Abstract
Hepatitis C virus (HCV) replication is dependent on microRNA 122 (miR-122), a liver-specific microRNA that recruits Argonaute 2 to the 5′ end of the viral genome, stabilizing it and slowing its decay both in cell-free reactions and in infected cells. Here we describe the RNA degradation pathways against which miR-122 provides protection. Transfected HCV RNA is degraded by both the 5′ exonuclease Xrn1 and 3′ exonuclease exosome complex, whereas replicating RNA within infected cells is degraded primarily by Xrn1 with no contribution from the exosome. Consistent with this, sequencing of the 5′ and 3′ ends of RNA degradation intermediates in infected cells confirmed that 5′ decay is the primary pathway for HCV RNA degradation. Xrn1 knockdown enhances HCV replication, indicating that Xrn1 decay and the viral replicase compete to set RNA abundance within infected cells. Xrn1 knockdown and miR-122 supplementation have equal, redundant, and nonadditive effects on the rate of viral RNA decay, indicating that miR-122 protects HCV RNA from 5′ decay. Nevertheless, Xrn1 knockdown does not rescue replication of a viral mutant defective in miR-122 binding, indicating that miR-122 has additional yet uncharacterized function(s) in the viral life cycle.
Keywords: host factor, RNA decay, translation, viral replicase
Hepatitis C virus (HCV) is a positive-strand RNA virus classified in the family Flaviviridae. It is highly hepatotropic and an important cause of human liver disease (1). Its replication is uniquely dependent on miR-122, which is the most abundant microRNA (miRNA) in the liver and accounts for >50% of mature miRNAs in human hepatocytes (2, 3). There are two conserved miR-122 binding sites (S1 and S2) located near the 5′ end of the positive-sense viral RNA genome, immediately upstream of an internal ribosome entry site (IRES) that mediates translation of the viral polyprotein. Direct interactions between the miR-122 seed sequence (nts 2–8) and these sites in the 5′ UTR are essential for amplification of the HCV genome (4, 5). Additional supplemental base-pairing upstream of S1 and S2 has also been demonstrated and is important for viral replication (6, 7).
Unlike typical interactions of miRNAs with mRNAs that involve binding within the 3′ UTR and promote translational repression and/or destabilization of the target RNA (8), binding of miR-122 to the 5′ UTR of HCV genomic RNA stimulates viral protein expression (4, 9) and, in association with Argonaute 2 (Ago2) protein, stabilizes HCV RNA (10). The rate of decay of either transfected synthetic genomic RNA or replicating viral RNA within infected cells is slowed when cells are transfected with synthetic duplex miR-122, thereby supplementing its endogenous abundance. Conversely, transfection of antisense RNA complementary to miR-122 enhances the rate with which HCV RNA decays in either context. HCV RNA is not thought to contain a 5′ cap structure, and the stabilizing action of miR-122 on synthetic RNA can be functionally substituted by addition of a 5′ cap analog (10). These observations point to the importance of RNA decay pathways in HCV replication and suggest that miR-122 is likely to prevent degradation of viral RNA from its 5′ end. However, the specific mechanisms involved in degradation of HCV RNA, or for that matter most positive-strand viral RNAs, have not been well studied and remain unclear. In addition, exactly how miR-122 stabilizes the viral RNA remains to be elucidated.
Because HCV genomic RNA is positive sense and serves directly as the mRNA for viral protein translation, it is reasonable to speculate that cellular mRNA decay pathways may also function in degradation of the viral RNA. In eukaryotic cells, bulk mRNA decay typically initiates with deadenylation, shortening the 3′ poly(A) tail, followed by degradation of the RNA molecule in a 5′→3′ or 3′→5′ direction (11). In the 5′ decay pathway, the monomethyl guanosine (m7G) cap is cleaved from mRNA by decapping enzymes (12, 13), exposing the 5′ monophosphorylated product to progressive 5′→3′ exoribonucleolytic degradation by Xrn1 (14, 15). In the 3′ decay pathway, deadenylated mRNA is degraded by the cytoplasmic RNA exosome complex, a multisubunit 3′→5′ exoribonuclease (16). The residual cap structure resulting from 3′ decay is hydrolyzed by the scavenger decapping enzyme DcpS (17). Because HCV genomic RNA contains neither a 3′ poly(A) tail nor 5′ cap, its degradation requires neither deadenylation nor decapping. Nonetheless, either or both of these two exoribonucleolytic pathways may potentially contribute to decay of HCV RNA in infected cells.
Here, we describe the roles played by Xrn1 and the exosome complex in HCV RNA decay. We show that viral RNA is degraded specifically by Xrn1 within infected cells. Our results reveal that miR-122 protects the genome against Xrn1-mediated decay but that it has additional functions beyond genome stabilization that are essential for viral replication.
Results
Xrn1 and the Exosome both Mediate Degradation of Transfected HCV RNA.
