Abstract
CD1d-restricted natural killer T (NKT) cells include two major subgroups. The most widely studied are Vα14Jα18+ invariant NKT (iNKT) cells that recognize the prototypical α-galactosylceramide antigen, whereas the other major group uses diverse T-cell receptor (TCR) α-and β-chains, does not recognize α-galactosylceramide, and is referred to as diverse NKT (dNKT) cells. dNKT cells play important roles during infection and autoimmunity, but the antigens they recognize remain poorly understood. Here, we identified phosphatidylglycerol (PG), diphosphatidylglycerol (DPG, or cardiolipin), and phosphatidylinositol from Mycobacterium tuberculosis or Corynebacterium glutamicum as microbial antigens that stimulated various dNKT, but not iNKT, hybridomas. dNKT hybridomas showed distinct reactivities for diverse antigens. Stimulation of dNKT hybridomas by microbial PG was independent of Toll-like receptor-mediated signaling by antigen-presenting cells and required lipid uptake and/or processing. Furthermore, microbial PG bound to CD1d molecules and plate-bound PG/CD1d complexes stimulated dNKT hybridomas, indicating direct recognition by the dNKT cell TCR. Interestingly, despite structural differences in acyl chain composition between microbial and mammalian PG and DPG, lipids from both sources stimulated dNKT hybridomas, suggesting that presentation of microbial lipids and enhanced availability of stimulatory self-lipids may both contribute to dNKT cell activation during infection.
Keywords: type II natural killer T cell, CD1 antigen presentation, innate immunity, adaptive immunity
Natural killer T (NKT) cells recognize lipid antigens presented by CD1d (1, 2). Two major subsets of NKT cells have been described in both mice and humans based on the utilization of T-cell receptor (TCR)-α and -β genes: the widely studied type I or invariant NKT (iNKT) cells express a semi-invariant Vα14Jα18 TCR α-chain in mice and Vα24Jα18 in humans paired with a limited set of β-chains (Vβ7, Vβ8, and Vβ2 in mice and Vβ11 in humans). iNKT cells can recognize the prototypical α-galactosylceramide (α-GalCer) antigen; several glycosylated microbial ceramides and diacylglycerol lipids; and the self-antigens β-glucosylceramide (β-GluCer), isoglobotrihexosylceramide, phospholipids, and lysophospholipids (3). iNKT cells display an effector/memory phenotype and rapidly secrete large amounts of cytokines following primary stimulation. In contrast, CD1d-restricted NKT cells that use diverse TCR α- and β-chains have also been identified, although recurrent use of certain germ-line segments has been observed (4–8). These NKT cells are referred to as type II or diverse NKT (dNKT) cells.
The TCR diversity of dNKT cells suggests antigen specificities that are distinct from those of iNKT cells. Indeed, dNKT cells do not recognize α-GalCer or any of the α-glycan–linked microbial lipid antigens that are recognized by iNKT cells, and the self-lipid sulfatide, which is recognized by a small subset of mouse dNKT cells, is not recognized by iNKT cells (9, 10). Furthermore, the self-antigen β-GluCer, which stimulates most or all human and mouse iNKT cells, stimulates only a small subset of mouse dNKT cells (11, 12). A number of studies have documented, or inferred, a function for dNKT cells during infection (13, 14), yet the antigens that stimulate dNKT cells during infection remain unknown. Studies using transgenic or sulfatide-specific dNKT cells revealed that a substantial fraction of dNKT cells have a naïve phenotype (CD44lo, CD62Lhi, CD69neg/lo) and express chemokine receptors, integrins and effector molecules distinct from iNKT cells (7, 15, 16). Thus, utilization of diverse TCRs by dNKT cells correlates with distinct antigen specificity, phenotype, and functional attributes compared with iNKT cells.
In humans, CD1a-, b-, and c-reactive T cells use diverse αβ TCRs. The pattern of intracellular trafficking and localization of human CD1b is strikingly similar to that of mouse CD1d (17). Human CD1b is known to present lipid antigens from Mycobacterium tuberculosis (Mtb), and CD1b-restricted T cells expand following Mycobacterium bovis bacillus Calmette–Guérin (BCG) immunization and Mtb infection in humans (14). Furthermore, immunization with Mtb lipids protected guinea pigs that harbor CD1b and CD1b-restricted T cells from subsequent Mtb infection (18). Based on the similarities between CD1b and CD1d and the utilization of diverse TCRs by both human CD1b-restricted T cells and mouse dNKT cells, we hypothesized that dNKT cells may also recognize mycobacterial lipid antigens.
Here, we found that Mtb-infected antigen-presenting cells (APCs) and several mycobacterial phospholipids stimulated dNKT hybridomas. dNKT hybridomas displayed distinct, albeit partially overlapping, antigen specificities for different lipid antigens. Interestingly, the mammalian counterparts of stimulatory microbial phospholipids, although composed of structurally distinct acyl chains, were also stimulatory for dNKT hybridomas, suggesting that the foreign-lipid reactive repertoire of dNKT cells is shaped by cross-reactivity with self-lipid antigens.
Results
Mtb Lipids Stimulate dNKT Cells.
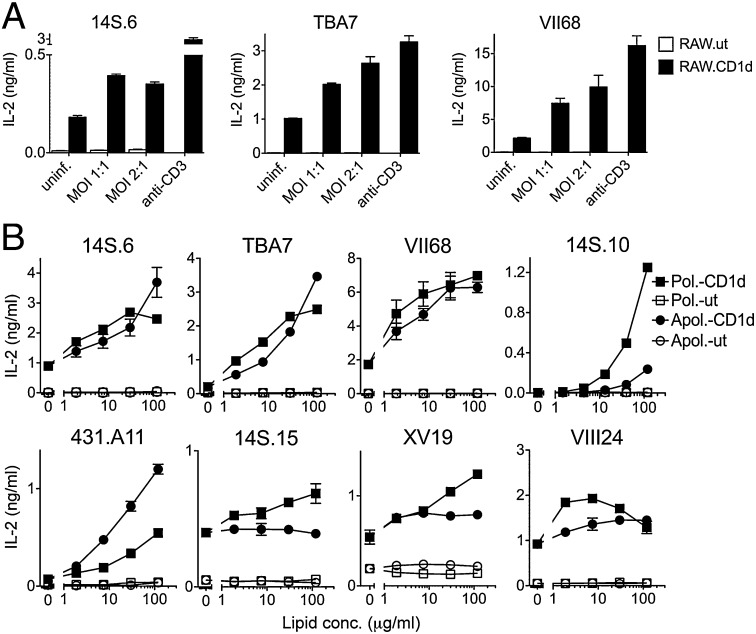
To investigate whether CD1d-restricted dNKT cells recognize microbial lipid antigens, we studied eight dNKT cell hybridomas that were previously identified based on their recognition of CD1d expressing APCs and the use of Vα14-negative TCRs. Of the eight hybridomas, the XV19 and 14S.15 hybridomas recognize sulfatide and the XV19 and VII68 hybridomas recognize β-GluCer. Seven of the hybridomas use diverse Vα and Jα TCR gene segments, whereas the 431.A11 hybridoma uses a recurrent Vα3.2/Jα9+ TCR. To investigate whether the dNKT cell hybridomas are activated by Mtb-infected APCs, we used an in vitro infection system with dNKT hybridomas and a CD1d-transfected macrophage cell line (RAW cells). After incubation with RAW cells infected with virulent Mtb H37Rv dNKT hybridomas produced IL-2 in a CD1d-dependent manner (Fig. 1A).
