Abstract
Epigenetic silencing in mammals involves DNA methylation and posttranslational modifications of core histones. Here we show that the H1 linker histone plays a key role in regulating both DNA methylation and histone H3 methylation at the H19 and Gtl2 loci in mouse ES cells. Some, but not all, murine H1 subtypes interact with DNA methyltransferases DNMT1 and DNMT3B. The interactions are direct and require a portion of the H1 C-terminal domain. Expression of an H1 subtype that interacts with DNMT1 and DNMT3B in ES cells leads to their recruitment and DNA methylation of the H19 and Gtl2 imprinting control regions. H1 also interferes with binding of the SET7/9 histone methyltransferase to the imprinting control regions, inhibiting production of an activating methylation mark on histone H3 lysine 4. H1-dependent recruitment of DNMT1 and DNMT3B and interference with the binding of SET7/9 also were observed with chromatin reconstituted in vitro. The data support a model in which H1 plays an active role in helping direct two processes that lead to the formation of epigenetic silencing marks. The data also provide evidence for functional differences among the H1 subtypes expressed in somatic mammalian cells.
Keywords: mouse embryonic stem cell, H1 histone triple knockout
Two major types of epigenetic marks occur in mammalian genomes, DNA methylation and posttranslational modifications of histones (1, 2). The methylation of cytosines in mammalian DNA occurs primarily at CpG dinucleotides and is essential for normal mammalian development (1, 3). Perturbations of DNA methylation patterns are thought to play a role in cancer development (4). There are two classes of mammalian DNA methyltransferases (DNMTs), one that functions primarily to establish DNA methylation de novo (DNMT3A and DNMT3B) and the other to maintain it (DNMT1) (3). DNA methylation can have profound effects on gene expression and is most often associated with transcriptional silencing (1, 2). Accumulating evidence indicates the existence of crosstalk between the DNA methylation and core histone modification systems (1, 2). For example, mouse ES cells deficient for the histone H3 lysine 9 (H3K9) methyltransferases Suv39h1/h2 exhibit hypomethylation of CpGs at a subset of repetitive DNA elements (5). Additionally, several lines of evidence indicate that the presence of unmethylated lysine 4 of histone H3 (H3K4) in chromatin favors de novo methylation of DNA (6–9). Despite these advances, the factors in chromatin that coordinate DNA methylation and histone H3 methylation have not been identified.
In this study, we investigated the role of the linker histone H1 in regulating DNA methylation and histone H3 methylation at the H19 and Gtl2 loci in mouse ES cells. H1 is the most distinct of the five histone proteins (H1, H2A, H2B, H3, and H4) in chromatin (10, 11). Mammals contain at least eight histone H1 subtypes or variants that differ in amino acid sequences and expression during development (12–14). Functional differences among these subtypes have been difficult to identify because they appear to act redundantly in development (14–17). Although the precise location of histone H1 within the chromatin fiber is uncertain, it is known that histone H1 resides outside the nucleosome core particle, where it associates with the DNA as it enters and exits the core particle and protects an additional ∼20 bp of DNA (linker DNA). In contrast to the core histones, H1 is highly mobile within the nuclei of living cells (18), consistent with its proposed role in dynamic regulation of chromatin structure and gene activity (19, 20). We show here that histone H1 is indeed a key player in regulating DNA methylation and histone H3 methylation at the H19 and Gtl2 loci in mouse ES cells. Our studies also reveal two mechanisms by which histone H1 acts to silence expression at these loci: by interacting directly with and recruiting DNMT1 and DNMT3B and by inhibiting SET7/9 binding and methylation of H3K 4. We also demonstrate functional differences in the silencing activity of histone H1 subtypes.
Results
Linker Histone H1 Mediates Reversible Changes in DNA Methylation and Gene Expression at the H19 and Gtl2 Loci.
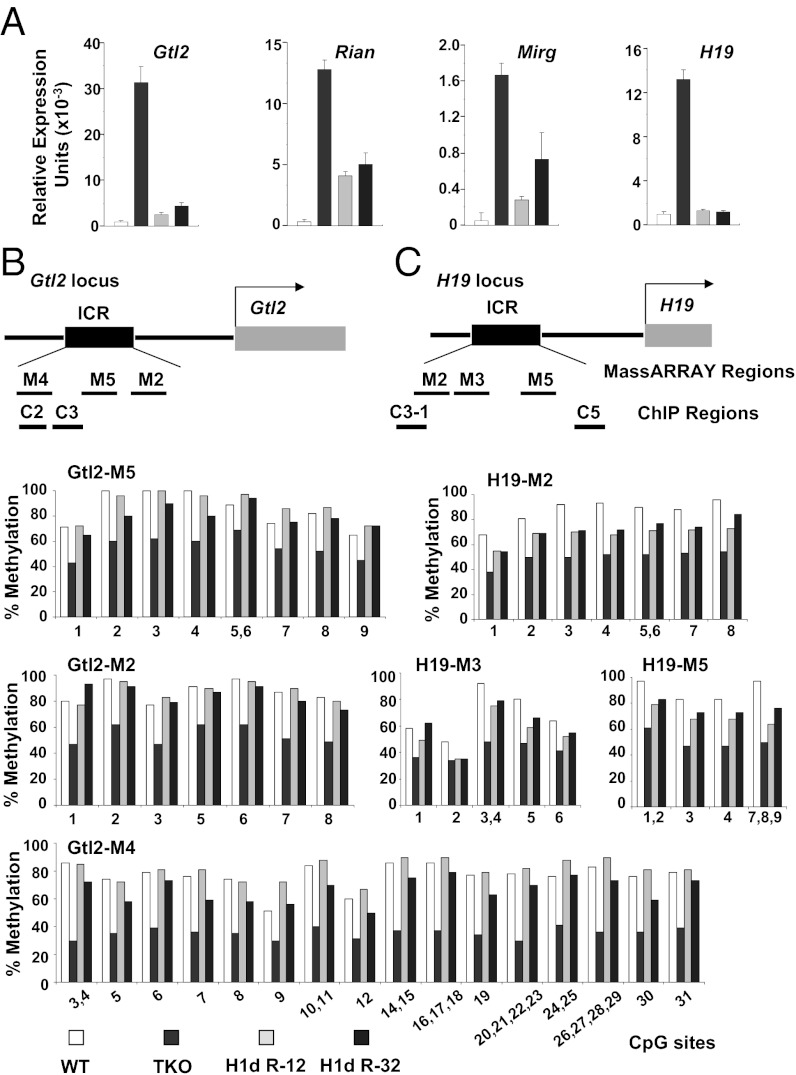