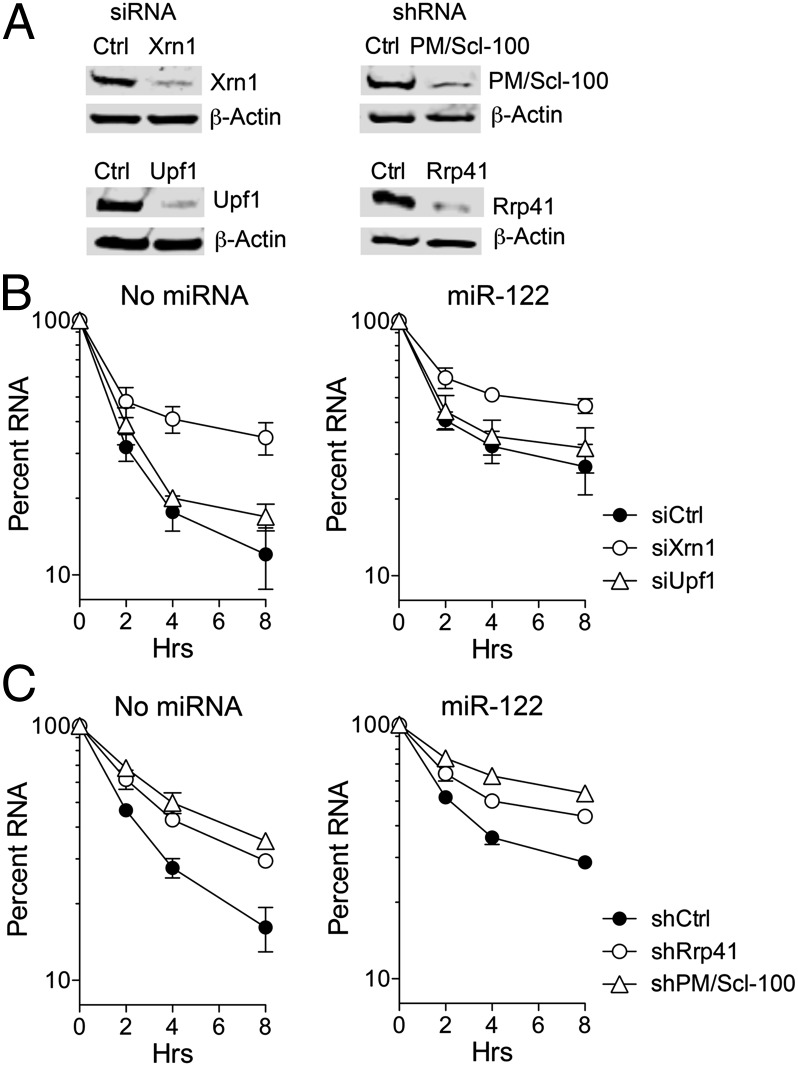
As a first step in determining whether mRNA decay pathways contribute to degradation of positive-strand HCV RNA, we examined the decay of HCV RNA transfected into HeLa cells that do not express endogenous miR-122. To assess the role of the 5′ exonuclease Xrn1 (XRN1), we reduced its expression by prior transfection of Xrn1-specific siRNA. We similarly knocked down expression of Upf1 (UPF1), which is a key factor in nonsense-mediated mRNA decay (18) and is also involved in degradation of histone mRNA and HIV RNA metabolism (19, 20). Immunoblots confirmed efficient knockdown of both RNA decay factors (Fig. 1A, Left). Synthetic replication-defective HCV RNA (H77S/AAG, which contains a lethal mutation in its RNA polymerase) (10) was electroporated into the cells with or without coelectroporation of duplex miR-122. This strategy eliminates any potentially confounding effects of replication on measurements of RNA stability. HCV RNA levels were determined by quantitative real-time RT-PCR (qRT-PCR) and normalized to β-actin mRNA. As shown in Fig. 1B, the transfected HCV RNA degraded rapidly, with a t1/2 of 1.4 h in cells transfected with a scrambled control siRNA (siCtrl) (Table S1). Knocking down Xrn1 significantly stabilized HCV RNA, increasing the t1/2 to 3.2 h, whereas Upf1 knockdown had little effect on its decay (t1/2 = 1.7 h) (Fig. 1B). As expected (10), coelectroporation of miR-122 significantly stabilized the RNA, leading to a t1/2 of 2.3 and 2.7 h in the siCtrl and siUpf1-transfected cells, respectively. miR-122 supplementation resulted in a proportionately similar increase in RNA stability after Xrn1 knockdown, boosting the t1/2 from 3.2 to 4.9 h. Maximum stabilization of HCV RNA was observed with combined Xrn1 knockdown and miR-122 supplementation (Fig. 1B). Collectively, these results indicate that Xrn1, but not Upf1, contributes to degradation of transfected HCV RNA.
Fig. 1.
Decay of transfected HCV RNA in HeLa cells. (A) Immunoblots of (Left) Xrn1 and Upf1 in HeLa cells following siRNA transfection and (Right) Rrp41 and PmScl100 in HeLa cells stably expressing the indicated shRNA. β-actin was a loading control. (B) HeLa cells were transfected with the indicated siRNAs for 48 h and electroporated with replication-deficient genotype 1a H77S-AAG RNA with or without miR-122 (1 μM). The percentage of HCV RNA remaining at each time point following electroporation without (Left) and with (Right) miR-122 supplementation was determined by qRT-PCR, relative to the abundance of β-actin mRNA. Results shown represent the means of three replicate experiments ± SEM. (C) HeLa cells expressing the indicated shRNAs were electroporated as in B. Percent HCV RNA remaining following electroporation without (Left) and with (Right) miR-122 supplementation was quantified by qRT-PCR relative to the abundance of β-actin mRNA. Results shown represent the means of three replicate experiments ± SEM.
To determine whether the exosome is also involved in degradation of transfected RNA, we knocked down two exosome components, Rrp41 (EXOSC4), a core structural component of the exosome, and PM/Scl-100 (EXOSC10), a 3′→5′ exonuclease, using lentiviruses expressing specific shRNAs (Fig. 1A, Right). Transfected HCV RNA was degraded somewhat more slowly in cells transduced with control lentivirus (shCtrl) than with siCtrl (t1/2 = 2.1 vs. 1.4 h) (Table S1), reflecting different conditions in these experiments (Materials and Methods). However, shRNA-mediated knockdown of either exosome component substantially stabilized the RNA (Fig. 1C; Table S1), indicating that the exosome is also involved in decay of transfected viral RNA. As with Xrn1, miR-122 supplementation further increased the stability of the transfected HCV RNA in the PM/Scl-100 and Rrp41 knockdown cells. We conclude from these results that both Xrn1 and the exosome contribute to degradation of transfected HCV RNA.
Degradation of HCV RNA in Cell-Free S10 Lysate Is Primarily Mediated by the Exosome.