Fig. 1.
Mtb infected APCs and Mtb lipids stimulate dNKT hybridomas. (A) CD1d-transfected or untransfected RAW cells were infected with virulent Mtb H37Rv and cocultured with dNKT hybridomas. (B) Mtb polar or apolar lipids stimulate dNKT hybridomas in the presence of CD1d-transfected or untransfected RAW cells. IL-2 in culture supernatants was measured by ELISA. Data show mean ± SEM and are representative of two independent experiments.
We next tested whether lipids isolated from Mtb bacteria were stimulatory for dNKT hybridomas. Lyophilized Mtb was extracted in different organic solvents to obtain lipid fractions extracted in apolar solvents (petroleum ether) or polar solvents (chloroform/methanol). All of the dNKT hybridomas tested were stimulated in cocultures with RAW.CD1d cells in the presence of either polar or apolar Mtb lipids (Fig. 1B). Four out of eight dNKT hybridomas (14S.10, 14S.15, XV19, VIII24) secreted more IL-2 in response to polar Mtb lipids compared with apolar Mtb lipids, whereas three hybridomas (14S.6, TBA7, VII68) showed comparable responses. Interestingly, the 431.A11 hybridoma responded more strongly to apolar than to polar Mtb lipids. In contrast, iNKT hybridomas, characterized by expression of the semi-invariant Vα14Jα18+ TCR, were not stimulated in the presence of Mtb lipids but secreted IL-2 following stimulation with α-GalCer (Fig. S1 A and B). Thus, dNKT hybridomas were stimulated by Mtb-infected APCs or Mtb lipids in a CD1d-dependent manner.
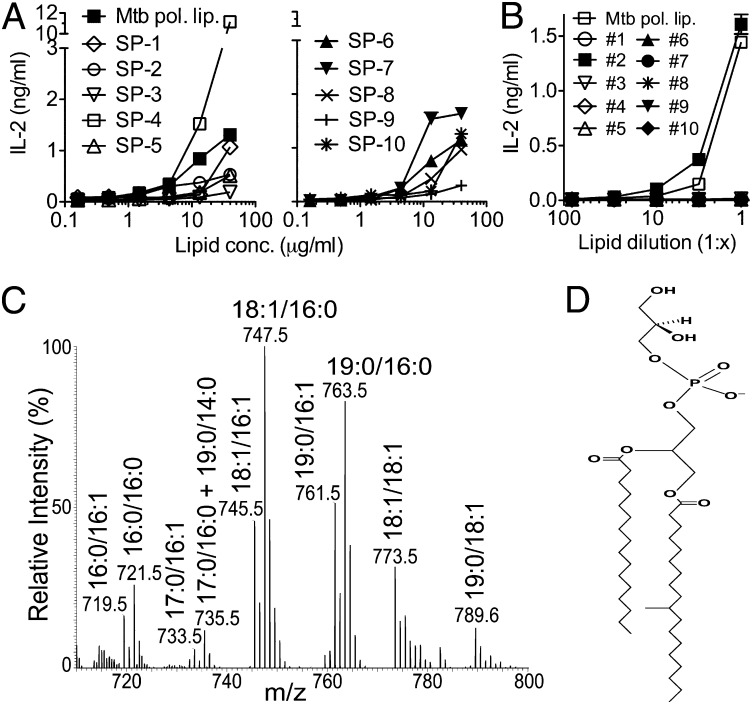
Next, to identify the Mtb lipid(s) that stimulated dNKT hybridomas, we performed extensive lipid separations and purifications and tested the purified lipids in our dNKT hybridoma coculture assay. To track the biological activity of the purified lipids, we used the 14S.10 dNKT hybridoma that displayed very low autoreactivity to CD1d-transfected RAW cells and preferentially recognized Mtb polar lipids (Fig. 1B). First, we separated Mtb polar lipids into 10 fractions (semipreps 1–10) using ion-exchange chromatography with diethylaminoethyl (DEAE) cellulose columns and salt gradient elution. Stimulation of the 14S.10 hybridoma with Mtb semipreps 1–10 in the presence of RAW.CD1d cells resulted in strong IL-2 secretion only with semiprep 4, whereas most other fractions did not simulate or only weakly simulated (Fig. 2A). To isolate the stimulatory lipid(s) from the mixture of lipids contained in semiprep 4, we further separated the lipids by preparative TLC into nine fractions (Fig. S2A). Stimulatory activity for the 14S.10 hybridoma was detected in fraction 4.3 (Fig. S2B). Based on the detection of multiple lipids in fraction 4.3 by one-dimensional (1D) and 2D TLC analysis (Fig. S2 A and C), we next separated 10 different lipids from fraction 4.3 by preparative 2D TLC (Fig. S2C). Stimulatory activity for the 14S.10 hybridoma was detected in fraction 4.3.2 (Fig. 2B). Phosphate staining was positive and α-naphthol staining was negative for lipid 4.3.2, indicating the presence of a phosphate moiety and the absence of carbohydrate moieties, respectively (Fig. S2C). Analysis of the highly purified 4.3.2 lipid by electrospray ionization–MS revealed dominant [M–H]− ions with a mass/charge (m/z) ratio of 763.5, 761.5, 747.5, and 745.5 (Fig. 2C) and was consistent with the lipid being phosphatidylglycerol (PG; Fig. 2D). Additional fatty acid analysis by tandem MS of the purified lipid confirmed the presence of molecular PG species containing C19:0/C16:0, C19:0/C16:1, C18:1/C16:0, and C18:1/C16:1 acyl chain combinations (Fig. 2C). Thus, our isolation and purification revealed Mtb PG to stimulate a dNKT hybridoma in a CD1d-dependent manner.