In a previous study, we used microarray analysis to identify gene-expression changes in mouse triple-knockout (TKO) ES cells partially depleted of H1 histones by homozygous inactivation of three H1 genes, H1c, H1d, and H1e (21). Surprisingly, we found that the TKO ES cells, which contain about 50% of the normal level of total histone H1, have very few differences in gene expression compared with WT ES cells. However, prominent among the small number of affected genes were H19 and Gtl2, genes whose expression is regulated by DNA methylation of their imprinting control regions (ICRs). Mouse ES cells normally silence both copies of the H19 gene on mouse chromosome 7 (21–26) as well as several genes (Gtl2, Rian, and Mirg) at the Gtl2 locus on mouse chromosome 12 (Fig. 1A). On the other hand, we found that the partially H1-depleted TKO ES cells express H19, Gtl2, Rian, and Mirg transcripts (Fig. 1A). We used bisulfite treatment of genomic DNA and analysis of specific PCR products with a Sequenom MassARRAY Epityper to quantify the extent of DNA methylation at a total of 47 CpG dinucleotides throughout the ICR regulatory regions at the H19 and Gtl2 loci. Consistent with the silencing of gene expression at the two loci in WT ES cells, we found that many CpG dinucleotides in the two ICRs are highly methylated in WT ES cells (Fig. 1 B and C). Moreover, derepression of the transcripts from the two loci observed in the H1-depleted TKO cells is accompanied by reductions in the methylation level at nearly all CpGs in the two ICRs (Fig. 1 B and C). The reduction of CpG methylation accompanying H1 depletion is specific for the H19 and Gtl2 ICRs, because it was not observed at highly methylated repetitive DNA sequences (21) or at clusters of CpGs located upstream and downstream of the two ICRs (Fig. S1 A and B). It also was not observed at a cluster of highly methylated CpGs in the upstream region of the β-actin gene (Fig. S1C), expression of which is unaffected by H1 depletion (21). Global DNA methylation also is not changed in the H1-depleted cells, as determined by the luminometric methylation assay (27) (Fig. S1D). Thus, DNA methylation at the H19 and Gtl2 ICRs is especially sensitive to the partial (50%) reduction in H1 histone levels in the TKO ES cells.
Fig. 1.
Changes in gene expression and DNA methylation caused by histone H1 depletion are reversed by restoration of H1d. (A) Transcript levels of the indicated genes were measured by quantitative RT-PCR in WT, H1 TKO, and two stably transfected TKO ES cell lines expressing exogenous H1d (R-12 and R-32). Transcript levels were normalized to the level of GAPDH mRNA and represent the average of at least three determinations. Similar results were obtained with several other ES cell lines of the same genotypes. The H1 histone subtype stoichiometries in the cell lines are shown in Fig. S2. (B and C) The extent of methylation of individual CpG’s within the indicated MassARRAY regions of the (B) Gtl2 and (C) H19 loci (Upper) in DNA from the cell lines described in A was determined by bisulfite treatment of genomic DNA and analysis of PCR products with a Sequenom MassARRAY Epityper. The results represent the average of analyses of at least two independent DNA preparations. The extent of methylation at all the indicated positions was significantly different between WT and TKO and between TKO and R-12 and R-32 cell lines, as determined by two-tailed Mann–Whitney tests (all P < 0.001).
To begin to understand the mechanism(s) by which H1 represses gene expression and promotes DNA methylation at the H19 and Gtl2 loci, we asked whether the effects of H1 depletion are reversible. Because the H1d gene was the last gene to be inactivated in the H1 TKO ES cells (16), we sought to restore its expression by stable transfection of the cells with an H1d expression vector. The vector consisted of the H1d gene and 7.1 kb and 1.9 kb of the 5′ and 3′ flanking sequences, respectively. Stably transfected clones expressing H1d were isolated, and their H1 composition was characterized by quantitative HPLC (Fig. S2A). Clones in which expression of H1d restored the level of total H1 (H1 per nucleosome) (Fig. S2B) nearly to levels in WT ES cells were chosen for further analysis. We found that restoration of H1d caused repression of the H19, Gtl2, Rian, and Mirg transcripts (Fig. 1A). Consistent with the renewed repression of the four genes at the two loci, DNA methylation analysis showed that most CpG dinucleotides in the Gtl2 and H19 ICRs were remethylated upon H1d restoration (Fig. 1 B and C). The observed differences in CpG methylation within the two ICRs in WT, TKO, and H1d-restored cells are highly significant, as determined by two-tailed Mann–Whitney tests (all P < 0.001). Thus, the gene-expression and DNA methylation changes at the H19 and Gtl2 loci caused by H1 depletion are reversible in ES cells.
