with folded ubiquitin at 300 K and 1 bar (4.1 ns) (28). This is in good agreement with a ratio of 3.64 for the water self-diffusion coefficients (D300K/1bar = 2.41 × 10−5 cm2/s, D258K/2,500bar = 6.60 × 10−6 cm2/s) (29). Apparently the Stokes–Einstein relation is approximately fulfilled, albeit at 258 K, 2,500 bar τc is about 11% larger than expected from the value at 300 K. This increase may be caused by an increase in the hydration shell of ubiquitin at low temperature and high pressure or by genuine differences in the pressure and temperature dependence of water rotational and translational diffusion (29). As noted before, a number of folded-state residues in the region D21–I44 showed signs of pressure-induced exchange broadening consistent with 15N T2 data at room temperature (17). These were excluded from the estimate of τc. Due to the low intensity of the resonances at 258 K, no quantitative analysis of the exchange was attempted.
In contrast to the folded state, the 15N backbone resonances of the denatured state at 258 K, 2,500 bar show strong variations in their relaxation behavior (Fig. S3B). Thus, the {1H}-15N NOE values are reduced from an average of 0.74 to 0.4, indicative of larger amplitude local backbone motions on the subnanosecond timescale. The NOE values are rather uniform and very similar to data (average 0.4) for the A-state at 300 K (19). Furthermore, T1 increases and T2 decreases along the sequence from residue Q2 to about Q40, indicating slower overall motions in the C-terminal part. An identical behavior is found for the A-state.
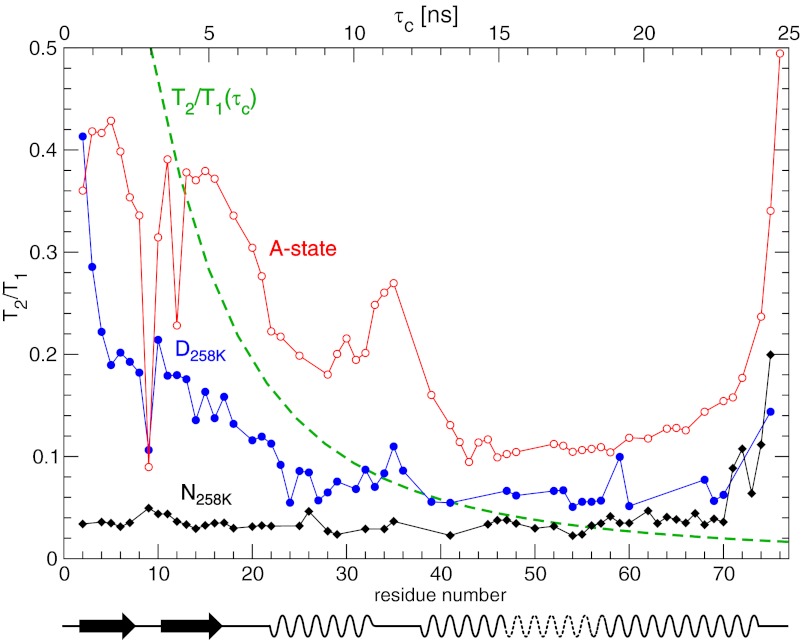
In the absence of exchange broadening, the 15N T2/T1 ratio is to good approximation independent of rapid internal motions and of the magnitude of the chemical shift anisotropy and depends only on the overall rotational diffusion tensor (28). Brutscher et al. have used this property to estimate effective tumbling times along the sequence of the A-state (19). Fig. 5 shows the 15N T2/T1 ratio for the denatured and the folded state at 258 K, 2,500 bar in comparison with the A-state at 300 K. In addition, also the expected T2/T1 ratio of an isotropic rotator is given as a function of the isotropic τc. For the cold-denatured state, the T2/T1 ratio decreases from about 0.4 at residue Q2 to about 0.06 in the region between residues Q40 and V70, corresponding to an increase in the effective rotational correlation time from about 4 ns to 13 ns. This profile is very similar to that of the A-state, albeit the A-state ratios are shifted to higher values due to the faster tumbling at 300 K. It has been noted previously (19) that the A-state T2/T1 ratios form plateaus with different values along the sequence, which roughly correspond to the first β-hairpin as well as to helices α and α′. It was thus concluded that these secondary structure elements move independently from each other. Similar plateaus are observed for the cold-denatured state, although the difference between helices α and α′ is not as pronounced, which may also be expected from the decreased slope of the T2/T1 curve at larger rotational correlation times. For both conditions, a similar increase in T2/T1 is observed around residue I35, which indicates increased mobility of the linker between helices α and α′. It is also evident that the N-terminal residues Q2–F4 of the cold-denatured state are more flexible than the rest of the β-hairpin, whereas no such N-terminal fraying is observed in the A-state. With the exception of this minor difference, the overall 15N relaxation behavior of both the cold-denatured and the A-state is very similar. It may thus be concluded that the cold-denatured state also consists of the three independently moving secondary structure elements β-hairpin, helix α, and helix α′.
