Abstract
Positron emission tomography (PET) reporter genes allow noninvasive whole-body imaging of transplanted cells by detection with radiolabeled probes. We used a human deoxycytidine kinase containing three amino acid substitutions within the active site (hdCK3mut) as a reporter gene in combination with the PET probe [18F]-L-FMAU (1-(2-deoxy-2-18fluoro-β-L-arabinofuranosyl)-5-methyluracil) to monitor models of mouse and human hematopoietic stem cell (HSC) transplantation. These mutations in hdCK3mut expanded the substrate capacity allowing for reporter-specific detection with a thymidine analog probe. Measurements of long-term engrafted cells (up to 32 wk) demonstrated that hdCK3mut expression is maintained in vivo with no counter selection against reporter-labeled cells. Reporter cells retained equivalent engraftment and differentiation capacity being detected in all major hematopoietic lineages and tissues. This reporter gene and probe should be applicable to noninvasively monitor therapeutic cell transplants in multiple tissues.
Keywords: gene therapy, molecular imaging
Genetically modifying cells can offer novel therapeutic strategies for currently untreatable diseases (1). Standard methods for monitoring the long-term viability of transplanted cells are inadequate. Improved methods to serially detect transplanted cells in several tissues throughout the body simultaneously and noninvasively are critical to measure therapeutic efficacy (2, 3).
Hematopoietic stem cell transplants (HSCT) from both autologous and allogeneic sources have been successfully used in regenerative medicine (4). Genetic engineering through viral vector integration repairs defects in HSCs expanding clinical applications (5). Effective transplantation requires the engraftment of HSCs followed by an expansion into mature hematopoietic lineages repopulating multiple organs and peripheral blood. Measurement of mature hematopoietic cells in the peripheral blood is the primary diagnostic method for evaluating transplant efficacy. The major limitation of this approach is the lack of information about the engraftment within hematopoietic tissues.
Cells engineered to express a positron emission tomography (PET) reporter gene can be serially imaged in vivo with a reporter-specific probe (2). Most studies have used variants of the herpes simplex virus type 1 thymidine kinase (HSV1-TK or HSV1-sr39TK) and a radiolabeled penciclovir analog (9-(4-[18F]-fluoro-3-hydroxymethylbutyl)guanine, [18F]-FHBG) to detect reporter-labeled cells (6, 7). However, HSV1-TK is immunogenic and cells expressing this enzyme are selectively cleared over time potentially causing therapeutic failure (8–10). This immunogenicity has prevented the routine use of PET reporter genes clinically (11, 12).
Alternative potentially nonimmunogenic PET reporter genes have been investigated (3). Human nucleoside kinases deoxycytidine kinase (dCK) and thymidine kinase 2 (TK2) have similar substrate specificity to HSV1-TK. Several studies demonstrated the specific detection of reporter-labeled cells in mouse models with these human nucleoside kinases as PET reporters. Two studies developed xenografts expressing truncated TK2 or a mutant TK2 demonstrating reporter-specific imaging when probed with [18F]-thymidine analogs (13, 14). Infiltrating tumor-specific human T cells expressing a mutant dCK (dCKDM: R104M, D133A) reporter were detected within lung lesions of mice after transplantation by 2′-[18F]fluoro-5-ethyl-1-beta-D-arabinofuranosyluracil ([18F]-FEAU) PET (15). These reporter-labeled T cells were tested for cytolytic activity in vitro against target cells demonstrating that expression of dCKDM did not alter their short-term function (15).
Further investigation of human dCK as a PET reporter was selected based on multiple factors. mRNA encoding DCK is ∼800 bp, smaller than HSV1-TK, causing a minimal size increase when inserted into therapeutic vectors. The biological function of dCK has been described in genetic knockout mice (16, 17). The enzyme structure and kinetics of dCK are well characterized (18) with known point mutations that shift substrate specificity (18–20). A previous study successfully demonstrated the use of an alternate mutant dCK reporter and probe (15). Endogenous dCK activity can be monitored with PET using an alternate radiolabeled nucleoside analog (21).
How human nucleoside kinase reporters affect long-term cell-based therapies remains uncertain. Specifically it is unknown if constitutive expression in reporter-labeled cells is maintained within the recipient with no perturbation on cell function. Knockout dCK mice have a significant reduction in the total quantity of T and B lymphocytes caused by cellular stress from imbalanced nucleotide pools (22). Ectopic expression of nucleoside kinases could cause similar imbalances. Potential complications may include growth defect, disadvantage, or counterselection resulting in the loss of engrafted cells over time.
Our study demonstrates that a hdCK PET reporter can successfully monitor transplanted cells long term with no toxicity or survival disadvantage in modified cells. Models of mouse and human hematopoietic reconstitution were used to compare our reporter tracking for monitoring engraftment to peripheral blood sampling. Reporter-labeled cells exhibited identical behavior to nonlabeled cells with no differences detected regarding cell cycle, lineage, or tissue location. Our data provide evidence that hdCK3mut is an optimal reporter gene for hematopoietic cell tracking with future applications in a broad range of therapeutic cell transplants.
Results
Mutant Human dCK Functions as a PET Reporter When Probed with Thymidine Analogs.