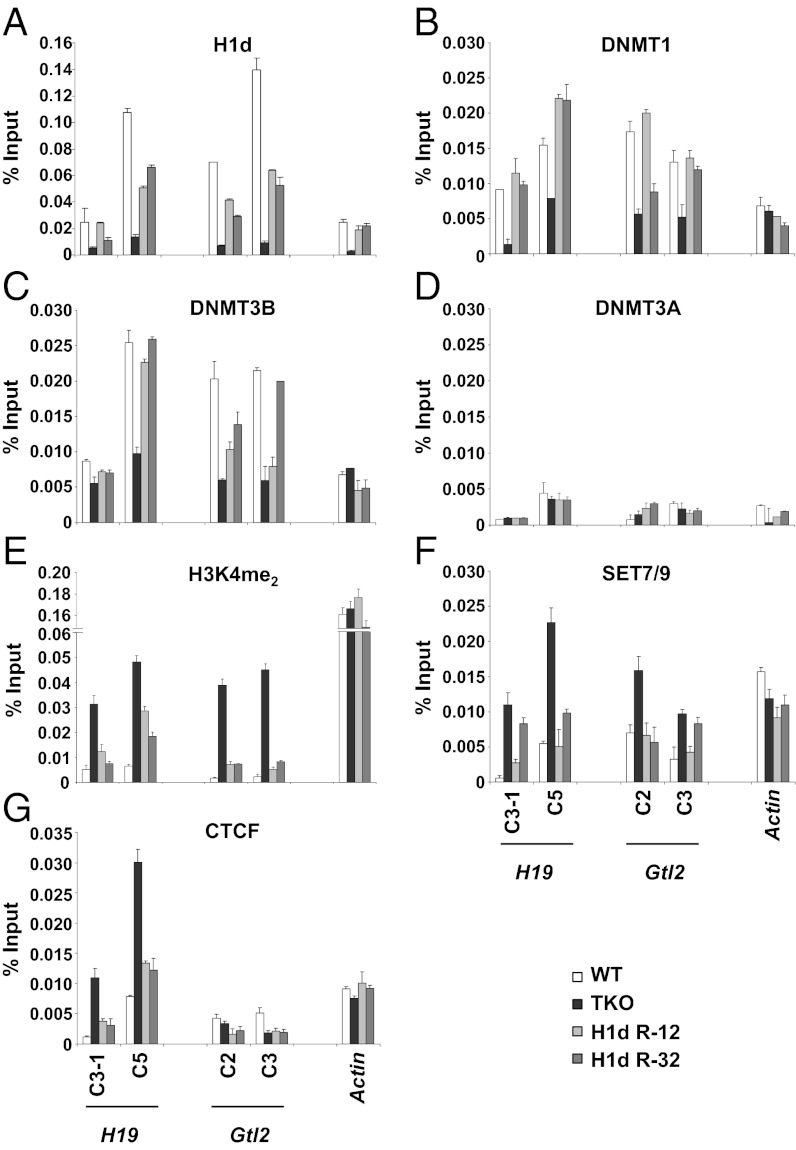
Regulation of H19 expression is mediated in part by the insulator-binding protein CTCF (28, 29). CTCF binds to multiple positions within the H19 ICR when CpG dinucleotides in its binding sites are unmethylated (28, 29). Consistent with the DNA methylation analysis, we found increased CTCF occupancy at the H19 ICR in the TKO cells, which have reduced ICR CpG methylation, compared with that in the WT and H1d-restored cells (Fig. 2G).
Fig. 2.
Histone H1 occupancy at ICRs leads to recruitment of DNMT1 and DNMT3B and interferes with SET7/9 binding and methylation of H3K4 in nucleosomes. (A–G) qChIP was performed with antisera specific for the indicated proteins and histone modifications on cross-linked chromatin from WT, H1 TKO, and two stably transfected TKO ES cell lines expressing exogenous H1d (R-12 and R-32). The bars indicate the percentage of input DNA fragment (see Fig. 1 B and C, Upper, ChIP regions) in specific immunoprecipitates after subtracting the percentage in immunoprecipitates using isotype-matched control antibodies. Error bars indicate the SD of triplicate determinations. Similar results were obtained in three repeat experiments. Specificity of the H1d antibody is demonstrated in Fig. S5.
H1 promotes recruitment of DNMT1 and DNMT3B and inhibits binding of SET7/9 methyltransferase and methylation of H3K4 in vivo and in chromatin reconstituted in vitro.
The finding that DNA hypomethylation at the ICRs caused by H1 depletion is readily reversible by restoration of H1 suggests that H1 directs a process leading to recruitment of DNMTs to these loci. To investigate this possibility, we carried out quantitative ChIP (qChIP) experiments to measure the level of occupancy of DNMTs at the H19 and Gtl2 ICRs. We observed that occupancy of DNMT1 and DNMT3B is highly correlated with the level of H1d occupancy at multiple positions within the two ICRs. The two DNA methyltransferases were bound at these loci in WT ES cells, in which H1d also is readily detectable in these regions (Fig. 2A), and they were much reduced at these positions in the H1 TKO cells lacking H1d (Fig. 2 B and C). Expression of exogenous H1d in the KO cells restored its binding to these loci (Fig. 2A) and also restored occupancy of DNMT1 and DNMT3B (Fig. 2 B and C). Note that the levels of DNMT1 and DNMT3B are very similar in the three types of ES cells (Fig. S3), indicating that the observed changes in DNMT1 and DNMT3B occupancy at the two ICRs are not caused by changes in the levels of expression of the proteins. In contrast, DNMT3A was not detectable at any of the studied positions in any condition (Fig. 2D), even though the DNMT3A2 isoform is well expressed in the three types of ES cells (Fig. S3). These results indicate that the presence of histone H1 at the H19 and Gtl2 ICRs in ES cells leads to recruitment of DNMT1 and DNMT3B.
Several reports indicate that CpG methylation throughout the genome correlates with the presence of unmethylated H3K4 (6–9) and that de novo methylation of DNA is inhibited by methylation of H3K4 (6, 8). Therefore, we studied H3K4 methylation, as well as occupancy of the SET7/9 H3K4 methyltransferase, at the H19 and Gtl2 ICRs in WT, H1-depleted, and H1d-restored ES cells. We found generally lower levels of H3K4me2 (Fig. 2E) and SET 7/9 occupancy (Fig. 2F) at several positions within the ICRs where DNA methylation and DNMT1 and DNMT3B occupancy is increased in WT and H1d-restored cells compared with H1-depleted cells. These results suggest that, in addition to promoting the binding of DNMT1 and DNMT3B, the presence of H1 at the ICRs interferes with SET7/9 binding and methylation of nucleosomes at the ICRs.