Fig. 2.
Isolation of stimulatory Mtb lipids. (A) Using DEAE cellulose columns, Mtb polar lipids were separated into 10 semipreps (SP) that were tested for stimulation of 14S.10 cells. (B) Lipids from fraction 4.3 were further separated by preparative 1D and 2D TLC, resulting in the stimulatory 4.3.2 Mtb lipid. (C and D) MS analysis revealed lipid 4.3.2 (C) to be PG with various molecular species (D). Data are representative of two independent experiments.
Corynebacterium glutamicum Lipids Stimulate dNKT Cells.
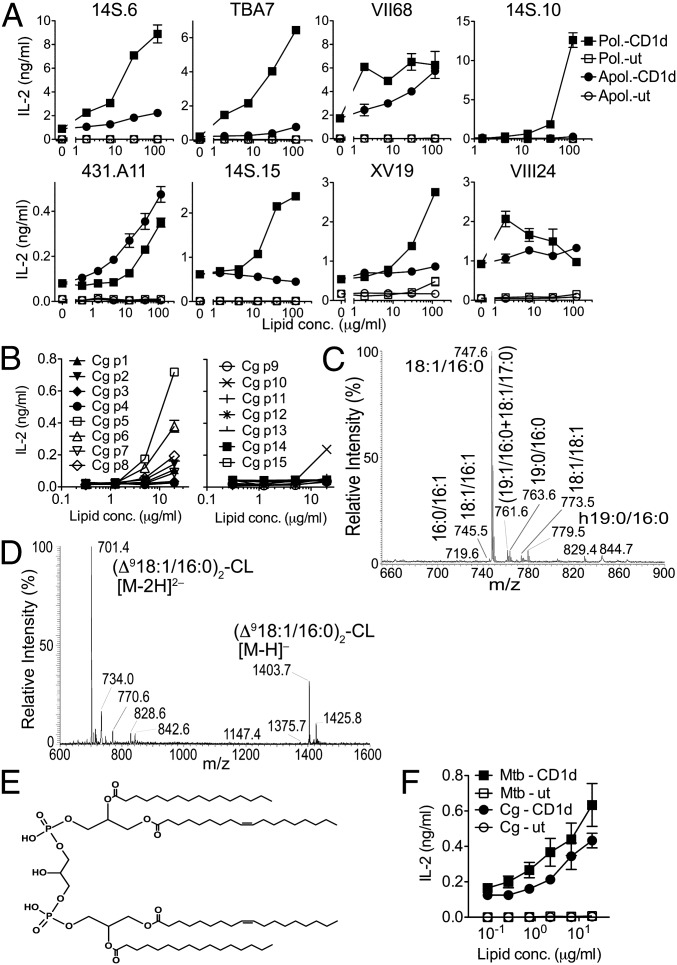
Because of the difficulty of generating large quantities of highly pure lipids from virulent Mtb, we established a system using Corynebacterium glutamicum (Cg) for the purification of large amounts of stimulatory mycobacterial lipids. Cg expresses a lipid profile that is similar, but less complex, than that of Mtb and is widely used to investigate mycobacterial lipid synthesis pathways. Following purification of Cg polar or apolar lipids, dNKT hybridomas were tested, and, similar to the responses observed with Mtb lipids, all hybridomas tested secreted IL-2 in response to Cg lipids in our in vitro RAW cell coculture assay (Fig. 3A). Seven of the hybridomas showed stronger responses to Cg polar lipids than to Cg apolar lipids, and similar to the responses seen with Mtb lipids, the 431.A11 hybridoma responded stronger to apolar than to polar Cg lipids. iNKT hybridomas tested did not respond to Cg lipids (Fig. S3A). Next, we separated and purified 15 individual polar Cg lipids by preparative 2D TLC (Fig. S3B). Cg lipids p6 and p5 were stimulatory in our 14S.10 hybridoma assay (Fig. 3B). Structural analyses by mass spectrometry and tandem mass spectrometry revealed the stimulatory Cg lipids p6 and p5 to be PG (Fig. 3C and Fig. S3 C and D) and diphosphatidylglycerol (DPG; or cardiolipin; Fig. 3 D and E), respectively. For Cg PG, the dominant ion at m/z 747.6 corresponds to C18:1/C16:0 and less abundant ions at m/z 763.6 and 773.5 represent C19:0/16:0 and C18:1/C18:1, respectively, in which the double bond of the C18:1 fatty acid substituent is located at Δ9 (Fig. S3D). When compared directly, PG isolated from Mtb and Cg stimulated the 14S.10 hybridoma in a similar manner (Fig. 3F). Cg DPG showed a dominant [M–H]− ion at m/z 1,403.7 corresponding to a (delta9 C18:1/C16:0)2-DPG (Fig. 3D). Thus, in addition to the isolation of PG from Mtb, independent purification of lipids from Cg polar lipids revealed Cg PG and the structurally related DPG as stimulatory lipids recognized by the 14S.10 dNKT hybridoma.
Fig. 3.
Cg lipids stimulate dNKT hybridomas. (A) Polar or apolar Cg lipids were tested for reactivity of dNKT hybridomas as in Fig. 1B. (B) Cg polar lipids were separated by 2D TLC and analyzed for reactivity with 14S.10 cells. (C–E) MS analysis revealed (C) the stimulatory lipid p6 to be PG (C) and the active lipid p5 (D) to be DPG (or cardiolipin; CL) (E). (F) Stimulation of 14S.10 cells in the presence of RAW.CD1d or RAW.ut cells and purified Mtb or Cg PG. Data are representative of two independent experiments.
Phosphatidylglycerophosphate Synthetase A-Deficient Cg Lacks Expression of PG and DPG.