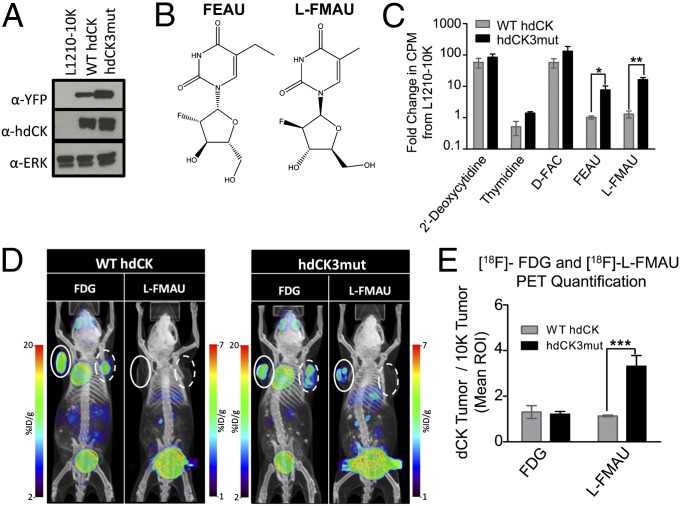
hdCK3mut contained three point mutations (A100V, R104M, and D133A) that were chosen based on a previous study that demonstrated a 1,100-fold increase in thymidine activity compared with wild-type dCK in enzyme kinetic assays (20). L1210-10K, a mouse leukemic cell line with no endogenous dCK was selected as a model cell line for in vitro studies (23). Stable expression of wild-type (WT) hdCK or hdCK3mut coexpressed with yellow fluorescent protein (YFP) through an internal ribosome entry site (IRES) in L1210-10K cells were generated to test probes for specific retention in hdCK3mut cells (Fig. 1A and vector maps in Fig. S1). Two thymidine analogs, 2′-fluoro-2′-deoxyarabinofuranosyl-5-ethyluracil (FEAU) and 1-(2-deoxy-2-fluoro-β-L-arabinofuranosyl)-5-methyluracil (L-FMAU), showed significant accumulation in reporter cell lines compared with wild type. Retention of the probe L-FMAU was 18-fold higher in hdCK3mut cells compared with WT hdCK (Fig. 1 B and C).
Fig. 1.
Development of a human thymidine selective PET reporter gene hdCK3mut. (A) Western blot analysis for equal expression of dCK and the linked fluorescent marker YFP in stable cell lines. (B) Chemical structure of two thymidine analogs FEAU and L-FMAU. (C) In vitro [3H]-nucleoside uptake assay. Results are displayed on a log10 scale as a fold change in counts per minute (cpm) from L1210-10K, a dCK-deficient cell line. (FEAU P = 0.027, L-FMAU P = 0.0052) (D) [18F]-FDG and [18F]-L-FMAU MicroPET scans of NSG mice with s.c. grafts. Right side is control L1210-10K (dotted line). Left side is L1210-10K cells with stable expression of WT dCK or hdCK3mut (solid line). (E) Region-of-interest quantification for [18F]-FDG and [18F]-L-FMAU (P = 0.0006).
Enzyme kinetic analysis further demonstrated high substrate affinity of hdCK3mut to L-FMAU with a measured Km of ∼13 µM. hdCK3mut had a fourfold lower Km for L-FMAU compared with the previously published dCKDM reporter (13 µM versus 56 µM). A high affinity PET reporter and probe combination is optimal because probes are administered at high specific activities with low concentrations of substrate. The decreased Km of hdCK3mut for L-FMAU demonstrates that it will achieve a higher velocity at a lower substrate concentration (Table S1).
Immune compromised NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were implanted with two s.c. grafts. The right side contained L1210-10K cells and the left side contained L1210-10k cell lines engineered to express WT hdCK or hdCK3mut. To determine tumor viability, animals were imaged by PET/CT with 2-deoxy-2-18fluoro-D-glucose ([18F]-FDG), a glucose analog used to measure glycolytic consumption. F-18 has a half-life of ∼110 min with probes decayed to undetectable levels within 24 h allowing for sequential scans with alternate probes. The following day [18F]-L-FMAU PET/CT scans detected hdCK3mut reporter expression with signal observed within hdCK3mut grafts (Fig. 1D). PET images were then quantified for total probe accumulation. Images were dose corrected to total radioactivity at the scan start time. Tumors were then selected in a region of interest (ROI) and the mean percent injected dose per gram (%ID/g) over the entire tumor was calculated. Tumor signal of the dCK transduced graft is compared as the fold change in probe retention to the nontransduced L1210-10K tumors of each animal. Tumors expressing hdCK3mut had a 3.3-fold increase in [18F]-L-FMAU retention (P = 0.0006) compared with WT hdCK grafts (Fig. 1E and Fig. S2). These results determined that hdCK3mut and L-FMAU make a suitable PET reporter gene and probe combination for in vivo studies.
Expression of hdCK3mut in Mouse HSCs Allow Noninvasive Detection of Reporter Cell Transplantation Before Normalization of Peripheral Blood Counts.
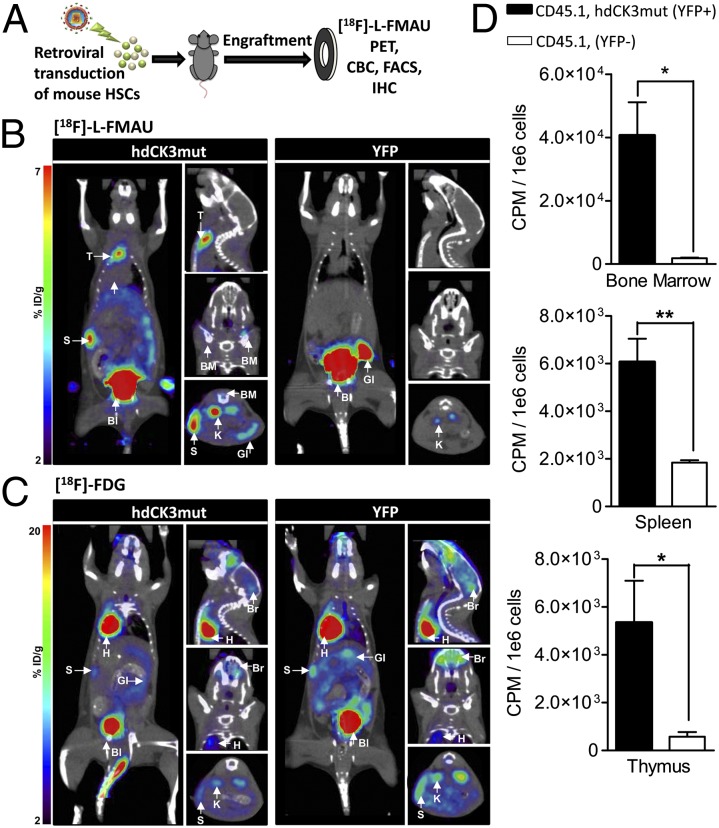
A competitive mouse bone marrow transplantation (BMT) study was chosen to test whether hdCK3mut can detect transplanted cells during early hematopoietic reconstitution (24–28).