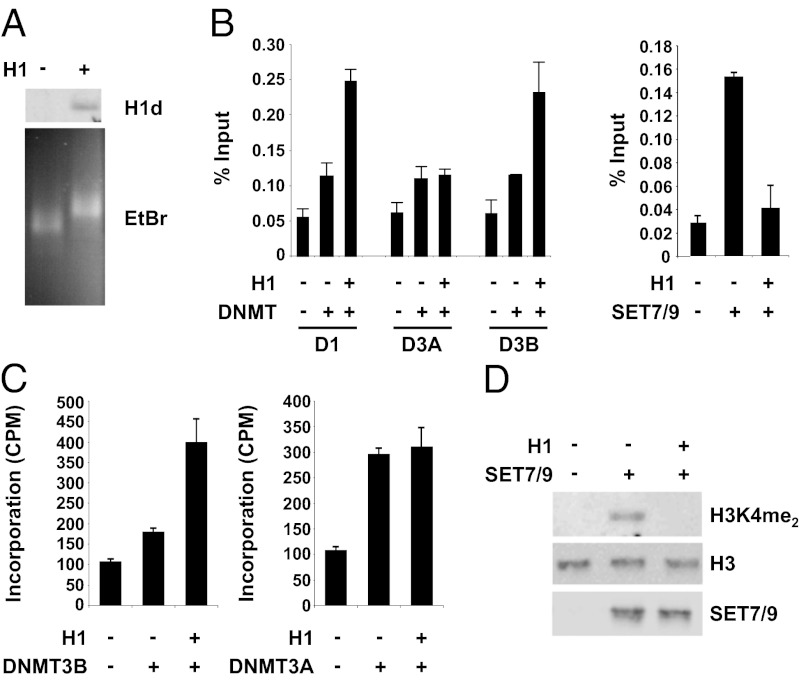
To investigate further the mechanism by which H1 stimulates DNMT1 and DNMT3B recruitment and interferes with SET7/9 binding, we purified recombinant forms of the three proteins (and DNMT3A) and analyzed their binding to chromatin reconstituted in vitro with and without histone H1. A DNA fragment containing two copies of a nucleosome-positioning sequence (30) was assembled into chromatin with purified core histones in the presence or absence of purified, recombinant H1d histone (Fig. 3A). The reconstituted chromatin then was incubated separately with each of the purified enzymes, and the binding to chromatin was measured by qChIP with specific antibodies. The results show that histone H1 stimulates the in vitro binding of DNMT1 and DNMT3B to the reconstituted, purified chromatin (Fig. 3B, Left). However, histone H1 markedly inhibits binding of SET7/9 to the chromatin (Fig. 3B, Right). Histone H1 did not affect DNMT3A in vitro binding to chromatin significantly, as is consistent with the in vivo results (Fig. 2D) and with the results of the H1–DNMT protein interaction studies described below. We also measured the effect of histone H1 incorporation in the chromatin on the de novo DNA methyltransferase activity of DNMT3B and DNMT3A, as well as on the lysine methyltransferase activity of SET7/9 toward the chromatin. We found that histone H1 stimulates DNMT3B-directed DNA methyltransferase activity (Fig. 3C) but does not affect DNMT3A activity. Importantly, histone H1 strongly inhibits SET7/9 lysine methyltransferase activity toward histone H3 in the chromatin (Fig. 3D). The results of these in vitro studies with purified components suggest that histone H1 acts directly to promote DNMT1 and DNMT3B recruitment and to block SET7/9 binding and methylation of H3K4 in chromatin.
Fig. 3.
H1 stimulates binding and DNA methyltransferase activity of DNMT1 and DNMT3B and interferes with SET7/9 binding and histone H3 methyltransferase activity in chromatin reconstituted in vitro. (A) Dinucleosomes were assembled with core histones from HeLa cells and purified as described in SI Materials and Methods. Purified dinucleosomes were incubated at 4 °C for 10 min without and with purified recombinant H1d prepared by thrombin cleavage of GST-H1d. Incorporation of H1 into dinucleosomes was demonstrated by nondenaturing gel electrophoresis and EtBr staining, which showed slower migration of dinucleosomes containing H1 (Lower). The reaction mixtures also were analyzed by SDS/PAGE and immunoblotting using an anti-H1d antibody (Upper). (B) In vitro binding of DNMTs and SET7/9 to dinucleosomes. Dinucleosomes lacking or containing H1, as indicated, were incubated with purified recombinant DNMT1, DNMT3A, DNMT3B, or SET7/9 as described in SI Materials and Methods. Protein binding to chromatin was analyzed by treating the reaction mixtures with formaldehyde followed by qChIP as described in SI Materials and Methods, using antibodies against DNMT1, DNMT3A, or DNMT3B (Left) or against SET7/9 (Right). Error bars indicate the SDs of duplicate experiments. (C) Dinucleosomes lacking or containing H1 were incubated with purified DNMT3B (Left) or DNMT3A (Right) in the presence of S-adenosyl-L-[methyl-3H] methionine, and the amount of radioactivity incorporated into DNA [reported as counts per minute (cpm) per reaction] was measured as described in SI Materials and Methods. Error bars indicate the SDs of duplicate experiments. (D) Dinucleosomes lacking or containing histone H1 were incubated with purified SET7/9 as described in SI Materials and Methods. The reaction mixtures were analyzed by SDS/PAGE and immunoblotting with the antisera indicated to the right of the figure.
The C-terminal domain of linker histone H1d interacts directly with DNMT1 and DNMT3B.
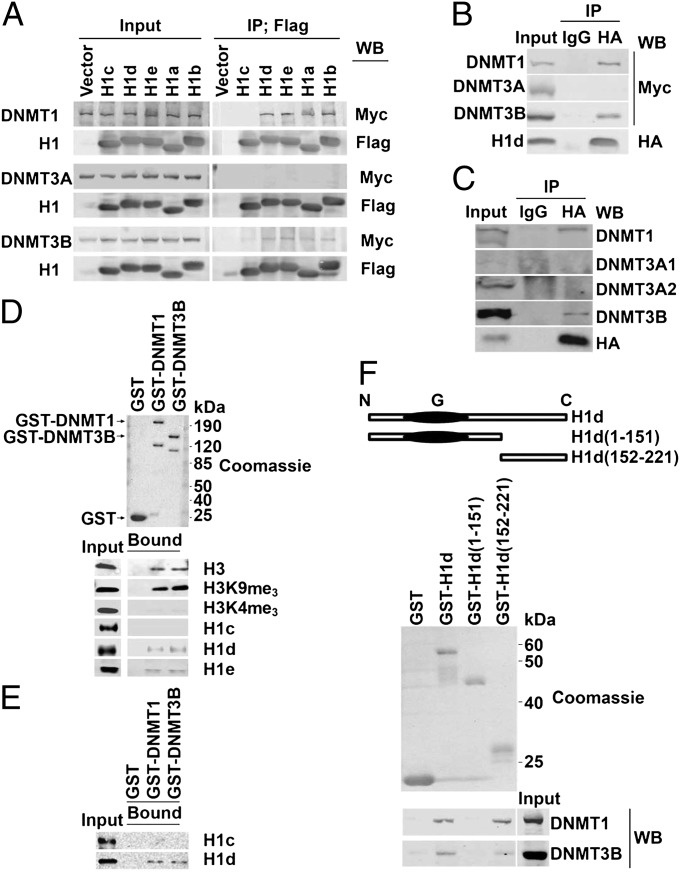
The results described in the preceding section raise the possibility that histone H1 may promote DNA methylation by interacting with DNMT1 and DNMT3B. We investigated this possibility by studying interactions between several histone H1 subtypes and the three DNMTs in several ways. First, Myc-tagged DNMT1, DNMT3A, and DNMT3B and Flag-tagged somatic mouse histone H1 subtypes H1a–H1e were coexpressed in 293T cells by transient transfection. Immunoprecipitation of cell lysates with an anti-Flag serum showed association of four H1 subtypes with DNMT1 and DNMT3B but not with DNMT3A (Fig. 4A). Interestingly, H1c did not interact with DNMT1 and showed only a very weak interaction with DNMT3B. As a control for possible artifacts caused by the Flag tag, which was at the N terminus of histone H1, similar experiments were carried out with H1d tagged with an HA epitope at the C terminus. Again, interaction was seen with DNMT1 and DNMT3B but not with DNMT3A, (Fig. 4B). The observed specificity for interaction of H1d with DNMT1 and DNMT3B but not with DNMT3A is entirely consistent with the results of the in vivo and in vitro ChIP experiments (Figs. 2 B–D and 3B). HA-tagged H1d expressed in 293T cells also coimmunoprecipitated with endogenous DNMT3B (Fig. S4A). Again, however, H1c, as well as the H1(0) subtype, interacted very poorly (Fig. S4A). To investigate interaction of histone H1 and DNMTs expressed at normal levels in ES cells, we prepared H1 TKO ES cells expressing HA-tagged H1d by stable transfection. Immunoprecipitation with anti-HA serum showed that H1d interacts with endogenous DNMT1 and DNMT3B but not with DNMT3A2 (Fig. 4C).