To confirm that PG and DPG indeed are stimulatory lipids produced by Cg, we generated mutant Cg bacteria that are deficient in PG and DPG synthesis. Phosphatidylglycerophosphate synthetase A (pgsA) is critical for the biosynthesis of PG and DPG in several bacterial species. We therefore deleted the pgsA gene in Cg (Fig. S4) and, as a control, generated pgsA-deficient strains with reconstituted pgsA expression. pgsA-deficient Cg was viable, and TLC analysis of polar lipids showed loss of expression of PG and DPG and increased abundance of a phospholipid that was not detected in WT Cg (Fig. S5A), whereas pgsA-deficient Cg that had complemented pgsA expression showed a lipid profile that was similar to WT Cg (Fig. S5B). Surprisingly, despite the absence of PG and DPG, total polar lipids isolated from pgsA-deficient Cg stimulated 14S.10 cells comparable to polar lipids from wild-type (WT) Cg or pgsA-deficient Cg with reconstituted pgsA expression (Fig. S5C). To determine whether pgsA-deficient Cg had specifically lost the stimulatory capacity from PG and DPG, we separated lipids isolated from pgsA-deficient Cg by 2D TLC from regions corresponding to the location of PG and DPG found with WT Cg lipids (Fig. S5A). We detected no stimulatory activity from the expected locations for both PG and DPG isolated from pgsA-deficient Cg (Fig. S5D). Thus, deletion of the pgsA enzyme in Cg clearly resulted in a loss of PG and DPG recognized by 14S.10.
To determine whether the apparent compensatory increase in production of the phospholipid by pgsA-deficient Cg might be responsible for stimulating the 14S.10 hybridoma when total polar lipids were used, we isolated this lipid. Our functional analysis revealed the lipid to be stimulatory for the 14S.10 hybridoma (Fig. S5E), and structural analysis by MS revealed this lipid to be phosphatidylinositol (PI) with a dominant ion with m/z of 835.5 corresponding to a C18:1/C16:0 fatty acid composition (Fig. S5 F and G). Together, these results demonstrate that Cg PG, DPG, and PI, in addition to Mtb PG, stimulate the 14S.10 dNKT hybridoma and suggest that an individual dNKT TCR can recognize structurally distinct microbial phospholipids.
Range of Reactivities to Microbial Lipids Among dNKT Cells.
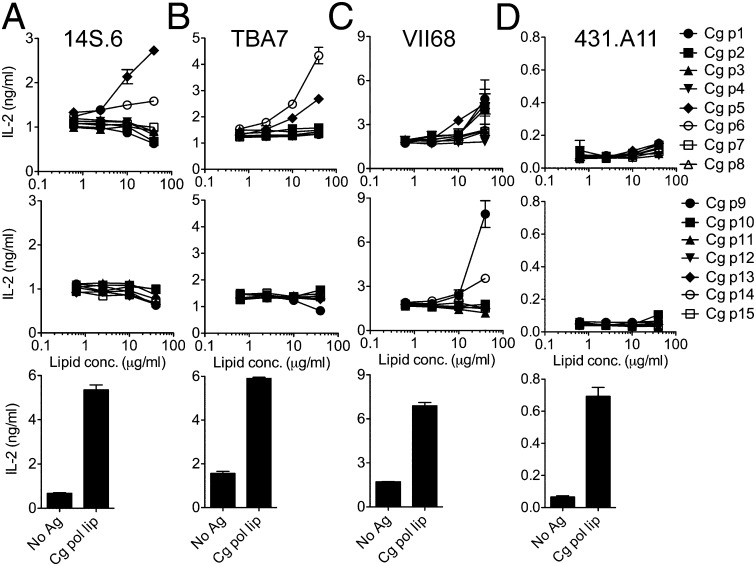
Our data suggested that several microbial phospholipids are recognized by the 14S.10 dNKT hybridoma, and, in addition, that several other dNKT hybridomas are stimulated by Cg polar lipids (Fig. 3A). To identify the lipids that specifically stimulate other dNKT hybridomas, we performed a large-scale preparative 2D TLC separation of Cg polar lipids (Fig. S3B). Then, we tested the reactivities to these purified lipids with additional dNKT hybridomas. Both 14S.6 and TBA7 hybridoma cells were stimulated by Cg PG (p6) and DPG (p5), with 14S.6 hybridoma cells responding to Cg DPG > PG, whereas TBA7 cells responded to Cg PG > DPG (Fig. 4 A and B). In contrast, the VII68 hybridoma recognized Cg PG, DPG, and several other lipid fractions only weakly, but strongly responded to Cg lipid p9, which failed to stimulate any of the other hybridomas (Fig. 4C). The identity of the lipid(s) contained in Cg fraction p9 remains to be determined. Interestingly, 431.A11, the hybridoma that was stimulated by Cg apolar > Cg polar lipids, did not respond to any of the 15 purified polar lipids (Fig. 4D), suggesting that this hybridoma recognizes distinct apolar lipid species. Together, these data suggest that the use of TCR α- and β-chains with extensive junctional and germ-line diversity by dNKT cells results in the recognition of distinct microbial lipids, with partially overlapping and occasionally unique antigen specificities.
Fig. 4.
Range of reactivities to microbial lipids among dNKT hybridomas. dNKT hybridomas were stimulated with lipids isolated from Cg polar lipids (as in Fig. 3B) in the presence of RAW.CD1d cells. (A) 14S.6. (B) TBA7. (C) VII68. (D) 431.A11. Cg lipid p6 is PG and p5 is DPG (Fig. 3). Table 1 shows TCR-α and -β gene utilization and summarizes reactivities. Data are representative of two independent experiments.
Antigen Uptake and/or Processing, but Not Toll-Like Receptor-Mediated Signaling, Is Required for dNKT Cell Activation by Microbial Lipids.