Donor cells were generated by treating mice with 5-flourouracil 5 d preharvest for HSC enrichment. Collected bone marrow was retrovirally infected with ∼40–60% transduction efficiency to express hdCK3mut (coexpressed with YFP through an IRES) or the control of IRES-YFP only (Fig. S1). Recipient mice then received a lethal irradiation dose of 900 rads to eliminate host bone marrow. Mice were transplanted with the mixed population of reporter/nonreporter HSC-enriched donor bone marrow (Fig. 2A).
Fig. 2.
hdCK3mut and [18F]-L-FMAU PET can track reporter-labeled mouse hematopoietic cells during early engraftment and expansion in bone marrow chimera mice. (A) Lethally irradiated C57BL/6 (CD45.2) mice were transplanted with retrovirally transduced 5-FU–enriched HSCs (CD45.1). Animals were monitored for hematopoietic reconstitution over their total lifespan. MicroPET scans shown from Left,coronal; Right Upper, sagittal; Center, coronal; and Lower, transverse. (B) [18F]-L-FMAU at 4 wk posttransplant. Reporter signal observed in hdCK3mut animals in spleen (S), thymus (T), and bone marrow (BM). Probe metabolism in both cohorts seen in gastrointestinal (GI), bladder (Bl), and kidney (K). (C) [18F]-FDG MicroPET scan at 4 wk posttransplant. Nonreporter-specific signal observed in both cohorts in heart (H), spleen (S), gastrointestinal (GI), brain (Br), with metabolism in kidneys (K) and bladder (Bl). (D) In vivo accumulation of [18F]-L-FMAU in sorted hematopoietic cells from hdCK3mut animals. Reporter positive: CD45.1+, YFP+ and reporter negative: CD45.1+, YFP−. (P < 0.05).
Under standard conditions mice will display normalized engraftment and complete blood counts (CBC) within ∼8 wk after BMT (27). We hypothesized that early engraftment and expansion could be monitored by reporter imaging before normalization of peripheral blood measurements. At 4 wk post-BMT, animals received PET/CT scans with [18F]-FDG and the following day [18F]-L-FMAU (Fig. 2 B and C).
Whole-body glucose consumption was measured by [18F]-FDG MicroPET and was indistinguishable between the two cohorts of animals. Weak signal was detected within the spleen and bone marrow indicating a similar glycolytic rate across all animals (Fig. 2C). The following day PET/CT with [18F]-L-FMAU detected only hdCK3mut cells engrafted within the spleen, thymus, and focal areas within the bone marrow of reporter-labeled animals (Fig. 2B and Fig. S3). Animals in the control YFP cohort had no hematopoietic signal observed with [18F]-L-FMAU (Fig. 2B). Visualization of [18F]-L-FMAU in hematopoietic tissues of hdCK3mut recipients verified that reporter imaging can monitor early engraftment after BMT.
To confirm that [18F]-L-FMAU accumulation was specific for hdCK3mut cells, in vivo reporter and nonreporter accumulation was measured. Donor hematopoietic cells from hdCK3mut recipients were sorted for reporter positive or negative and then were counted for total radioactivity in counts per minute (cpm) normalized to cpm/1e6 cells. Cells expressing hdCK3mut had a significantly (P < 0.05) higher accumulation of [18F]-L-FMAU compared with unlabeled cells in all hematopoietic tissues (Fig. 2D).
Reporter Labeled Mouse HSCs Retain Expression of hdCK3mut with Equivalent Engraftment and Differentiation Capacity.
Overexpression of enzymes or reporter genes can potentially cause cellular stress, developmental defects during differentiation, growth disadvantage, or transformation (29, 30). The long-term effects from forced expression of hdCK3mut on mouse HSC’s engraftment and differentiation capacity was investigated. Reconstituted chimeric mice were evaluated to confirm that hdCK3mut expression was preserved and that mouse HSCs expressing the reporter maintained normal function after transplantation.
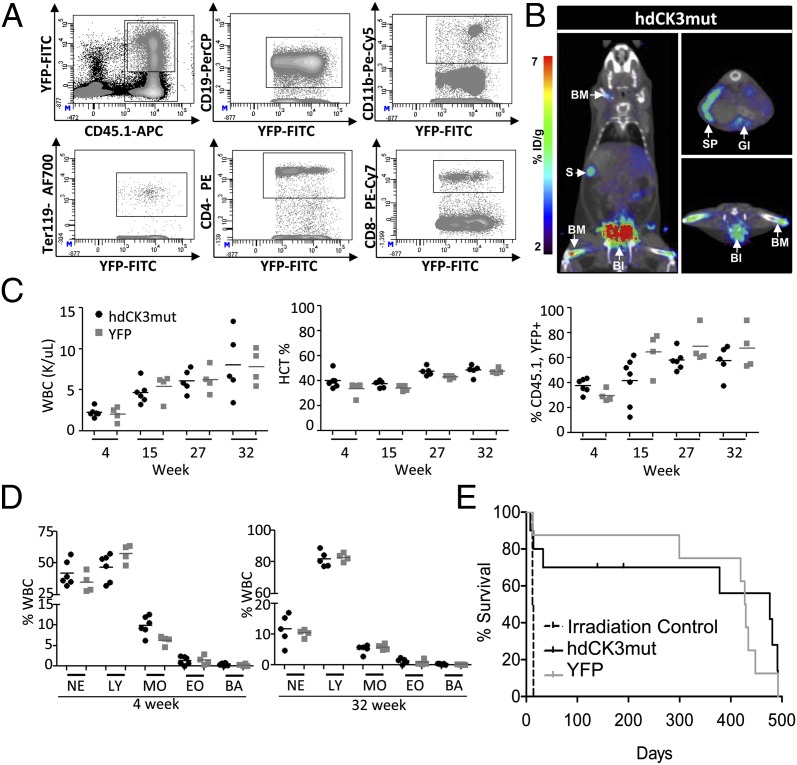
Reconstituted chimeric mice 6–8 wk post-BMT were analyzed by flow cytometry and immunohistochemical (IHC) analysis (Fig. 3A and Figs. S4 and S5). Flow cytometry analysis evaluated the spleen, thymus, bone marrow, and peripheral blood for total donor engraftment by lineage, reporter expression (YFP expression), and cell cycle. A representative fluorescent-activated cell sorting (FACS) plot of hdCK3mut engraftment within the spleen is displayed (Fig. 3A). No significant difference in reporter engraftment based on tissue location, lineage distribution, or cell cycle profiles from nonreporter labeled cells or in comparison with the YFP cohort was observed (Fig. S4).