Fig. 5.
T2/T1 ratios calculated from 600-MHz 15N relaxation data for ubiquitin. Black curve: folded-state (258 K, 2,500 bar) T2/T1 ratios as a function of residue number (bottom horizontal axis). Blue curve: cold-denatured state (258 K, 2,500 bar). Red curve: A-state (300 K, 1 bar) replotted from Brutscher et al. (19). The dashed green curve shows the T2/T1 ratio for an isotropic rotator at 600 MHz as a function of τc (top horizontal axis). Secondary structure elements of the A-state are drawn at the bottom.
Pressure-Induced Unfolding in Methanol.
Cold denaturation is caused by a reduction in the energetic cost of exposing hydrophobic groups to water, i.e., by a reduction of the hydrophobic effect at low temperatures (Discussion). Because methanol also reduces the strength of the hydrophobic effect, it seems not surprising that the A-state at 60% methanol and pH 2 has strong similarities to the pressure-assisted cold-denatured state. This observation prompted us to test whether a combination of methanol and pressure alone would also induce an A-state–like structural ensemble of ubiquitin.
This is indeed the case. At pH 4.6, 45% methanol, and 308 K, predominantly native-state resonances are observed in the 1H-15N HSQC at 1 bar (Fig. 6A). However, also a second set of very weak resonances is visible, which according to their intensities corresponds to about 14% population. When pressure is increased gradually toward 2,500 bar, the native state resonances become weaker at the expense of the second set of resonances. The transition is almost complete at 2,500 bar (Fig. 6A), where the second state has a population of about 93%. An assignment of the backbone resonances shows that the 13Cα, 13C′, and 15N secondary shifts of this pressure-induced state (Fig. 6C) are almost identical to those of the A-state at 298 K (Fig. 3). These results clearly show that pressure has a very similar effect to that of methanol on the conformation of ubiquitin.
Fig. 6.
(A) Pressure-induced unfolding of ubiquitin in 45% methanol at 308 K 1H-15N HSQC recorded on a 600-MHz spectrometer at pressures of 1 (dark blue), 500 (blue), 1,000 (green), 1,500 (magenta), 2,000 (orange), and 2,500 bar (red), respectively. (B) Expanded region of A showing the transition in the glycine region. Resonances are labeled with assignment information corresponding to the native state (dark blue) or the pressure-induced A-state (red). (C) Secondary chemical shift analysis (13Cα, 13C′, 15N) of the pressure-induced unfolded state of ubiquitin in 45% methanol (308 K, 2,500 bar). The secondary shifts are almost identical to those of the A-state (Fig. 3). Secondary structure elements of the A-state are drawn at the top.
A few general conclusions follow from a closer look at the behavior of individual resonances during the pressure variation from 1 to 2,500 bar (Fig. 6B): (i) because distinct sets of resonances are observed for native and A-state, the exchange time between both states is significantly longer than the inverse of their chemical shift separation, i.e., tens of milliseconds. (ii) From the intensity-derived population ratios it follows that the free energy of the native state is about 4.5 kJ/mol lower than that of the A-state at 1 bar, whereas at 2,500 bar, it is about 6.7 kJ/mol higher. (iii) The chemical shift change of resonances for both states during the pressure variation shows that their conformations vary to some extent with pressure. Thus, pressure changes not only the depth of the free energy wells for both states, but also their shape.