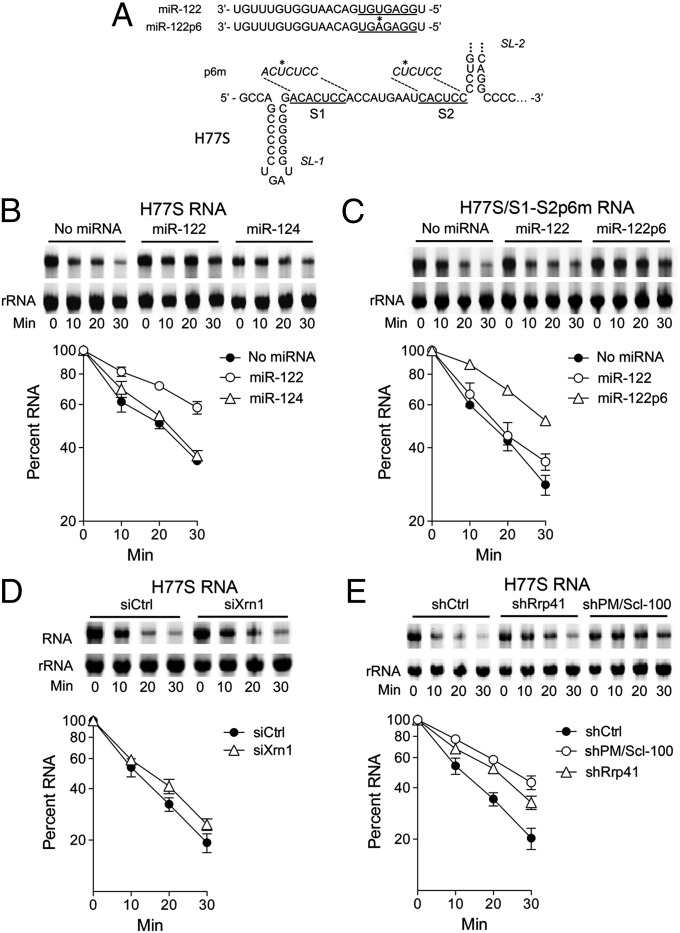
We previously demonstrated that miR-122 stabilizes a chimeric poliovirus RNA that contains the HCV 5′ UTR in lieu of the poliovirus 5′UTR when added with it to HeLa S10 lysate (10). Using SYTO 62 infrared fluorescence to quantify the RNA (Fig. S1A), we found that full-length genotype 1a HCV RNA (H77S) (21) is similarly stabilized by miR-122 when incubated in S10 lysate. HCV RNA degrades rapidly in HeLa S10 lysate (Fig. 2B). However, prior addition of duplex miR-122, but not miR-124 (a brain-specific miRNA), to the lysate doubled the half-life of the RNA (t1/2 increasing from 18.6 to 39 min; Table S2). In contrast, a related viral RNA mutant with single base substitutions in S1 and S2 that ablate miR-122 binding (H77S/S1-S2p6m; Fig. 2A) (4) was not stabilized by miR-122, but was stabilized by the complementary miR-122 mutant (miR-122p6) (Fig. 2C; Table S2), thereby confirming that HCV RNA is physically stabilized in S10 lysate as a result of miR-122 binding to the 5′ UTR. The addition of a 5′ cap also stabilized HCV RNA in S10 lysate (Fig. S1B), and the stability of the capped RNA was further enhanced by miR-122 supplementation (Fig. S1C).
Fig. 2.
Decay of HCV RNA in HeLa S10 lysate. (A) (Upper) miR-122 and mutant miR-122p6 guide strand sequence; (Lower) 5′ terminal sequence of HCV RNA (H77S.3/AAG) with S1 and S2 miR-122 seed sequence-binding sites underlined. Point mutations (*) in the related S1-S2p6m mutant are shown above. (B) H77S.3 RNA was incubated with HeLa S10 lysate containing the indicated duplex miRNA (1 μM). RNAs were extracted at indicated time intervals, stained with SYTO 62, and resolved in 1% agarose. Percent HCV RNA remaining was quantified by the Odyssey Infrared Imaging System relative to the 28S rRNA. Results are the means of three experiments ± SEM. (C) Decay assays were carried out as in B with the H77S.3/S1-S2p6m mutant RNA. (D and E) Decay assays were carried out as in B with lysates from (D) HeLa cells transfected with control or Xrn1 siRNA (mean ± SEM from two replicate experiments) or (E) lysates from HeLa cells stably expressing the indicated shRNA (mean ± SEM from three replicate experiments).
To investigate how HCV RNA is degraded in this cell-free system, we carried out similar assays using S10 lysates from Xrn1 and exosome knockdown cells. In contrast to what we observed with transfected RNA, Xrn1 knockdown minimally affected the HCV RNA decay rate (Fig. 2D; Table S3), whereas knocking down either of the exosome components, Rrp41 and PM/Scl-100, slowed degradation of HCV RNA substantially (Fig. 2E). Thus, the 3′ exosome-mediated decay pathway is mainly responsible for degradation of HCV RNA in HeLa S10 lysate. This finding is surprising, because miR-122 stabilizes the RNA by binding near its 5′ end (Fig. 2 A and B) and is considered further in Discussion.
Degradation of Replicating HCV RNA Is Mediated by Xrn1.