Fig. 3.
hdCK3mut mouse HSCs persist in vivo allowing long-term monitoring of therapeutic cell transplantation. (A) Representative FACS plot for hdCK3mut engraftment within the spleen. Cells were monitored for CD45.1 (donor) and YFP (reporter) positive. Further gating demonstrates that reporter positive (YFP+) cells can be found in all major lineages. (B) [18F]-L-FMAU MicroPET at 32 wk post-BMT. (C) Serial monitoring of peripheral blood. Animals were monitored for total white blood cell (WBC), hematocrit (HCT), and reporter-labeled donor engraftment (CD45.1+, YFP+). (D) Distribution of white blood cells at early and late engraftment are indistinguishable between YFP and hdCK3mut animals. NE, neutrophils; LY, lymphocytes; MO, monocyte; EO, eosinophil; BA, basophil. (E) No survival disadvantage seen in hdCK3mut reporter animals.
Tissue architecture of the spleen and thymus was examined by hematoxylin and eosin staining (H&E) with normal morphology in both hdCK3mut and YFP mice. hdCK3mut engraftment was then detected although IHC of anti-dCK with no staining seen in YFP recipients. Anti-YFP IHC detected the linked fluorescent marker in both tissues and cohorts of animals confirming the flow cytometry data. Both dCK and YFP IHC identified the same engrafted hematopoietic cells in hdCK3mut animals, demonstrating the specificity of reporter detection using a newly developed monoclonal antibody generated in our laboratory (Fig. S5).
Mice received HSC-enriched bone marrow that was retrovirally transduced to express hdCK3mut or YFP in ∼50% of cells. This enrichment technique also contains residual committed short-term progenitor cells that can express the reporter. Transplantation of these progenitor cells is necessary for animal survival but these cells confound analysis of HSC differentiation at early time points. Measurements of mature hematopoietic cells from HSCs and progenitor cells are indistinguishable in peripheral blood analysis. A methylcellulose (MC) colony forming assay measured the expansion and differentiation capacity of reporter-labeled bone marrow 6 wk posttransplantation. Recipient animals’ bone marrow was harvested, sorted, and placed in MC for 12 d (Fig. S6A) (25). Quantification of the MC assay determined that cells from hdCK3mut recipient mice were equivalent to YFP animals and nonchimeric bone marrow in colony forming capacity (CFC) as well as colony type distribution (Fig. S6 B–F). Sorted hdCK3mut or YFP positive cells were comparable in CFC and colony type, demonstrating that expression of hdCK3mut does not cause a disadvantage during in vitro differentiation (Fig. S6E). Sorted cells from both YFP and hdCK3mut recipients retained reporter expression during in vitro differentiation detected through flow cytometry, confirming the continued expression of hdCK3mut throughout cell development (Fig. S6F).
Together these experiments demonstrate that expression of the hdCK3mut reporter gene has no observable selective disadvantage on hematopoietic cell engraftment and expansion capacity in a mouse HSC transplantation model.
hdCK3mut Mouse HSCs Persist in Vivo Allowing Long-Term Monitoring of Therapeutic Cell Transplantation.
Long-term effects from the expression of human nucleoside reporters are poorly defined. Potential concerns include selective vector silencing or counterselection of reporter-labeled cells over time.
We examined whether reporter cells in BMT-recipient mice retained the PET reporter function through serial imaging in vivo. Consecutive scans and peripheral blood analysis were obtained at 4, 15, 27, and 32 wk post-BMT allowing for detection of both short- and long-lived reporter HSCs (27, 28).
At 32 wk post-BMT hdCK3mut reporter-specific signal was detected within the spleen and bone marrow, demonstrating long-term engraftment capability of reporter-labeled hematopoietic cells (Fig. 3B). Previous scans of the same animals demonstrated similar signal at 15 and 27 wk (Fig. S7). Serial detection of hdCK3mut through PET/CT reveals that the reporter gene functions through hematopoiesis. It is hypothesized that each sequential scan is detecting different hematopoietic cells that have homed to the spleen.
Peripheral blood was collected at each time point and analyzed for CBC and reporter engraftment by flow cytometry. No significant difference was observed in the retention of circulating reporter positive hematopoietic cells between hdCK3mut and YFP recipients. Comparison of the total white blood cell (WBC) count and hematocrit were normal and equivalent between both hdCK3mut and YFP after BMT (Fig. 3C). Peripheral WBC differential demonstrated that at both early and late engraftment the distribution of WBC subtypes were consistent between both groups (Fig. 3D).
Reporter-labeled bone marrow transduced with YFP or hdCK3mut was able to successfully rescue lethally irradiated recipient animals. Long-term monitoring determined there was no survival disadvantage for hdCK3mut recipients over YFP as indicated in a Kaplan–Meier survival curve (Fig. 3E). Collectively these experiments demonstrate that expression of hdCK3mut is an inert reporter gene capable of long-term noninvasive tracking method throughout the recipients’ lifespan.
hdCK3mut Allows for Noninvasive Detection of Human HSC Engraftment.