The pressure-induced transition is completely reversible and will allow a detailed analysis of the transition from ubiquitin’s native to its A-state. Such an analysis is currently in progress. The methanol/pressure transition can also be induced in other proteins. Fig. S4 shows the continuous pressure-induced unfolding for protein G at 50% methanol and 303 K, pH 2.5. Again a second distinct set of resonances becomes visible at high pressure, which is populated to about 45% at 2,500 bar. Such an A-state of protein G has not been observed before under normal pressure conditions. The backbone assignment and analysis of secondary shifts (Figs. S5 and S6) reveals that the A-state of protein G mainly consists of one central helix that is in a similar position to that of the native-state helix. However, its N-terminal end is extended by about one helical turn to residue 17 and its population is reduced to about 50% as indicated by a respective reduction in the size of secondary shifts. In addition, small propensities for β-conformations are observed in the A-state at the positions of the native-state N-terminal and C-terminal β-strands 1–4.
Discussion
Here we have presented a detailed structural characterization of ubiquitin under pressure-assisted cold-denaturing conditions, which is based on extensive assignments of the backbone resonances at 258 K and 2,500 bar and the analysis of secondary chemical shifts and 15N relaxation data. Under these conditions, ubiquitin exists in slow exchange equilibrium between a folded and a disordered state. The folded state closely resembles the native-state structure of ubiquitin as determined in crystal or in solution, although as shown previously for high pressure at room temperature (17) enhanced mobility of many residues, in particular in the C-terminal part, is evident from the 15N relaxation data.
The disordered state is not purely random coil, but has propensities for a native-like N-terminal β-hairpin and α-helix as well as a nonnative C-terminal α-helix that are very similar to those described previously for the A-state (19). The stabilization of a helical form for the C-terminal part is in line with theoretical (30) and FTIR studies (31) on helical peptides that indicate only minor destabilization or even stabilization under pressure. Throughout the sequence, the observed structural propensities are on the order of 20% of fully formed secondary structure elements. Similar to that in the A-state, 15N relaxation data indicate an independent segmental motion of these three secondary structure elements with limited mobility of the backbone N-H vectors on the subnanosecond timescale. A certain similarity also exists to ubiquitin’s urea-denatured state, where propensities for a native-like N-terminal β-hairpin and α-helix have been reported (22, 23). However, the latter state is considerably more flexible on the nanosecond timescale as evident from its strongly reduced {1H}-15N NOE values (32).
The observed structural propensities of the pressure-assisted cold-denatured state should be put in the context of proposed mechanisms for cold and pressure denaturation. Cold denaturation is a consequence of the positive ΔCp, which is commonly attributed to the ordering of water molecules around unfolded hydrophobic residues, i.e., the hydrophobic effect (6, 7). Lowering the temperature reduces the free energy penalty of the hydrophobic ordering and thus favors unfolding. Strong evidence for this relation to the hydrophobic effect comes from the proportionality of ΔCp to the buried hydrophobic surface area (33). In contrast, the cause of pressure denaturation is less clear. Theoretical studies indicate that insertion of water between two hydrophobic molecules is more favorable at high pressure (34). Protein unfolding at high pressure may then be understood as transfer of water into the hydrophobic protein interior rather than the transfer of nonpolar residues to water. Other simulations show that water in the vicinity of exposed hydrophobic groups has much higher compressibility than that of hydrophilic surface water, bulk water, or the protein interior (35–37). This would reduce the strength of the hydrophobic effect at high pressure and hence favor unfolding. In contrast, experimental data show that the loss of internal cavities under unfolding significantly reduces the total volume of the system and hence favors unfolding at high pressure (38, 39).
Penetration of water molecules into the interior of ubiquitin at high pressure has been observed in simulations (40). However, experimental studies on dynamics (41) and hydrogen exchange (42) have so far failed to detect clear effects for the native state under high pressure. The secondary structure propensities in the pressure-assisted cold-denatured state of ubiquitin are consistent with the notion that water has penetrated into the hydrophobic protein interior and pried apart the protein. Because this is achieved by cold and pressure, a combination of all of the above-mentioned mechanisms may be responsible for reducing the free energy penalty of opening the hydrophobic core. It is remarkable, however, that native as well as nonnative secondary structures persist to about 25% in populations under these conditions. This means that these subglobal units are energetically so well defined that they still dominate the energy landscape despite many of the more long-range hydrophobic contacts being broken. The similarity of pressure/cold-, alcohol-, and urea-denatured states attests to the strong stabilization of these subunits. This observation is also consistent with NMR data on the cold denaturation of ubiquitin in reverse micelles at 253 K (14), where resonances of different regions of the protein were found to disappear at different temperatures, indicating higher stability for the α-helix and parts of the first β-hairpin.