Fig. 4.
H1 histones interact directly with DNMT1 and DNMT3B. (A) Coimmunoprecipitation of five Flag-tagged H1 subtypes with Myc-tagged DNMT1 and DNMT3B expressed in 293T cells. Lysates of transfected cells were immunoprecipitated with anti-Flag serum, and the immunoprecipitates were analyzed by SDS/PAGE and immunoblotting with either anti-Myc or anti-Flag serum. Anti-Flag immunoblots demonstrate approximately equal abundance of the five H1 proteins in the immunoprecipitates. Lysates of cells transfected with an empty expression vector (Vector) served as negative controls. Coimmunoprecipitation was not detected in lysates from cells expressing exogenous DNMT3A or H1c that served as additional negative controls. Input lanes are 5% of the extract used for immunoprecipitation. (B) Coimmunoprecipitation of HA-tagged H1d and Myc-tagged DNMT1 and DNMT3B expressed in 293T cells. The input lane is 5% of the extract used for immunoprecipitation. (C) Coimmunoprecipitation of endogenous DNMT1 and DNMT3B with HA-tagged H1d expressed at normal levels by stable transfection of H1 TKO ES cells. The input lane is 5% of the extract used for immunoprecipitation. HPLC analyses like those shown in Fig. S2 showed that HA-tagged H1d is expressed at levels similar to that of H1d in WT ES cells. (D) GST-DNMT1 and GST-DNMT3B interact with purified H1d and H1e, as well as with histone H3 lacking lysine 4 trimethylation. Purified histones were prepared from mouse ES cell chromatin by extraction with 0.2 N H2SO4 and mixed with GST-DNMT1 or GST-DNMT3B or GST bound to Glutathione-Sepharose 4B. Bound proteins were collected by centrifugation and analyzed by SDS/PAGE and immunoblotting with the antisera indicated at the right of the figure. The input lane is 5% of the extract used for interaction. Specificity of the histone H1 antibodies is shown in Fig. S5. (E) GST-DNMT1 and GST-DNMT3B interact directly with H1d but not with H1c. Recombinant H1c and H1d were produced in bacteria as GST fusion proteins. Purified GST-H1c and GST-H1d were treated with 2 units of thrombin (Sigma) at 22 °C overnight, and the remaining thrombin was removed by incubation with pAminobenzamidine-Agarose (Sigma) at 22 °C for 30 min and centrifugation. The resulting GST-free H1 proteins were mixed with GST-DNMT1 or GST-DNMT3B or GST bound to Glutathione-Sepharose 4B and analyzed as described in D. (F) DNMT1 and DNMT3B bind to the CTD of histone H1. GST fusion proteins containing the indicated regions of H1d (Upper) were bound to Glutathione-Sepharose 4B and mixed with extracts of 293T cells expressing Myc-tagged DNMT1 or DNMT3B. Bound proteins were collected as in D (Lower) and analyzed by immunoblotting with an anti-Myc serum.
To investigate further H1 interaction with DNMTs, we carried out in vitro interaction studies with GST-DNMT fusion proteins and purified histones prepared by acid extraction of ES cell chromatin. Using specific antisera for the H1c, H1d, and H1e subtypes (Fig. S5), we observed that GST-DMNT1 and GST-DMNT3B interact with H1d and H1e but do not interact with H1c (Fig. 4D). Interaction of DNMT1 and DNMT3B with purified histone H1 was confirmed by far-Western blotting experiments (Fig. S6).
The in vitro interaction studies with H1 histones purified from mammalian cells suggest that some histone H1 subtypes may interact directly with DNMT1 and DNMT3B. To determine whether the interactions are indeed direct, we used purified H1 proteins produced in bacteria. We found that GST-DNMT1 and GST-DNMT3B bound recombinant H1d but did not bind H1c (Fig. 4E). These results demonstrate that H1d interacts directly with DNMT1 and DNMT3B. Combined with the in vivo and in vitro ChIP data (Figs. 2 B and C and 3B), these results indicate that H1d, and probably certain other H1 subtypes, can promote DNA methylation by interacting directly with and recruiting DNMT1 and DNMT3B to chromatin.
H1 linker histones consist of three domains, a short N-terminal region, a central, highly structured globular region, and a basic C-terminal domain (CTD) (31). To determine which of these regions interact with DNMTs, we prepared three fusion proteins in which GST was joined to full-length H1d or to the first 151 or the last 70 residues of H1d. We found that DNMT1 and DNMT3B bound to the full-length fusion protein and to the fusion protein containing the last 70 residues of the CTD but did not bind appreciably to the fusion protein containing residues 1–151 (Fig. 4F). The importance of the histone H1 CTD for binding DNMT1 and DNMT3B was demonstrated further by showing that substituting this region from H1d for the corresponding region in H1c, which binds these DNMTs only very weakly if at all (Fig. 4 A, D, and E), greatly increases binding of the two DNMTs (Fig. S4B). These results indicate that the last 70 residues of the H1d CTD are necessary and sufficient to bind both DNMT1 and DNMT3B.