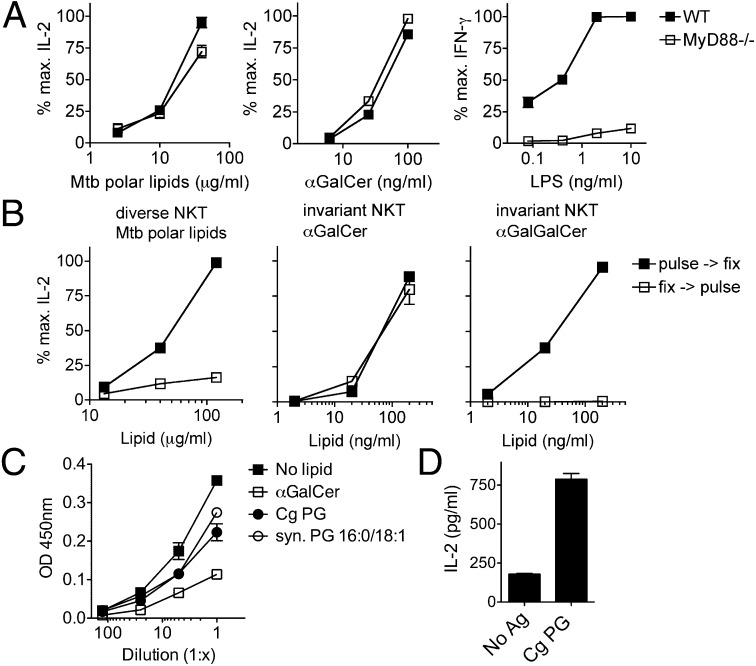
iNKT cells can be stimulated by microbes either by direct recognition of microbial antigens or indirectly through APC-mediated activation that results in secretion of stimulatory cytokines such as IL-12, -18, or type I IFNs and CD1d presentation of stimulatory self-lipids. To bias our in vitro coculture assay toward the discovery of microbial lipids that stimulate dNKT cells directly and to allow possible processing of lipids, we used dNKT hybridomas in combination with RAW.CD1d cells as APCs. IL-2 production by T-cell hybridomas is expected to be primarily the result of TCR-mediated rather than cytokine-mediated activation. To confirm that activation of 14S.10 dNKT hybridomas was not dependent on Toll-like receptor (TLR)-mediated signaling by APCs to induce cytokine secretion or self-lipid synthesis, we stimulated the 14S.10 hybridoma with Mtb polar lipids in the presence of MyD88-deficient bone marrow-derived dendritic cells (BMDCs) that are defective in TLR-mediated signaling. IL-2 production by the 14S.10 hybridoma in response to Mtb polar lipids in the presence of MyD88-deficient BMDCs was not different from stimulation with WT BMDCs (Fig. 5A). As a control, stimulation of an iNKT hybridoma with α-GalCer also was comparable in the presence of WT and MyD88-deficient BMDCs (Fig. 5A). In contrast, stimulation of a primary iNKT cell line with the TLR agonist LPS resulted in production of IFN-γ only in the presence of WT, but not MyD88-deficient, BMDCs, showing that, unlike hybridomas, primary iNKT cells are cytokine responsive (Fig. 5A and Fig. S6). Thus, stimulation of the 14S.10 dNKT hybridoma did not require TLR-mediated APC activation, suggesting that this dNKT hybridoma may directly recognize CD1d-presented Mtb lipids.
Fig. 5.
Microbial lipids activate dNKT cells independent of APC TLR signaling and require uptake and/or processing. (A) 14S.10 dNKT hybridoma cells (Left) and 24.9 iNKT hybridoma cells (Center) or an iNKT cell line (Right) were stimulated with Mtb polar lipids, α-GalCer, or LPS, respectively, in the presence of WT or MyD88−/− BMDCs. (B) RAW.CD1d cells were either pulse→fix or fix→pulse treated with lipids and cultured with 14S.10 dNKT or 24.9 iNKT hybridoma cells. (C) α-GalCer, Cg PG (contains C18:1/16:0), and synthetic C16:0/18:1 PG (representing mammalian PG) compete with biotin–PE binding to CD1d. (D) Stimulation of 14S.10 cells with Cg PG presented by plate-bound CD1d in the absence of APCs. Data are representative of two independent experiments.
Table 1.
Reactivity of dNKT hybridomas to Cg lipids
| Hybridoma | Source | Vα/Jα genes | Vβ/Jβ genes | Cg polar | Cg lipid | Cg apolar |
| 14S.10 | Ref. 5 | Vα11.3/Jα4 | Vβ8.1/Jβ2.7 | ++ | DPG, PG, PI | 0 |
| 14S.6 | Ref. 5 | Vα17/Jα32 | Vβ14.1/Jβ2.1 | +++ | DPG, PG | + |
| TBA7 | Ref. 6 | Vα1/Jα18* | Vβ8.2/Jβ2.6 | +++ | PG, DPG | 0/+ |
| VII68 | Ref. 4 | Vα4/Jα32* | Vβ11/Jβ2.5 | +/++ | p9 | +/++ |
| 14S.15 | Ref. 5 | Vα4/Jα38* | Vβ5.1/Jβ2.4 | +/++ | None | 0 |
| 431.A11 | Ref. 6 | Vα3.2/Jα9 | Vβ8.2/Jβ16* | + | None | ++ |
| XV19 | Ref. 4 | Vα1/Jα26 | Vβ16/Jβ2.1 | +++ | NT | 0 |
| VIII24 | Ref. 4 | Vα3.2/Jα26* | Vβ9/Jβ1.4 | 0/+ | NT | 0 |
TCR-α and -β gene use was determined for dNKT hybridomas used in this study. +++Very strong; ++robust; +weak; 0, SI < 3; NT, not tested.
*Gene use differs from reference.
We next determined whether antigen uptake and processing by APCs was required for activation of the 14S.10 hybridoma following stimulation with Mtb polar lipids. RAW.CD1d cells were either pulsed with Mtb polar lipids for 2 h and subsequently fixed with glutaraldehyde (“pulse→fix”) or fixed before incubation with Mtb lipids (“fix→pulse”) and then cocultured with 14S.10 hybridomas. Pulse→fix-treated, but not fix→pulse-treated, RAW cells stimulated IL-2 secretion by 14S.10 cells (Fig. 5B). Control experiments were performed with α-GalCer, an iNKT cell antigen known to be able to load onto CD1d molecules on the cell surface, and α-GalGalCer, an iNKT cell antigen known to require intracellular cleavage of the second galactose to result in generation of the stimulatory α-GalCer (19). Pulse→fix and fix→pulse treatment was similar for stimulation of an iNKT hybridoma with α-GalCer, but only pulse→fix treatment resulted in stimulation of the iNKT hybridoma with α-GalGalCer (Fig. 5B). Thus, stimulation of the 14S.10 hybridoma with Mtb lipids required lipid uptake and/or processing by APCs, and together, these results suggest that the 14S.10 dNKT TCR directly recognizes Mtb lipid antigen(s) presented by CD1d.
Formation of CD1d–Lipid Complexes and Their Recognition by dNKT Cells.