The steady-state data on the pressure/cold-denatured state of ubiquitin do not provide direct information on the folding pathway at room temperature and normal pressure. Such information must come from kinetic experiments under the latter conditions. Nevertheless, it is tempting to speculate that the residual native-like structure in the pressure/cold-denatured state of ubiquitin is related to a hierarchical order of protein folding under normal conditions. In the hierarchical model larger entities of native structure form from smaller native-like pieces (43). Evidence for hierarchical folding has been given in certain cases, e.g., for cytochrome c from the heterogeneity of hydrogen exchange rates under mild unfolding conditions (44) and for a destabilized mutant of L9 that allowed observation of a cold-denatured state containing native and nonnative structural elements at ambient temperatures (9). For ubiquitin, intermediates in folding have been identified in kinetic experiments (45) and have previously been postulated from heterogeneous line broadening of the cold-denatured state in a reverse micelle system (14). Extrapolation of our data to folding under normal conditions may indicate the following complex folding landscape: The structural propensities in the N-terminal part of the cold-denatured state are native-like, but they are nonnative in the C-terminal part. Thus, the formation of the C-terminal α-helix is off-pathway. This off-pathway structure first needs to unfold before the native C-terminal β-sheet can form and close the structure via the parallel connection from the C-terminal strand β5 to the N-terminal strand β1. The latter connection has the largest sequence separation, i.e., contact order in ubiquitin. Its formation is thus the most entropically disfavored of all. Because average contact order and folding speed of single-domain proteins are inversely correlated (46), it is expected that the formation of the antiparallel sheet β1/β5 is the rate-limiting final step in the folding of ubiquitin. The concomitant presence of the on-pathway N-terminal β-hairpin and α-helix and the off-pathway C-terminal α-helix in the cold-denatured state is consistent with such an order of folding events. Furthermore, also the inverse process of unfolding has been shown to start by breaking sheet β1/β5 in the case of thermal (47) and pressure (42) denaturation. We conclude therefore that an A-state–like conformational ensemble is the major thermodynamic and presumably also kinetic intermediate nearest to the native state in the ubiquitin folding pathway.
The observation of cold or pressure denaturation is severely limited by the low temperatures and high pressures required. The cold denaturation temperature of ubiquitin at ambient pressure is below 238 K (12), whereas at room temperature pressure denaturation occurs at 5.4 kbar (48). Similar cold denaturation temperatures at ambient pressure and unfolding pressures at ambient temperature are required for many other proteins (3). The simultaneous application of low temperature and pressure allowed us to observe about 50% of denatured ubiquitin at 258 K and the current maximal pressure of 2,500 bar suitable for high-sensitivity NMR experiments. The close similarity of the pressure-assisted cold-denatured state and the methanol-denatured state is consistent with the notion that in both cases the destabilization results from the weakening of the hydrophobic effect. We could show here that the combination with methanol shifts the unfolding transition farther into the easily observable pressure and temperature range. Indeed a complete transition from native state to unfolded ensemble occurs at 308 K, 45% methanol in the pressure range from 1 to 2,500 bar. The unfolded ensemble under these conditions is highly similar to both the pressure-assisted cold and the methanol-denatured states of ubiquitin. A similar transition is observable for protein G. The methanol-assisted pressure transitions are completely reversible. This opens the way to follow the unfolding of proteins in a continuous and very detailed manner by NMR experiments.
Materials and Methods
Sample Preparation.