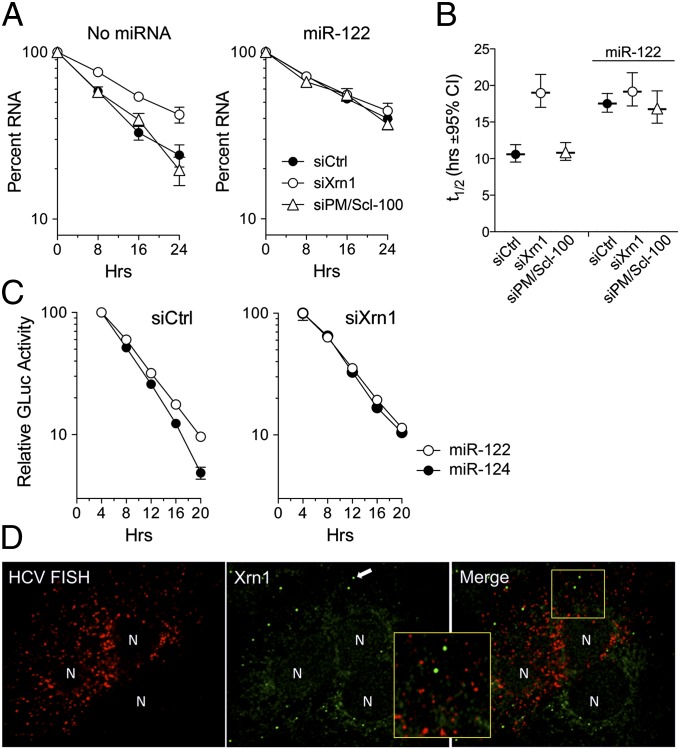
The experiments described above examined decay of synthetic HCV RNA after transfection into cells or when added to cell-free lysates and may not recapitulate the RNA decay pathways acting on replicating viral genomes. To assess this, we treated Huh-7.5 cells infected with HCV (H77S.3 virus) with PSI-6130, a potent and specific nucleoside inhibitor of the HCV NS5B RNA-dependent RNA polymerase that blocks viral RNA synthesis (22). As observed previously following the arrest of new viral RNA synthesis (10), replicating RNA degraded much more slowly than transfected RNA (t1/2 of 10.6 h in siCtrl-transfected cells) (Fig. 3A, Left, compare with Fig. 1A). Knocking down Xrn1 slowed this rate of HCV RNA decay impressively (t1/2 = 19.0 h). In contrast, knocking down the exosome component PM/Scl-100 had no effect (t1/2 of 10.8 h) (Fig. 3 A, Right, and B). Thus, replicating HCV RNA is degraded by the 5′ exonuclease Xrn1, but not the exosome complex. The viral RNA was significantly stabilized by miR-122 supplementation in siCtrl and PM/Scl-100 knockdown cells, reaching a half-life approximating that in the Xrn1 knockdown cells (t1/2 = 16.8–17.5 h; Fig. 3B). However, miR-122 supplementation resulted in no additional increase in RNA stability in the Xrn1 knockdown cells (t1/2 of 19.2 h). Thus, Xrn1 knockdown and miR-122 supplementation have equal and redundant effects on stability of the RNA, from which we infer that miR-122 enhances HCV RNA stability by protecting it from Xrn1-mediated 5′→3′ degradation.
Fig. 3.
Decay of replicating HCV RNA in Xrn1- and PM/Scl-100–depleted cells following PSI-6130 arrest of viral RNA synthesis. (A) Huh-7.5 cells were transfected with replication-competent genotype 1a H77S.3 RNA, and then retransfected 48 h later with siRNAs specific for Xrn1 or PM/Scl-100 or scrambled siCtrl. After an additional 48-h incubation (time = 0), the cells were treated with 10 μM PSI-6130, with (Right) or without (Left) simultaneous miR-122 supplementation. Data shown represent percent HCV RNA remaining following addition of PSI-6130. RNA was quantified by qRT-PCR relative to the abundance of actin mRNA. Results are the means of three experiments ± SEM. (B) Estimated half-life (t1/2) of HCV RNA ± 95% CI under the conditions shown in A. The data were fit to a one-phase decay model (R2 = 0.946–0.983). While the decay constant, k, differed significantly between siXrn1-, siPmScl-100–, and siCtrl-transfected cells in the absence of miR-122 supplementation (P < 0.0001 by the extra sum-of-squares F test), there was no significant difference in cells supplemented with miR-122 (P = 0.19). (C) GLuc expression from cells transfected with H77S.3/GLuc2A RNA, followed by transfection of siXrn1 or siCtrl and PSI-6130 arrest of new viral RNA synthesis as in A. Cells were supplemented with miR-122 or miR-124 at the time of addition of PSI-6130, and media were replaced at 4-h intervals thereafter. Data shown are the mean GLuc activity ± SD from four replicate cultures and are representative of multiple experiments. (D) Confocal microscopy demonstrating the absence of colocalization of HCV RNA with Xrn1. Huh-7.5 cells were transfected with H77S.3 RNA for 4 d and then subjected to FISH for detection of HCV RNA (red). Xrn1 was visualized by subsequent immunostaining (green) and is concentrated in P bodies (arrow in center panel). Nuclei (N) are marked by the absence of HCV RNA and Xrn1. Inset represents an enlarged view of a portion of the merged image. An uninfected cell in the lower right quadrant provides an internal control for FISH.
We also assessed the effect of miR-122 on HCV protein expression under these conditions. For this, we measured Gaussia princeps luciferase (GLuc) secreted into the media by cells supporting replication of a viral RNA (H77S.3/GLuc2A) that expresses GLuc as part of its polyprotein (23). GLuc production, assessed at 4-h intervals after the arrest of viral RNA synthesis with PSI-6130, declined with a t1/2 of about 8 h (Fig. 3C). miR-122 supplementation resulted in a small but reproducible increase in GLuc production in siCtrl-transfected cells (Fig. 3C, Left), consistent with stabilization of the viral RNA. No such increase was observed in Xrn1-depleted cells (Fig. 3C, Right), indicating that miR-122 supplementation does not enhance viral protein translation under these conditions.