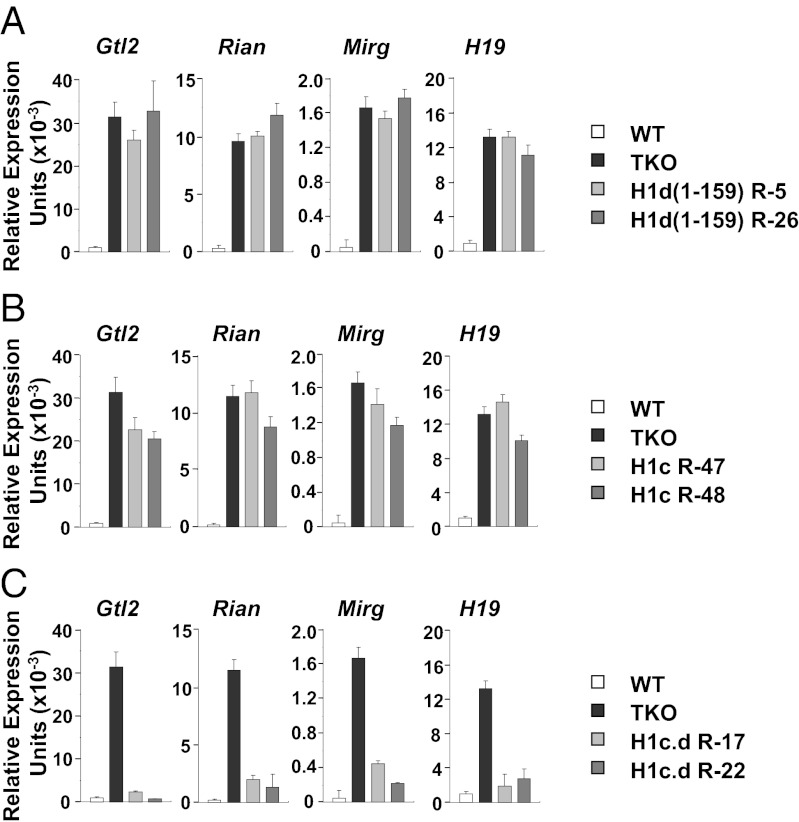
The preceding findings imply that residues 152–221 of the H1d CTD are crucial for the ability of H1d to recruit DNMT1 and DNMT3B to the H19 and Gtl2 ICRs and to repress expression of the H19, Gtl2, Rian, and Mirg transcripts. To test this prediction, we expressed an H1d protein (H1d 1–159) lacking the C-terminal 62 residues in the H1-depleted TKO ES cells. We found that the truncated H1d is unable to repress expression of the four genes (compare Figs. 5A and 1A). Using ChIP, we found that the truncated H1d also is unable to recruit DNMT1 and DNMT3B to the H19 and Gtl2 ICRs (Fig. S7). To prove that residues 152–221 of the H1d CTD are essential for gene silencing and recruitment of the two DNMTs, we made use of the fact that the H1c subtype interacts very poorly with the two DNMTs (Fig. 2A and Fig. S4). We found that when H1c is expressed in the TKO ES cells, it is much less effective than H1d in repressing expression of the four transcripts (Fig. 5B). It also does not promote recruitment of DNMT1 and DNMT3B to the ICRs in ES cells (Fig. S7). Therefore, we substituted residues 167–221 of the H1d CTD for the corresponding region of H1c and expressed the H1c-H1d chimeric protein in the TKO ES cells. As described above, the chimeric protein binds DNMT1 and DNMT3B (Fig. S4). We found that residues 167–221 of the H1d CTD conferred on the chimeric H1 the ability to repress expression of the four transcripts (Fig. 5C). Moreover, ChIP experiments showed that expression of the chimeric H1 leads to recruitment of DNMT1 and DNMT3B to the ICRs (Fig. S7). Taken together, these results clearly demonstrate the importance of the C-terminal region of the H1d CTD for H1-mediated effects at the ICRs in vivo.
Fig. 5.
The H1d CTD is required for silencing expression of the H19, Gtl2, Rian, and Mirg transcripts. (A) Levels of the indicated transcripts were measured by quantitative RT-PCR in WT, H1 TKO, and two stably transfected TKO ES cell lines expressing an exogenous H1d mutant lacking the C-terminal 62 residues [H1d(1–159) R-5 or H1d(1–159) R-26]. (B) Levels of the indicated transcripts were measured by quantitative RT-PCR in WT, H1 TKO, and two stably transfected TKO ES cell lines expressing exogenous H1c (H1c R-47 and H1c R-48). (C) Levels of the indicated transcripts were measured by quantitative RT-PCR in WT, H1 TKO, and two stably transfected TKO ES cell lines expressing an exogenous chimeric H1 (H1c.d R-17 and H1c.d R-22). The H1 histone subtype stoichiometries in these cell lines are shown in Fig. S8. Other details are as described in Fig. 1.
Interestingly, we found that both GST-DNMT1 and GST-DNMT3B also interact with histone H3 (Fig. 4D). Interaction of DNMT3B with the N-terminal tail of histone H3 was reported recently (32). Consistent with this report, we found that interaction of DNMT3B with histone H3 is strongly inhibited by H3K4 trimethylation but not by trimethylation of H3K9 (Fig. 4D). We also observed that H3K4 trimethylation inhibited the interaction of DNMT1 with histone H3 (Fig. 4D). Interactions of DNMT1 and DNMT3B with purified histone H3 were confirmed by far-Western blotting experiments (Fig. S6).
In summary, DNMT1 and DNMT3B interact with two of the major building blocks of chromatin, histones H1 and H3. Moreover, the interactions with histone H3 depend on the state of H3K4 methylation, which also is regulated by histone H1 through its ability to block binding and H3K4 methylation by the SET7/9 methyltransferase. We propose that histone H1 is a key player in regulating epigenetic silencing at the H19 and Gtl2 loci through a dual mechanism involving direct interaction and recruitment of DNMT1 and DNMT3B and also by inhibiting binding of SET7/9 and methylation of H3K4 (Fig. S9 and Discussion).