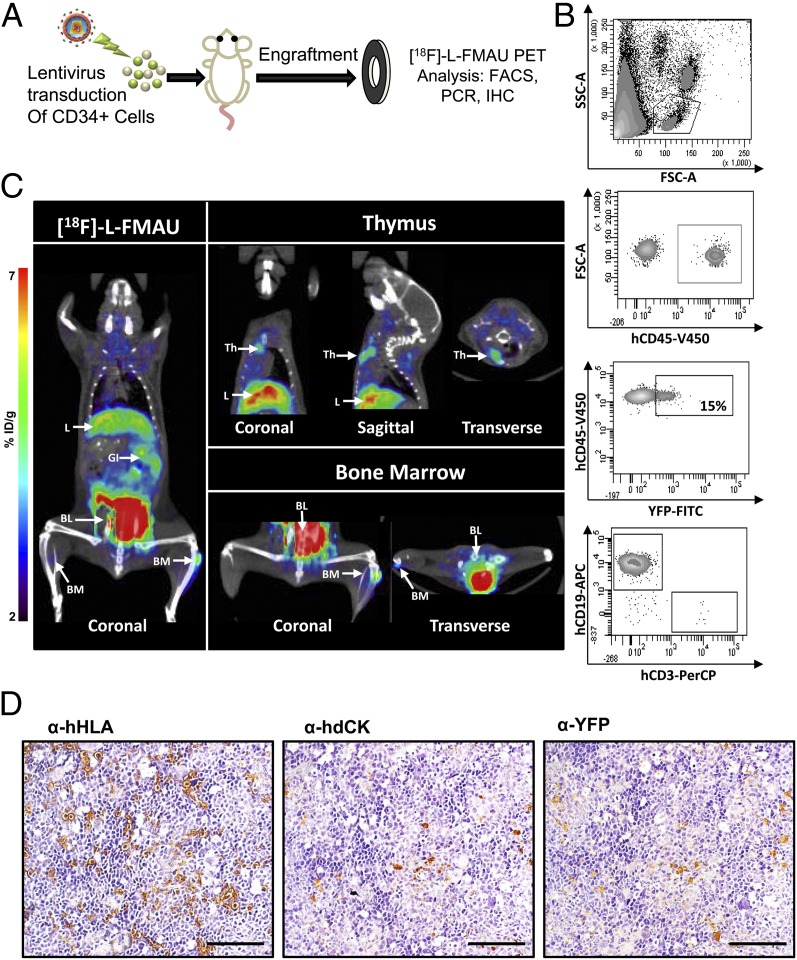
Probe retention in reporter-labeled cells is dependent upon transport of the radio-labeled probe intracellularly with sequential phosphorylation by hdCK3mut. Although mice and humans have similar nucleoside transporters, total expression or variation based on cell lineage may differ between species (31, 32). A humanized xenotransplantation model was used to validate that hdCK3mut would function as a reporter in human HSCs. Human hematopoiesis occurs within the spleen, bone marrow, and thymus of NSG mice when transplanted as neonates providing a tool to study in vivo human HSC differentiation (33). Isolated CD34+ cord blood HSCs were transduced with hdCK3mut or control lentiviral vectors (Fig. S1B) and transplanted into sublethally irradiated neonate NSG recipient animals (Fig. 4A). When transplanted as neonates CD34+ cells engraft and expand within multiple tissues and can home to the empty NSG thymus. These mice develop partial human hematopoietic systems with mature human myeloid, T-, and B cells.
Fig. 4.
hdCK3mut allows for noninvasive detection of human HSC engraftment. (A) Schematic of human HSC xenotransplantation. CD34+ cells from cord blood donors are transduced with lentivirus. Sublethally irradiated neonate NSG recipients are intrahepatically transplanted. (B) FACS plots for total human engraftment at 8 wk posttransplantation. hCD45 denotes total human cells, reporter cells are detected by YFP+. (C) [18F]-L-FMAU MicroPET detects human hematopoietic cells expressing hdCK3mut within the bone marrow (BM) and thymus (Th). Background signal from probe metabolism is seen in liver (L), gastrointestinal (GI) with probe clearance through the bladder (BL). NSG mice displayed higher probe background compared with C57Bl6 animals seen by increased nonspecific liver signal. (D) IHC detects hdCK3mut-labeled human hematopoietic cells within the spleen. Total engraftment detected with α-hHLA, reporter cells detected by α-hdCK and α-YFP. (Scale bar, 100 μm.)
At 8 wk post human HSC transplantation peripheral blood analysis detected hdCK3mut reporter human engraftment by flow cytometry (Fig. 4B). Peripheral blood mononuclear cells (PBMCs) were stained for human CD45 to detect total human engraftment. Additional markers were used to detect human myeloid, B-cell, T-cell, and YFP for reporter-labeled cells. MicroPET scans with [18F]-L-FMAU detected hdCK3mut cells within the thymus and bone marrow of chimeric recipient mice (Fig. 4C). This demonstrates that human cells labeled with hdCK3mut retain expression of the reporter and are capable of engrafting after transplantation.
IHC analysis of human HSC engraftment in the spleen and thymus was performed at the experimental endpoint (Fig. 4D and Fig. S8). Total human engraftment was detected with human-specific HLA staining. Sequential sections verified reporter positive cells by anti-dCK and anti-YFP staining. Anti-dCK IHC in the spleen stained a fraction of the total engrafted human cells, consistent with the peripheral blood FACS (Fig. 4B), which revealed ∼15% of human cells that were reporter positive based on YFP expression (Fig. 4D). This supports the hypothesis that human hematopoietic cell maturation and homing is retained in cells expressing hdCK3mut.
Overlapping Integration Sites in hdCK3mut-Labeled Human Hematopoietic Cells Defines a Common Cell of Origin with Multilineage Differentiation Capacity in Vivo.
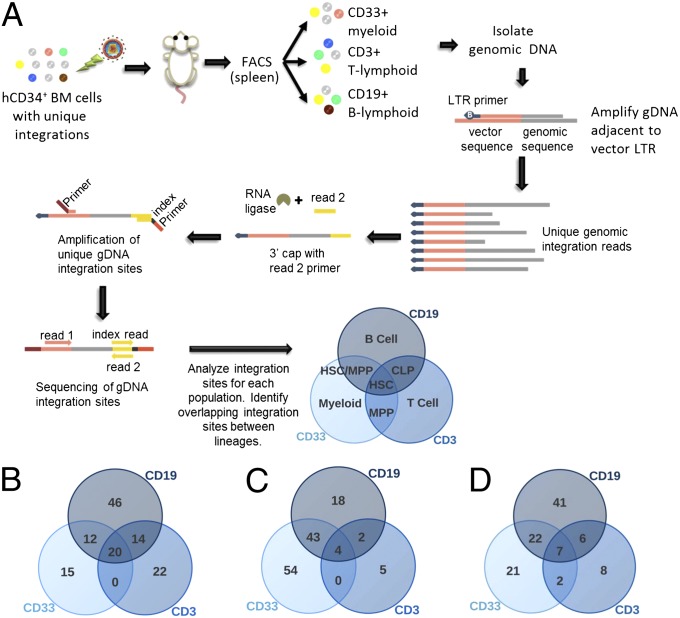
A concern of gene therapy trials for the correction of inherited diseases is the potential for insertional mutagenesis that has been observed in rare cases (34). Vector integration within tumor suppressors, near the transcriptional start site of oncogenes, or at sites that alter cell function are potential complications when using viral integration methods. Integration of lentiviral vectors is less likely to cause oncogenic transformation that was previously seen with other retroviral vectors (35). Recent studies have focused on identifying integration sites of modified hHSCs to detect potential problems such as dominant clonal expansion or lineage restriction (36, 37). Integration site analysis on long-term engrafted human chimeric mice was used to determine if expression and integration of hdCK3mut resulted in an abnormal event.