Xrn1 is expressed within the cytosol but enriched within P bodies, sites of mRNA storage and decay (24). We previously demonstrated that double-stranded RNA (dsRNA), an intermediate in the HCV replication cycle, is not localized to P bodies, the morphology of which is well preserved within infected cells with only a minimal reduction in their number (25). To ascertain whether single-stranded positive-sense viral RNA colocalizes with Xrn1, we used a sensitive FISH method to detect HCV RNA in infected cells, counterstaining with antibody specific for Xrn1 (Fig. 3D). FISH confirmed the absence of HCV RNA in P bodies. A quantitative pixel analysis of confocal microscopic images revealed 0.67 ± 0.35% SD overlap of the RNA signal with Xrn1 in P bodies vs. 22.3 ± 5.4% SD overlap with Xrn1 in the cytosol. Thus, although HCV RNA localizing to P bodies could be degraded by Xrn1 (and hence not detectable by FISH), it is possible that HCV RNA is degraded by cytosolic Xrn1. Importantly, we observed no differences in the cellular localization of Xrn1 in infected vs. uninfected cells. Similar studies using an antibody specific for the decapping factor DCP1a as a marker of P bodies also showed no association of HCV RNA with P bodies or changes in DCP1a localization (Fig. S2).
Identification of HCV RNA Degradation Intermediates.
To directly identify HCV RNAs that have been partially degraded, we adapted a circularization RT-PCR (cRT-PCR) strategy to capture HCV RNAs with 5′ monophosphate (5′P). HCV RNAs with 5′P can be ligated with the 3′OH to form a circular RNA, and the region containing 5′ and 3′ ends is subsequently amplified by RT-PCR (Fig. S3A). HCV RNAs with 5′ triphosphate are incompetent for ligation and can only be detected if treated first with RNA polyphosphatase. We applied this method to total RNA isolated from Huh-7 cells stably infected with HJ3-5 virus (Fig. S3B). Full-length HCV RNA with intact 5′ and 3′ ends was identified only after polyphosphatase treatment (Fig. S3C), confirming that the RNA possesses a 5′ triphosphate RNA in cells. Importantly, without pretreatment with RNA polyphosphatase, we could identify HCV RNA degradation intermediates containing 5′P (Fig. S3D). When amplified and sequenced, these intermediates all contained truncated 5′ ends while retaining intact 3′ ends. This finding provides strong evidence that 5′ decay is the primary pathway for HCV RNA degradation in cells and is consistent with Xrn1 being the major contributor to degradation of replicating HCV RNA.
Xrn1 Knockdown Enhances HCV Replication.
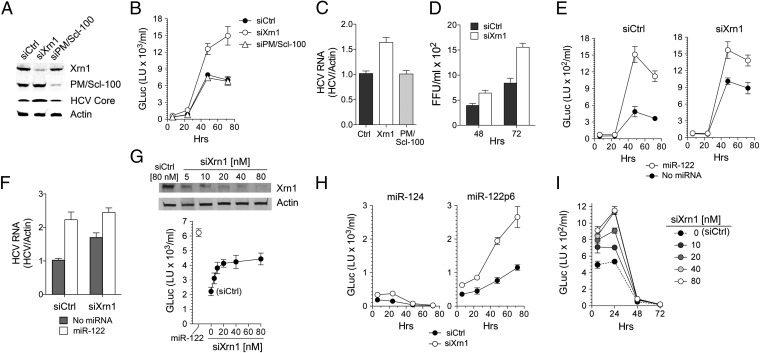
We next asked whether the increased HCV RNA stability in Xrn1 knockdown cells would enhance viral replication. If so, this would indicate that the 5′ Xrn1 decay pathway competes with viral RNA synthetic machinery to set the level to which HCV RNA replicates in cells. To test this hypothesis, we transfected Huh-7.5 cells with siRNAs specific for Xrn1 or PM/Scl-100, resulting in efficient knockdown of these proteins (Fig. 4A). The cells were then retransfected with H77S.3/GLuc2A (23) RNA, and secreted GLuc activity was assessed thereafter as a measure of its replication. Xrn1 knockdown resulted in a twofold increase in GLuc activity at 48–72 h compared with control siRNA, whereas PM/Scl-100 knockdown had no effect (Fig. 4B). Intracellular HCV RNA was similarly increased in Xrn1 knockdown cells (Fig. 4C and Fig. S4), as was the abundance of HCV core protein (Fig. 4A). Xrn1 knockdown also increased the yield of infectious virus released into supernatant fluids (Fig. 4D). Collectively, these data show that Xrn1-mediated decay competes with RNA synthesis directed by the HCV replicase to set the level of viral RNA in infected cells. Previous efforts to investigate how Xrn1 influences HCV replication have produced conflicting results, with two studies showing no effects of Xrn1 depletion (26, 27). Our results are consistent with those of Jones et al. (28) and Ruggieri et al. (29) and provide a mechanism for why Xrn1 depletion promotes HCV replication.
Fig. 4.
Xrn1 knockdown enhances HCV replication in Huh-7.5 cells. Huh-7.5 cells were transfected with siRNAs specific for Xrn1 or PM/Scl-100 or scrambled siCtrl and then 24 h later retransfected with H77S.3/GLuc2A RNA. (A) Immunoblots of Xrn1, PM/Scl-100, and HCV core protein 72 h after HCV RNA transfection, with β-actin as a loading control. (B) GLuc activity in supernatant fluids from Huh-7.5 cells transfected with HCV RNA and the indicated siRNAs. (C) HCV RNA was quantified by qRT-PCR 72 h after HCV RNA transfection relative to β-actin mRNA. (D) Infectious virus titer of supernatant fluids from Huh-7.5 cells transfected with HCV RNA for 48 or 72 h, determined by a fluorescent focus formation assay. (E) GLuc assays were carried out as in B with or without cotransfection of miR-122 (50 nM) and the indicated siRNAs. (F) Cells in E at 72 h posttransfection were harvested, and HCV RNA was quantified by qRT-PCR relative to β-actin mRNA. (G) Graded knockdown of Xrn1 (Upper) by transfection of increasing concentrations of siXrn1 results in proportionate increases in GLuc expression (Lower) that plateaus at the highest siXrn1 concentrations significantly below the level of GLuc expression resulting from miR-122 supplementation. Cells shown as transfected with 0 nM siXrn1 received 80 nM siCtrl. (H) Huh-7.5 cells were transfected with HCV RNA containing p6m mutations in both miR-122 binding sites (HJ3-5/GLuc2A-S1-S2p6m) with cotransfection of the indicated miRNAs. GLuc activity in supernatant fluids was analyzed at indicated time intervals. (I) Graded knockdown of Xrn1 (see G) results in increased transient expression of GLuc from HJ3-5/GLuc2A-S1-S2p6m (due to stabilization of the transfected RNA) but fails to rescue replication of the miR-122 binding mutant. Results shown in G and I represent the mean ± range from replicate cultures, whereas all other results represent the means of three replicate experiments ± SEM.