Discussion
The results reported here demonstrate that H1 linker histone promotes epigenetic silencing at the H19 and Gtl2 loci in mouse ES cells in two ways (Fig. S9). Histone H1 interacts directly with DNMT1 and DNMT3B, leading to their recruitment and DNA methylation of the ICRs. Histone H1 also interferes with binding of the SET7/9 methyltransferase to chromatin, inhibiting methylation of H3K4 in nucleosomes. These dual activities of histone H1 were observed both in vivo and with nucleosomes reconstituted in vitro. Hsitone H1 also was reported to inhibit histone H3 acetylation by PCAF in vitro (33). Low levels of the methylated H3K4 chromatin marker are generally associated with reduced transcription (34). Genome-wide measurements indicate that CpG methylation is highly correlated with unmethylated H3K4 (7, 9). Binding of DNMT-containing complexes to nucleosomes also is inhibited by methylation of H3K4 (6, 8, 32). Thus, the ability of histone H1 to interfere with H3K4 methylation contributes to its silencing activity and may further favor binding of DNMTs (Fig. S9). Recent work indicates that the methylation status of CpGs in mammals is highly dynamic because of both methylation and active demethylation reactions (35, 36). Because histone H1 can bind both DNMT3B and DNMT1, it is able to promote both the establishment and maintenance of methylated CpGs. Interestingly, we did not detect protein–protein interaction between histone H1 and DNMT3A. We also did not observe histone H1-mediated recruitment of DNMT3A to the H19 and Gtl2 ICRs. Thus, the effect of histone H1 appears to be specific for DNMT1- and DNMT3B-directed DNA methylation.
Our results also indicate that recruitment of DNMT1 and DNMT3B occurs via direct physical contact between the C-terminal end of the histone H1 CTD and the DNA methyltransferases. The evidence includes protein–protein interaction studies with histone H1 and DNMT1 and DNMT3B produced in bacteria. Although the histone H1 CTD is highly basic, this property alone is unlikely to account for binding of DNMT1 and DNMT3B. The CTDs of the H1c and H1(0) subtypes have a density of basic residues similar to that of other histone H1 subtypes, and they bind DNMT1 and DNMT3B much more weakly. The lack of interaction between histone H1 and DNMT3A also indicates a high degree of specificity for the interaction and is entirely consistent with our ChIP data showing that DNMT3A is not present at the H19 and Gtl2 ICRs. The histone H1 CTD has been implicated in chromatin condensation, and recent in vitro studies indicate this property localizes to two subdomains (37). Histone H1 has been reported to bind a number of other proteins, but in many cases the binding regions and functional consequences of binding have not been explored (31). However, two examples of functionally important interactions between the histone H1 CTD and other proteins have been studied. An and coworkers (38) reported that human H1.2 forms a complex with several other proteins that can repress p53-dependent transcription. Repression by the complex is dependent on direct interaction of H1.2 with p53, which requires the histone H1 CTD. Garrard and coworkers (39) have studied a functional interaction between the histone H1 CTD and the DNA fragmentation factor DFF40 that cleaves the linker DNA between nucleosomes during apoptosis-induced release of chromatin. Interestingly, it was shown that portions of the histone H1 CTD could promote DFF40 DNA binding and cleavage, independent of the histone H1 N-terminal and globular domains. Along with these other reports, the work described here significantly strengthens the view that the histone H1 CTD functions not only as a key structural component of chromatin but also as an adaptor module that enables certain other proteins to access chromatin.
We also found differences in the DNMT binding and silencing activity among the six H1 subtypes expressed in the ES cells. The H1c and H1(0) subtypes bound DNMT1 and DNMT3B much more weakly than the four other tested histone H1 subtypes (H1a, H1b, H1d, and H1e). H1c also was unable to silence expression at the H19 and Gtl2 loci. We localized the difference in silencing activity to the CTD by showing that expression of an H1c.d chimeric protein in which residues 167–221 from the C-terminal end of H1d replaced the corresponding region of H1c can silence expression. Expression of the chimeric H1 also led to recruitment of DNMT1 and DNMT3B to the H19 and Gtl2 ICRs, whereas expression of H1c and a truncated H1d lacking this region of the CTD did not lead to recruitment of the two DNMTs. Gene inactivation studies in mice have not revealed essential functions for any of these histone H1 subtypes, including H1c and H1(0) (15–17). Interestingly however, we reported previously that the genes encoding H1(0) and H1c exhibit differences in their regulation compared with the genes encoding the four other subtypes (40–42). These differences are attributable both to differences in transcriptional control and to the formation of H1(0) and H1c polyadenylated mRNAs, which are distinct from the cell cycle-dependent, nonpolyadenylated mRNAs formed by the other four genes. The functional differences among the histone H1 subtypes described here may provide a basis for further understanding the diversity in the mammalian histone H1 gene family.
Materials and Methods
Quantitative RT-PCR analyses of total RNA was performed as described previously (43). qChIP was carried out as described in our previous publications (21, 43–46) with specific modifications described in Sl Materials and Methods. Procedures for protein immunoprecipitation, Western blotting, and protein interactions with GST fusion proteins were described previously (47). Far-Western blotting experiments were carried out with histones extracted from chromatin with 0.2 N sulfuric acid, followed by SDS/PAGE, transfer to Immobilon-FL PVDF membranes (Millipore), and denaturation and renaturation as described previously (48). Procedures for DNA methylation analysis, in vitro chromatin assembly, histone methyltransferase and DNA methyltransferase assays, and in vitro qChIP with reconstituted chromatin are described in Sl Materials and Methods. Details of plasmid constructions, antibodies, and cell culture are described in Sl Materials and Methods. PCR primers used to assay specific mRNAs are listed in Table S1. PCR primers used to assay ChIP products are listed in Table S2. PCR primers used for MassARRAY are listed in Table S3.
Supplementary Material
Acknowledgments
We thank Yuhong Fan, Emilie Brasset, and Wouter de Laat for preliminary studies of CTCF occupancy; Dmitry Fyodorov, Matthew Gamble, John Greally, and Sandeep Wontakal for critical reading of this manuscript; and Keiichi Nakayama and Kunio Shiota for kindly providing plasmids. This work was supported by National Institutes of Health Grant CA079057. A.I.S. also received support from National Cancer Institute Cancer Center Grant 2P30CA13330.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213266110/-/DCSupplemental.