15N/13C-labeled human ubiquitin was prepared as described (49). NMR samples for the pressure-assisted cold-denatured state contained 1.0 mM 15N/13C-labeled protein in 10 mM potassium phosphate buffer, 0.02% NaN3, 90/10% (vol/vol) H2O/D2O, pH 6.5. For measurements under methanol-destabilizing conditions, a further ubiquitin sample contained 0.6 mM 15N/13C-labeled protein, 30 mM sodium acetate, 45% methanol, 45% H2O, 10% D2O, pH 4.6.
15N/13C-labeled protein G was prepared as described in ref. 50. For measurements under methanol-destabilizing conditions, the sample contained 0.25 mM 15N/13C-labeled protein G, 25 mM potassium phosphate, 50% methanol, 40% H2O, 10% D2O, pH 2.5.
High-Pressure Measurements.
All experiments were carried out using a commercial high-pressure NMR cell (Daedalus Innovations LLC) with an inner diameter of 3 mm and an active volume of 120 μL. The tube was rated to 2,500 bar and used in an aluminum alloy static pressure cell connected to a high-pressure generator (High Pressure Equipment Company) via a pressure line. Both line and pressure generator were filled with extralow viscosity paraffin wax (Sigma-Aldrich; 95369).
NMR Experiments.
All NMR spectra were recorded on Bruker Avance DRX800 or DRX600 spectrometers equipped with triple-resonance pulsed-field gradient TXI probes. Backbone assignments were achieved using standard 3D CBCA(CO)NH, HNCA, HNCO, and 15N-edited 1H-1H nuclear Overhauser effect spectroscopy (NOESY) experiments. 15N relaxation measurements (T1, T2, {1H}-15N NOE) for the cold-denatured state of ubiquitin were recorded at 600 MHz. All spectra were processed with NMRPipe (51) and evaluated with NMRView (52) or PIPP (53).
Analysis of Relaxation Data.
Resonance intensities of NMR relaxation spectra were extracted using the program nlinLS contained in NMRPipe (51). T1 and T2 decay curves were then fitted to the intensities by an in-house written routine implemented in Matlab (MathWorks), using Monte Carlo estimation of errors.
Determination of Secondary Shifts.
Chemical shifts for native-state ubiquitin (293 K, 1 bar, pH 6.6) are from BioMagResBank (BMRB) entry 6457 and those for urea-denatured ubiquitin (8 M urea, 10 mM glycine, 90%/10% (vol/vol) H2O/D2O, pH 2.5, 298 K) are from BMRB entry 16626. For the latter, an offset of 0.2 ppm was added to account for an uncorrected shift of the water line used for indirect referencing in the presence of urea. Chemical shifts of the ubiquitin’s A-state (60% methanol, 30% H2O, 10% D2O, pH 2, 298 K) were obtained as described in ref. 20.
Secondary chemical shifts were obtained by subtracting random coil shifts generated by the University of Kopenhagen web server (http://www1.bio.ku.dk/english/research/pv/sbin_lab/staff/MAK/randomcoil/script/), which uses protein sequence, pH, and temperature corrections (54–56).
Supplementary Material
Acknowledgments
We thank Marco Rogowski and Klara Rathgeb-Szabo for protein preparation and Florence Cordier as well as Shin Isogai for valuable discussions. This work was supported by SNF Grant 31-132857 (to S.G.) and by a stipend from the Boehringer Ingelheim Fonds (to L.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Chemical shifts have been deposited in the BioMagResBank (BMRB) for ubiquitin in water at 258 K, 2,500 bar (ID 18610) and for ubiquitin in 45% methanol at 308 K, 2,500 bar (ID 18611).
See Author Summary on page 1578 (volume 110, number 5).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212222110/-/DCSupplemental.