Cells were sorted from the spleens of engrafted animals into three lineages based on human CD33, CD3, or CD19 expression. Total genomic DNA was isolated and sequences flanking the vector integration sites were amplified by using common primers within the LTRs. Short primers were then ligated to the 3′ end of all amplified DNA allowing uniform ends of all fragments. A second PCR amplification was then performed to attach unique barcode sequences to the 3′ end. This allows multiple samples to be sequenced together and each sample to be precisely identified. Pooled samples underwent paired-end 50-nt Illumina sequencing to identify the unique integration site. Results were then aligned against genomic DNA to identify the exact integration location. Comparison of myeloid, B- and T-cell integrations were analyzed for each animal identifying individual and overlapping integrations (Fig. 5A).
Fig. 5.
Overlapping integration sites in hdCK3mut-labeled human hematopoietic cells defines a common cell of origin with multipotent lineage capacity in vivo. (A) Schematic of integration site analysis. Cells from the spleens of engrafted animals are sorted into three populations. DNA is amplified from the LTR and a 3′ cap is attached. Unique index sequence is placed on all samples in a second amplification. Samples are sequenced, and genomic integration sites are determined. Overlapping integration sites from multiple lineages determines a common cell of origin differentiated into multiple hematopoietic lineages. (B) YFP-engrafted animal. (C and D) hdCK3mut-engrafted animals.
Lineage-specific integrations identified committed progenitor cells. Integration sites found in all three populations are derived from a common transduced cell of origin (HSC) which then differentiated into all lineages (Fig. 5 B–D). Previous vector copy number per cell was determined by PCR to be ∼0.5 (0.485, hdCK3mut; 0.494, YFP) after transduction. It is estimated that each HSC integration site represents a single engrafted clone.
A comparison of the total number of integrations from each sample in hdCK3mut and YFP animals confirms that the number of integrations detected is similar between vectors. This demonstrates that the expression of hdCK3mut does not prevent hHSCs from differentiating into all major hematopoietic lineages within the humanized mouse model. hdCK3mut also had no effect on long-term engraftment and was detected up to 5 mo post-HSC transplant with no lineage restriction due to gene toxicity or clonal expansion due to growth advantage.
Discussion
We have demonstrated that an alternate dCK mutant (hdCK3mut) is well-tolerated, highly sensitive, and capable of monitoring long-term HSC engraftment.
Expression of HSV1-TK After Gene Transfer Provides a Safety Mechanism in Aberrant Reporter Cell Populations Through Reporter-Specific Cytoxicity (38, 39).
hdCK3mut provides an alternative PET reporter gene to sr39TK. One concern for alternate reporters is the loss of suicide gene function, which is gained when HSV1-TK and mutants are used (40). Gene transfer of HSV1-TK into select cell populations allowed for targeted cytotoxicity when treated with FDA-approved acycloguanosine-based antiviral drugs such as ganciclovir (41). This eliminates all cells expressing the reporter with limited off-target cytotoxicity (38). A limitation in HSV1-TK expression is that prophylaxis treatment with acycloguanosine antivirals to minimize cytomegalovirus infection in immunocompromised individuals cannot be administered without potentially eliminating the donor cells (42).
In previous studies, hdCK3mut also exhibited higher substrate specificity for several chemotherapeutic and antiviral nucleoside analogs in comparison with wild-type dCK (18–20). hdCK3mut had a higher activity with gemcitabine (2′,2′-difluorodeoxycytidine, dFdC) with little activity toward acycloguanosine drugs. dFdC is an FDA-approved chemotherapeutic that works through self-potentiation in the diphosphate and triphosphate forms (43). The triphosphate form is then incorporated into DNA, causing chain termination. Treatment with drugs that have a higher affinity for hdCK3mut may allow a targeted suicide gene therapy. Proper in vivo models will need to be tested to validate hdCK3mut enhanced sensitivity and to evaluate the off-target effects from treatment.
Evaluating the Potential Immunogenicity of hdC3Kmut in Short-Term Therapeutic Cell Transplants.
Before transitioning hdCK3mut into a clinical PET reporter for stem cell or long-term therapeutic transplants experimental short-lived transplanted cells need to be investigated. The immunogenicity of HSV1-TK was found when reporter-labeled lymphocytes caused a CD8 immune response selectively killing reporter cells (8–10). A similar study could determine if expression of hdCK3mut will cause an immune response.
hdCK3mut contains only three point mutations and is expected to not cause an immunogenic epitope for MHC class 1 presentation. Using a predictive software for MHC class 1 presentation (44), hdCK3mut has only one additional peptide fragment, which is predicted to be different from WT hdCK. This peptide, which incorporates the mutated amino acid methionine at position 104, could use the methionine as an anchoring residue within the MHC (45). All other amino acids that are displayed in the MHC are natural, and therefore the peptide fragments displayed from hdCK3mut are predicted to be recognized as “self” avoiding immune cell detection.
Summary.
We demonstrate how long-term follow-up of transplanted cells can be managed noninvasively by using reporter PET imaging. This study provides a comprehensive analysis on the inert biological effect of hdCK3mut on hematopoietic cells. Therefore, it is anticipated that hdCK3mut monitoring will provide a safe and effective mechanism for longitudinal monitoring of a broad range of transplanted cells.