References
- 1.Klose RJ, Bird AP. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci. 2006;31(2):89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Cedar H, Bergman Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 3.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehnertz B, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13(14):1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 6.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448(7154):714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu JL, et al. The N-terminus of histone H3 is required for de novo DNA methylation in chromatin. Proc Natl Acad Sci USA. 2009;106(52):22187–22192. doi: 10.1073/pnas.0905767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laurent L, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20(3):320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Holde KE. Chromatin. New York: Springer; 1989. [Google Scholar]
- 11.Wolffe A. Chromatin: Structure and Function. London: Academic; 1998. [Google Scholar]
- 12.Izzo A, Kamieniarz K, Schneider R. The histone H1 family: Specific members, specific functions? Biol Chem. 2008;389(4):333–343. doi: 10.1515/BC.2008.037. [DOI] [PubMed] [Google Scholar]
- 13.Brown DT. 2001 Histone variants: Are they functionally heterogeneous? Genome Biol 2(7): REVIEWS 0006. [Google Scholar]
- 14.Happel N, Doenecke D. Histone H1 and its isoforms: Contribution to chromatin structure and function. Gene. 2009;431(1-2):1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Fan YH, Sirotkin A, Russell RG, Ayala J, Skoultchi AI. Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol Cell Biol. 2001;21(23):7933–7943. doi: 10.1128/MCB.21.23.7933-7943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan YH, et al. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol. 2003;23(13):4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sirotkin AM, et al. Mice develop normally without the H1(0) linker histone. Proc Natl Acad Sci USA. 1995;92(14):6434–6438. doi: 10.1073/pnas.92.14.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catez F, Ueda T, Bustin M. Determinants of histone H1 mobility and chromatin binding in living cells. Nat Struct Mol Biol. 2006;13(4):305–310. doi: 10.1038/nsmb1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown DT. Histone H1 and the dynamic regulation of chromatin function. Biochem Cell Biol. 2003;81(3):221–227. doi: 10.1139/o03-049. [DOI] [PubMed] [Google Scholar]
- 20.Bustin M, Catez F, Lim JH. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17(5):617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 21.Fan Y, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123(7):1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 22.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366(6453):362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 23.Tucker KL, et al. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 1996;10(8):1008–1020. doi: 10.1101/gad.10.8.1008. [DOI] [PubMed] [Google Scholar]
- 24.Dean W, et al. Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: Association with aberrant phenotypes. Development. 1998;125(12):2273–2282. doi: 10.1242/dev.125.12.2273. [DOI] [PubMed] [Google Scholar]
- 25.Warnecke PM, Biniszkiewicz D, Jaenisch R, Frommer M, Clark SJ. Sequence-specific methylation of the mouse H19 gene in embryonic cells deficient in the Dnmt-1 gene. Dev Genet. 1998;22(2):111–121. doi: 10.1002/(SICI)1520-6408(1998)22:2<111::AID-DVG1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 26.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 27.Karimi M, Luttropp K, Ekström TJ. Global DNA methylation analysis using the Luminometric Methylation Assay. Methods Mol Biol. 2011;791:135–144. doi: 10.1007/978-1-61779-316-5_11. [DOI] [PubMed] [Google Scholar]
- 28.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405(6785):482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 29.Hark AT, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405(6785):486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 30.Martin C, Cao R, Zhang Y. Substrate preferences of the EZH2 histone methyltransferase complex. J Biol Chem. 2006;281(13):8365–8370. doi: 10.1074/jbc.M513425200. [DOI] [PubMed] [Google Scholar]
- 31.McBryant SJ, Lu X, Hansen JC. Multifunctionality of the linker histones: An emerging role for protein-protein interactions. Cell Res. 2010;20(5):519–528. doi: 10.1038/cr.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010;38(13):4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrera JE, West KL, Schiltz RL, Nakatani Y, Bustin M. Histone H1 is a specific repressor of core histone acetylation in chromatin. Mol Cell Biol. 2000;20(2):523–529. doi: 10.1128/mcb.20.2.523-529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 35.Bhutani N, et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463(7284):1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popp C, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463(7284):1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu X, Hansen JC. Identification of specific functional subdomains within the linker histone H10 C-terminal domain. J Biol Chem. 2004;279(10):8701–8707. doi: 10.1074/jbc.M311348200. [DOI] [PubMed] [Google Scholar]
- 38.Kim K, et al. Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J Biol Chem. 2008;283(14):9113–9126. doi: 10.1074/jbc.M708205200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Widlak P, et al. The histone H1 C-terminal domain binds to the apoptotic nuclease, DNA fragmentation factor (DFF40/CAD) and stimulates DNA cleavage. Biochemistry. 2005;44(21):7871–7878. doi: 10.1021/bi050100n. [DOI] [PubMed] [Google Scholar]
- 40.Dong Y, Liu D, Skoultchi AI. An upstream control region required for inducible transcription of the mouse H1(zero) histone gene during terminal differentiation. Mol Cell Biol. 1995;15(4):1889–1900. doi: 10.1128/mcb.15.4.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng GH, Nandi A, Clerk S, Skoultchi AI. Different 3′-end processing produces two independently regulated mRNAs from a single H1 histone gene. Proc Natl Acad Sci USA. 1989;86(18):7002–7006. doi: 10.1073/pnas.86.18.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng GH, Skoultchi AI. Rapid induction of polyadenylated H1 histone mRNAs in mouse erythroleukemia cells is regulated by c-myc. Mol Cell Biol. 1989;9(6):2332–2340. doi: 10.1128/mcb.9.6.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J. 2005;24(21):3712–3723. doi: 10.1038/sj.emboj.7600834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rekhtman N, et al. PU.1 and pRB interact and cooperate to repress GATA-1 and block erythroid differentiation. Mol Cell Biol. 2003;23(21):7460–7474. doi: 10.1128/MCB.23.21.7460-7474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choe KS, Ujhelly O, Wontakal SN, Skoultchi AI. PU.1 directly regulates cdk6 gene expression, linking the cell proliferation and differentiation programs in erythroid cells. J Biol Chem. 2010;285(5):3044–3052. doi: 10.1074/jbc.M109.077727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wontakal SN, et al. A large gene network in immature erythroid cells is controlled by the myeloid and B cell transcriptional regulator PU.1. PLoS Genet. 2011;7(6):e1001392. doi: 10.1371/journal.pgen.1001392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: Functional antagonism in erythroid cells. Genes Dev. 1999;13(11):1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Y, Li Q, Chen XZ. Detecting protein-protein interactions by Far western blotting. Nat Protoc. 2007;2(12):3278–3284. doi: 10.1038/nprot.2007.459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.