References
- 1.Hawley SA. Reversible pressure—temperature denaturation of chymotrypsinogen. Biochemistry. 1971;10(13):2436–2442. doi: 10.1021/bi00789a002. [DOI] [PubMed] [Google Scholar]
- 2.Privalov PL, Griko YuV, Venyaminov SYu, Kutyshenko VP. Cold denaturation of myoglobin. J Mol Biol. 1986;190(3):487–498. doi: 10.1016/0022-2836(86)90017-3. [DOI] [PubMed] [Google Scholar]
- 3.Smeller L. Pressure-temperature phase diagrams of biomolecules. Biochim Biophys Acta. 2002;1595(1–2):11–29. doi: 10.1016/s0167-4838(01)00332-6. [DOI] [PubMed] [Google Scholar]
- 4.Kauzmann W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 5.Tanford C. Contribution of hydrophobic interactions to the stability of the globular conformation of proteins. J Am Chem Soc. 1962;84:4240–4247. [Google Scholar]
- 6.Privalov PL. Stability of proteins: Small globular proteins. Adv Protein Chem. 1979;33:167–241. doi: 10.1016/s0065-3233(08)60460-x. [DOI] [PubMed] [Google Scholar]
- 7.Baldwin RL. Temperature dependence of the hydrophobic interaction in protein folding. Proc Natl Acad Sci USA. 1986;83(21):8069–8072. doi: 10.1073/pnas.83.21.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kauzmann W. Protein stabilization - Thermodynamics of unfolding. Nature. 1987;325:763–764. [Google Scholar]
- 9.Shan B, McClendon S, Rospigliosi C, Eliezer D, Raleigh DP. The cold denatured state of the C-terminal domain of protein L9 is compact and contains both native and non-native structure. J Am Chem Soc. 2010;132(13):4669–4677. doi: 10.1021/ja908104s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szyperski T, Mills JL. NMR-based structural biology of proteins in supercooled water. J Struct Funct Genomics. 2011;12(1):1–7. doi: 10.1007/s10969-011-9111-5. [DOI] [PubMed] [Google Scholar]
- 11.Szyperski T, Mills JL, Perl D, Balbach J. Combined NMR-observation of cold denaturation in supercooled water and heat denaturation enables accurate measurement of deltaC(p) of protein unfolding. Eur Biophys J. 2006;35(4):363–366. doi: 10.1007/s00249-005-0028-4. [DOI] [PubMed] [Google Scholar]
- 12.Skalicky J, Sukumaran D, Mills J, Szyperski T. Toward structural biology in supercooled water. J Am Chem Soc. 2000;122:3230–3231. [Google Scholar]
- 13.Davidovic M, Mattea C, Qvist J, Halle B. Protein cold denaturation as seen from the solvent. J Am Chem Soc. 2009;131(3):1025–1036. doi: 10.1021/ja8056419. [DOI] [PubMed] [Google Scholar]
- 14.Babu CR, Hilser VJ, Wand AJ. Direct access to the cooperative substructure of proteins and the protein ensemble via cold denaturation. Nat Struct Mol Biol. 2004;11(4):352–357. doi: 10.1038/nsmb739. [DOI] [PubMed] [Google Scholar]
- 15.Kitahara R, Yamada H, Akasaka K. Two folded conformers of ubiquitin revealed by high-pressure NMR. Biochemistry. 2001;40(45):13556–13563. doi: 10.1021/bi010922u. [DOI] [PubMed] [Google Scholar]
- 16.Kitahara R, Akasaka K. Close identity of a pressure-stabilized intermediate with a kinetic intermediate in protein folding. Proc Natl Acad Sci USA. 2003;100(6):3167–3172. doi: 10.1073/pnas.0630309100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitahara R, Yokoyama S, Akasaka K. NMR snapshots of a fluctuating protein structure: Ubiquitin at 30 bar-3 kbar. J Mol Biol. 2005;347(2):277–285. doi: 10.1016/j.jmb.2005.01.052. [DOI] [PubMed] [Google Scholar]
- 18.Kitahara R, et al. Cold denaturation of ubiquitin at high pressure. Magn Reson Chem. 2006;44(Spec No):S108–S113. doi: 10.1002/mrc.1820. [DOI] [PubMed] [Google Scholar]
- 19.Brutscher B, Brüschweiler R, Ernst RR. Backbone dynamics and structural characterization of the partially folded A state of ubiquitin by 1H, 13C, and 15N nuclear magnetic resonance spectroscopy. Biochemistry. 1997;36(42):13043–13053. doi: 10.1021/bi971538t. [DOI] [PubMed] [Google Scholar]
- 20.Cordier F, Grzesiek S. Quantitative comparison of the hydrogen bond network of A-state and native ubiquitin by hydrogen bond scalar couplings. Biochemistry. 2004;43(35):11295–11301. doi: 10.1021/bi049314f. [DOI] [PubMed] [Google Scholar]