We next determined whether miR-122 supplementation enhances HCV replication in Huh-7.5 cells following knockdown of Xrn1. If miR-122 acts only to stabilize the viral RNA, we reasoned that there should be relatively little enhancement of replication, as Xrn1 knockdown was as effective as miR-122 supplementation in stabilizing a nonreplicating viral RNA (Fig. 3 A and B). We tested this hypothesis by supplementing cells with miR-122 before transfection of the H77S/GLuc2A RNA. Although Xrn1 knockdown diminished the magnitude of the effect, miR-122 still effectively boosted HCV replication, as determined by measurements of GLuc expression (Fig. 4E, compare Left and Right) and viral RNA abundance (Fig. 4F). To confirm that this was not due to incomplete knockdown of Xrn1, we transfected cells with increasing concentrations of siXrn1. This resulted in increasing depletion of Xrn1, which was near maximal at 20 nM (Fig. 4G, Upper) and was matched by increasing HCV RNA replication (GLuc expression), which plateaued at the same siXrn1 concentration (Fig. 4G, Lower). Despite this evidence for maximal knockdown of Xrn1 expression, miR-122 supplementation resulted in a greater increase in HCV replication (Fig. 4G, Lower). This increase was associated with an increase in the number of large, NS5A-containing replication complexes, which were visualized in microscopic images of cells supporting replication of an HCV RNA in which enhanced yellow fluorescent protein (EYFP) was fused in frame with NS5A (30) (Fig. S5). Thus, whereas the enhanced level of replication in Xrn1 knockdown cells confirms that Xrn1 mediates viral RNA degradation in competition with RNA synthesis, the continued ability of miR-122 to enhance replication in Xrn1-depleted cells suggests that miR-122 plays a role in replication beyond its ability to protect viral RNA from Xrn1 exonuclease.
Xrn1 Knockdown Does Not Rescue Replication of a miR-122 Binding Mutant.
To further assess this Xrn1-independent function of miR-122 in HCV RNA replication, we asked whether Xrn1 knockdown could rescue replication of a viral RNA containing the p6m mutation in both S1 and S2 sites (Fig. 2A) that ablates miR-122 binding (HJ3-5/GLuc2A-S1-S2p6m RNA) (7). As expected (4), this mutant RNA did not replicate when transfected into Huh-7.5 cells, generating only diminishing GLuc activity over the ensuing 72 h (Fig. 4H, Left). Also as expected, cotransfection of the complementary miR-122p6 mutant (Fig. 2A) rescued its replication (Fig. 4H, Right), confirming that the lack of replication is due to its inability to bind miR-122 (4). Significantly, although GLuc expression was increased about twofold (consistent with greater RNA stability), there was no evidence of replication of the mutant RNA when it was transfected into Xrn1 knockdown cells in the absence of miR-122p6m (Fig. 4 H, Left, and I). Thus, although Xrn1 knockdown is able to fully substitute for miR-122 in stabilizing the viral RNA in infected Huh-7.5 cells (Fig. 3 A and B), it is not able to rescue replication of a miR-122 binding mutant. Consistent with the absence of an effect of miR-122 on viral translation after PSI-6130 arrest of RNA synthesis (Fig. 3C, Right), polysome analysis demonstrated no differences in the loading of the WT and miR-122 binding mutant RNAs on ribosomes (Fig. S6). The absence of a difference in ribosome loading is strong evidence that miR-122 has additional function(s) in viral genome amplification beyond stabilization and translation of HCV RNA.
Discussion
Recognition that miR-122 slows the decay of HCV RNA through a process involving recruitment of Ago2 to its 5′ UTR (10), coupled with the importance of miR-122 in the overall viral life cycle (3, 4), led us to characterize how HCV RNA is degraded in cells. We found the decay rates of transfected HCV RNA, HCV RNA added to HeLa S10 lysate, and viral RNA replicating within infected cells were substantially different (Figs. 1–3). These results suggest different decay mechanisms or possibly differential access of the RNA to decay pathways, despite the fact that miR-122 stabilizes the RNA in each of these experimental systems (10). Transfected RNA is subject to both Xrn1 and exosome-mediated decay, with a t1/2 of ∼1.8 h, whereas RNA added to S10 lysate has a t1/2 of ∼17 min and is mainly degraded by the exosome complex and not Xrn1. It is interesting that miR-122 protects HCV RNA from decay in HeLa lysate, as it does this by binding near the end of the 5′UTR (Fig. 2C). One possible explanation is that HCV RNA may be degraded by an unknown 5′ exonuclease (not Xrn1) present in S10 lysate. A second possibility is that miR-122, or the Ago2 protein it recruits to the 5′ UTR (10, 31), may prevent 3′ exosome degradation by promoting circularization of HCV RNA, similar to how poly(rC) binding protein 2 (PCBP2) promotes circularization and stabilization of the poliovirus genome (32, 33). Such a hypothesis could also explain the increased stability conferred on capped HCV RNA in S10 lysate by miR-122 (Fig. S1C).