Materials and Methods
Detailed information on animals, constructs and cloning, cell lines, uptake assays, enzyme kinetic assay, grafts, mouse HSC transplant, human HSC transplant, MicroPET imaging, peripheral blood analysis, FACS, methylcellulose assay, antibodies, Western blot, IHC, integration site analysis, and statistics can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Nagichettiar Satyamurthy and Jeffery Collins for the radiosynthesis of [18F]-FDG and [18F]-L-FMAU and Dr. Waldemer Lando, Dr. David Stout, and Darrin Williams in the Crump Institute for Molecular Imaging facility for their technical help with PET/CT scans. M.N.M. is supported by California Institute for Regenerative Medicine Training Grant TG2-01169. E.H.G. is supported by Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA. A.R.C. is supported by a Philip Whitcome training grant. C.N. is supported by the California Institute for Regenerative Medicine Bridges Training Program. O.N.W. is supported by a California Institute for Regenerative Medicine Tools/Technology Award RT1-01126. O.N.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221840110/-/DCSupplemental.
References
- 1.Sng J, Lufkin T. Emerging stem cell therapies: Treatment, safety, and biology. Stem Cells Int. 2012;2012:521343. doi: 10.1155/2012/521343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herschman HR. PET reporter genes for noninvasive imaging of gene therapy, cell tracking and transgenic analysis. Crit Rev Oncol Hematol. 2004;51(3):191–204. doi: 10.1016/j.critrevonc.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Serganova I, Ponomarev V, Blasberg R. Human reporter genes: Potential use in clinical studies. Nucl Med Biol. 2007;34(7):791–807. doi: 10.1016/j.nucmedbio.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Gyurkocza B, Rezvani A, Storb RF. Allogeneic hematopoietic cell transplantation: The state of the art. Expert Rev Hematol. 2010;3(3):285–299. doi: 10.1586/ehm.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohn DB. Update on gene therapy for immunodeficiencies. Clin Immunol. 2010;135(2):247–254. doi: 10.1016/j.clim.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gambhir SS, et al. A mutant herpes simplex virus type 1 thymidine kinase reporter gene shows improved sensitivity for imaging reporter gene expression with positron emission tomography. Proc Natl Acad Sci USA. 2000;97(6):2785–2790. doi: 10.1073/pnas.97.6.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tjuvajev JG, et al. Imaging herpes virus thymidine kinase gene transfer and expression by positron emission tomography. Cancer Res. 1998;58(19):4333–4341. [PubMed] [Google Scholar]
- 8.Berger C, Flowers ME, Warren EH, Riddell SR. Analysis of transgene-specific immune responses that limit the in vivo persistence of adoptively transferred HSV-TK-modified donor T cells after allogeneic hematopoietic cell transplantation. Blood. 2006;107(6):2294–2302. doi: 10.1182/blood-2005-08-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riddell SR, et al. T-cell mediated rejection of gene-modified HIV-specific cytotoxic T lymphocytes in HIV-infected patients. Nat Med. 1996;2(2):216–223. doi: 10.1038/nm0296-216. [DOI] [PubMed] [Google Scholar]
- 10.Traversari C, et al. The potential immunogenicity of the TK suicide gene does not prevent full clinical benefit associated with the use of TK-transduced donor lymphocytes in HSCT for hematologic malignancies. Blood. 2007;109(11):4708–4715. doi: 10.1182/blood-2006-04-015230. [DOI] [PubMed] [Google Scholar]
- 11.Mercier-Letondal P, et al. Early immune response against retrovirally transduced herpes simplex virus thymidine kinase-expressing gene-modified T cells coinfused with a T cell-depleted marrow graft: An altered immune response? Hum Gene Ther. 2008;19(9):937–950. doi: 10.1089/hum.2007.156. [DOI] [PubMed] [Google Scholar]
- 12.Oliveira G, Greco R, Lupo-Stanghellini MT, Vago L, Bonini C. Use of TK-cells in haploidentical hematopoietic stem cell transplantation. Curr Opin Hematol. 2012;19(6):427–433. doi: 10.1097/MOH.0b013e32835822f5. [DOI] [PubMed] [Google Scholar]
- 13.Campbell DO, et al. Structure-guided engineering of human thymidine kinase 2 as a positron emission tomography reporter gene for enhanced phosphorylation of a non-natural thymidine analog reporter probe. J Biol Chem. 2012;287(1):446–454. doi: 10.1074/jbc.M111.314666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ponomarev V, et al. A human-derived reporter gene for noninvasive imaging in humans: Mitochondrial thymidine kinase type 2. J Nucl Med. 2007;48(5):819–826. doi: 10.2967/jnumed.106.036962. [DOI] [PubMed] [Google Scholar]
- 15.Likar Y, et al. A new pyrimidine-specific reporter gene: A mutated human deoxycytidine kinase suitable for PET during treatment with acycloguanosine-based cytotoxic drugs. J Nucl Med. 2010;51(9):1395–1403. doi: 10.2967/jnumed.109.074344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi O, et al. A deficiency in nucleoside salvage impairs murine lymphocyte development, homeostasis, and survival. J Immunol. 2012;188(8):3920–3927. doi: 10.4049/jimmunol.1102587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toy G, et al. Requirement for deoxycytidine kinase in T and B lymphocyte development. Proc Natl Acad Sci USA. 2010;107(12):5551–5556. doi: 10.1073/pnas.0913900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabini E, Ort S, Monnerjahn C, Konrad M, Lavie A. Structure of human dCK suggests strategies to improve anticancer and antiviral therapy. Nat Struct Biol. 2003;10(7):513–519. doi: 10.1038/nsb942. [DOI] [PubMed] [Google Scholar]