- 21.Wilkinson KD, Mayer AN. Alcohol-induced conformational changes of ubiquitin. Arch Biochem Biophys. 1986;250(2):390–399. doi: 10.1016/0003-9861(86)90741-1. [DOI] [PubMed] [Google Scholar]
- 22.Meier S, Strohmeier M, Blackledge M, Grzesiek S. Direct observation of dipolar couplings and hydrogen bonds across a β-hairpin in 8 M urea. J Am Chem Soc. 2007;129(4):754–755. doi: 10.1021/ja067522k. [DOI] [PubMed] [Google Scholar]
- 23.Huang J-R, Grzesiek S. Ensemble calculations of unstructured proteins constrained by RDC and PRE data: A case study of urea-denatured ubiquitin. J Am Chem Soc. 2010;132(2):694–705. doi: 10.1021/ja907974m. [DOI] [PubMed] [Google Scholar]
- 24.Spera S, Bax A. Empirical correlation between protein backbone conformation and C.alpha. and C.beta. 13C nuclear magnetic resonance chemical shifts. J Am Chem Soc. 1991;113:5490–5492. [Google Scholar]
- 25.Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol. 1991;222(2):311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 26.Markley JL, et al. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the Standardization of Data Bases of Protein and Nucleic Acid Structures Determined by NMR Spectroscopy Recommendations for the presentation of NMR structures of proteins and nucleic acids. J Biomol NMR. 1998;12(1):1–23. doi: 10.1023/a:1008290618449. [DOI] [PubMed] [Google Scholar]
- 27.Huang J-R, Gabel F, Jensen MR, Grzesiek S, Blackledge M. Sequence-specific mapping of the interaction between urea and unfolded ubiquitin from ensemble analysis of NMR and small angle scattering data. J Am Chem Soc. 2012;134(9):4429–4436. doi: 10.1021/ja2118688. [DOI] [PubMed] [Google Scholar]
- 28.Tjandra N, Feller SE, Pastor RW, Bax A. Rotational diffusion anisotropy of human ubiquitin from 15N NMR relaxation. J Am Chem Soc. 1995;117:12562–12566. [Google Scholar]
- 29.Prielmeier F, Lang E, Speedy R, Lüdemann H. Diffusion in supercooled water to 300 MPa. Phys Rev Lett. 1987;59(10):1128–1131. doi: 10.1103/PhysRevLett.59.1128. [DOI] [PubMed] [Google Scholar]
- 30.Paschek D, Gnanakaran S, García AE. Simulations of the pressure and temperature unfolding of an alpha-helical peptide. Proc Natl Acad Sci USA. 2005;102(19):6765–6770. doi: 10.1073/pnas.0408527102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takekiyo T, Imai T, Kato M, Taniguchi Y. Understanding high pressure stability of helical conformation of oligopeptides and helix bundle protein high pressure FT-IR and RISM theoretical studies. Biochim Biophys Acta. 2006;1764(3):355–363. doi: 10.1016/j.bbapap.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 32.Wirmer J, Peti W, Schwalbe H. Motional properties of unfolded ubiquitin: A model for a random coil protein. J Biomol NMR. 2006;35(3):175–186. doi: 10.1007/s10858-006-9026-9. [DOI] [PubMed] [Google Scholar]
- 33.Spolar RS, Ha JH, Record MT., Jr Hydrophobic effect in protein folding and other noncovalent processes involving proteins. Proc Natl Acad Sci USA. 1989;86(21):8382–8385. doi: 10.1073/pnas.86.21.8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hummer G, Garde S, García AE, Paulaitis ME, Pratt LR. The pressure dependence of hydrophobic interactions is consistent with the observed pressure denaturation of proteins. Proc Natl Acad Sci USA. 1998;95(4):1552–1555. doi: 10.1073/pnas.95.4.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smolin N, Winter R. A molecular dynamics simulation of SNase and its hydration shell at high temperature and high pressure. Biochim Biophys Acta. 2006;1764(3):522–534. doi: 10.1016/j.bbapap.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Sarupria S, Garde S. Quantifying water density fluctuations and compressibility of hydration shells of hydrophobic solutes and proteins. Phys Rev Lett. 2009;103(3):037803. doi: 10.1103/PhysRevLett.103.037803. [DOI] [PubMed] [Google Scholar]