Replicating viral RNA appears to be subject to very different decay pathways than transfected RNA or RNA added to HeLa lysate. siRNA-mediated depletion of Xrn1 could have unintended consequences on cellular mRNA abundance, thereby potentially altering the abundance of host cell proteins. Despite this caveat, our data suggest that the decay of replicating RNA in infected cells is mediated specifically by Xrn1 with no contribution from the 3′ exosome decay pathway (Fig. 3A). The slow rate of decay under these conditions (t1/2 ∼11 h) is likely to reflect limited access of Xrn1 to HCV RNA within the membranous web (34), a membrane-associated complex containing multiple viral nonstructural proteins that directs new viral RNA synthesis. If the membranous web represents a sanctuary within which newly synthesized viral RNA is protected from cytosolic Xrn1, it would explain the longer t1/2 observed for replicating viral RNA than transfected, replication incompetent RNA (1.8 vs. 11 h). Larger replication complexes constituting the membranous web are relatively static and stable over many hours (35), consistent with the t1/2 we observed for replicating viral RNA. Viral RNA may become subject to Xrn1 degradation during movement from the membranous web to sites of virion assembly on the surface of cytoplasmic lipid droplets (36). The decay rate may thus be more indicative of the turnover and release of viral RNA from these complexes, rather than the rate of Xrn1-mediated 5′→3′ exoribonucleolytic digestion per se.
Alternatively, the slow rate of decay of replicating RNA may reflect the need for removal of a 5′ terminal triphosphate from HCV RNA, as efficient degradation of RNA by Xrn1 requires a 5′ monophosphate (37). Proteins with pyrophosphatase activity on 5′ triphosphate RNA have been reported, including RppH in Escherichia coli and Rai1p in yeast (38, 39). Whether proteins with similar activity are necessary for HCV RNA degradation remains to be determined. Other viruses in the family Flaviviridae express subgenomic RNAs (sfRNAs) that are produced by Xrn1 degradation of the viral genome and are important for pathogenicity (40, 41). Unlike HCV, however, which does not express sfRNA, these flavivirus genomes possess a 5′ cap, and details of their decay pathway are thus likely to differ from HCV.
Although we show that miR-122 protects the viral RNA from Xrn1-mediated decay (Fig. 3 A and B), our data indicate that there is an additional role for miR-122 in the viral life cycle beyond stabilization of the RNA. The critical role that it plays as an HCV host factor is demonstrated by the lack of replication of HCV RNA with single base substitutions in S1 and S2 that ablate miR-122 binding (Fig. 4H). We found that the binding of miR-122 to the 5′ UTR is essential for RNA replication even after knockdown of Xrn1 (Fig. 4 H and I), conditions under which miR-122 supplementation has no influence on the stability of the RNA (Fig. 3 A and B). Similarly, it does not appear that the requirement for miR-122 is related to viral protein translation or the activity of the HCV IRES. Previous studies indicate that miR-122 promotes viral protein expression (4, 9), but it seems likely that this effect is due to its ability to stabilize HCV RNA and increase its abundance (10). Importantly, we observed no increase in viral protein expression following miR-122 supplementation in Xrn1-depleted cells (Fig. 3C, Right), and no defects in ribosome loading by a mutant HCV RNA defective in miR-122 binding (Fig. S6). Collectively, these data argue strongly for a primary role of miR-122 in HCV replication that is independent of its effects on RNA stability or viral protein expression.
What other role might miR-122 play in the viral life cycle? Although it is required for replication of infectious virus (4, 42), miR-122 is also required for amplification of HCV RNA replicons (3) and thus has an essential role independent of viral entry or assembly and release. Absent an essential effect on viral protein translation or RNA stability in Xrn1-depleted cells, our data point strongly toward a primary requirement for miR-122 in HCV RNA synthesis. Despite this, previous efforts to demonstrate direct involvement of miR-122 in viral RNA synthesis have not succeeded. Short-term pulse-labeling experiments failed to show a diminution in HCV RNA synthesis in vivo following transfection of an antisense RNA targeting miR-122 (43), and we found that altering the concentration of miR-122 had no effect on ex vivo synthesis of viral RNA by isolated membrane-bound replicase complexes (44). However, a requirement for miR-122 in initiation of viral RNA synthesis, or for recruitment of host replication factors to the replication complex, might be missed in short-term pulse-labeling studies or in ex vivo studies of isolated replicase complexes (43, 44). Such models are consistent with the clear-cut separation of the effects of miR-122 on viral RNA stability and protein expression versus viral genome amplification that we demonstrated in the studies described herein and suggest new directions for future efforts to elucidate the role of miR-122 in the HCV life cycle.
Materials and Methods
Viral RNA Stability in Transfected and Infected Cells.
For transfection experiments, RNA was transcribed in vitro from pH77S/GLuc2A-AAG (complete genotype 1a HCV sequence with GLuc2A placed in-frame within the polyprotein-coding region and a lethal GDD to AAG mutation in NS5B) and electroporated into cells together with duplex miRNA. Total RNA was harvested at intervals, and HCV RNA was measured by qRT-PCR. Stability of replicating viral RNAs was assessed following arrest of new viral RNA synthesis with PSI-6130 as described (10).
RNA Stability in HeLa S10 Lysate.
Viral RNAs were incubated in HeLa S10 lysates, prepared as previously described except for the deletion of RNase treatment (32, 45), and preincubated with duplex miRNA as described previously. Reactions were stopped at intervals, and RNA was extracted, labeled with SYTO 62 dye, and then resolved by electrophoresis through a 1% Tris Borate EDTA-agarose gel.
Statistical Methods.
qRT-PCR data were fit to a one-phase decay model, and decay constants were compared using the extra sum-of-squares F test.
Additional details can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. William Marzluff, Angela Lam, Phil Furman, and Charles Rice for kindly providing reagents, and the Michael Hooker Microscopy Facility and University of North Carolina In Situ Hybridization Core Facility for technical support. This work was supported by National Institutes of Health Grants R01-AI095690 and P20-CA150343 and the University Cancer Research Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 1571.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213515110/-/DCSupplemental.