To determine whether the stimulatory microbial phospholipids bind directly to CD1d molecules, we used an established competition assay (11). We incubated biotin–phosphatidylethanolamine (PE)/CD1d antigen complexes with microbial Cg PG and, as controls, synthetic C16:0/C18:1 PG (representing mammalian PG) and α-GalCer. Both microbial Cg PG and synthetic mammalian PG displaced biotin–PE, as did the control iNKT cell antigen α-GalCer (Fig. 5C), suggesting that microbial as well as mammalian PG can bind to CD1d and form lipid–CD1d complexes. Stimulation of NKT cells by CD1d–antigen complexes in the absence of APCs is considered strong evidence that a lipid is directly recognized as a cognate antigen. Indeed, recombinant CD1d molecules loaded with Cg PG stimulated 14S.10 cells (Fig. 5D). Together, these results suggest that Cg PG binds to CD1d and that dNKT cells directly recognize Cg PG/CD1d complexes to result in their activation.
Microbial and Mammalian PG and DPG Stimulate dNKT Cells.
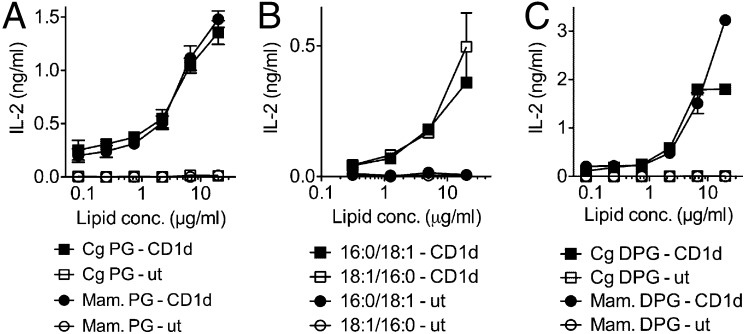
We next examined the structural features of phospholipid antigens that determine dNKT cell activation. The dominant ions detected in the stimulatory Mtb and Cg PG corresponded to C19:0/C16:0 and C18:1/C16:0 (Figs. 2C and 3C). In contrast to microbes, mammalian cells express C16:0/18:1 PG that differs from microbial PG in the position of the fatty acids at sn-1 and -2 of the glycerol backbone (compared to C18:1/C16:0 and C19:0/C16:0 in Cg and Mtb PG). To elucidate the structural requirements for stimulating the 14S.10 hybridoma, we next examined the ability of a synthetic C16:0/18:1 PG (representing the mammalian sn-1/sn-2 fatty acid orientation) to stimulate the 14S.10 hybridoma. For iNKT cell lipid antigens, subtle lipid tail modifications, such as the sn-1/sn-2 orientation of two different fatty acids or the position of a double bond, can profoundly impact iNKT cell activation (20, 21). Surprisingly, the synthetic C16:0/18:1 PG, similar to the microbial Cg PG, stimulated 14S.10 cells in a CD1d-dependent manner (Fig. 6A). We complemented these experiments with a synthetic chemistry approach and synthesized two PG molecules that only differed in the sn-1/sn-2 position of the C18:1 and C16:0 fatty acids at the glycerol backbone. The synthetic C18:1/16:0 (microbial) as well as the C16:0/18:1 (mammalian) PG stimulated 14S.10 hybridoma cells in a CD1d-dependent manner (Fig. 6B). Next, we examined whether mammalian DPG, in addition to microbial Cg DPG, stimulated 14S.10 hybridoma cells. Bovine DPG, containing dominantly (C18:2/C18:2)2 species, as well as microbial DPG, containing (C18:1/C16:0)2 species, stimulated 14S.10 cells in a CD1d-dependent manner (Fig. 6C). Together, these data suggest that microbial as well as mammalian PG and DPG are recognized by and stimulate CD1d-restricted dNKT hybridomas.
Fig. 6.
Microbial and mammalian phospholipids stimulate dNKT cells. (A) Microbial Cg PG (containing C18:1/16:0) or mammalian PG (isolated from murine skin, containing C16:0/18:1) was added to RAW.CD1d or RAW.ut cocultures with 14S.10 hybridoma cells. (B) Synthetic versions representing the dominant microbial PG (C18:1/16:0) and mammalian PG (C16:0/18:1) species stimulated 14S.10 cells. (C) Cg and bovine DPG stimulated 14S.10 cells. Data are representative of two independent experiments.
Discussion
dNKT cells play an increasingly appreciated role in antimicrobial immunity, yet the antigens they recognize remain poorly defined. Here, we tested lipids from Mtb and Cg, closely related bacterial species, for their ability to stimulate a panel of dNKT cell hybridomas that exhibited a spectrum of TCR Vα and Vβ use, and we identified several microbial phospholipids and other lipid fractions that activated dNKT cells. dNKT cell activation by microbial lipids was CD1d-dependent, occurred in the absence of TLR or IL-12 signaling by APCs, and required lipid uptake and/or intracellular processing by APCs. Furthermore, plate-bound microbial PG/CD1d complexes were sufficient to stimulate dNKT cells. Together, these findings demonstrate that the stimulatory microbial lipids activate dNKT cells through direct cognate interaction between the dNKT TCR and lipid antigen/CD1d complexes.