- 37.Grigera JR, McCarthy AN. The behavior of the hydrophobic effect under pressure and protein denaturation. Biophys J. 2010;98(8):1626–1631. doi: 10.1016/j.bpj.2009.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouget J-B, et al. Unique features of the folding landscape of a repeat protein revealed by pressure perturbation. Biophys J. 2010;98(11):2712–2721. doi: 10.1016/j.bpj.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roche J, et al. Cavities determine the pressure unfolding of proteins. Proc Natl Acad Sci USA. 2012;109(18):6945–6950. doi: 10.1073/pnas.1200915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Day R, García AE. Water penetration in the low and high pressure native states of ubiquitin. Proteins. 2008;70(4):1175–1184. doi: 10.1002/prot.21562. [DOI] [PubMed] [Google Scholar]
- 41.Fu Y, et al. Coupled motion in proteins revealed by pressure perturbation. J Am Chem Soc. 2012;134(20):8543–8550. doi: 10.1021/ja3004655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nisius L, Grzesiek S. Key stabilizing elements of protein structure identified through pressure and temperature perturbation of its hydrogen bond network. Nat Chem. 2012;4(9):711–717. doi: 10.1038/nchem.1396. [DOI] [PubMed] [Google Scholar]
- 43.Fitzkee NC, et al. Are proteins made from a limited parts list? Trends Biochem Sci. 2005;30(2):73–80. doi: 10.1016/j.tibs.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 44.Bai Y, Sosnick TR, Mayne L, Englander SW. Protein folding intermediates: Native-state hydrogen exchange. Science. 1995;269(5221):192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khorasanizadeh S, Peters ID, Roder H. Evidence for a three-state model of protein folding from kinetic analysis of ubiquitin variants with altered core residues. Nat Struct Biol. 1996;3(2):193–205. doi: 10.1038/nsb0296-193. [DOI] [PubMed] [Google Scholar]
- 46.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277(4):985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 47.Cordier F, Grzesiek S. Temperature-dependence of protein hydrogen bond properties as studied by high-resolution NMR. J Mol Biol. 2002;317(5):739–752. doi: 10.1006/jmbi.2002.5446. [DOI] [PubMed] [Google Scholar]
- 48.Herberhold H, Winter R. Temperature- and pressure-induced unfolding and refolding of ubiquitin: A static and kinetic Fourier transform infrared spectroscopy study. Biochemistry. 2002;41(7):2396–2401. doi: 10.1021/bi012023b. [DOI] [PubMed] [Google Scholar]
- 49.Sass J, et al. Purple membrane induced alignment of biological macromolecules in the magnetic field. J Am Chem Soc. 1999;121:2047–2055. [Google Scholar]
- 50.Vajpai N, Gentner M, Huang J-R, Blackledge M, Grzesiek S. Side-chain chi(1) conformations in urea-denatured ubiquitin and protein G from (3)J coupling constants and residual dipolar couplings. J Am Chem Soc. 2010;132(9):3196–3203. doi: 10.1021/ja910331t. [DOI] [PubMed] [Google Scholar]
- 51.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 52.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4(5):603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 53.Garrett D, Powers R, Gronenborn A, Clore G. A common sense approach to peak picking in two-, three-, and four dimensional spectra using automatic computer analysis of contour diagrams. J Magn Reson. 1991;95:214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 54.Kjaergaard M, Brander S, Poulsen FM. Random coil chemical shift for intrinsically disordered proteins: Effects of temperature and pH. J Biomol NMR. 2011;49(2):139–149. doi: 10.1007/s10858-011-9472-x. [DOI] [PubMed] [Google Scholar]
- 55.Kjaergaard M, Poulsen FM. Sequence correction of random coil chemical shifts: Correlation between neighbor correction factors and changes in the Ramachandran distribution. J Biomol NMR. 2011;50(2):157–165. doi: 10.1007/s10858-011-9508-2. [DOI] [PubMed] [Google Scholar]
- 56.Schwarzinger S, et al. Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc. 2001;123(13):2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]