References
- 1.Lemon SM, Walker C, Alter MJ, Yi M. Hepatitis C viruses. In: Knipe DM, et al., editors. Fields Virology. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1253–1304. [Google Scholar]
- 2.Chang J, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1(2):106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 3.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309(5740):1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 4.Jangra RK, Yi M, Lemon SM. miR-122 regulation of hepatitis C virus translation and infectious virus production. J Virol. 2010;84:6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jopling CL, Schütz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4(1):77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Machlin ES, Sarnow P, Sagan SM. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci USA. 2011;108(8):3193–3198. doi: 10.1073/pnas.1012464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimakami T, et al. Base pairing between hepatitis C virus RNA and microRNA 122 3′ of its seed sequence is essential for genome stabilization and production of infectious virus. J Virol. 2012;86(13):7372–7383. doi: 10.1128/JVI.00513-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: A look under the hood of miRISC. Nat Struct Mol Biol. 2012;19(6):586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 9.Henke JI, et al. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27(24):3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimakami T, et al. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci USA. 2012;109(3):941–946. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8(2):113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 12.Song M-G, Li Y, Kiledjian M. Multiple mRNA decapping enzymes in mammalian cells. Mol Cell. 2010;40(3):423–432. doi: 10.1016/j.molcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Kiledjian M. Regulation of mRNA decapping. Wiley Interdiscip Rev RNA. 2010;1(2):253–265. doi: 10.1002/wrna.15. [DOI] [PubMed] [Google Scholar]
- 14.Hsu CL, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13(8):4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones CI, Zabolotskaya MV, Newbury SF. The 5′ → 3′ exoribonuclease XRN1/Pacman and its functions in cellular processes and development. Wiley Interdiscip Rev RNA. 2012;3(4):455–468. doi: 10.1002/wrna.1109. [DOI] [PubMed] [Google Scholar]
- 16.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127(6):1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Rodgers ND, Jiao X, Kiledjian M. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 2002;21(17):4699–4708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103(7):1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 19.Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol. 2005;12(9):794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- 20.Ajamian L, et al. Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA. 2008;14(5):914–927. doi: 10.1261/rna.829208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci USA. 2006;103(7):2310–2315. doi: 10.1073/pnas.0510727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stuyver LJ, et al. Inhibition of hepatitis C replicon RNA synthesis by β-D-2′-deoxy-2′-fluoro-2′-C-methylcytidine: a specific inhibitor of hepatitis C virus replication. Antivir Chem Chemother. 2006;17(2):79–87. doi: 10.1177/095632020601700203. [DOI] [PubMed] [Google Scholar]
- 23.Shimakami T, et al. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology. 2011;140(2):667–675. doi: 10.1053/j.gastro.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balagopal V, Parker R. Polysomes, P bodies and stress granules: States and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21(3):403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jangra RK, Yi M, Lemon SM. DDX6 (Rck/p54) is required for efficient hepatitis C virus replication but not IRES-directed translation. J Virol. 2010;84:6810–6824. doi: 10.1128/JVI.00397-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheller N, et al. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc Natl Acad Sci USA. 2009;106(32):13517–13522. doi: 10.1073/pnas.0906413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ariumi Y, et al. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J Virol. 2011;85(14):6882–6892. doi: 10.1128/JVI.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones DM, Domingues P, Targett-Adams P, McLauchlan J. Comparison of U2OS and Huh-7 cells for identifying host factors that affect hepatitis C virus RNA replication. J Gen Virol. 2010;91(Pt 9):2238–2248. doi: 10.1099/vir.0.022210-0. [DOI] [PubMed] [Google Scholar]
- 29.Ruggieri A, et al. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe. 2012;12(1):71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Y, et al. Hepatitis C virus NS2 protein serves as a scaffold for virus assembly by interacting with both structural and nonstructural proteins. J Virol. 2011;85(1):86–97. doi: 10.1128/JVI.01070-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson JA, Zhang C, Huys A, Richardson CD. Human Ago2 is required for efficient miR-122 regulation of HCV RNA accumulation and translation. J Virol. 2011;85:2342–2350. doi: 10.1128/JVI.02046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray KE, Roberts AW, Barton DJ. Poly(rC) binding proteins mediate poliovirus mRNA stability. RNA. 2001;7(8):1126–1141. doi: 10.1017/s1355838201010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herold J, Andino R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol Cell. 2001;7(3):581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gosert R, et al. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77(9):5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wölk B, Büchele B, Moradpour D, Rice CM. A dynamic view of hepatitis C virus replication complexes. J Virol. 2008;82(21):10519–10531. doi: 10.1128/JVI.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyanari Y, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9(9):1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 37.Stevens A. Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5′-mononucleotides by a 5′ leads to 3′ mode of hydrolysis. J Biol Chem. 1980;255(7):3080–3085. [PubMed] [Google Scholar]
- 38.Xiang S, et al. Structure and function of the 5′→3′ exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458(7239):784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451(7176):355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 40.Pijlman GP, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4(6):579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Silva PA, Pereira CF, Dalebout TJ, Spaan WJ, Bredenbeek PJ. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J Virol. 2010;84(21):11395–11406. doi: 10.1128/JVI.01047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Randall G, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci USA. 2007;104(31):12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norman KL, Sarnow P. Modulation of hepatitis C virus RNA abundance and the isoprenoid biosynthesis pathway by microRNA miR-122 involves distinct mechanisms. J Virol. 2010;84(1):666–670. doi: 10.1128/JVI.01156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villanueva RA, et al. miR-122 does not modulate the elongation phase of hepatitis C virus RNA synthesis in isolated replicase complexes. Antiviral Res. 2010;88(1):119–123. doi: 10.1016/j.antiviral.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barton DJ, Morasco BJ, Flanegan JB. Assays for poliovirus polymerase, 3D(Pol), and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 1996;275:35–57. doi: 10.1016/s0076-6879(96)75005-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.