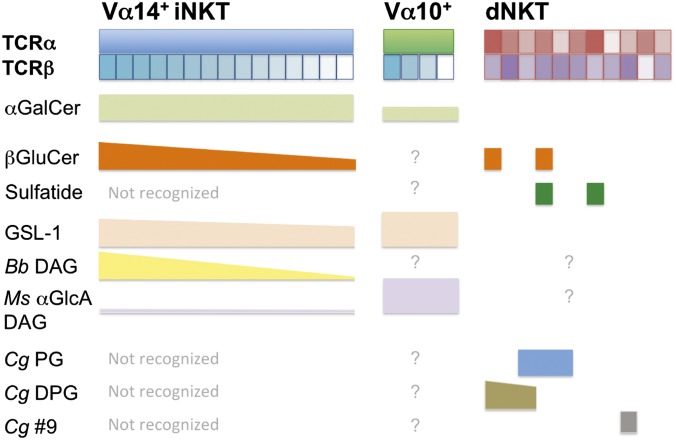
Characterization of the stimulatory lipids revealed a unique pattern of distinct and overlapping reactivities for microbial antigens among several dNKT cell hybridomas. Three dNKT cell hybridomas recognized microbial PG, DPG, and PI, with differential hierarchies in their responses to these antigens. Other dNKT hybridomas did not recognize those lipids, but instead had unique antigen specificities. For example, the VII68 hybridoma recognized Cg lipid p9, that has not yet been identified, and the 431.A11 hybridoma did not react to any of the isolated Cg lipid species and instead appeared to recognize apolar Cg and Mtb lipids. Of note, none of the lipid antigens from Mtb or Cg that activated dNKT cells stimulated iNKT cells. This pattern of dNKT cell antigen specificity is distinct from both the uniform and pattern recognition receptor-like TCR specificity observed for iNKT cells and the high degree of diversity and antigen specificity observed for MHC-restricted T cells (3) (antigen specificities of NKT cell subsets are summarized in Fig. 7). The α-GalCer antigen and its derivatives, the α-glycan–linked microbial iNKT cell antigens described so far, and the β-GluCer self-antigen are stimulatory for most or all iNKT cells. Although there is some hierarchy in affinity of different mouse iNKT cell TCRs based on TCR β-chain use (22, 23), stimulation with iNKT cell antigens generally results in immediate, innate-like responses of the entire iNKT cell population. iNKT cells, and the recently described semi-invariant Vα10+ CD1d-restricted NKT cells, use a unique mode of TCR recognition of lipid/CD1d complexes in which recognition of the lipid antigen is mediated primarily by the semi-invariant TCR α-chain, and the TCR β-chain appears to only modulate affinity (24–26). In contrast, MHC-restricted T cells, as well as human CD1a-, b-, and c-restricted T cells, show a high degree of specificity for unique antigens and stimulation with their cognate antigens results in clonal expansion and adaptive T-cell responses (27). Thus, the ability to discriminate between several lipid antigens as observed here for dNKT cells suggests a different mode for TCR-mediated recognition of lipid/CD1d complexes compared with iNKT cells, which indeed was recently confirmed by structural analysis of a dNKT TCR in complex with CD1d/sulfatide (28, 29).
Fig. 7.
Antigen specificities of Vα14+ iNKT, Vα10+ NKT, and dNKT cells. GSL-1 is found in Sphingomonas spp., Bb DAG in Borrelia burgdorferi, and Ms α-GlcA DAG in Mycobacterium smegmatis (3).
In humans, CD1b molecules present Mtb lipids to CD1b-restricted, microbial lipid-specific T cells that expand following vaccination, and Mtb lipid-specific CD1b-restricted T cells elicited by lipid vaccination provided protection in a guinea pig model of tuberculosis (14). In addition, iNKT cells, although not required for optimum immunity to Mtb infection in mice, can contribute to the protective immune response during Mtb infection and suppress bacterial replication in vitro (30). Here, dNKT cells were activated by Mtb-infected APCs, making it tempting to speculate that dNKT cells may also contribute to protective immunity during Mtb infection. Although there is no evidence that iNKT cells are capable of generating adaptive, antigen-specific responses, the partially naïve phenotype observed on some dNKT cells, together with their distinct antigen specificities, suggests that they may be capable of generating memory responses to these microbial antigens.
A striking feature of the observed dNKT cell specificity for microbial lipid antigens is the unexpected cross-reactivity for microbial phospholipids and the homologous mammalian phospholipids, because for several iNKT cell antigens, structural features unique to microbial lipids are critical for antigenicity (21, 31, 32). Both PG and DPG isolated from Cg stimulated dNKT cells comparable to their mammalian counterparts, despite apparent structural differences such as the reversal of the 16:0/18:1 fatty acid chains in the sn-1/sn-2 positions. The description here of three DPG/cardiolipin-reactive dNKT cells demonstrates that CD1d-restricted NKT cells that recognize cardiolipin can also use αβ TCRs, in addition to the cardiolipin-specific γδ T cells observed in mice (33). Overall, our findings suggest that the cross-reactivity between microbial and self-lipid antigens may be a central feature of dNKT cell function, allowing both presentation of microbial lipids and enhanced availability of stimulatory self-lipids to contribute to dNKT cell activation during infection. Furthermore, it is possible that dNKT cell activation during infection requires TLR- and cytokine-mediated costimulation in addition to the TCR-mediated signals induced by recognition of weak self-lipid or microbial lipid antigens, as has been observed for iNKT cells (34, 35).
In summary, our results show a pattern of antigen specificity for dNKT cells characterized by cross-reactivity between mammalian and microbial phospholipid antigens, and distinct from both the uniform and pattern recognition receptor-like antigen specificity observed for iNKT cells and the high degree of diversity and antigen specificity observed for MHC-restricted T cells. With this example of microbial antigens for dNKT cells, it will be intriguing to determine the role of their recognition in microbial infection and host defense.
Materials and Methods
dNKT hybridomas (4–6) were cocultured with CD1d-transfected or untransfected RAW cells, or BMDCs from WT or Myd88-deficient mice, in the presence of various concentration of lipids, or following infection of RAW cells with Mtb H37Rv. Cytokines were measured in culture supernatants by ELISA. Lipids were extracted from Mtb or Cg by using organic solvents and isolated by using HPLC and TLC, and structural analysis was performed as described (35). Synthesis of PG will be described elsewhere, and other lipids were from Avanti Polar Lipids. pgsA-deficient Cg was generated by targeting NCgl1889 with a construct based on pk19mobsacB and plasmid pEKEx2-1889 served for complementation studies. CD1d binding of lipids and CD1d plate assays used biotinylated CD1d proteins as described (11). For details of experimental conditions and analyses, see SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants AI077795 (to M.B.), AI028973 and AI063428 (to M.B.B.), and RR000954, DK034388, DK020579 (Mass Spectrometry Center, Washington University); and the Australian National Health and Medical Research Council (D.I.G., C.F.A., and J.R.). C.F.A. is supported by a PhD scholarship from Fundaçao para a Ciencia e Tecnologia Portugal; D.I.G. is supported by an Australian National Health and Medical Research Council (NHMRC) Senior Principal Research Fellowship, and J.R. is supported by an NHMRC Australia Fellowship. G.S.B. is supported by a Personal Research Chair from Mr. James Bardrick, The Wellcome Trust (084923/B/08/Z), and the Medical Research Council. NMR spectrometers used in this research were funded in part through Birmingham Science City: Innovative Uses for Advanced Materials in the Modern World (West Midlands Centre for Advanced Materials Project 2), with support from Advantage West Midlands and the European Regional Development Fund.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220601110/-/DCSupplemental.