- 19.Hazra S, Sabini E, Ort S, Konrad M, Lavie A. Extending thymidine kinase activity to the catalytic repertoire of human deoxycytidine kinase. Biochemistry. 2009;48(6):1256–1263. doi: 10.1021/bi802062w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iyidogan P, Lutz S. Systematic exploration of active site mutations on human deoxycytidine kinase substrate specificity. Biochemistry. 2008;47(16):4711–4720. doi: 10.1021/bi800157e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radu CG, et al. Molecular imaging of lymphoid organs and immune activation by positron emission tomography with a new [18F]-labeled 2′-deoxycytidine analog. Nat Med. 2008;14(7):783–788. doi: 10.1038/nm1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Austin WR, et al. Nucleoside salvage pathway kinases regulate hematopoiesis by linking nucleotide metabolism with replication stress. J Exp Med. 2012;209(12):2215–2228. doi: 10.1084/jem.20121061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordheim LP, et al. Characterization of a gemcitabine-resistant murine leukemic cell line: Reversion of in vitro resistance by a mononucleotide prodrug. Clin Cancer Res. 2004;10(16):5614–5621. doi: 10.1158/1078-0432.CCR-04-0506. [DOI] [PubMed] [Google Scholar]
- 24.Baum C, et al. Side effects of retroviral gene transfer into hematopoietic stem cells. Blood. 2003;101(6):2099–2114. doi: 10.1182/blood-2002-07-2314. [DOI] [PubMed] [Google Scholar]
- 25.Drize N, Chertkov J, Sadovnikova E, Tiessen S, Zander A. Long-term maintenance of hematopoiesis in irradiated mice by retrovirally transduced peripheral blood stem cells. Blood. 1997;89(5):1811–1817. [PubMed] [Google Scholar]
- 26.Stewart FM, Crittenden RB, Lowry PA, Pearson-White S, Quesenberry PJ. Long-term engraftment of normal and post-5-fluorouracil murine marrow into normal nonmyeloablated mice. Blood. 1993;81(10):2566–2571. [PubMed] [Google Scholar]
- 27.Kang E, et al. In vivo persistence of retrovirally transduced murine long-term repopulating cells is not limited by expression of foreign gene products in the fully or minimally myeloablated setting. Hum Gene Ther. 2001;12(13):1663–1672. doi: 10.1089/10430340152528156. [DOI] [PubMed] [Google Scholar]
- 28.Zavidij O, et al. Stable long-term blood formation by stem cells in murine steady-state hematopoiesis. Stem Cells. 2012;30(9):1961–1970. doi: 10.1002/stem.1151. [DOI] [PubMed] [Google Scholar]
- 29.Gambhir SS, et al. Imaging transgene expression with radionuclide imaging technologies. Neoplasia. 2000;2(1–2):118–138. doi: 10.1038/sj.neo.7900083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hallek M, Wanders L, Strohmeyer S, Emmerich B. Thymidine kinase: a Tumor marker with prognostic value for non-Hodgkin’s lymphoma and a broad range of potential clinical applications. Ann Hematol. 1992;65(1):1–5. doi: 10.1007/BF01715117. [DOI] [PubMed] [Google Scholar]
- 31.Baldwin SA, et al. The equilibrative nucleoside transporter family, SLC29. Pflugers Arch. 2004;447(5):735–743. doi: 10.1007/s00424-003-1103-2. [DOI] [PubMed] [Google Scholar]
- 32.Gray JH, Owen RP, Giacomini KM. The concentrative nucleoside transporter family, SLC28. Pflugers Arch. 2004;447(5):728–734. doi: 10.1007/s00424-003-1107-y. [DOI] [PubMed] [Google Scholar]
- 33.Park CY, Majeti R, Weissman IL. In vivo evaluation of human hematopoiesis through xenotransplantation of purified hematopoietic stem cells from umbilical cord blood. Nat Protoc. 2008;3(12):1932–1940. doi: 10.1038/nprot.2008.194. [DOI] [PubMed] [Google Scholar]
- 34.Corrigan-Curay J, et al. Challenges in vector and trial design using retroviral vectors for long-term gene correction in hematopoietic stem cell gene therapy. Mol Ther. 2012;20(6):1084–1094. doi: 10.1038/mt.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biffi A, et al. Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood. 2011;117(20):5332–5339. doi: 10.1182/blood-2010-09-306761. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez-Murillo A, Lozano ML, Montini E, Bueren JA, Guenechea G. Unaltered repopulation properties of mouse hematopoietic stem cells transduced with lentiviral vectors. Blood. 2008;112(8):3138–3147. doi: 10.1182/blood-2008-03-142661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ronen K, et al. Distribution of lentiviral vector integration sites in mice following therapeutic gene transfer to treat β-thalassemia. Mol Ther. 2011;19(7):1273–1286. doi: 10.1038/mt.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonini C, et al. HSV-TK gene transfer into donor lymphocytes for control of allogeneic graft-versus-leukemia. Science. 1997;276(5319):1719–1724. doi: 10.1126/science.276.5319.1719. [DOI] [PubMed] [Google Scholar]
- 39.Ciceri F, et al. Antitumor effects of HSV-TK-engineered donor lymphocytes after allogeneic stem-cell transplantation. Blood. 2007;109(11):4698–4707. doi: 10.1182/blood-2006-05-023416. [DOI] [PubMed] [Google Scholar]
- 40.Lupo-Stanghellini MT, et al. Clinical impact of suicide gene therapy in allogeneic hematopoietic stem cell transplantation. Hum Gene Ther. 2010;21(3):241–250. doi: 10.1089/hum.2010.014. [DOI] [PubMed] [Google Scholar]
- 41.Tiberghien P, et al. Ganciclovir treatment of herpes simplex thymidine kinase-transduced primary T lymphocytes: An approach for specific in vivo donor T-cell depletion after bone marrow transplantation? Blood. 1994;84(4):1333–1341. [PubMed] [Google Scholar]
- 42.Hébrard C, Dumontet C, Jordheim LP. Development of gene therapy in association with clinically used cytotoxic deoxynucleoside analogues. Cancer Gene Ther. 2009;16(7):541–550. doi: 10.1038/cgt.2009.25. [DOI] [PubMed] [Google Scholar]
- 43.Burris HA, 3rd, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: A randomized trial. J Clin Oncol. 1997;15(6):2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 44.Hakenberg J, et al. MAPPP: MHC class I antigenic peptide processing prediction. Appl Bioinformatics. 2003;2(3):155–158. [PubMed] [Google Scholar]
- 45.Reche PA, Glutting JP, Reinherz EL. Prediction of MHC class I binding peptides using profile motifs. Hum Immunol. 2002;63(9):701–709. doi: 10.1016/s0198-8859(02)00432-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.