References
- 1.Brigl M, Brenner MB. CD1: Antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 2.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 3.Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d-restricted antigens by natural killer T cells. Nat Rev Immunol. 2012;12(12):845–857. doi: 10.1038/nri3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardell S, et al. CD1-restricted CD4+ T cells in major histocompatibility complex class II-deficient mice. J Exp Med. 1995;182(4):993–1004. doi: 10.1084/jem.182.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behar SM, Podrebarac TA, Roy CJ, Wang CR, Brenner MB. Diverse TCRs recognize murine CD1. J Immunol. 1999;162(1):161–167. [PubMed] [Google Scholar]
- 6.Park SH, et al. The mouse CD1d-restricted repertoire is dominated by a few autoreactive T cell receptor families. J Exp Med. 2001;193(8):893–904. doi: 10.1084/jem.193.8.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arrenberg P, Halder R, Dai Y, Maricic I, Kumar V. Oligoclonality and innate-like features in the TCR repertoire of type II NKT cells reactive to a beta-linked self-glycolipid. Proc Natl Acad Sci USA. 2010;107(24):10984–10989. doi: 10.1073/pnas.1000576107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhost S, Sedimbi S, Kadri N, Cardell SL. Immunomodulatory type II natural killer T lymphocytes in health and disease. Scand J Immunol. 2012;76(3):246–255. doi: 10.1111/j.1365-3083.2012.02750.x. [DOI] [PubMed] [Google Scholar]
- 9.Jahng A, et al. Prevention of autoimmunity by targeting a distinct, noninvariant CD1d-reactive T cell population reactive to sulfatide. J Exp Med. 2004;199(7):947–957. doi: 10.1084/jem.20031389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blomqvist M, et al. Multiple tissue-specific isoforms of sulfatide activate CD1d-restricted type II NKT cells. Eur J Immunol. 2009;39(7):1726–1735. doi: 10.1002/eji.200839001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennan PJ, et al. Invariant natural killer T cells recognize lipid self antigen induced by microbial danger signals. Nat Immunol. 2011;12(12):1202–1211. doi: 10.1038/ni.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhost S, et al. Identification of novel glycolipid ligands activating a sulfatide-reactive, CD1d-restricted, type II natural killer T lymphocyte. Eur J Immunol. 2012;42(11):2851–2860. doi: 10.1002/eji.201142350. [DOI] [PubMed] [Google Scholar]
- 13.Tupin E, Kinjo Y, Kronenberg M. The unique role of natural killer T cells in the response to microorganisms. Nat Rev Microbiol. 2007;5(6):405–417. doi: 10.1038/nrmicro1657. [DOI] [PubMed] [Google Scholar]
- 14.Cohen NR, Garg S, Brenner MB. Antigen presentation by CD1 lipids, T cells, and NKT cells in microbial immunity. Adv Immunol. 2009;102:1–94. doi: 10.1016/S0065-2776(09)01201-2. [DOI] [PubMed] [Google Scholar]
- 15.Sköld M, Faizunnessa NN, Wang CR, Cardell S. CD1d-specific NK1.1+ T cells with a transgenic variant TCR. J Immunol. 2000;165(1):168–174. doi: 10.4049/jimmunol.165.1.168. [DOI] [PubMed] [Google Scholar]
- 16.Rolf J, et al. Molecular profiling reveals distinct functional attributes of CD1d-restricted natural killer (NK) T cell subsets. Mol Immunol. 2008;45(9):2607–2620. doi: 10.1016/j.molimm.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 17.Moody DB, Porcelli SA. Intracellular pathways of CD1 antigen presentation. Nat Rev Immunol. 2003;3(1):11–22. doi: 10.1038/nri979. [DOI] [PubMed] [Google Scholar]
- 18.Dascher CC, et al. Immunization with a mycobacterial lipid vaccine improves pulmonary pathology in the guinea pig model of tuberculosis. Int Immunol. 2003;15(8):915–925. doi: 10.1093/intimm/dxg091. [DOI] [PubMed] [Google Scholar]
- 19.Prigozy TI, et al. Glycolipid antigen processing for presentation by CD1d molecules. Science. 2001;291(5504):664–667. doi: 10.1126/science.291.5504.664. [DOI] [PubMed] [Google Scholar]
- 20.Kinjo Y, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7(9):978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- 21.Kinjo Y, et al. Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat Immunol. 2011;12(10):966–974. doi: 10.1038/ni.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mallevaey T, et al. T cell receptor CDR2 beta and CDR3 beta loops collaborate functionally to shape the iNKT cell repertoire. Immunity. 2009;31(1):60–71. doi: 10.1016/j.immuni.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pellicci DG, et al. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity. 2009;31(1):47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallevaey T, et al. A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity. 2011;34(3):315–326. doi: 10.1016/j.immuni.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pellicci DG, et al. Recognition of β-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat Immunol. 2011;12(9):827–833. doi: 10.1038/ni.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uldrich AP, et al. A semi-invariant Vα10+ T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties. Nat Immunol. 2011;12(7):616–623. doi: 10.1038/ni.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Godfrey DI, Rossjohn J, McCluskey J. The fidelity, occasional promiscuity, and versatility of T cell receptor recognition. Immunity. 2008;28(3):304–314. doi: 10.1016/j.immuni.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Girardi E, et al. Type II natural killer T cells use features of both innate-like and conventional T cells to recognize sulfatide self antigens. Nat Immunol. 2012;13(9):851–856. doi: 10.1038/ni.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel O, et al. Recognition of CD1d-sulfatide mediated by a type II natural killer T cell antigen receptor. Nat Immunol. 2012;13(9):857–863. doi: 10.1038/ni.2372. [DOI] [PubMed] [Google Scholar]
- 30.Sada-Ovalle I, Chiba A, Gonzales A, Brenner MB, Behar SM. Innate invariant NKT cells recognize Mycobacterium tuberculosis-infected macrophages, produce interferon-gamma, and kill intracellular bacteria. PLoS Pathog. 2008;4(12):e1000239. doi: 10.1371/journal.ppat.1000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinjo Y, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434(7032):520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 32.Mattner J, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434(7032):525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 33.Dieudé M, et al. Cardiolipin binds to CD1d and stimulates CD1d-restricted γδ T cells in the normal murine repertoire. J Immunol. 2011;186(8):4771–4781. doi: 10.4049/jimmunol.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brigl M, Bry L, Kent SC, Gumperz JE, Brenner MB. Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat Immunol. 2003;4(12):1230–1237. doi: 10.1038/ni1002. [DOI] [PubMed] [Google Scholar]
- 35.Brigl M, et al. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J Exp Med. 2011;208(6):1163–1177. doi: 10.1084/jem.20102555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.